Abstract
Rad50 ABC-ATPase complex with Mre11 nuclease is essential for dsDNA break repair, telomere maintenance, and ataxia telangiectasia mutated kinase checkpoint signaling. How Rad50 affects Mre11 functions, and how ABC-ATPases communicate nucleotide binding and ligand states across long distances and among protein partners, has remained obscure. Here, structures of Mre11–Rad50 complexes define the Mre11 2-helix Rad50 binding domain (RBD) that forms a 4-helix interface with Rad50 coiled-coils adjoining the ATPase core. Newly identified effector and basic-switch helix motifs extend the ABC-ATPase signature motif to link ATP-driven Rad50 movements to coiled-coils binding Mre11, implying a ~30 Å pull on the linker to the nuclease domain. Both RBD and basic-switch mutations cause clastogen sensitivity. Collective results characterize flexible ATP-dependent Mre11 regulation, defects in cancer-linked RBD mutations, conserved superfamily basic-switches and motifs effecting ATP-driven conformational change, and a unified comprehension of ABC-ATPase activities.
Mre11–Rad50 complex, or Mre11–Rad50–Nbs1 (MRN) in higher eukaryotes, coordinates detection, signaling, and repair of cytotoxic and mutagenic DNA double-strand breaks (DSBs). Mre11 3′-5′ exonuclease and single stranded DNA (ssDNA) endonuclease activities are regulated by Rad50 ATP binding and hydrolysis within the MRN complex1,2. Combined structural, biochemical, and cell biology results show MRN serves as a DNA damage sensor, enzymatic effecter, and a transducer of cell-cycle checkpoint signals for DNA double-stand break repair (DSBR)3,4. Critical MRN tumor suppressor functions are underscored by the facts that NBS1 mutations cause a radiosensitive and chromosome instability disorder Nijmegen breakage syndrome (NBS)5, MRE11 mutations cause ataxia telangiectasia-like disorder, and RAD50 mutations result in an NBS-like syndrome6. Other MRE11 variants (including L473F) are linked to colorectal cancer7, yet the molecular basis for such defects remain undefined.
The Mre11–Rad50 core complex has critical DNA end-bridging and ATP-regulated endonucleolytic activities for initiation of homologous recombination (HR) repair of DSBs8,9; yet, high-resolution structures of this Mre11–Rad50 complex and its critical interfaces have eluded characterizations. Crystal structures showed an 80 kDa Mre11 dimer can directly bridge DNA ends9, characterized Rad50 ABC-ATPase monomer with and without adjacent coiled-coil regions10,11, and defined a nucleotide-bound Rad50 dimer lacking coiled-coil regions11 (see Supplementary Fig. 1). Also, hints on quaternary assembly come from electron micrographs (EM) of Mre11–Rad50 that revealed a ~100 Å diameter four-lobed Mre11–Rad50 head (M2R2-head) with ~500 Å long Rad50 coiled-coil protrusions9,10,12,13. Yet, no Mre11–Rad50 co-complex structures exist in either nucleotide-bound or free states. So how Mre11 is physically linked to Rad50, how Rad50 subunits assemble within the M2R2-head, and how nucleotide binding to the ABC-ATPase may regulate Mre11–Rad50 structure and functions remain mysteries3.
Here we use Pyrococcus furiosus proteins and Schizosaccharomyces pombe genetics to define the critical, conserved Mre11–Rad50 interface, the molecular basis of this interaction, and the importance of this interface for DSBR in vivo. To elucidate Rad50 conformational changes that would impact Mre11, we solved four new structures of Rad50 containing critical coiled-coil regions in complex with either the Mre11 Rad50 binding domain (RBD), AMP:PNP, or both. Combined structural and mutational results define the ABC-ATPase signature helix motif and key basic-switch residues that drive and coordinate Rad50 domain rotations by toggling between specific, distinct salt bridge networks. Collective results help explain defects in cancer-linked Mre11 mutations, and identify an underlying molecular basis, conserved across the ABC-ATPase superfamily, for coupling ATPase nucleotide state to biological outcomes through conformational changes impacting interfaces and attached functional domains.
RESULTS
The Rad50 binding domain (RBD) of Mre11
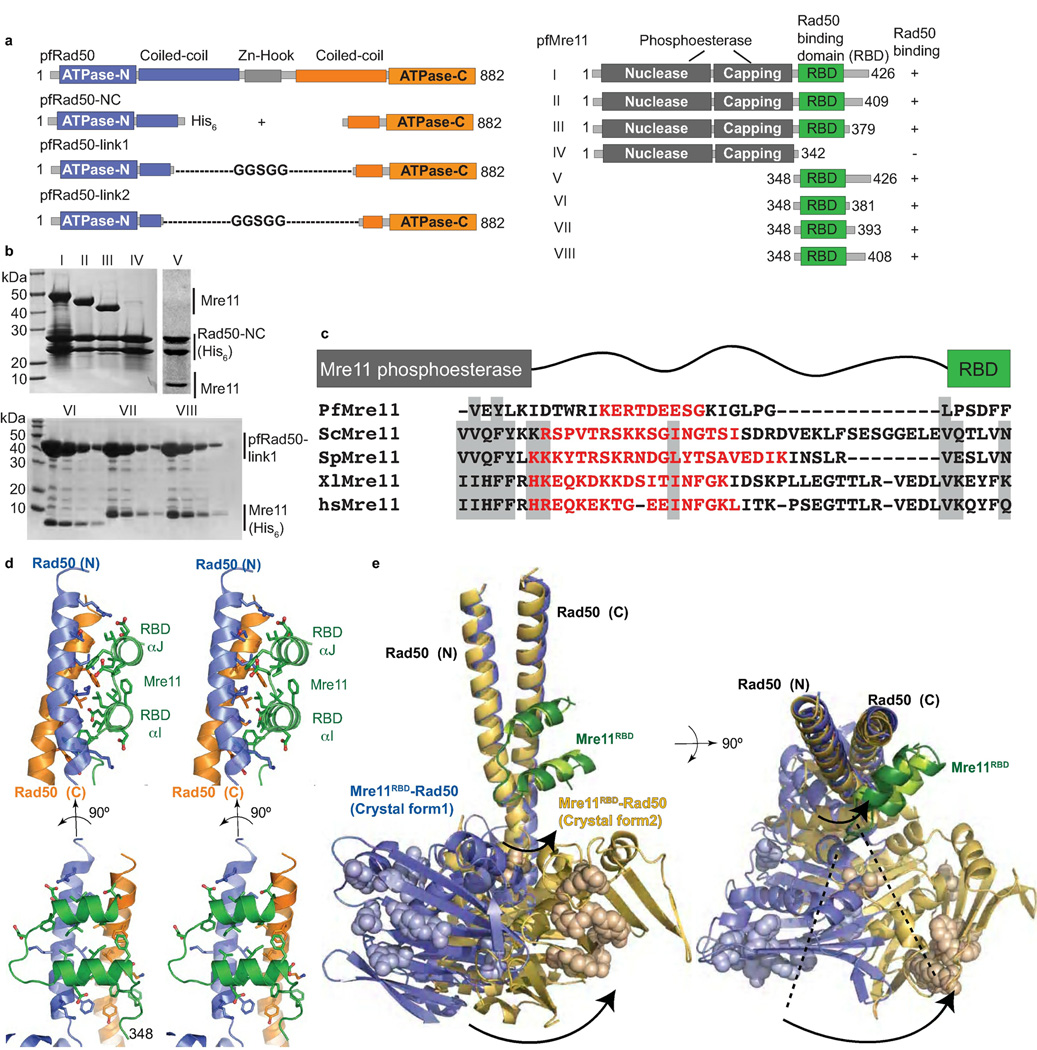
To map the Mre11 RBD, we generated a series of P. furiosus Mre11 (pfMre11) deletion constructs and tested their ability to co-express and co-purify with coiled-coil truncated pfRad50 constructs (Fig. 1a,b). Mre11 C-terminal truncations, which left the N-terminal core nuclease domain intact, revealed that the 342–379 region contained residues essential for binding histidine-tagged pfRad50 (pfRad50-NC). A C-terminal Mre11 construct (residues 348–426) also bound Rad50 (Fig. 1b). To finely map the Mre11 RBD, we expressed and co-purified Mre11–Rad50 complexes of untagged Rad50 with shortened coiled-coils connected by intramolecular "Gly-Gly-Ser-Gly-Gly" sequences (pfRad50-link1) with predicted minimal Mre11 RBD regions containing a histidine-tag. Our shortest Mre11 construct, residues 348–381 (Mre11RBD), bound tightly to and co-purified with pfRad50-link1. Collectively, these data delineate a major physical Mre11–Rad50 interaction for Mre11 residues 348–379 and a corresponding Rad50 binding site within the first coiled-coil ~6 heptad repeats, proximal to the ATPase core.
Figure 1. The Mre11RBD–Rad50 interface.
(a) pfRad50 and pfMre11 construct schematics for domain mapping and crystallizations. pfRad50-link constructs contain Gly-Ser repeat sequences to intramolecularly link Rad50 N- and C-lobes. (b) Mapping of Mre11 RBD. Top: His6-tagged Rad50 was co-expressed with Mre11 variants (I–V) shown in (a). Bottom: His6-tagged Mre11 variants (V–VIII) were co-expressed with untagged pfRad50-link1. Minimal pfMre11 polypeptide (Mre11RBD, residues 348–381) interacts with Rad50-link1. (c) Sequence alignment of the linker region connecting the Mre11 RBD to the nuclease capping domain in pfMre11, S. cerevisiae (scMre11), S. pombe (spMre11), Xenopus laevis (xlMre11) and human (hsMre11). Shaded regions show well-conserved residues. Disordered residues are shown in red as seen in pfMre11 crystal structures or as predicted by Disopred2. (d) Mre11RBD–Rad50 interface, shown in orthogonal stereo views. The hydrophobic Mre11RBD–Rad50 interaction core is augmented by four flanking complementary salt bridge interactions with acidic residues from Mre11 RBD interacting with 4 positively-charged Rad50 surface residues. (e) Superimposition of two nucleotide-free Mre11RBD–Rad50 crystal forms. The core Mre11RBD–Rad50 interface is maintained, but a ~35° rotation about the base of the Rad50 coiled-coil identifies a flexible linkage to Rad50 ATPase domains. Residues equivalent to those mutated in Rad50S yeast phenotypes are shown in space fill representations.
The four-helix architecture of the Mre11–Rad50 interface
To define the Mre11–Rad50 interface structure, we solved two independent X-ray crystal structures, to 2.1 Å and 3.4 Å resolution, of pfMre11RBD bound to pfRad50-NC (Table 1), which reveal the same interface. Our 2.1 Å structure provides high-resolution details about this interface (Fig. 1). The Mre11 RBD consists of two helices (RBD-αI and RBD-αJ, named sequentially from nuclease core labeling10) that interact with the Rad50 coiled-coil base through a conserved hydrophobic surface patch. This 4-helix interaction differs from classical 4-helix bundle interfaces, such as in human Mn superoxide dismutase14 and typical coiled-coil packing such as in bacterial pili15,16, as Mre11 helices pack almost orthogonally to the two Rad50 coiled-coil helices. The Mre11RBD–Rad50 interface includes 72% of the 32 Mre11RBD residues and has a ~970 Å2 buried surface area (BSA). Ten Mre11RBD hydrophobic core residues account for 75% of the total BSA, and this strong interface spans ~20 Å across and ~30 Å up the Rad50 coiled-coils. The conserved interface in human Mre11–Rad50 likely involves Mre11 RBD residues Gln435–Lys475, and Rad50 coiled-coil regions around Arg184–Lys204 and Lys1,098–Asp1,129.
Table 1.
Data collection and refinement statistics
| Rad50-link1- AMP:PNP-Mg2 |
Mre11RBD- Rad50 crystal form 1 |
Mre11RBD- Rad50 crystal form 2 |
Mre11RBD- Rad50-link2- AMP:PNP- Mg2+ |
|
|---|---|---|---|---|
| Data collection | ||||
| Space group | P212121 | P212121 | P61 | P6522 |
| Cell dimensions | ||||
| a, b, c (Å) | 83.39, 108.52, 150.37 | 55.78, 91.23, 107.33 | 116.04, 116.04, 109.85 | 177.4, 177.4, 130.4 |
| α, β, γ (°) (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50.0-1.90 (1.92-1.90)* | 50.0-2.10 (2.17-2.10) | 50.0-3.4 (3.49-3.40) | 50.0-3.3 (3.39-3.30) |
| Rsym or Rmerge | 5.6 (48.0) | 5.1 (45.7) | 10.9 (33.5) | 4.8 (66.0) |
| // I /σ/ | 25.4 (1.9) | 27.5 (2.8) | 14.2 (2.0) | 35.7 (3.6) |
| Completeness (%) | 97.9 (81.8) | 97.0 (85.6) | 92.0 (59.3) | 99.8 (100.0) |
| Redundancy | 5.3 (2.6) | 4.6 (4.2) | 6.6 (2.2) | 9.8 (10.1) |
| Refinement | ||||
| Resolution (Å) | ||||
| No. reflections | 105157 (8694) | 31661 (2748) | 10679 (682) | 18667 (1840) |
| Rwork / Rfree | 19.2/24.4 | 21.0/25.6 | 21.1/27.4 | 25.8/30.8 |
| No. atoms | ||||
| Protein | 10535 | 3237 | 3354 | 5567 |
| Ligand/ion | 128 | 0 | 10 | 64 |
| Water | 935 | 244 | 7 | 0 |
| B-factors | ||||
| Protein | 41.8 | 61.2 | 100.0 | 124.3 |
| Ligand/ion | 20.68 | 107.3 | ||
| Water | 46.40 | 63.0 | 77.6 | |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.006 | 0.003 | 0.006 | 0.01 |
| Bond angles (°) | 1.091 | 0.679 | 1.031 | 1.223 |
One crystal was used for each data set. Values in parentheses are for highest-resolution shell.
Mre11 RBD is flexibly linked to the Rad50 coiled-coils
Our sequence alignments and Disopred2 (ref.17) disorder predictions show residues spanning from 333 at the end of previous Mre11 nuclease coordinates9,10 to reside 348 of our interface structure have high sequence divergence, suggesting intrinsic disorder (Fig. 1c). In fact, residues 334–342 were present and disordered in our previous structures (PDB codes 3DSC, 3DSD, 1II7)9,10, but we missed the significance of this observation. Thus, combined results reveal that a flexible tether links the Mre11 nuclease and RBD domains.
Comparison of Mre11RBD–Rad50 structures from two different crystal forms furthermore reveals that the Rad50 coiled-coils can adopt dramatically variable orientations relative to the ATPase domains in the nucleotide free form (Fig. 1e). These changes are highlighted by core Mre11RBD–Rad50 superimpositions; this region superimposes well, but the Rad50 ATPase domain can rotate substantially with respect to the coiled-coils. These domain motions uncover intrinsic flexibility in the hinge region at the base of the coiled-coil N-terminal α-helix, which is adjacent to the Mre11RBD–Rad50 interface. In our structures this region has limited contacts with the N-terminal half (N-lobe) of the Rad50 ATPase domain and adopts dramatically different conformations, imparting a 30° twist and 15 Å shift relative to the coiled-coils. Conversely, the base of the C-terminal helix of the coiled-coil is rigid, and makes extensive contacts with the C-terminal half (C-lobe) of the Rad50 ATPase core.
Mre11 RBD mutations disrupt Mre11–Rad50 interactions in vivo
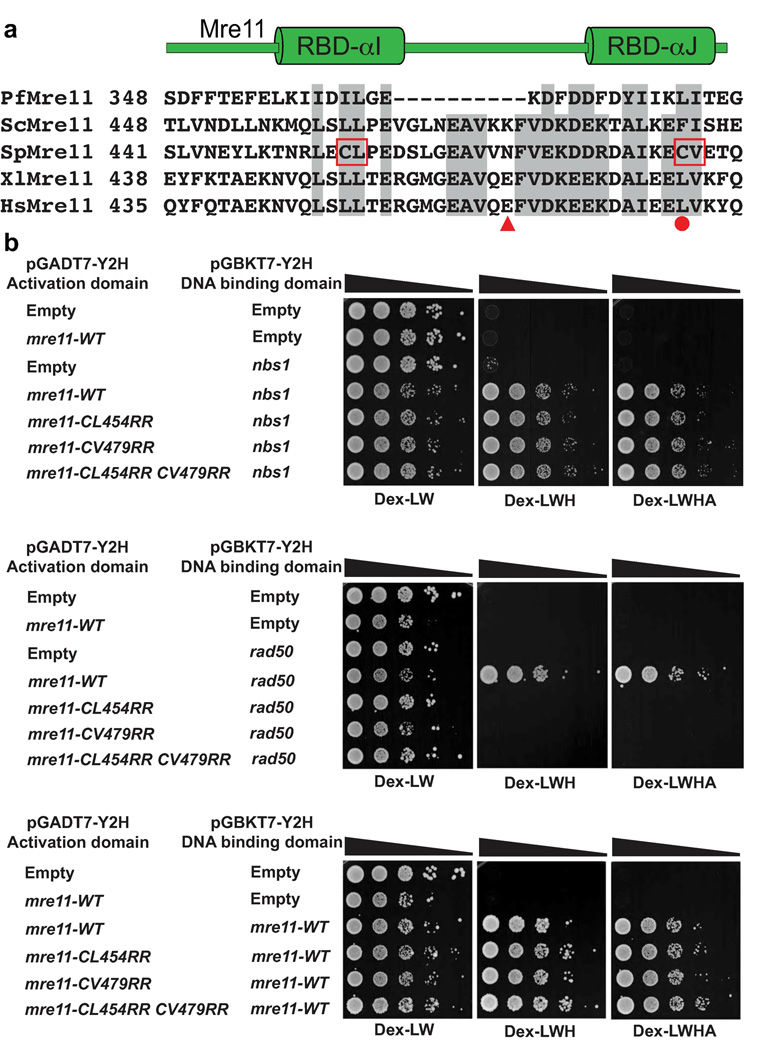
Based on sequence alignments of Mre11 orthologs (Fig. 2a), mutations were introduced into S. pombe Mre11 (also known as Rad32) to test the functional significance of the Mre11 RBD. Hydrophobic Cys-Leu (CL) residues in RBD-αI and Cys-Val (CV) residues in RBD-αJ were changed to charged Arg-Arg (RR) residues, either separately or in combination. Collectively, these residues at the Mre11 RBD core mediate hydrophobic interactions to both of the Rad50 coiled-coil α-helices and also between RBD-αI and RBD-αJ. Thus, their substitution to charged Arg residues should disrupt the interface. Indeed, two-hybrid analyses showed that Mre11 and Rad50 have a robust interaction, but this was severely diminished by the RR mutation in either RBD-αI or RBD-αJ (Fig. 2b). Importantly, our Mre11 RBD mutations did not impair Mre11 homodimeric or Nbs1 interactions, indicating specific disruption of the major Mre11–Rad50 interface.
Figure 2. A conserved interface links eukaryotic Mre11 and Rad50.
(a) Multiple sequence alignment of Mre11 RBD from pfMre11, scMre11, spMre11, xlMre11 and hsMre11. Shaded regions show well-conserved residues. CL→RR and CV→RR mark hydrophobic to charged surface substitutions introduced into S. pombe Mre11 RBD. HsMre11 mutations identified in somatic colorectal cancers are highlighted (solid red circle, point mutation; triangle, truncation7). (b) S. pombe Mre11 RBD variant interactions with Rad50, Nbs1 and the Mre11 homodimeric interaction analyzed by two-hybrid. Growth on Dex-WL plates (minimal glucose media lacking tryptophan and leucine) indicates the reporter strain transformed with plasmids pGADT7 (Gal4 activating domain) and pGBKT7 (Gal4 DNA binding domain) fused to the respective proteins. Growth on Dex-LWH (less stringent; lacking histidine) and Dex-LWHA (more stringent; lacking histidine and adenine) indicates a positive two-hybrid interaction. We have previously shown that pGBKT7-mre11-WT alone does not autoactivate9 and this was not repeated here. Mre11 RBD mutants fail to interact with Rad50 yet retain homodimerization and Nbs1 interactions. Strains used are detailed in Supplementary Table 1.
The Mre11 RBD is critical for DSBR in fission yeast
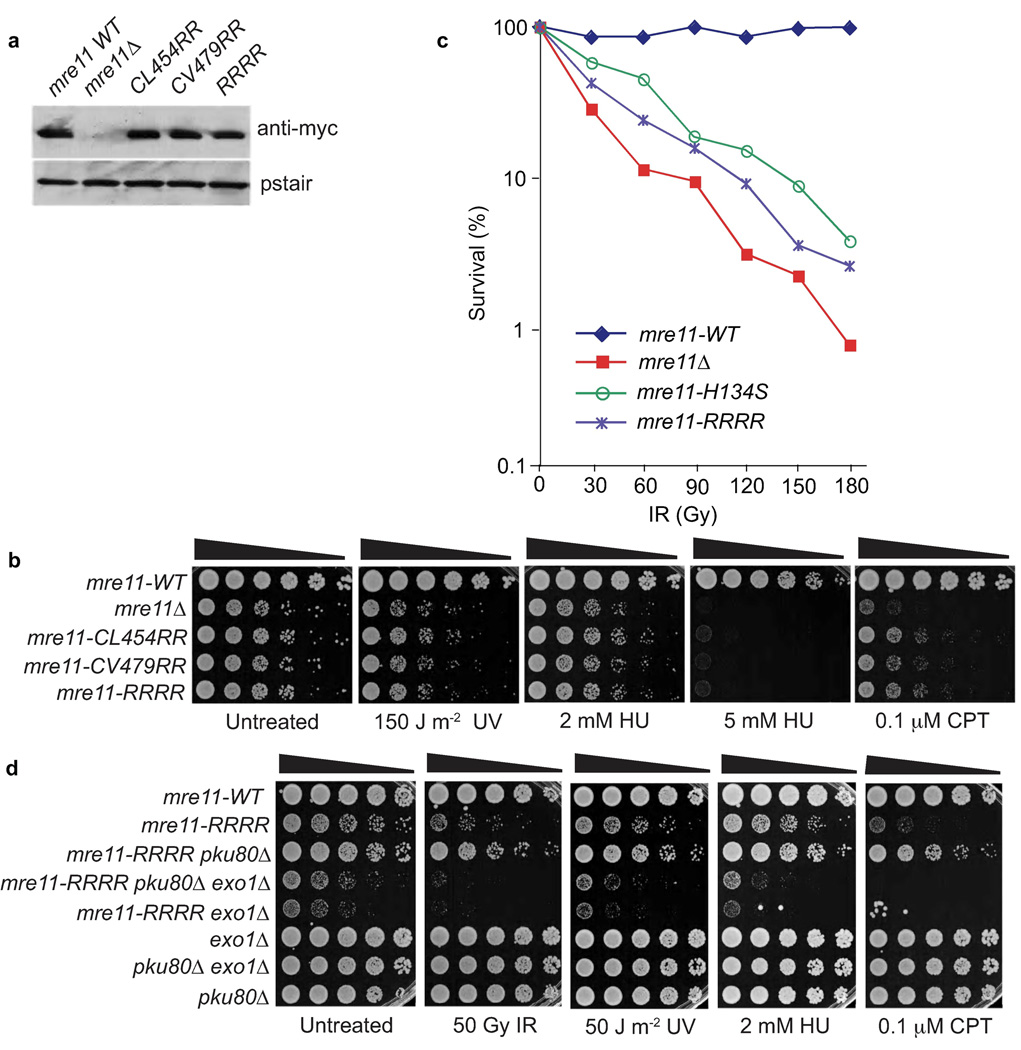
To test whether the Mre11 RBD mutants show increased DNA damage sensitivity, we examined responses to four genotoxins: 1) ionizing radiation (IR), which directly makes DSBs; 2) UV light, which creates DNA photoproducts that can be processed into DSBs; 3) camptothecin (CPT), a topoisomerase inhibitor that causes replication fork breakage when the replisome encounters a topoisomerase–CPT complex; and 4) hydroxyurea (HU), which stalls replication forks by inhibiting ribonucleotide reductase required for dNTP synthesis. The mre11 alleles replace genomic mre11 (mre11-WT) and encode a C-terminal myc-tag. These strains were compared to mre11Δ and myc-tagged mre11-WT control strains. This myc-tag does not noticeably impair Mre11 function9. Immunoblotting showed that the Mre11 RBD mutants were expressed at levels comparable to wild type (Fig. 3a).
Figure 3. The Mre11–Rad50 interaction interface coordinates DSBR in S. pombe.
(a) Expression levels of myc-tagged Mre11 variants. (b) Mre11 RBD variant UV, HU, and CPT genotoxin sensitivity. Mre11–Rad50 interaction interface disruption causes clastogen sensitivity. Five-fold serial dilutions of cells on rich media plates were photographed following 2–3 days at 30°C. (c) Mre11 RBD variants are IR sensitive. This plot is representative of two independent experiments (Supplementary Fig. 2). (d) The IR, UV, HU, and CPT survival defects of mre11-RRRR are suppressed by Ku80 elimination. This rescue depends on Exo1. Strains used are detailed in Supplementary Table 1.
In agreement with their poor abilities to interact with Rad50 in two-hybrid assays (Fig. 2b), the Mre11 RBD-αI (mre11-CL454RR) and RBD-αJ (mre11-CV479RR) mutants resembled mre11Δ in being very sensitive to IR, UV, CPT and HU (Fig. 3b,c). These mutants also formed smaller colonies than wild type, indicating defects in repair of spontaneous DNA damage. Serial dilution assays performed with UV, HU or CPT shows that the Mre11 RBD mutants are slightly more resistant than mre11Δ cells. This small difference might be because the Mre11 RBD-αI and RBD-αJ mutants retain residual interactions with Rad50 that were not detected by yeast two-hybrid analysis; thus we did survival assays on an mre11-RRRR allele that should completely disrupt the interface. This allele appeared identical to the mre11-CL454RR and mre11-CV479RR alleles in serial dilution assays (Fig. 3b) and in IR survival assays the mre11-RRRR strain is slightly more resistant than mre11Δ (Fig. 3c and Supplementary Fig. 2). Collectively, these data show that the Mre11 RBD forms an interface that is critical for DSBR, although weak function is maintained when it is mutated.
ExoI nuclease can compensate for Mre11 RBD mutant phenotypes
To see if Mre11 interface mutants impact DNA end-processing in fission yeast, we tested if ExoI nuclease can compensate for Mre11. In the IR survival assays the mre11-RRRR strain is slightly more sensitive than the previously characterized mre11-H134S mutant (Fig. 3c). Genetic and biochemical studies indicate that the mre11-H134S genotoxin sensitivity is caused by a defect in ssDNA endonuclease activity9 that is suppressed by inactivating the Ku70–Ku80 complex, which can bind and block ends. This rescue requires Exo1 exonuclease, indicating that Mre11 endonuclease activity is critical for generating single-strand overhangs that are competent for HR repair18,19. To assess whether the mre11-RRRR mutant is defective in DNA end-processing, we created mre11-RRRR strains lacking Ku80 and/or Exo1. We found that the pku80Δ mutation suppressed the slow growth phenotype as well as the IR, CPT, UV and HU sensitivities of mre11-RRRR cells (Fig. 3d). This supports a model in which Ku promotes non-homologous end-joining by binding to DSB ends and inhibits Exo1-dependent resection19. Accordingly, the extreme genotoxin sensitivity of the mre11-RRRR pku80Δ exo1Δ strain showed that this suppression was dependent on Exo1 activity. Indeed, the exo1Δ mutation substantially exacerbated the mre11-RRRR phenotypes. These results indicate that the mre11-RRRR phenotypes primarily reflect an inability of Mre11–Rad50 to process DNA ends for DSBR by HR.
Architecture of the M2R2-head
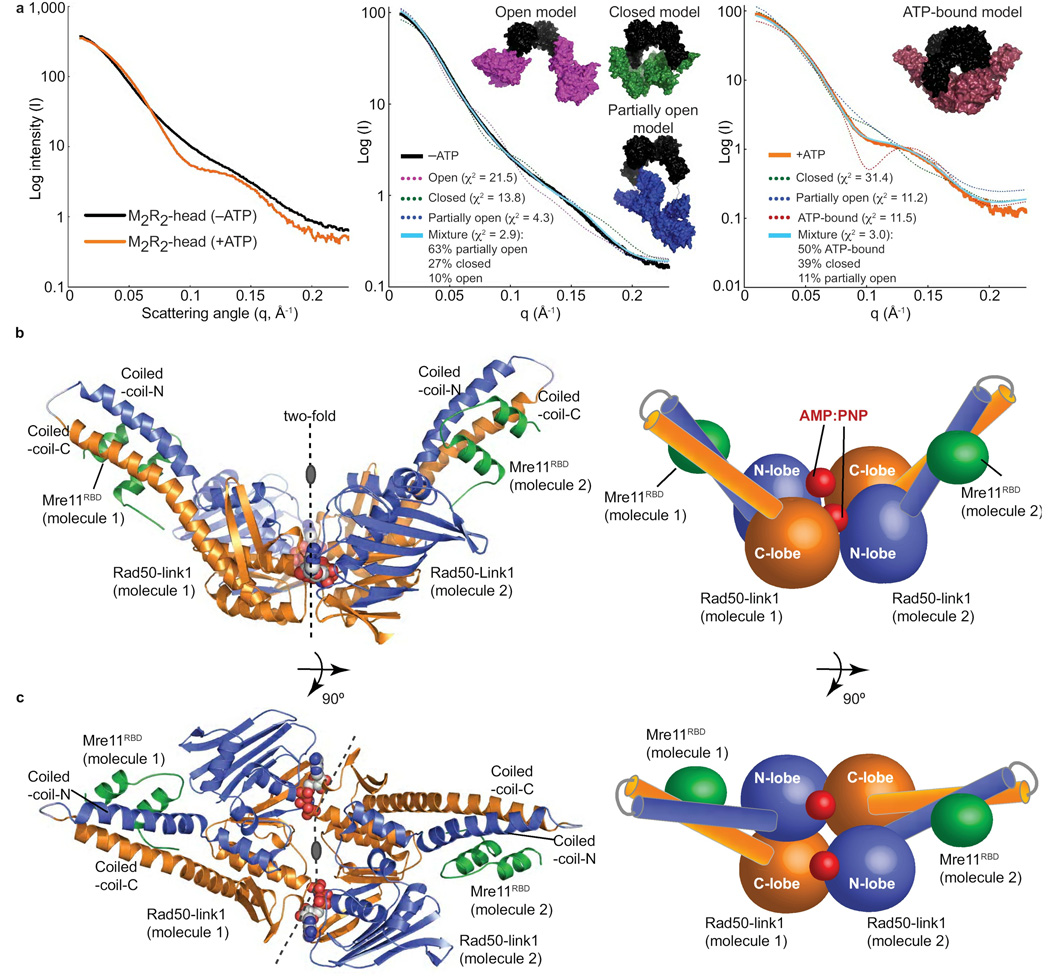
To test the Mre11RBD–Rad50 complex flexibility implied from crystal structures, we examined M2R2-head solution conformations with small angle X-ray scattering (SAXS). SAXS combined with crystal structure restraints can accurately define flexible conformations and ensembles in solution, and can also identify existing structures that most closely match the measured experimental scattering20. Experimental SAXS curves of M2R2-head preparations (Fig. 4a) show dramatic scattering curve changes supporting a flexible to more ordered transition: featureless without ATP (−ATP) to defined peaks and troughs with ATP (+ATP). Further, the radius of gyration decreases from 46.5 to 41.0 Å upon ATP binding, dimensions resembling M2R2-head regions within intact pfMre11–Rad50 EM images12. These results, along with a compaction observed in the pair distribution (p(r)) plot (Supplementary Fig. 3), show that the M2R2-head transitions from a more open to compacted states upon ATP binding. This supports our hypothesis that ATP binding in the M2R2-head leads to Rad50 dimerization, which would close the M2R2-head to form a globular, toroidal structure.
Figure 4. The M2R2-head assembly.
(a) SAXS analysis of the M2R2-head reveals a transition from a conformationally flexible, open complex to a globular, closed complex upon ATP binding. Left: experimental SAXS curves of the M2R2-head without (−ATP) and with (+ATP) nucleotide. Fits of single and MES models of M2R2-heads to −ATP (middle panel) and +ATP (right panel) data. Models are shown as surfaces with Mre11 core dimer colored black and Rad50 domains with attached Mre11 RBD colored for open (magenta), partially open (blue), closed (green) and ATP-bound (red) conformations. Fits to the experimental data are shown for single models (dashed line) and the MES ensemble (cyan line) with quality of fit shown by χ2. (b) Mre11RBD–Rad50-link1–AMP:PNP–Mg2+ complex architecture. Left: Mre11RBD (green) binds to Rad50 coiled-coil base. Right: Schematic of the structure. (c) Orthogonal views of the complex as in (b). See Table 1 for data processing and refinement statistics.
To model the conformational flexibility implied for the −ATP data by the featureless curve and overall architecture of the M2R2-head in the absence and presence of ATP, we used molecular dynamics (MD) and minimal ensemble searches (MES)21 to find M2R2-head structural models that best fit the data. We find the predominant M2R2-head architecture without ATP is a partially open state; yet, improved fit to the data by a mixture of open, partially open and closed conformations shows the inherent flexibility of the complex without nucleotide (Fig. 4a). With ATP we expected to see mixed ATP-bound and free states in solution. So to accurately model M2R2-head complex with ATP, we used MES with closed ATP-bound M2R2-head models, based on our crystal structures described below, combined with models identified for the −ATP data (Fig. 4a). The results suggest that 89% of the M2R2-head is in a closed state, with the Mre11 dimer and ATP-induced Rad50 forming a toroidal, globular structure.
The M2R2-head toroidal structure from our SAXS analyses is independently supported through direct comparisons of M2R2-head scattering curves with SAXS curves calculated from the Protein Data Bank, using the database for rapid search of structural neighbors (DARA)22. DARA analysis shows that the M2R2-head quaternary assembly is most structurally conserved with Topoisomerase-II and the DNA mismatch repair protein MutS, another ABC-ATPase superfamily member (Supplementary Fig. 3). Both of these proteins form toroidal-like structures, supporting the accuracy of M2R2-head architecture observed from our MD and MES modeling. Notably, the architectural similarity of M2R2-head with MutS reveals a striking convergent evolution of quaternary assembly modes for divergent members of the ABC-ATPase superfamily.
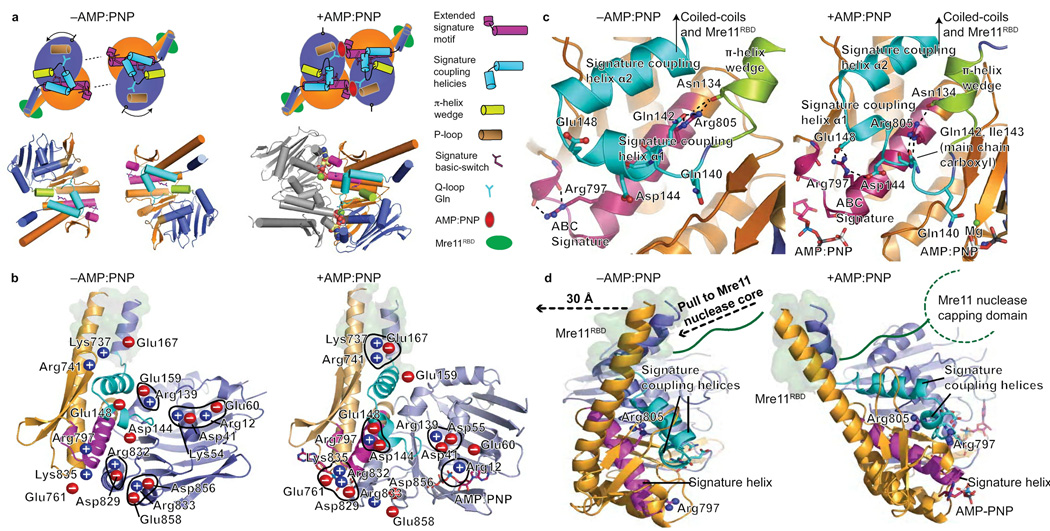
ABC signature helices couple nucleotide state to domain movements
To test if this Mre11RBD–Rad50 interface is impacted upon nucleotide binding in the Rad50 ABC-ATPase core, we used our Rad50 constructs that intramolecularly link the ABC-ATPase N- and C-lobes with adjacent coiled-coil regions (Fig. 1a): these constructs facilitated ternary structure solution of Mre11RBD–Rad50-link1–AMP:PNP–Mg2+ to 3.3 Å (Fig. 4b,c) and Rad50-link2–AMP:PNP–Mg2+ to 1.9 Å resolution (Table 1). Superimposition of our nucleotide-free and -bound structures of Rad50 containing coiled-coil regions and morphing between crystallographically-defined states reveals both global conformational changes (Supplementary Movies 1–3) and their underlying basis (Fig. 5). As Rad50 AMP:PNP structures with and without bound Mre11RBD superimpose well, we used the higher resolution structure for most analyses.
Figure 5. Rad50 nucleotide-binding induced conformational changes.
(a) Nucleotide free (−AMP:PNP) and bound (AMP:PNP) Rad50 conformations. Nucleotide binding is coordinated by the signature motif, Q-loop, P-loop, and induces a ~35° subdomain rotation. Rad50 N-lobe rotation drives the π-helix wedge into the signature coupling helices, dramatically altering signature coupling helix conformation relative to ATPase subdomain interactions. Motifs are colored as in key. (b) Twenty salt bridge switches rearrange upon nucleotide binding and coordinate domain rotations, see Supplementary Movie 1. Blue (positive) and red (negative) circles highlight charged residues. The Mre11 RBD is highlighted by a green surface representation of Rad50 residues involved in the interface. (c) Signature helix Arg797 and Arg805 rearrangements link nucleotide binding with domain rotations, conformational change of the signature-coupling helices and Q-loop, and motions in the Rad50 coiled-coils (see Supplementary Movie 2). The Mre11 RBD is highlighted as in (b). (d) Nucleotide-binding induced Rad50 ATPase C-lobe rotation relative to the N-lobe drives coiled-coil repositioning to impact bound Mre11 RBD, highlighted as in (b). The Rad50 domain rotation is transduced through coiled-coil repositioning (see Supplementary Movie 3), into a linear pull on the linker between the Mre11 RBD and nuclease capping domain as depicted by dashed arrows.
Our Mre11RBD–Rad50-link1–AMP:PNP–Mg2+ structure defines the nucleotide bound state of the unknown half of the M2R2-head, with molecular dimensions of 120 × 74 × 62 Å. Globally, the Rad50 ABC-ATPase core dimerizes with AMP:PNP–Mg2+ sandwiched at the crystallographic two-fold interface (Fig. 4b), inducing a ~35° rotation of the C-lobe relative to the N-lobe, supporting and extending changes proposed from core structures lacking all coiled-coil regions11. However, our new nucleotide-bound structures reveal novel positions of two helices, which we term the signature coupling helices, immediately C-terminal to the Rad50 Q-loop. These helices, which are absent from the original nucleotide-bound Rad50 structure, connect the Q-loop to the base of the N-terminal helix of the coiled-coil. Upon nucleotide binding, Rad50 N-lobe rotation drives a π-helix element (π-helix wedge) between the signature coupling helices to splay them apart (Fig. 5a–c and Supplementary Fig. 4). The signature coupling helices movement, resembling the opening of an arm at the elbow, acts as a lever exerting force on the base of the N-terminal coiled-coil helix, which our nucleotide-free structures show is a point of flexion. This force repositions the coiled-coils with respect to the ATPase core, impacting the Mre11 RBD position. As shown by structure-based animation (Supplementary Movie 3), the ATP driven domain rotation is transduced to a ~30 Å linear pull on the Mre11 linker by the Rad50 coiled-coil movement at the Mre11 interface (Fig. 5d).
Basic-switches and alternating salt bridges control ATPase rotations
Underlying the global conformational changes described above is a striking, extensive network of >20 charge pairs that switch upon nucleotide binding (Fig. 5b). These changes provide a mechanism to physically couple ATPase conformational rotation to coiled-coil and attached Mre11 RBD repositioning. Intriguingly, basic-switch residues (Arg797 and Arg805 in pfRad50) immediately adjacent to the conserved signature motif, which defines the ABC-ATPases superfamily, occupy a conserved helix. We term this the signature helix as it encodes a molecular conformational switch that links signature motif nucleotide recognition to subdomain rotation.
Arg797 hydrogen bonds to main chain signature motif atoms in the nucleotide-free state. Upon nucleotide binding, the signature motif moves to contact the nucleotide. As a consequence, Arg797 detaches from the signature motif and moves to form interaction networks with Glu148 and Asp144 on signature coupling helix-α1 (Fig. 5c and Supplementary Fig. 4). These new Arg797 interactions can only form after opening and translation of the signature coupling helices following N-lobe rotation and limit further rotation of the first helix. Arg797 movements thus directly link nucleotide recognition by the signature motif to movements of the signature coupling helices that control coiled-coil positioning.
Signature helix Arg805 integrates the signature motif, Q-loop, and domain rotations. Nucleotide binding breaks Arg805 hydrogen bonds to the Asn134 main chain. Arg805 then moves towards the protein surface with concomitant rearrangements of the Q-loop. Arg805 rotates into the signature coupling helices and guides the π–helix to wedge open signature coupling helix-α1 and form new hydrogen bonds to Gln142 and Ile143 main chain carboxyl atoms (Fig. 5c). This switch at the junction between the N-lobe, C-lobe, and coiled-coil base, implicates Arg805 as a key coordinator of Rad50 ATPase lobe rotation and its coupling to coiled-coil rearrangements.
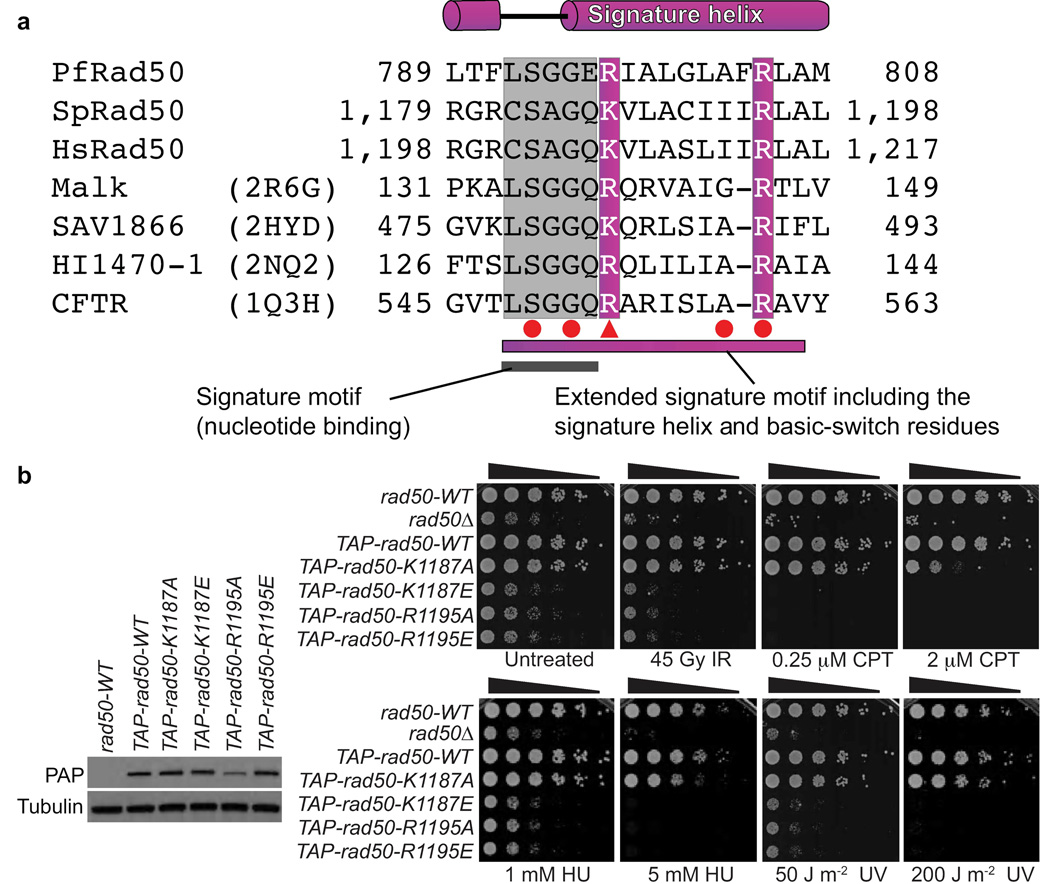
Rad50 basic-switches are critical for DSBR in fission yeast
Rad50 ortholog sequence alignments reveal the conservation of basic residues corresponding to pfRad50 Arg797 and Arg805 (Fig. 6a). To test the functional significance of these basic-switch residues for DSBR in vivo, we made K1187A, K1187E (pfRad50 Arg797 equivalent), R1195A and R1195E (pfRad50 Arg805 equivalent) mutations in S. pombe Rad50. The rad50 alleles replace genomic rad50 (rad50-WT) and encode a TAP-tag. These strains were compared to rad50Δ and TAP-tagged-rad50-WT control strains. This TAP-tag does not noticeably impair Rad50 function and immunoblotting showed that the Rad50 basic-switch mutants were expressed at wild type levels, with the exception of R1195A that has reduced expression (Fig. 6b).
Figure 6. ABC-ATPase superfamily conserved basic-switch residues in Rad50 coordinate DSBR in S. pombe.
(a) Rad50 sequence alignment with ABC transporters shows the extended signature motif, with well-conserved residues shaded. Red circles (point mutations) and triangles (truncations) denote cystic fibrosis causing CFTR mutations. (b) S. pombe Rad50 basic-switch variants on the signature helix are defective for DSBR. Left: Expression levels of TAP-tagged Rad50 variants, as probed by PAP antibody. Right: IR, CPT, HU and UV sensitivity of Rad50 basic-switch variants. Five-fold serial dilutions of cells on rich media plates were photographed following 2–3 days at 30°C. Strains used are detailed in Supplementary Table 1.
To test whether the Rad50 basic-switch mutants show increased DNA damage sensitivity, we examined their responses to genotoxins. Serial dilution assays show that Rad50 basic-switch mutants are more sensitive to the clastogen agents than the rad50-WT control strain (Fig. 6b). The K1187A variant phenotype is seen mainly with higher doses of clastogens. In contrast, the K1187E and both R1195A and R1195E variants are strikingly sensitive to clastogen agents and are as deleterious as rad50Δ. Thus, these assays reveal the importance of Rad50 signature helix basic-switch residues for DSBR in vivo.
DISCUSSION
Rad50 conformations, Mre11 interface, and Mre11–Rad50 functions
Long-range allostery in Mre11–Rad50 is implied by coupling Rad50 nucleotide states to differential impacts on Mre11 endonuclease and exonuclease activities1, Rad50 Zn-hook region mutations that disrupt binding to Mre11 over a distance of ~500 Å12, and DNA binding at the M2R2-head that straightens Rad50 coiled-coils3,23. This allostery has been enigmatic, but collective results presented here illuminate a chemo-mechanical conduit coupling Rad50 state to Mre11–DNA interactions. Combined crystal structures and SAXS solution results show the Mre11–Rad50 complex undergoes open to closed conformational changes upon ATP binding, appropriate to load Mre11–Rad50 onto DNA ends blocked by covalent adducts or Ku binding. Notably, the linkage between the Mre11 RBD and Rad50 coiled-coils is essential for DSBR in vivo, and nucleotide-binding driven Rad50 conformation change is suitable to mediate communication within and between MRN complexes.
The core Mre11 dimer adopts different conformations to symmetrically synapse DNA ends at two-ended breaks, or asymmetrically bind one-ended DSBs9. These conformational changes are linked to rotations of the nuclease capping domain, with this ratchet action controlling DNA access to the Mre11 active site. Our results reveal that the conserved Mre11 RBD flexibly extends from the Mre11 nuclease capping domain to interact with the Rad50 coiled-coil base. This connection suggests that rotation and flexure of the Mre11 capping domain in response to DNA binding is positioned to signal DNA occupancy state via the flexible linker to Rad50 through symmetry to asymmetry transitions in the Mre11 DNA binding cleft (Supplementary Fig. 5).
Similarly, ATP-driven movement in Rad50 could not only control Mre11–Rad50 loading onto DNA but also be transmitted via the Mre11 RBD interface to effect positioning of Mre11 nuclease capping domains, to regulate DNA access and nuclease activities at the active site. Nucleotide induced dimerization of Rad50 ABC-ATPase within the M2R2-head has key architectural consequences (Fig. 4,5; Supplementary Movies 1–3). 1) The M2R2-head loses degrees of freedom, moving from a conformationally flexible, open state to a closed, toroidal architecture. 2) Nucleotide binding fixes subdomain rotation within each Rad50 ABC-ATPase, resulting in Rad50 coiled-coil and attached Mre11 RBD repositioning, which pulls on the nuclease capping domain (Fig. 5d; Supplementary Fig. 5). Interestingly, Saccharomyces cerevisiae mutations resulting in rad50S phenotypes, which show persistent DNA damage signaling and defects in processing covalent protein–DNA adducts24, map primarily to the Rad50 N-lobe surface both distal from and, in the nucleotide-free state, flexible with respect to the Mre11 RBD (Fig. 1e). Our results show these sites undergo flexible to fixed conformational switching upon nucleotide binding, suggesting Rad50S mutations either affect this locked conformation or disrupt transmission of conformational changes to partners.
The Mre11–Rad50 mechanical linkage provides an elegant mechanism to couple Rad50 nucleotide states to Mre11 endonuclease and exonuclease activities, and helps explain results from the S. cerevisiae mre11-6 allele. mre11-6 lacks linkage between the Mre11 capping domain and RBD through the deletion of the RL6 DNA binding loop, yet retains the RBD and Rad50 binding24. Notably, mre11-6 impacts Mre11–Rad50 activities in vivo and in vitro as observed by deficiencies in meiotic DSB processing, moderate sensitivity to the DNA alkylating agent MMS, and decreases in DNA binding, 3′–5′ exonuclease and ssDNA endonuclease activities. Thus, the flexible Mre11 capping domain connection to the RBD appears critical for multiple Mre11–Rad50 catalytic and DNA binding functions. Significantly, this key Mre11–Rad50 linkage is evidently impaired in human cancers. Both a truncation between the MRE11 RBD-αI and RBD-αJ helices (Glu460) as well as a point mutation mapping to the RBD-αJ helix (L473F) in human MRE11 (Fig. 2a) were identified as somatic mutations in colorectal cancers7, underlining the importance of this evolutionarily critical Mre11–Rad50 interface in mediating MRN tumor suppressor functions. Collective MRN complex results reveal that Mre11 uses flexible linkers that can pull on the nuclease for both Rad50 and Nbs1 (ref.13). Thus, functional Mre11 nuclease interactions with diverse DNA damaged ends require a tethered but flexible linkage to its protein partners. The unexpected flexible attachment of Mre11 to Rad50 elucidated here arguably enables Mre11 interactions with diverse DNA substrates and protein partners required for effective orchestration of DNA end-processing and repair during HR, telomere maintenance, and non-homologous plus microhomology-mediated end-joining.
Implications for the ABC-ATPase superfamily
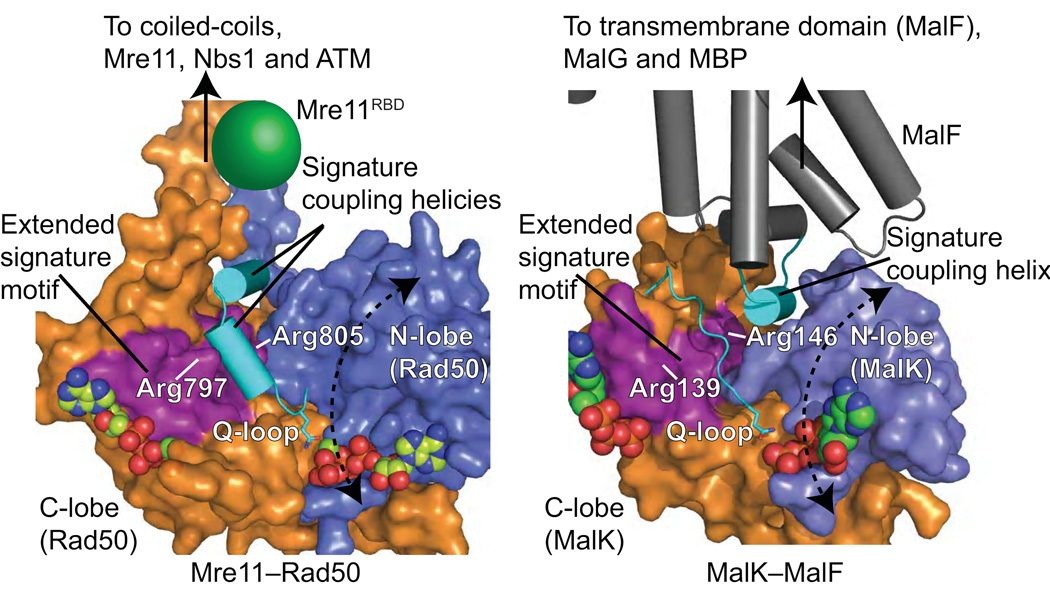
ABC-ATPases function in DNA repair and chromosome segregation through Rad50, MutS and SMC proteins, and in ABC transporters, where nucleotide binding regulates transmembrane domain opening and closing to control cellular import and export25. ABC-ATPase superfamily members are also associated with human disease and bacterial pathogenicity, and are thus of extreme biomedical interest26. For example, dysfunction of the cystic fibrosis transmembrane conductance regulator (CFTR) results in cystic fibrosis, P-glycoprotein ABC transporter acts in multidrug resistance of cancer cells, and inherited mutations in MRN or MutS result in cancer-predisposition diseases.
All ABC-ATPase machines have a strikingly similar heterotetrameric assembly, composed of attached ABC-ATPase and substrate- or function-specific dimers. Rad50 structures provided prototypical information on the ABC-ATPase superfamily by revealing that the signature motif acts in trans across the dimer11. However, an unanswered fundamental scientific question has been how ABC-ATPases communicate nucleotide binding and ligand states across long distances and among protein partners to effect diverse functions. We show here that nucleotide sensing by the signature motif in Rad50 is connected to the ABC-ATPase subdomain rotation through basic-switch residues encoded on the adjacent α-helix, which we call the signature helix. In ABC transporters similar movements occur between nucleotide states with the α-helical subdomain, equivalent to Rad50 C-lobe, rotating ~15° with respect to the RecA-like subdomain (Supplementary Fig. 6). The importance of the signature helix as a common denominator for ABC-ATPases is supported by its structural conservation in the ABC-ATPase superfamily and sequence conservation of basic-switch residues equivalent to pfRad50 Arg797 and Arg805. This is corroborated by mapping of causative cystic fibrosis mutations to this region in CFTR. The major cause of cystic fibrosis is deletion of CFTR Phe508, which maps to a region topologically equivalent to the Mre11–Rad50 interface and acts at the interface between CFTR ABC-ATPase and transmembrane domains. Yet, other disease-causing CFTR mutations map to the signature motif and signature helix, including two point mutations within the signature motif (Ser549 and Gly551), two more at the end of the signature helix (Ala559 and Arg560), and a truncation at Arg55327,28 (Fig. 6a). CFTR Arg553 and Arg560 are equivalent to pfRad50 Arg797 and Arg805 respectively.
Comparison of Rad50 structures with those of ABC transporters including maltose transporter MalK–MalF29, the multidrug exporter SAV186630 and a metal-chelate-type transporter31 also reveal critical conserved features of the ABC-ATPase domain important for propagating the subdomain rotation to conformational changes in the divergent functional domain. An α-helix from the transmembrane domain that inserts into the interface of the ATPase lobes is topologically equivalent to the Rad50 signature coupling helix-α2 (Fig. 7). In ABC transporters, this transmission helix is important for the concerted conformational changes that occur within the intact transporter between different states, undergoing a rotation and translation upon the rotation of the ABC-ATPase domain (also called the nucleotide binding domain)29. In contrast to ABC transporters, the Rad50 ABC-ATPase subdomain rotation is much larger, ~35° vs. ~15°. While this results in a similar rotation and translation of the transmission interface equivalent helix, its transmission to the attached functional domain, Mre11, is indirectly propagated through repositioning the coiled-coils.
Figure 7. Topologically equivalent signature helices connect nucleotide binding to conformational changes in the ABC-ATPase superfamily.
Rad50 ABC-ATPase molecular surface with attached coiled-coil and mapped Mre11 RBD compared to MalK ABC-ATPase with interacting MalF transmembrane protein. The extended signature helix (purple) and signature coupling helices or helix (cyan) connect nucleotide binding to movements of attached functional domains and proteins.
Direct observation and quantitation of Rad50 structures here show the importance of Rad50 basic switches for DSBR and their conservation within ABC transporters. Yet, divergence has likely resulted in adaptations to couple conformational changes to diverse functional domains. For example, in CFTR there is an essential role for signature helix Arg555, which falls between the basic switches identified here, in coordinating dimer interactions during ATP-driven conformational changes32. Also, mutation of CFTR Arg1303, present on the signature helix of the second ABC-ATPase domain but mapping past pfRad50 Arg805, results in misregulation of ATP-mediated allostery, promoting spontaneous, ATP-independent opening of CFTR33.
Collectively, our results redefine a structurally and functionally relevant extended ABC-ATPase superfamily signature motif (Fig. 6a). This novel loop-helix motif encodes the means for Rad50 and ABC transporters to utilize ATP-regulated inter-subunit domain rotation to modulate orientations of attached but structurally diverged functional domains through an analogous transmission interface (Fig. 7). Thus, Rad50 results now provide a prototypical molecular framework and unifying hypothesis for how ABC-ATPase superfamily proteins couple nucleotide binding in the ATPase core to attached functional domain movements to perform diverse biological roles. As many ABC transporters are antibiotic targets34,35 and as inhibition of DNA repair pathways, including molecular disruption of Rad50, can sensitize cancer cells to traditional treatments36–38, development of small molecules targeting these allosteric controls would be an exciting prospect. In fact, ligands targeting protein conformational states, such as those defined here, can activate as well as inhibit protein functions39. Parts of the Rad50 basic-switch sites are surface accessible, so their movements will provide pockets for water and ligand binding40. Thus, designing small-molecules targeting Rad50 arginine switches should be feasible, as high affinity inhibitors to the arginine binding site in nitric oxide synthase were designed that employ initial binding to push open larger binding pockets41. So the experimentally defined interface, ATP-dependent conformational changes, basic-switches, and signature coupling helical motifs characterized here for Rad50 provide an informed basis to broadly interrogate and control functions of ABC-ATPase superfamily members in cell biology.
METHODS
Protein expression and purification
Recombinant P. furiosus Mre11 constructs I–V (Fig. 1a and 1b) were co-expressed and co-purified with pfRad50-NC as previously described10,11. The His6-tagged Mre11RBD variants (constructs VI–VIII, Fig. 1a) were cloned with a thrombin-cleavable His6-tag and expressed from pET15b. Untagged intramolecularly linked Rad50 constructs were made by PCR to introduce a 'Gly-Gly-Ser-Gly-Gly' bridge between pfRad50 residues Lys177 and Thr726 (pfRad50-link2) or between residues Tyr187 and Ile716 (PfRad50-link1). His6-Mre11RBD constructs (VI–VIII) were co-expressed with either pfRad50-link1 or pfRad50-link2 in E. coli Rosetta2 (DE3) cells (Novagen) grown in Terrific Broth plus 0.4% (v/v) glycerol and induced with IPTG at 16°C overnight.
For co-expressed constructs, co-purification of thermostable pfMre11–Rad50 complexes was achieved by sequential Ni-affinity chromatography, heat denaturation of E. coli proteins at 65°C, Superdex 200 (GE Healthcare) gel filtration, and cation exchange chromatography. For crystallization, the N-terminal His6-tag of pfMre11RBD (construct VI, Mre11 residues 348–381) was removed by thrombin digestion. Untagged pfRad50-link2 was purified by heat denaturation of E. coli proteins at 65°C, Superdex 200 (GE Healthcare) gel filtration, and cation exchange chromatography. Proteins concentrated to 10 mg mL−1 in protein buffer 1 (50 mM Tris pH 7.5, 500 mM NaCl) were buffer exchanged into protein buffer 2 (20 mM Tris pH 7.5, 150 mM NaCl) prior to crystallization.
Crystallization
Crystals were grown by hanging-drop vapor diffusion. Crystals of the Rad50-link2–AMP:PNP–Mg2+ complex were grown by mixing 1 µl of 10 mg mL−1 Rad50-link2 in protein buffer 3 (20 mM Tris pH 7.5, 150 mM NaCl, 10 mM MgCl2, 2.5 mM AMP:PNP (Sigma)) with 1 µl of crystallization solution 1 (100 mM Tris pH 9.0, 100 mM NaCl, 16–18% (w/v) PEG 550 MME). Mre11RBD–Rad50-link1–AMP:PNP–Mg2+ complex crystals were grown by mixing 1 µl of protein at 10 mg mL−1 in protein buffer 3 with 1 µl of crystallization solution 2 (100 mM Tris pH 8.5, 200–300 mM LiSO4, 12–13% (w/v) PEG 3350). Nucleotide free Mre11RBD–Rad50-NC crystals were grown at 20°C in crystallization solution 3 (100 mM Tris pH 7.5, 200 mM NaCl, 2 M ammonium sulfate; crystal form 2) or at 4°C in crystallization solution 4 (100 mM Tris pH 7.5, 200 mM MgCl2, 20% (w/v) PEG 3350; crystal form 1). Crystal cryoprotection was done by rapid soaks in Paratone-N (Hampton Research) for all crystal forms except Mre11RBD–Rad50-link1–AMP:PNP–Mg2+ complex crystals which were gradually soaked into crystallization solution 2 supplemented with 26% (v/v) ethylene glycol prior to flash cooling in liquid nitrogen.
X-ray diffraction data collection, structure determination and processing
Data were collected and processed in HKL2000. Structures were solved by molecular replacement with PHASER and refined in REFMAC with rounds of manual rebuilding in O and COOT as detailed in Supplementary Methods. Refined models of Mre11RBD–Rad50 crystal form 1 (2.1 Å), Mre11–Rad50 crystal form 2 (3.4 Å), Mre11RBD–Rad50-link1–AMP:PNP–Mg2+ (3.3 Å) and Rad50-link2–AMP:PNP–Mg2+ (1.9 Å) all have good statistics and geometry (Table 1).
Protein pull-down assays
Constructs detailed in Fig. 1a and 1b were co-expressed as described above and, following lysis in 50 mM Tris pH 7.5, 500 mM NaCl, soluble extracts incubated with Ni-NTA beads (Qiagen). Bound protein was eluted with lysis buffer containing 300 mM imidazole after washing with 50 mM imidazole and analyzed by SDS PAGE.
Small angle X-ray scattering data collection and processing
M2R2-heads were purified following co-expression and purification of Rad50-NC and Mre11 (1–379) constructs (Fig. 1) and SAXS data collected and analyzed at the Advanced Light Source SIBYLS beamline (BL12.3.1) as described42. Briefly, data were collected at a wavelength of 1.0 Å and sample-to-detector distance of 1.5 m. Purified M2R2-head protein at 6 mg mL−1 was dialyzed into SAXS buffer (50 mM Tris 7.5, 150 mM NaCl). Protein was diluted 1:1 in SAXS buffer (−ATP data) or SAXS buffer with 2.5 mM ATP and 2.5 mM MgCl2 (for +ATP). Following heating at 55°C, designed to trap ATP-bound states, short (0.5 s) and long (2 s) SAXS exposures were collected at 20°C for protein and relevant buffer. Scattering profiles were generated by subtracting buffer from sample exposures followed by merging of short and long exposures in PRIMUS43 to generate SAXS data including the entire scattering spectrum. Guinier analysis (Supplementary Fig. 3) revealed the absence of aggregates. SAXS scattering data analysis using MD and MES was done using BILBOMD21 and FoxS software44, as described in Supplementary Methods.
Strain construction, survival assays and yeast two-hybrid analysis
Strain genotypes are listed in Supplementary Table 1. Growth media and methods for S. pombe were performed as described45. Spot assays were performed by plating 5-fold serial dilutions of exponentially growing cells onto rich media plates in the absence or presence of the indicated DNA damaging agents. Plates were incubated at 30°C and scanned after 2–3 days of growth. The IR survival was assayed by counting cells plated in triplicate onto rich medium after exposure to indicated IR doses. Normalization was to untreated samples. Yeast two-hybrid analysis was done as described with S. cerevisiae reporter strain AH109 (Clontech Matchmaker system)9,18.
Immunoblotting
Anti-Myc (9E10: Santa Cruz Biotechnology), Pstair and PAP (Sigma) antibodies were used for western blotting as described9,46.
Supplementary Material
ACKNOWLEDGMENTS
This MRN research is supported by National Cancer Institute grants CA117638 (J.A.T and P.R), CA92584 (J.A.T), CA77325 (P.R) and in part by the NIH Intramural Research program 1Z01ES102765-01 (R.S.W). Microbial complex efforts are supported by the ENIGMA Program of the Department of Energy, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 with Lawrence Berkeley National Laboratory (J.A.T). The SIBYLS beamline (BL12.3.1) at the Advanced Light Source is supported by United States Department of Energy program Integrated Diffraction Analysis Technologies DE-AC02-05CH11231 (J.A.T). We thank Greg Hura (Lawrence Berkeley National Laboratory) for expert SAXS data collection assistance.
Footnotes
Accession numbers
Coordinates and structure factors for Mre11RBD–Rad50 crystal form 1 (3QKS), Mre11–Rad50 crystal form 2 (3QKR), Mre11RBD–Rad50-link1–AMP:PNP–Mg2+ (3QKU) and Rad50-link2–AMP:PNP–Mg2+ (3QKT) have been deposited in the Protein Data Bank, with accession codes in parentheses,.
AUTHOR CONTRIBUTIONS
G.J.W. analyzed results, did SAXS experiments and wrote manuscript. J.S.W and O.L. did S. pombe experiments and analysis. G.M. and R.S.W. solved crystal structures. A.A. refined structures. S.S. and G.G purified proteins. M.H. assisted SAXS analyses. R.S.W., J.S.W., P.R. and J.A.T. designed research, analyzed results and helped write the manuscript.
REFERENCES
- 1.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50-Mre11 complex. J. Biol. Chem. 2001;276:35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- 3.Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 2010;9:1299–1306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 5.Carney JP, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 6.Waltes R, et al. Human RAD50 Deficiency in a Nijmegen Breakage Syndrome-like Disorder. Am. J. Hum. Genet. 2009;84:605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, et al. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res. 2004;64:2998–3001. doi: 10.1158/0008-5472.can-04-0587. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins BB, Paull TT. The P. furiosus Mre11/Rad50 Complex Promotes 5′ Strand Resection at a DNA Double-Strand Break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams RS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopfner KP, et al. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 11.Hopfner KP, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 12.Hopfner KP, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 13.Williams RS, et al. Nbs1 Flexibly Tethers Ctp1 and Mre11-Rad50 to Coordinate DNA Double-Strand Break Processing and Repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borgstahl GE, et al. The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell. 1992;71:107–118. doi: 10.1016/0092-8674(92)90270-m. [DOI] [PubMed] [Google Scholar]
- 15.Craig L, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell. 2003;11:1139–1150. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 16.Craig L, et al. Type IV Pilus Structure by Cryo-Electron Microscopy and Crystallography: Implications for Pilus Assembly and Functions. Mol. Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Limbo O, et al. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomita K, et al. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell Biol. 2003;23:5186–5197. doi: 10.1128/MCB.23.15.5186-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 21.Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen. Physiol. Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sokolova AV, Volkov VV, Svergun DI. Prototype of a database for rapid protein classification based on solution scattering data. J Appl. Crystallogr. 2003;36:865–868. [Google Scholar]
- 23.Moreno-Herrero F, et al. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 24.Usui T, Petrini JH, Morales M. Rad50S alleles of the Mre11 complex: questions answered and questions raised. Exp. Cell Res. 2006;312:2694–2699. doi: 10.1016/j.yexcr.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopfner KP, Tainer JA. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 27.Bobadilla JL, Macek M, Fine JP, Farrell PM. Cystic fibrosis: a worldwide analysis of CFTR mutations--correlation with incidence data and application to screening. Hum. Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 28.Lewis HA, et al. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450:515–521. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 30.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 31.Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC. An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science. 2007;315:373–377. doi: 10.1126/science.1133488. [DOI] [PubMed] [Google Scholar]
- 32.Vergani P, Lockless SW, Nairn AC, Gadsby DC. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature. 2005;433:876–880. doi: 10.1038/nature03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szollosi A, Vergani P, Csanády L. Involvement of F1296 and N1303 of CFTR in induced-fit conformational change in response to ATP binding at NBD2. J. Gen. Physiol. 2010;136:407–423. doi: 10.1085/jgp.201010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garmory HS, Titball RW. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect. Immun. 2004;72:6757–6763. doi: 10.1128/IAI.72.12.6757-6763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lou H, Dean M. Targeted therapy for cancer stem cells: the patched pathway and ABC transporters. Oncogene. 2007;26:1357–1360. doi: 10.1038/sj.onc.1210200. [DOI] [PubMed] [Google Scholar]
- 36.Abuzeid WM, et al. Molecular disruption of RAD50 sensitizes human tumor cells to cisplatin-based chemotherapy. J. Clin. Invest. 2009;119:1974–1985. doi: 10.1172/JCI33816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dupré A, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat. Chem. Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 39.Zorn JA, Wells JA. Turning enzymes ON with small molecules. Nat. Chem. Biol. 2010;6:179–188. doi: 10.1038/nchembio.318. [DOI] [PubMed] [Google Scholar]
- 40.Kuhn LA, et al. The interdependence of protein surface topography and bound water molecules revealed by surface accessibility and fractal density measures. J. Mol. Biol. 1992;228:13–22. doi: 10.1016/0022-2836(92)90487-5. [DOI] [PubMed] [Google Scholar]
- 41.Garcin ED, et al. Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nat. Chem. Biol. 2008;4:700–707. doi: 10.1038/nchembio.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat. Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. App. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 44.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 46.Werler PJH, Hartsuiker E, Carr AM. A simple Cre-loxP method for chromosomal N-terminal tagging of essential and non-essential Schizosaccharomyces pombe genes. Gene. 2003;304:133–141. doi: 10.1016/s0378-1119(03)00402-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.