Abstract
Metagenomics sequencing projects have dramatically increased our knowledge of the protein universe and provided over one-half of currently known protein sequences; they have also introduced a much broader phylogenetic diversity into the protein databases. The full analysis of metagenomic datasets is only beginning, but it has already led to the discovery of thousands of new protein families, likely representing novel functions specific to given environments. At the same time, a deeper analysis of such novel families, including experimental structure determination of some representatives, suggests that most of them represent distant homologs of already characterized protein families, and thus most of the protein diversity present in the new environments are due to functional divergence of the known protein families rather than the emergence of new ones.
Introduction
The number of possible protein sequences is astronomically large. A back-of-the-envelope calculation suggests that there are 20150 or the order of 10195 possible proteins with 150 amino acids length alone. Only a very small percentage of them exists, or ever existed, in nature. The size of the set of actual proteins, called protein hyperspace, or protein universe, is more difficult to estimate. One way to do it is to look at the number of existing species. For eukaryotes, about 2 million species have already been described; the total count is believed to be between 5 and 100 million. Given that an average eukaryotic genome has between 10- to 20-thousand genes, we can safely assume that the size of the eukaryotic protein universe exceeds 1012, not counting intra-species variations, such as SNPs or splicing variants. The number of prokaryotic species is even more difficult to estimate—not in the least because of the difficulties in defining species—but estimates between 107–109 have been discussed in literature 1, and lower numbers, between 35 and 350 thousand have been suggested using a broader definition of species 2. Given the few thousand genes in a typical prokaryote (or hundred of thousands in a pan-genome 3,4 of broadly defined species), these estimates suggest that the prokaryotic protein universe may be similar in size to the eukaryotic one. This count does not include viral proteins, which may be actually the largest contributors to the protein universe, nor account for the possibility that most of the diversity may reside in rare species that are not seen in current surveys 5.
There is a lot of interest in the structure and properties of the protein universe, as it provides us with a different perspective about the evolution of life on earth compared to a standard, species-based phylogeny. We know that proteins can be grouped in families; the number of currently classified protein families ranges from 12 6 to 14 thousand 7. However, currently existing libraries of protein families cover only about 60% of all known proteins, and they are likely to expand further even if no new proteins were ever sequenced. The next level of organization of the protein universe is provided by three-dimensional structures of proteins; at this level, the protein universe is clearly more coarsely grained, with about 1,300 protein folds identified today 8 and the estimates of the number of possible folds range from 5,000 to 20,000. The structural view of the protein universe is interesting because similarity between 3D structures may be interpreted as a distant evolutionary relationship between proteins and their families and the small number of folds as compared to the number of protein families may suggest that evolution played a major role in shaping the protein universe. At the same time, the number of possible protein folds is obviously limited by the constraints of compact packing in three-dimensional space, and thus it is possible that the same overall structures were discovered independently by convergent evolution.
In this review we specifically look at insights into the structure of protein universe brought about by the emergence of a new strategy in DNA sequencing; namely, metagenomics. Metagenomics is a term used to describe both a technique of sequencing of DNA purified directly from a natural environment and the research field focusing on studying microbial communities in their natural state 9,10. Metagenomics holds an enormous promise in many areas of science, from environmental processes to human health. Here we analyze one very specific aspect of metagenomics, its role in mapping the protein universe.
Our growing knowledge of the protein universe
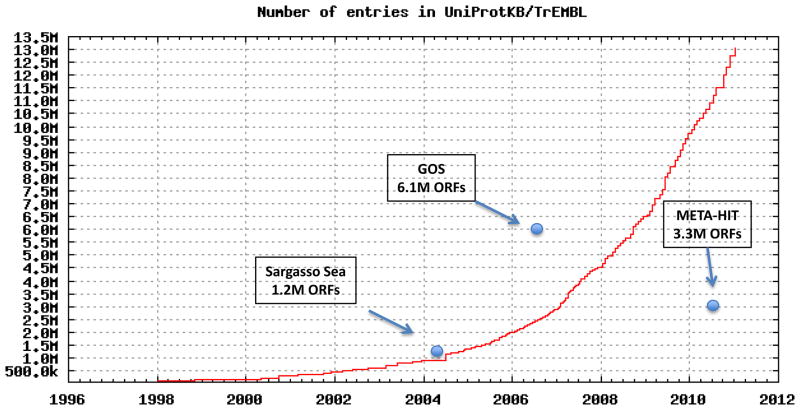
We know, mostly through prediction of open reading frames (ORFs) from genomic DNA, the sequences of millions of proteins. The latest count lists over 13-million proteins in automatically generated databases, such as TrEMBL 11 or NCBI NR 12. Based on the previous estimates, this represents only a small fraction of the entire protein universe; less than 1/100 of 1%. As DNA sequencing technology is breaking one technological barrier after another, and new protein sequencing techniques are possibly emerging 13, we can be sure this percentage will grow rapidly (see Box 1).
Box 1.
Growth of TrEMBL protein database 11 from its inception in 1996 and for comparison, timing and size of the largest metagenomics datasets. The largest single deposit to protein sequence databases, Global Ocean Survey dataset, quadrupled the then known protein universe 17*. An earlier Sargasso Sea set doubled it 16*, while the more recent set from human gut microbiome sequencing increased it by 25% 20**.
Still, in the foreseeable future, we would be obviously limited to sparse sampling, rather than covering, of the protein universe. All our current observations about internal structure of the protein universe—about the number and size distribution of protein families, number of folds, etc.—are based on this very limited sampling. The critical question about the validity of insights gained from the sampling depends on how representative is the selected sample.
In this context, it is interesting to note that until recently, only genes that were already a subject of direct experiments were ever sequenced. This introduced strong biases; the early datasets were depleted for example in genes coding for transmembrane and low-complexity proteins. Expansion of DNA sequencing technology to sequence entire genomes removed some of the biases and lead to the discovery of classes of novel, never-before-seen proteins. Complete sequences of over 1,000 prokaryotic and 100 eukaryotic genomes are now known but, still, biases exist. 80% of all sequenced bacterial genomes come from only three bacterial phyla 14; of the over 70 identified to date, over one-half do not have a single sequenced representative 15. Only 13 of the 36 known eukaryotic phyla have sequenced representatives. Such “phylogenetic bias”, reflecting our biased research interests, also translates into a bias in protein universe coverage.
Metagenomics enters the stage
Improvements in DNA sequencing technology in recent years not only have increased the speed of sequencing, but also have allowed a more fundamental breakthrough: metagenomics, or unbiased study of complete microbial communities. The impact of this approach for protein universe mapping was dramatically demonstrated with the first large scale metagenomic project of a microbiologically rich environment, where over 1.2 million protein coding ORFs were identified in samples from the Sargasso sea 16*. In a single project, the size of the then-known protein universe was doubled. Over 95% of proteins identified in this study proved to be unique, i.e., never seen before, and most of them came from species that had never been studied before. The next study, the Global Ocean Survey, in which the ocean metagenome was studied systematically in an around-the-world trip, tripled the size of the already larger protein databases with the release of 6.1 million predicted protein sequences identified during the voyage 17*.
This pattern continues as metagenomics is applied to new environments. Most notable in this respect were studies of the human gut environment. Following the first study of only two individuals 18* that identified sequences of about 50,000 new proteins, the next study 19 provided over 600,000 sequences; the latest in this series 20** over 3.3 million, again significantly increasing the protein universe coverage in a single experiment. It is important to note that these large metagenomic studies are part of large, multivel projects using both metagenomic and genomic approaches; mapping of the ocean (http://camera.calit2.net/microgenome/) and the human microbiome 21.
Of course metagenomics, while addressing some earlier biases, introduces new ones. The most important one is related to the procedures (or lack thereof) for the elimination of errors in sequencing. In genome sequencing, error correction is achieved by multiple coverage of the genome with repeated reads. Every genome fragment is sequenced many times, automatically correcting errors from individual reads. Metagenomic studies of diverse communities typically produces very few read overlaps, and most ORFs are predicted based on single reads. This leads to a large and difficult-to-estimate number of errors, from fragmentary ORFs to completely wrong gene calls (and hence, wrong ORFs) and biases – for instance by relative underrepresentation of long genes.
The problems with ORF definitions are one of the reasons that sets of ORFs derived in metagenomics projects are not integrated with other databases. NCBI and UniProt maintain separate (and very incomplete) databases of proteins from “environmental samples”. There are dedicated public resources devoted to data from the Global Ocean Sampling (CAMERA) 22 and the Human Microbiome Project Data Analysis and Coordination Center [DACC], http://www.hmpdacc.org/, and some data, especially on the smaller projects are available from sequencing centers (JGI, http://img.jgi.doe.gov/cgi-bin/m/main.cgi). Data from some of other projects, such as MetaHIT 20** can be downloaded in raw form. But overall, so far there is no single resource that provides uniform data and analyses across all metagenomics datasets. This unfortunate situation makes the analysis of these sets and especially the comparison between them quite difficult.
Another way to address the “phylogenetic bias” in protein sequence databases was proposed in the Genomic Encyclopedia of Bacteria and Archaea (GEBA) project, which specifically selects phylogenetically diverse bacteria for genome sequencing based on the tree-of-life coverage with completely sequenced genomes (http://www.jgi.doe.gov/programs/GEBA/index.html). Analysis of 56 genomes sequenced in the pilot phase of the project were reported recently 23**. Even though strictly a genomics project, GEBA, like metagenomics projects, specifically targets novel regions of protein sequence space and thus was included in this review.
What is there—or what have we learned from the metagenomics-driven expansion of the protein universe?
While driven by their specific scientific goals, projects such as the Global Ocean Survey, the Human Microbiome Project, MetaHIT or GEBA, all resulted in dramatic, one-step increases in our knowledge of the protein universe. Their contributions were remarkable not only by their size, but also by the fact that they addressed some of the biases of the previous sequencing strategies. There are many other metagenomics projects that collectively have also significantly increased our knowledge of the protein universe, but here we focus on these four, since their authors extensively analyzed their datasets for novelty and internal structure.
The follow-up study for the Global Ocean Survey (GOS) focused specifically on the estimates of the number of new protein families identified in this dataset 24**. Using massive computer resources, the set of all protein sequences identified at that time was clustered based on sequence similarity. Excluding singletons and small protein clusters, over 17,000 protein clusters were identified, with 4,000 of them consisting entirely of novel proteins identified in this study, and almost one-half (1,700) showing no recognizable sequence similarity to clusters containing non-GOS proteins. A vast majority (95%) of clusters are small (<20 proteins), and more than one-half of all predicted ORFs are singletons, illustrating both the diversity of the set and the likely problems with ORF calling..
The first human gut metagenomics projects mentioned a very high number of unannotated “hypothetical proteins: but didn’t analyze them in any detail 18. The next study 19 added over 600,000 ORFs identified in their study to a number of other metagenomics datasets and performed clustering to identify novel families in metagenomics sets; however, the list of these families was not made public. Data from the several human metagenomic studies were analyzed for the presence of novel families specific to this particular environment, with over 1,800 novel protein clusters identified in automated analysis and almost 200 new families curated by hand 25*. Families from this study are now being added to the PFAM database and will be a part of the next release. The MetaHIT study performed clustering of the over 3 million ORFs identified over 6000 novel gene families with more than 20 members 20**, but again the list of these families was not releases.
Between the three largest metagenomics projects, almost 10,000 novel protein families were reported. Unfortunately, none of the definitions of new families was released and at this point we don’t know how much these lists overlap. At the same time, all studies used automated clustering procedures. Groups of proteins indentified in such approaches, after more careful human curation can often be linked to previously known families and/or combined with each other, therefore the final count of new families is likely to be smaller. Effects of different thresholds in identifying can be best illustrated by the differences between the GEBA study 23** which didn’t use a size threshold in defining novel families and identified over 60,000 of them in set much smaller than any of the three metagenomics sets discussed here. All this makes it very difficult to describe the really novel part of protein universe identified in these studies. As analyses of these datasets progresses we may arrive at more precise numbers, but undoubtedly there are thousands on new protein families to be found. The question of possible relations between these families on the higher level of organization of the protein universe, that of superfamilies or fold groups is of course completely open and is awaiting more detailed studies, but preliminary results (see the next paragraph) support current observations of possible saturation or at least slowing down the pace of discovery of really new protein families 26* 27.
Characterizing (at least some of) the new families
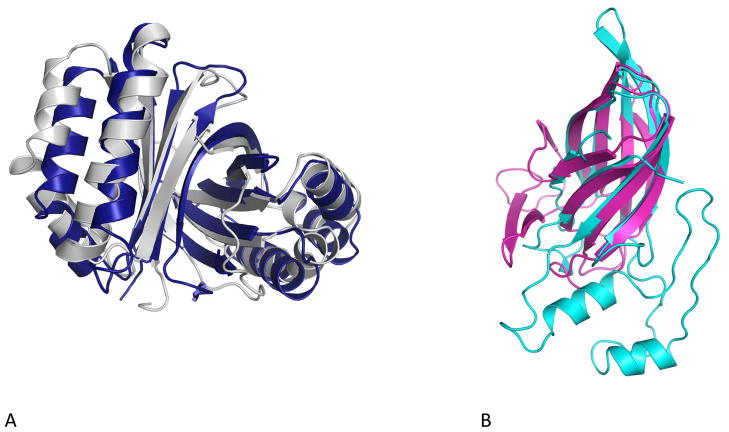
The challenge of characterizing new, completely uncharacterized groups of proteins prompted NIH Protein Structure Initiative centers, which were charged to use the newly developed techniques of high throughput protein structure determination to provide structural coverage of protein universe 28 to perform pilot studies targeting proteins from uncultured bacteria. As of today, structures of eight proteins from the Sargasso Sea, six from the GOS and three from the GEBA protect were solved (http://www.rcsb.org). Several of them are homologs of well studied enzymes, including deaminases 29 and Zn-dependent carboxypeptidases 30,31, and their comparison to their homologs from cultured bacteria clearly identified their similarities. Uncharacterized proteins from the GOS set were selected from novel families identified in the automated clustering study 32, but rather unexpectedly, all six were shown to be distant homologs of already characterized proteins. One of them, a first representative of a large (>600 homologs) protein family, was shown to be distantly related to Sm/LSm RNA-binding proteins 33*. Four others belong to a dimeric ferrodoxin superfamily, with the closest characterized homologs annotated as monooxygenases from the PFAM ABM (antibiotic biosynthesis monooxygenase) family 34. Interestingly, both cases represent extremely distant homologies with sequence identity on the level of 10–12% (see 33* and Box 2, Figure A). Structures of three proteins from the GEBA project all were shown to represent previously known protein families, despite very low sequence similarity (see Box 2, Figure B for an example). This pattern—recognizing apparently novel families as distant homologs of previously characterized ones only after experimental structure determination—may seem surprising, but in a recent analysis, it was shown to be fairly typical for proteins from other uncharacterized families: out of 250 examples, over 70% were shown to represent known folds and are most likely divergent variants of already characterized protein families 35.
Box 2.
Many proteins from the new environments are highly divergent members of known protein families, displaying high structural (and possibly functional) similarity despite highly divergent, sometimes beyond recognition, amino acid sequences. A. An uncharacterized protein identified in global ocean survey (PDB code:2pgc, gray) is highly similar (3 Å RMSD over 193 amino acids) to putative monooxygenase from Lactobacillus acidophilus (PDB code: 2f44) with sequence identity of the structural alignment of 11% seq id, i.e. close to the random level. Both structures were solved by the JCSG center as part of the coverageof the protein structural space project 37. B. An uncharacterized protein from Sulfurospirillum deleyianum, (PDB code 3nkg, cyan) part of the GEBA project shows strong structural similarity to carbohydrate binding module from Saccharophagus degradans (PDB code 2cdo, magenta) 38. The S. delevianum protein was solved by the MCSG center as part of the structural survey of novel organisms 28.
Conclusions
Metagenomics, and especially large projects, such as the Global Ocean Survey for the ocean and MetaHIT for the human microbiome, have opened new regions of protein space for direct analysis. The detailed analysis of these and other metagenomics datasets is still ongoing, but we can already say that, despite their novelty and diversity, the analyses have mostly confirmed our previous picture of the protein universe since the vast majority of proteins in the metagenomics sets are related, albeit sometimes very distantly, to previously identified protein families. Thousands of new protein families already have been identified, and thousands more undoubtedly will be, but detailed analyses generally show that even such novel families are built around already known structural themes. The protein universe appears to be shaped to a large extent by divergence, sometimes extreme, of large, but limited, number of protein families. This is especially visible at the level of protein three-dimensional structures, where previously known folds are increasingly found for apparently novel protein families. This doesn’t diminish the novelty, especially in terms of functions, of the proteins in these families; nor does it diminish the challenges in identifying these functions 36. Nature has built an incredible diversity of molecular functions by modifying and adapting existing solutions and while we may be coming close to identifying the core number of these, we can look forward to new insights and discoveries that will be found in the remaining 99.99% of the protein universe that we still haven’t mapped.
Acknowledgments
The author’s research is supported by NIH grants R01 GM087218 (FFAS) and U54 GM094586 (JCSG). Discussions with colleagues in JCSG and other PSI projects, and especially in the JCSG Bioinformatics Core are gratefully acknowledged. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci U S A. 2002;99:10494–9. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schloss PD, Handelsman J. Status of the microbial census. Microbiol Mol Biol Rev. 2004;68:686–91. doi: 10.1128/MMBR.68.4.686-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102:13950–5. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mira A, Martin-Cuadrado AB, D’Auria G, Rodriguez-Valera F. The bacterial pan-genome:a new paradigm in microbiology. Int Microbiol. 2010;13:45–57. doi: 10.2436/20.1501.01.110. [DOI] [PubMed] [Google Scholar]
- 5.Sogin ML, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A. 2006;103:12115–20. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–22. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter S, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–5. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreeva A, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–25. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–85. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Research Council (U.S.). Committee on Metagenomics: Challenges and Functional Applications. & National Academies Press (U.S.). . The new science of metagenomics : revealing the secrets of our microbial planet. National Academies Press; Washington, DC: 2007. p. xii.p. 158. [Google Scholar]
- 11.The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–8. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bandeira N, Clauser KR, Pevzner PA. Shotgun protein sequencing: assembly of peptide tandem mass spectra from mixtures of modified proteins. Mol Cell Proteomics. 2007;6:1123–34. doi: 10.1074/mcp.M700001-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Hugenholtz P, Kyrpides NC. A changing of the guard. Environ Microbiol. 2009;11:551–3. doi: 10.1111/j.1462-2920.2009.01888.x. [DOI] [PubMed] [Google Scholar]
- 15.Pace NR. Mapping the tree of life: progress and prospects. Microbiol Mol Biol Rev. 2009;73:565–76. doi: 10.1128/MMBR.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. the first large scale metagenomics project, which popularized the concept of metagenomics and introduced the first of its type, massive set of functionaly connected proteins. [DOI] [PubMed] [Google Scholar]
- 17*.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. the largest single metagenomics project to date, impressive by its goals and its scope. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. doi: 10.1126/science.1124234. first application of metagenomics to a most important environment for us all - the human body. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–81. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. concepts and techniques of metagenomics applied to the human distal gut, first study on a scale sufficiently large to identify saturation of human gut environment with protein families and to define a central, conserved gut proteome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson J, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–23. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun S, et al. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: the CAMERA resource. Nucleic Acids Res. 39:D546–51. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Wu D, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–60. doi: 10.1038/nature08656. a study presenting a different approach to battle the “phylogenetic bias”, systematic exploration of neglected corners of the tree of life. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. follow-up study of the largest metagenomic project ever, focusing on the internal structure of this set. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Ellrott K, Jaroszewski L, Li W, Wooley JC, Godzik A. Expansion of the protein repertoire in newly explored environments: human gut microbiome specific protein families. PLoS Comput Biol. 2010;6:e1000798. doi: 10.1371/journal.pcbi.1000798. study identifying protein families specific to a given environment (here, human gut). Also first comparison of automated vs. curated protein families and the only (so far) study that released the list of novel families. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Levitt M. Nature of the protein universe. Proc Natl Acad Sci U S A. 2009;106:11079–84. doi: 10.1073/pnas.0905029106. a broad analysis of the protein universe in terms of protein families, showing first signs of saturation in terms of pace of discovery of new families. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chubb D, Jefferys BR, Sternberg MJ, Kelley LA. Sequencing delivers diminishing returns for homology detection: implications for mapping the protein universe. Bioinformatics. 2010;26:2664–71. doi: 10.1093/bioinformatics/btq527. [DOI] [PubMed] [Google Scholar]
- 28.Joachimiak A. High-throughput crystallography for structural genomics. Curr Opin Struct Biol. 2009;19:573–84. doi: 10.1016/j.sbi.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall RS, et al. The hunt for 8-oxoguanine deaminase. J Am Chem Soc. 2010;132:1762–3. doi: 10.1021/ja909817d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang DF, et al. Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes. Biochemistry. 2009;48:3730–42. doi: 10.1021/bi900111q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang DF, et al. Functional annotation of two new carboxypeptidases from the amidohydrolase superfamily of enzymes. Biochemistry. 2009;48:4567–76. doi: 10.1021/bi900453u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Wooley JC, Godzik A. Probing metagenomics by rapid cluster analysis of very large datasets. PLoS One. 2008;3:e3375. doi: 10.1371/journal.pone.0003375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Das D, et al. Crystal structure of a novel Sm-like protein of putative cyanophage origin at 2.60 A resolution. Proteins. 2009;75:296–307. doi: 10.1002/prot.22360. discovery of a surprising connection between an apparently novel, ocean specific protein family and well known SM-like proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sciara G, et al. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 2003;22:205–15. doi: 10.1093/emboj/cdg031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaroszewski L, et al. Exploration of uncharted regions of the protein universe. PLoS Biol. 2009;7:e1000205. doi: 10.1371/journal.pbio.1000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dessailly BH, Redfern OC, Cuff AL, Orengo CA. Detailed analysis of function divergence in a large and diverse domain superfamily: toward a refined protocol of function classification. Structure. 2010;18:1522–35. doi: 10.1016/j.str.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elsliger MA, et al. The JCSG high-throughput structural biology pipeline. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1137–42. doi: 10.1107/S1744309110038212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henshaw J, et al. Family 6 carbohydrate binding modules in beta-agarases display exquisite selectivity for the non-reducing termini of agarose chains. J Biol Chem. 2006;281:17099–107. doi: 10.1074/jbc.M600702200. [DOI] [PubMed] [Google Scholar]