Abstract
Women with stage III/IV versus stage I/II endometriosis have lower implantation and pregnancy rates in natural and assisted reproduction cycles. To elucidate potential molecular mechanisms underlying these clinical observations, herein we investigated the transcriptome of eutopic endometrium across the menstrual cycle in the setting of severe versus mild endometriosis. Proliferative (PE), early secretory (ESE), and mid-secretory (MSE) endometrial tissues were obtained from 63 participants with endometriosis (19 mild and 44 severe). Purified RNA was subjected to microarray analysis using the Gene 1.0 ST Affymetrix platform. Data were analyzed with GeneSpring and Ingenuity Pathway Analysis and subsequently validated. Comparison of differentially regulated genes, analyzed by cycle phase, revealed dysregulation of progesterone and/or cyclic adenosine monophosphate (cAMP)-regulated genes and genes related to thyroid hormone action and metabolism. Also, members of the epidermal growth factor receptor (EGFR) signaling pathway were observed, with the greatest upregulation of EGFR in severe versus mild disease during the early secretory phase. The extracellular matrix proteoglycan versican (VCAN), which regulates cell proliferation and apoptosis, was the most highly expressed gene in severe versus mild disease. Upregulation of microRNA 21 (MIR21) and DICER1 transcripts suggests roles for microRNAs (miRNAs) in the pathogenesis of severe versus mild endometriosis, potentially through regulation of gene silencing and epigenetic mechanisms. These observed differences in transcriptomic signatures and signaling pathways may result in poorly programmed endometrium during the cycle, contributing to lower implantation and pregnancy rates in women with severe versus mild endometriosis.
Keywords: severe endometriosis, mild endometriosis, eutopic endometrium, microarray, transcriptome
Introduction
Endometriosis is a benign gynecologic disease characterized by endometrial-like tissue (epithelium and stroma) outside the uterus. It affects primarily women of reproductive age and presents with pelvic pain and infertility.1,2 Endometriosis is diagnosed mainly by visualization at surgery, and the revisedAmerican Society for Reproductive Medicine (ASRM) staging system recognizes minimal, mild, moderate, and severe (I-IV) stages of disease, based on the number and character of peritoneal lesions, ovarian and other organ involvement, and presence, type, and extent of adhesions.3,4 Although peritoneal, ovarian and rectovaginal endometriotic lesions are considered distinct entities with different pathogenesis,5–7 mild and severe stages of peritoneal endometriosis may also be distinct disorders, though the supporting data are limited. However, there is clinical evidence that embryonic implantation rates differ in women with severe versus mild endometriosis (see below), suggesting that the eutopic endometrium is different functionally and biochemically in these 2 types of endometriosis.
Women with moderate–severe endometriosis have more difficulty conceiving, compared to those with minimal–mild disease.8 Also, women with stage III/IV endometriosis have significantly lower implantation rates (13.7% vs 28.3%, respectively; P < .05) and pregnancy rates (22.6% vs 40.0%, respectively; P < .01) but not fertilization or miscarriage rates, compared to women with stage I/II endometriosis.9 A meta-analysis of 22 published studies on endometriosis and in vitro fertilization (IVF) outcomes showed that IVF pregnancy rates are significantly lower in women with severe versus mild endometriosis (13.84% vs 21.12%, respectively; P < .001),10 underscoring a potential endometrial origin of these differences. Also, participants with advanced disease demonstrate diminished ovarian response and higher cancellation rates in IVF cycles, but improved implantation, pregnancy, miscarriage, and delivery rates, after surgery, similar to those for women with tubal factor infertility,11 suggesting that removal of disease improves endometrial receptivity.
We have previously compared the transcriptome of eutopic endometrium from women with minimal/mild disease with the endometrium from women without disease during the window of implantation (mid-secretory endometrium [MSE])12 and also the endometrial transcriptome from women with moderate/severe disease compared with no disease in proliferative (PE), early secretory endometrium (ESE), and MSE.13 Based on these and other studies,14 endometrium from women with endometriosis appears to differ from that of disease-free women.12,13,15 Herein, we compared the transcriptome of eutopic endometrium from women with severe versus mild endometriosis at different times in the menstrual cycle, in an attempt to understand the differences and their potential roles contributing to the pathophysiology of infertility in women with endometriosis.
Materials and Methods
Study Participants
The study was approved by the Committee on Human Research of the University of California−San Francisco (UCSF) and the Stanford University Committee on the Use of Human Subjects in Medical Research. Samples were obtained from the National Institute of Health Specialized Cooperative Centers Program in Reproduction and Infertility Research (NIH SCCPRR) Human Endometrial Tissue and DNA Bank at UCSF. Endometrial tissue was obtained from 12 participants without endometriosis undergoing endometrial biopsy or hysterectomy for benign disorders not related to endometrial pathology (used in immunohistochemistry experiments) and from 63 participants with endometriosis undergoing endometrial biopsy for infertility evaluation or hysterectomy for treatment of severe pelvic pain and extensive endometriosis (Table 1 ). All participants were documented not to be pregnant and not to have had hormonal treatment for at least 3 months before surgery. Staging of endometriosis was performed according to the revised American Fertility Society classification system.3,4 Of the 63 participants, 19 had mild and 44 had severe endometriosis. The majority of endometriosis samples were obtained by endometrial biopsy, whereas the majority of control (no endometriosis) samples were obtained after hysterectomy (Table 1). (Note our previous studies demonstrated that sampling technique does not affect the endometrial transcriptome.16) The mean ages (years) of patients were: mild endometriosis group, 35.7 ± 1.4; severe endometriosis group, 35.2 ± 1.2l (P > .05). Menstrual cycle phase was determined based on histological evaluation of the tissue by 3 independent readers and according to the Noyes' criteria.17
Table 1.
Characteristics of Participants and Endometrial Tissue Samples in the Study
| Participant ID | Cycle Phase | Experiment | How Endometrium was Obtained | Age | Ethnicity |
|---|---|---|---|---|---|
| Whole tissue biopsy used for microarray analysis and validations | |||||
| Mild endometriosis | |||||
| 609 | PE | Microarray, IHC | Endometrial biopsy | 27 | Mixed |
| 621 | PE | Microarray, IHC | Hysterectomy | 37 | Caucasian |
| 658 | PE | Microarray, QPCR, IHC | Endometrial biopsy | 36 | Caucasian |
| 660 | PE | Microarray, QPCR | Endometrial biopsy | 46 | Caucasian |
| ST-007 | PE | Microarray, QPCR | Endometrial biopsy | 41 | Unknown |
| ST-012 | PE | Microarray, QPCR, | Endometrial biopsy | 41 | Unknown |
| ST-042 | PE | Microarray | Endometrial biopsy | 34 | Caucasian |
| ST-071 | PE | Microarray | Endometrial biopsy | 42 | Caucasian |
| ST-082 | PE | Microarray, IHC | Endometrial biopsy | 32 | Caucasian |
| ST-50 | PE | Microarray, QPCR | Endometrial biopsy | 39 | Caucasian |
| ST-080 | ESE | Microarray, QPCR, IHC | Endometrial biopsy | 43 | Unknown |
| ST-089 | ESE | Microarray, QPCR, IHC | Endometrial biopsy | 33 | Caucasian |
| ST-113 | ESE | Microarray, QPCR, IHC | Endometrial biopsy | 27 | Caucasian |
| 550 | MSE | Microarray | Endometrial biopsy | 38 | Mixed |
| ST-009 | MSE | Microarray, QPCR, IHC | Endometrial biopsy | 31 | Caucasian |
| ST-014 | MSE | Microarray, QPCR, IHC | Endometrial biopsy | 28 | Black |
| ST-033 | MSE | Microarray, QPCR | Endometrial biopsy | 42 | Caucasian |
| ST-038 | MSE | Microarray, QPCR | Endometrial biopsy | 36 | Caucasian |
| ST-121 | MSE | Microarray, QPCR, IHC | Endometrial biopsy | 25 | Caucasian |
| Severe endometriosis | |||||
| 26Aa | PE | Microarray | Endometrial biopsy | 31 | Caucasian |
| 508a | PE | Microarray, QPCR | Endometrial biopsy | 25 | Caucasian |
| 511 | PE | Microarray | Endometrial biopsy | 42 | Caucasian |
| 575a | PE | Microarray, IHC | Endometrial biopsy | 26 | Unknown |
| 587a | PE | Microarray, QPCR | Endometrial biopsy | 37 | Caucasian |
| 589 | PE | Microarray, IHC | Hysterectomy | 48 | Asian |
| 594a | PE | Microarray | Endometrial biopsy | 38 | Caucasian |
| 595 | PE | Microarray | Endometrial biopsy | 37 | Asian |
| 647a | PE | Microarray, QPCR | Endometrial biopsy | 39 | Caucasian |
| 651a | PE | Microarray, QPCR | Endometrial biopsy | 37 | Caucasian |
| ST-049 | PE | Microarray | Endometrial biopsy | 29 | Caucasian |
| ST-076 | PE | Microarray | Endometrial biopsy | 30 | Hispanic |
| ST-084 | PE | Microarray, QPCR, IHC | Endometrial biopsy | 37 | Caucasian |
| ST-090 | PE | Microarray, QPCR, IHC | Endometrial biopsy | 42 | Caucasian |
| ST-70 | PE | Microarray | Endometrial biopsy | 22 | Caucasian |
| 489a | ESE | Microarray, IHC | Hysterectomy | 39 | Asian |
| 496a | ESE | Microarray, QPCR, IHC | Endometrial biopsy | 37 | Caucasian |
| 517a | ESE | Microarray | Endometrial biopsy | 35 | Asian |
| 599a | ESE | Microarray, IHC | Endometrial biopsy | 35 | Black |
| 607 | ESE | Microarray, IHC | Endometrial biopsy | 24 | Asian |
| 684 | ESE | Microarray, QPCR, IHC | Hysterectomy | 36 | Caucasian |
| 27Aa | ESE | Microarray | Endometrial biopsy | 22 | Caucasian |
| ST-036 | ESE | Microarray | Endometrial biopsy | 45 | Caucasian |
| ST-065 | ESE | Microarray, QPCR | Endometrial biopsy | 34 | Asian |
| ST-112 | ESE | Microarray, QPCR, IHC | Endometrial biopsy | 38 | Caucasian |
| ST-127 | ESE | Microarray, IHC | Endometrial biopsy | 43 | Caucasian |
| ST-130 | ESE | Microarray, QPCR, IHC | Endometrial biopsy | 35 | Caucasian |
| 516a | MSE | Microarray | Endometrial biopsy | 34 | Asian |
| 526 | MSE | Microarray, QPCR, IHC | Hysterectomy | 48 | Unknown |
| 540a | MSE | Microarray | Endometrial biopsy | 37 | Caucasian |
| 543a | MSE | Microarray | Endometrial biopsy | 38 | Caucasian |
| 544 | MSE | Microarray, QPCR, IHC | Endometrial biopsy | 46 | Caucasian |
| 645a | MSE | Microarray | Endometrial biopsy | 39 | Asian Indian |
| 678a | MSE | Microarray | Hysterectomy | 44 | Asian |
| 33Aa | MSE | Microarray | Endometrial biopsy | 27 | Caucasian |
| 72Aa | MSE | Microarray | Endometrial biopsy | 31 | Caucasian |
| 73Aa | MSE | Microarray | Endometrial biopsy | 26 | Caucasian |
| 97Aa | MSE | Microarray | Endometrial biopsy | 35 | Unknown |
| ST-037 | MSE | Microarray, QPCR, IHC | Hysterectomy | 32 | Caucasian |
| ST-039 | MSE | Microarray, QPCR | Endometrial biopsy | 44 | Caucasian |
| ST-078 | MSE | Microarray | Endometrial biopsy | 32 | Caucasian |
| ST-091 | MSE | Microarray, QPCR | Endometrial biopsy | 20 | Caucasian |
| ST-096 | MSE | Microarray, QPCR, IHC | Endometrial biopsy | 31 | Caucasian |
| ST-119 | MSE | Microarray, IHC | Endometrial biopsy | 41 | Asian |
| No endometriosis | |||||
| 455b | PE | IHC | Hysterectomy | 39 | Caucasian |
| 469 | PE | IHC | Hysterectomy | 42 | Caucasian |
| 604 | PE | IHC | Hysterectomy | 44 | Caucasian |
| 693 | PE | IHC | Hysterectomy | 46 | Caucasian |
| UC-24 | ESE | IHC | Hysterectomy | 45 | Black |
| UC-26 | ESE | IHC | Hysterectomy | 34 | Caucasian |
| 629 | ESE | IHC | Hysterectomy | 46 | Caucasian |
| 680 | ESE | IHC | Hysterectomy | 34 | Caucasian |
| 463 | MSE | IHC | Hysterectomy | 48 | Caucasian |
| 501 | MSE | IHC | Hysterectomy | 49 | Caucasian |
| 610b | MSE | IHC | Hysterectomy | 50 | Caucasian |
| 626b | MSE | IHC | Hysterectomy | 42 | Caucasian |
Isolation of RNA and Preparation for Hybridization
Each endometrial tissue specimen was processed individually for microarray hybridization, as described earlier.13 Briefly, total RNA was extracted from whole-tissue specimens using the Trizol reagent (Invitrogen, Carlsbad, California), subjected to DNase treatment, and purified using the RNeasy Plus Kit (QIAGEN, Valencia, California). RNA purity was assessed by the A260/A280 ratio, and quality and integrity were assessed using the Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, California), with all samples having high-quality RNA (RNA Integrity Number (RIN) = 9.7-10).
RNA samples were prepared for microarray analysis according to the Affymetrix protocol (Affymetrix, Inc, Santa Clara, California), as described earlier.13,16 Briefly, for each sample, 5 µg of total RNA were reverse transcribed to complementary DNA (cDNA). Second strand DNA was generated using DNA polymerase, followed by overnight in vitro transcription to generate cRNA. After chemical fragmentation, biotinylated cRNAs were ready for hybridization. Quality of the final product was assessed in the Agilent Bioanalyzer. Each sample was hybridized to HU133 Plus 2.0 high-density oligonucleotide array (Affymetrix), with 54 600 genes and expressed sequence tags (ESTs), at the UCSF Genomic Core Facility. The data were scanned according to the protocol described in Assay Manual from Affymetrix.
Microarray Data Analysis
The .cel data files were imported into GeneSpring GX 10.0 software (Agilent Technologies) and processed using the robust multiarray analysis (RMA) algorithm for background adjustment, normalization, and log2-transformation of perfect match (PM) values.16 The data during each menstrual cycle phase (PE, ESE, and MSE were compared between the severe and mild endometriosis groups. The generated gene lists included only genes with >2.0-fold change (FC) and P < .05 by 1-way analysis of variance (ANOVA) with Tukey post hoc test and Benjamini-Hochberg multiple testing correction for false discovery rate.
Principal Component Analysis and Hierarchical Clustering
Principal component analysis (PCA) and hierarchical clustering were performed as described.15,16 Principal component analysis is an unbiased analysis performed in GeneSpring with all samples, using all 42 203 genes and 12 397 ESTs on Affymetrix Human HU133 Plus 2.0 arrays to look for similar expression patterns and underlying cluster structures. Hierarchical cluster analysis of differentially expressed genes from all samples was conducted using the smooth correlation distance measure algorithm (GeneSpring) to identify samples with similar patterns of gene expression. Compared to PCA, hierarchical clustering uses only informational genes—that is are differentially expressed among all experimental conditions.
Ingenuity Pathway Analysis
Gene symbols and FCs of the up- and downregulated genes in each pairwise comparison were imported into Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, California), as described earlier.15 For each comparison, associated top significantly regulated molecular and biological networks and canonical molecular pathways were identified. Only networks with the highest score were selected for the analysis. This was followed by functional analysis on the data set level and canonical pathway analysis. The significance of the association between the genes from the data set and the canonical pathway (in the IPA library) was presented as a ratio of the number of genes from the data set in a given pathway divided by the total number of molecules that make up the canonical pathway (Fisher exact test was used to calculate a P value). Pathways with P < .05 and ratio >0.05 were considered significant.
Microarray Validation by Real-Time Reverse Transcriptase−Polymerase Chain Reaction
Real-time reverse transcriptase−polymerase chain reaction (RT-PCR) was performed in duplicate using the SYBR Green PCR Mix (Fermentas Inc, Glen Burnie, Maryland), according to the manufacturer’s instructions. The housekeeping gene RPL19 was used as the normalizer. Numbers of mild endometriosis samples used for validation were n = 5, n = 3, and n = 5 for PE, ESE, and MSE, respectively, and in the severe endometriosis group, n = 6, n = 5, and n = 6 for PE, ESE, and MSE, respectively (Table 1 ). The following primer sequences were used: thyroxine deiodinase 2 (DIO2) sense 5′- TTGTACTTACTCTAAATTTCCCAAGG-3′ and antisense 5′-CATTGCCACTGTTGT CACCT-3′; insulin-like growth factor binding protein 5 (IGFBP5) sense 5′-TGCACCTGAGATGAGACAGG-3′ and antisense 5′-GCTTCATCCCGTACTTGTCC-3′; somatostatin (SST) sense 5′-CCCAGACTCCGTCAGTTTCT-3′ and antisense 5′-ATCATTCTCCGTCTGGTTGG-3′; transgelin (TAGLN) sense 5′-TTAGCTTTCCCCAGACATGG-3′ and antisense 5′-CGGTAGTGCCCATCATTCTT-3′; versican (VCAN) sense 5′-CCAGCCCCCTGTTGTAGAAA-3′ and antisense 5′’-ATTGAATTGTCCTTT GCTGATG-3′; solute carrier family 1, member 1 (SLC1A1) sense 5′-AACACTGCCTGTCACCTTCC-3′ and antisense 5′-GCACTCAGCACAATCACCAT-3′; epidermal growth factor receptor (EGFR) sense 5′-GAATGCATTTGCCAAGTCCT-3′ and antisense 5′-CGTCTATGCTGTCCTCAGTCA-3′; and RPL19 sense 5′-GCAGAT AATGGGAGGAGCC-3′ and antisense 5′- GCCCATCTTTGATGAGCTTC-3′. Polymerase chain reactions were run on the Mx4000 and Mx3005 quantitative real-time reverse transcriptase−polymerase chain reaction (QPCR) Stratagene systems (Agilent Technologies), using thermal cycling conditions, as described.15,18 Statistical analysis for the QRT-PCR results was performed using the nonparametric Mann-Whitney test. Significance was determined at P ≤ .05.
Immunohistochemistry
Immunostaining was performed for VCAN and EGFR using 4 µm thick paraffin-embedded endometrial tissue sections from women with mild and severe endometriosis: PE, n = 4 and n = 5, respectively; ESE, n = 3 and n = 7, respectively; and MSE, n = 3 and n = 5, respectively), as well as women without endometriosis (n = 4 in all phases). The samples were de-paraffinized in Xylene (Sigma-Aldrich, St Louis, Missouri) and rehydrated in decreasing concentrations of ethanol. All slides were incubated for 15 minutes in H2O2 (3% in methanol) to block endogenous peroxidase activity after antigen retrieval by boiling slides in citrate buffer (pH = 6.0). Thereafter the slides were blocked with normal horse serum for 45 minutes, followed by incubation with the primary antibody: overnight 4°C incubation with the rabbit polyclonal anti-VCAN antibody (Versican V0/V2 Neo, ThermoScientific, Waltham, Massachusetts) at 5 µg/mL concentration, and 1 hour at room temperature for the rabbit anti-human EGFR antibody (Santa Cruz Biotechnology, Inc, Santa Cruz, California, kind gift from Dr M Hsieh, UCSF) at 1:50 dilution.
In negative control slides, the primary antibody was replaced with nonimmune immunoglobulin G (IgG) of equivalent concentration from the same species. All slides were incubated with universal goat anti-rabbit/mouse secondary antibodies (Vector Laboratories Inc, Burlingame, California) for 30 minutes at room temperature. A freshly prepared diaminobenzidine-hydrogen peroxide solution (ImmPACT DAB kit, Vector Laboratories) was added to the slides, which were thereafter rinsed with distilled water. The slides were counterstained with haematoxylin (Vector Laboratories) and mounted with Clarion mounting medium (SigmaAldrich). A Leica microscope was used to visualize the immunostaining and to photograph the results. Sections of mouse ovarian and lung tissue (a kind gift from Dr Marco Conti, UCSF) were used as positive controls for VCAN immunostaining.19,20 Sections of 12-week human placental tissue served as a positive control for EGFR staining21; myometrium served as an internal positive control.22,23
Results
Cluster Analysis
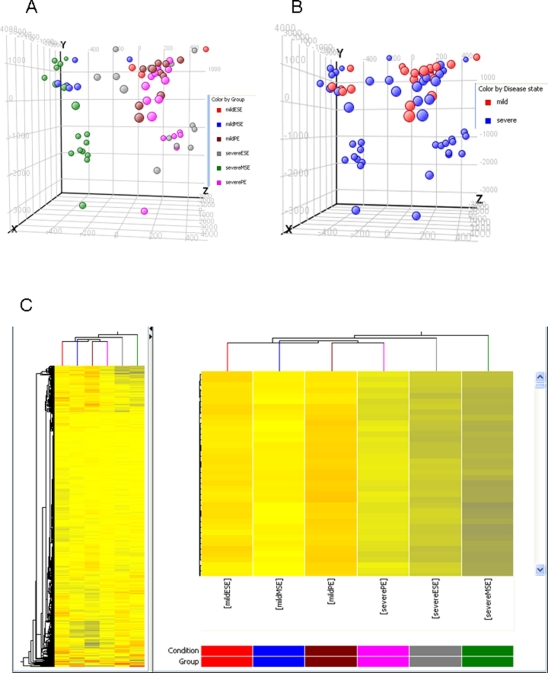
Principal component analysis of all genes showed that mild and severe endometriosis samples cluster according to their cycle phase rather than the disease stage (Figure 1A), confirming previous observations of phase-dependent segregation when analyzing endometrial tissue or isolated cells.13,15 However, PCA followed by subsequent analysis of disease stage demonstrated that severe endometriosis samples cluster separately from mild endometriosis, regardless of cycle phase, although there was some overlap (Figure 1B).
Figure 1.
Clustering analyses of samples from participants with mild and severe endometriosis. Panels A and B, Principal component analysis (PCA) of samples. A, Analyzed by menstrual cycle phase; and B, Analyzed by disease stage. PCA was applied to all endometrial samples that were characterized by the gene expression of all probes on the Affymetrix Gene 1.0 ST platform. C, Hierarchical clustering analysis of no-endometriosis and mild and severe endometriosis samples throughout the menstrual cycle, using the profiles of significantly regulated genes. PE indicates proliferative phase endometrium; ESE, early secretory phase endometrium; MSE, mid-secretory phase endometrium; m, mild endometriosis; s, severe endometriosis.
Unsupervised hierarchical clustering analysis was conducted using the profiles of significantly regulated genes in each study group (Figure 1C, clusterogram). Severe endometriosis samples clustered together and separately from mild endometriosis samples. Early secretory endometriosis from the mild endometriosis group clustered close to the mild PE group. Remarkably, even though clustering analysis of mild and severe endometriosis samples showed that they clustered separately from each other, signifying the difference between these 2 stages of endometriosis, PE as well as ESE and MSE samples from all groups demonstrated branching from the same stem, supporting the conclusion that cycle phase has greater impact than disease stage in sample clustering (Figure 1C).
Endometrial Transcriptome
Severe versus mild endometriosis
Comparison of severe versus mild endometriosis samples in the proliferative phase revealed 380 differentially regulated genes (P < .05, FC =2; Supplement Table 1), with 120 up- and 260 downregulated. Transcripts for several extracellular matrix (ECM) proteins and their receptors, such as VCAN, laminin-β1, fibrillin 1, and integrin-β1 (fibronectin receptor), were upregulated in severe endometriosis PE samples, as were heat shock proteins, DIO2 (the enzyme that converts thyroxine T4 to triiodothyronine T3), thioredoxin interacting protein (TXNIP), relaxin/insulin-like family peptide receptor 1, EGFR, microRNA 21 (MIR21), interferon-γ receptor 1, neuropilin, and others (Supplement Table 1).
Comparison of severe versus mild endometriosis samples in the early secretory phase revealed 817 differentially regulated genes (166 up- and 651 downregulated; Supplement Table 2 ). Although dysregulation of some genes persisted from the proliferative phase, some new genes were revealed, including upregulation of CYP26A1, IGF1, DICER1, DUSP1, KLF9, PAPPA, FOXO1A, neurotrophic tyrosine kinase receptor type 3, transducer of ERBB2 (TOB), and sulfatase 2 and downregulation of thyrotropin-releasing hormone (TRH), SST, lactotransferrin (LTF), TAGLN, Indian hedgehog homolog (IHH), BMP7, CXCL14, and others (Supplement Table 2). Some of the upregulated genes are progesterone and/or estradiol dependent, although some known progesterone-regulated genes (eg, IGFBP6, secretoglobin family 3A1, complement D, and glutathioine peroxidase 3 [GPX3]) were downregulated. These data suggest that the steroid hormone response and intracellular programs are disordered in both severe and mild forms of endometriosis in the early secretory phase.
Table 2.
The Most Represented Gene Ontology (GO) Categories in Severe Endometriosis
| GO Biological Process | GO Cellular Component | GO Molecular Function |
|---|---|---|
| PE vs mild endometriosis PE | ||
| Transcription | Nucleus | Nucleotide binding |
| Transport | Intracellular | Protein binding |
| Cell adhesion | Cytoplasm | DNA binding |
| Nuclear mRNA splicing, via spliceosome | Extracellular region | Receptor activity |
| Proteolysis | Membrane | Nucleic acid binding |
| Translation | Mitochondrion | Catalytic activity |
| Signal transduction | Plasma membrane | Actin binding |
| Negative regulation of transcription from RNA polymerase II promoter | Membrane fraction | Binding |
| Lipid metabolic process | Integral to plasma membrane | Structural constituent of ribosome |
| Protein folding | Golgi membrane | Signal transducer activity |
| Regulation of transcription, DNA-dependent | Endoplasmic reticulum | Structural molecule activity |
| Skeletal system development | Integral to membrane | Zinc ion binding |
| Mesoderm formation | Golgi apparatus | Magnesium ion binding |
| Cell cycle | Ruffle | Receptor binding |
| Protein amino acid phosphorylation | Cytosol | Insulin receptor binding |
| Ubiquitin-dependent protein catabolic process | Cytoskeleton | RNA binding |
| Carbohydrate metabolic process | Extracellular space | Transporter activity |
| Mitochondrial electron transport, NADH to ubiquinone | Filopodium | NADH dehydrogenase activity |
| Multicellular organismal development | Mitochondrial inner membrane | Hydrolase activity |
| RNA processing | Eukaryotic translation initiation factor 4F complex | Ion channel activity |
| mRNA processing | Nucleolus | Calcium ion binding |
| Organ morphogenesis | Cornified envelope | Metalloendopeptidase activity |
| Cell fate determination | Endosome | Transcription factor activity |
| Angiogenesis | Inner acrosomal membrane | Ubiquitin-protein ligase activity |
| DNA repair | Microfibril | Iron ion binding |
| ESE vs mild endometriosis ESE | ||
| Transport | Nucleus | Protein binding |
| Transcription | Extracellular region | Nucleotide binding |
| Cell adhesion | Cytoplasm | DNA binding |
| Proteolysis | Membrane | Nucleic acid binding |
| Cell cycle | Intracellular | Catalytic activity |
| Signal transduction | Mitochondrion | Receptor activity |
| Translation | Plasma membrane | Actin binding |
| Lipid metabolic process | Membrane fraction | Signal transducer activity |
| Skeletal system development | Endoplasmic reticulum | Structural molecule activity |
| Immune response | Golgi membrane | Transporter activity |
| Nuclear mRNA splicing, via spliceosome | Integral to plasma membrane | Calcium ion binding |
| Carbohydrate metabolic process | Lysosome | Cytokine activity |
| Apoptosis | Extracellular space | RNA binding |
| Ubiquitin-dependent protein catabolic process | Cytosol | Ion channel activity |
| Protein amino acid phosphorylation | Golgi apparatus | Magnesium ion binding |
| Metabolic process | Soluble fraction | Binding |
| Protein folding | Ubiquitin ligase complex | Structural constituent of ribosome |
| Protein modification process | Nucleosome | Zinc ion binding |
| Regulation of cell growth | Endosome | Receptor binding |
| rRNA processing | Chromatin | Hydrolase activity |
| tRNA processing | Ruffle | Endopeptidase inhibitor activity |
| Ossification | Mediator complex | Metalloendopeptidase activity |
| DNA repair | Cytoskeleton | Ubiquitin-protein ligase activity |
| Acute-phase response | Integral to membrane of membrane fraction | Phosphoprotein phosphatase activity |
| Regulation of transcription, DNA-dependent | Exosome (RNase complex) | Antigen binding |
| MSE vs mild endometriosis MSE | ||
| Transcription | Nucleus | Nucleotide binding |
| Transport | Cytoplasm | Protein binding |
| Signal transduction | Extracellular region | DNA binding |
| Cell adhesion | Intracellular | Nucleic acid binding |
| Translation | Membrane | Receptor activity |
| Nuclear mRNA splicing, via spliceosome | Mitochondrion | Catalytic activity |
| Proteolysis | Plasma membrane | Signal transducer activity |
| Lipid metabolic process | Endoplasmic reticulum | Binding |
| Protein amino acid phosphorylation | Membrane fraction | Actin binding |
| Ubiquitin-dependent protein catabolic process | Golgi membrane | Transporter activity |
| Negative regulation of transcription from RNA polymerase II promoter | Integral to plasma membrane | RNA binding |
| Cell cycle | Lysosome | Calcium ion binding |
| Multicellular organismal development | Golgi apparatus | Structural molecule activity |
| DNA repair | Cytosol | Magnesium ion binding |
| Angiogenesis | Soluble fraction | Structural constituent of ribosome |
| Immune response | Integral to membrane | Zinc ion binding |
| Skeletal system development | Ubiquitin ligase complex | Ion channel activity |
| Protein folding | Proteasome complex | Ubiquitin-protein ligase activity |
| Apoptosis | Nucleosome | Iron ion binding |
| Carbohydrate metabolic process | Ruffle | Endopeptidase inhibitor activity |
| Metabolic process | Endosome | Phosphoprotein phosphatase activity |
| Regulation of cell growth | Chromatin | Receptor binding |
| Protein modification process | Telomeric region | Iron ion transmembrane transporter activity |
| Regulation of transcription, DNA-dependent | Mitochondrion | Enzyme inhibitor activity |
| mRNA processing | Coated pit | hydrolase activity |
Abbreviations: PE, proliferative endometrium; ESE, early secretory endometrium; MSE, mid-secretory endometrium.
Comparison of severe versus mild endometriosis samples in the mid-secretory phase revealed 1286 differentially regulated genes (Supplement Table 3 ), with 377 and 909genes being up- and downregulated, respectively. These data are consistent with the hierarchical clusterogram and indicate that the greatest differences between severe and mild endometriosis occur in the window of implantation (Figure 1C). Interestingly, some progesterone-regulated genes such as DKK1, MAOA, MAOB, CXCL14, IL15, IL1R1, IDO1, and CD55 were upregulated in this comparison group, although other progesterone-regulated genes, for example, KLF-13, IGFBP6, and members of the Notch-signaling pathway were downregulated (Supplemental Table 3). These data are consistent with dysregulation in the response to progesterone in MSE in both forms of endometriosis, as observed in ESE.
Table 3.
Top Networks Regulated in Endometriuum from Severe Versus Mild Endometriosis
| Top 5 networks regulated in proliferative phase endometrium (PE) from severe endometriosis versus mild PE | ||||
| ID | Molecules in Network | Score | Focus Molecules | Top Functions |
| 1 | ADAMTS9, Caspase, CDKN2A, Cyclin A, DBP, E2f, EN2, ERCC1, FOXL2, GAS2L1, GNA11, GSPT1, GTF2H4, GTSE1, HNRNPA2B1, HNRNPH1, Ifn gamma, IFNGR1, IL32, KRT17, LIN37, MAFF, MAP3K7IP2, NFkB (complex), NLRP1, PYCARD, RBCK1, RRAS, RRM1 (includes EG:6240), SUZ12, TANK, TFDP1, TFIIH, UHMK1, WTAP | 52 | 29 | Cell Death, Hair and Skin Development and Function, Organ Development |
| 2 | ANXA1, BAX, C12ORF10, Calpain, CBLC, DDX3X, DNAJA1, EGFR, FANCA, Fibrinogen, GNRH, HSP, Hsp70, Hsp90, HSP90AA1, HSP90AB1, HSP90B1, HSPA8, HSPH1, IFN Beta, IL1, LAMB1, LARP1, LRRFIP1, MACF1, MYOF, PAFAH1B3, PCM1, PI3K, PLA2, Proteasome, REEP6, SFRP1, SSB, YWHAZ | 39 | 24 | Cellular Compromise, Post-Translational Modification, Protein Folding |
| 3 | 14-3-3, ADD3, Akt, ASAH1, BAD, CCNL1, Cdc2, CEL, CP, Ctbp, CTBP1, Cyclin E, DCN, DDX42, EXOSC4, FZD2, GADD45GIP1, HISTONE, HMG20B, HNRNPR, JDP2, MAP2K1/2, NFIC, p70 S6k, PDGF BB, PDPK1, PNN, PP2A, RBM5, RBMX, RPL13, SF3A2, SF3B1, SFRS1, SFRS7 | 39 | 25 | RNA Post-Transcriptional Modification, Lipid Metabolism, Small Molecule Biochemistry |
| 4 | ATP2B4, C1QTNF2, CTTN, DIO2, EPHA2, ERK, FAK, FBN1, Fgf, Fgfr, FGFR3, FOSL2, GDI2, IL27RA, ITGB1, JAK, JAK1, KLF13, LMO4, NRTN, NUFIP1, Pak, Pdgf, Pdgfr, PI3K p85, PLC gamma, PSMD7, PTPN11, RAB1A, Raf, RLIM, Shc, STAT, STAT5a/b, VCAN | 32 | 21 | Cellular Development, Skeletal and Muscular System Development and Function, Cellular Movement |
| 5 |
ARAP1, BRD4, Calmodulin, Ck2, CRIM1, CSDC2, GGA1, Histone h3, Ikb, IKK (complex), Insulin, MATR3, MIR21 (includes EG:406991), MRPL12, MSX2, MTUS1, MYCN, NOP2, NUDT1, PIK3R1, PSPC1, PTEN, RBM14, RNA polymerase II, RPL13, SFPQ, SLC39A4, SMC4, SORD, UBE2G2, UBE2I, Ubiquitin, XIST, ZNF146, ZNF451 |
27 |
19 |
Cell Cycle, Embryonic Development, Cancer |
| Top 5 networks regulated in early secretory phase endometrium (ESE) from severe endometriosis versus mild ESE | ||||
| ID | Molecules in Network | Score | Focus Molecules | Top Functions |
| 1 | ADAMTS9, AP1S1, BCR, BEX2, BGN, C1q, C1QA, C1QB, C1S, CARD10, Complement component 1, CXCL16, ENPP1, FMOD, G0S2, IgG, IGKC, IGL@, Igm, IL32, KRT7, KRT13, KRT17, NFkB (complex), NFKBIZ, PIGR, PTPLAD1, RBCK1, SERINC3, SERPING1, SLC16A1, SLC3A1, SLC7A1, STAP2, TNFRSF18 | 42 | 29 | Skeletal and Muscular System Development and Function, Amino Acid Metabolism, Dermatological Diseases |
| 2 | ALP, ALPP, ASS1, CCNO, CFD, DDX3X, DIO2, DKK1, DNMT3A, Fgf, FGF18, FOXL2, FOXS1, Frizzled, FRZB, FXYD5, FZD2, FZD8, FZD10, GAS1, HES1, MAFF, MIB2, MSX2, MUC4, NEDD4L, P38 MAPK, PDGF BB, PORCN, SLC30A5, SMAD6, SOX4, TOB1, Wnt, WNT4 | 42 | 29 | Cell Development, Connective Tissue Development and Function, Skeletal and Muscular System Development |
| 3 | ADD3, AGTRAP, ATP1A2, BMP7, C1QTNF2, CCL5, CTSG, ERK, Fibrin, FXYD4, Growth hormone, Igf, Igfbp, IGFBP3, IGFBP5, IGFBP6, IL27RA, LCN2, Mmp, MMP7, Na+,K+ -ATPase, NADPH oxidase, NAMPT, NRTN, POSTN, PRPF4, RARRES2, SERPINA1, SERPINA3, Smad2/3-Smad4, STRA13, Tgf beta, TIMP1, VCAN, VitaminD3-VDR-RXR | 30 | 24 | Cellular Movement, Cancer, Gastrointestinal Disease |
| 4 | ABP1, Adaptor protein 2, ADRB2, ALDOA, Angiotensin II receptor type 1, Beta Arrestin, C12ORF10, CAV1, Clathrin, CMTM8, Creatine Kinase, DAB2, Dynamin, EGFR, FBP1, GFER, HDL, HSP90AB1, HSPA8, MACF1, MAT2A, Mek, MIR21 (includes EG:406991), NCK, NFIB, NUMA1, PLTP, PTPRS (includes EG:5802), SNX9 (includes EG:51429), Sos, SPDEF, TRAK1, TRAK2, TRH, Vegf | 30 | 24 | Cellular Assembly and Organization, Cardiac Hypertrophy, Cardiovascular Disease |
| 5 |
ADCYAP1R1, AGR2, BLVRB, Cbp/p300, CYP26A1, DHRS13, FJX1, FOXO1, hCG, Histone h3, Histone h4, HMOX1, HOXB8, KLF6, LBH, LSR, MAF, MGMT, MTUS1, Nfat (family), NPTX2, NPTXR, Oxidoreductase, PDGFA, Pkc(s), PURA, Rxr, SFRP4, SP3, TAGLN, TBL1X, Thyroid hormone receptor, TNFRSF4, TSPO, XIST |
30 |
26 |
Developmental Disorder, Genetic Disorder, Neurological Disease |
| Top 5 networks regulated in mid-secretory phase endometrium (MSE) from severe endometriosis versus mild MSE | ||||
| ID | Molecules in Network | Score | Focus Molecules | Top Functions |
| 1 | AHNAK, ARID5B, B3GNT1, BGN, CAND2, CHI3L1, COL16A1, COL6A2, EHD2, ELN, FOXS1, FXYD6, H1FX, HNRPDL, KCNG1, KRT7, MFAP2, MPHOSPH9, MYOF, NPTX2, NPTXR, PDLIM4, PDZK1IP1, PKIG, PLOD2, PMEPA1, RAB25, RAMP2, RBPMS, SCG5, SPAG4, STARD10, TGFB1, WFS1, ZNF581 | 51 | 35 | Cancer, Cell-To-Cell Signaling and Interaction, Skeletal and Muscular Disorders |
| 2 | ANXA11, APBA3, APEX2, AQP1, BIK, C12ORF10, CBLC, CCT2, DOCK5, EFEMP2, EGFR, FAM107A, GAS5, GPC1, HNRNPH1, HSP90AB1, HSPH1, LDOC1, LRRFIP1, MACF1, MBNL1, MPG, PAFAH1B3, PGK1, PHLDA1, PSMD7, PTPRS (includes EG:5802), RAB1A, RABAC1, SCAMP1, SFRP1, TRAK2, TUBB2A, WWP1, ZNF638 | 51 | 35 | Cardiovascular System Development and Function, Cellular Development, Cell Growth and Proliferation |
| 3 | BCLAF1, C19ORF43, CAMK2N1, CCNL2, CHD1 (includes EG:1105), CLK1, DDX42, DEAF1, ERAL1, ERK, GNRH, HMG20B, HNRNPA2B1, HNRNPL, IL27RA, IL6ST, KLF13, KLHL22, LMO4, LOXL1, PRPF4, RBM5, RBMX, RLIM, SEMA3F, SF3A2, SF3B1, SFRS1, SFRS5, SFRS7, SFRS11, SFRS2IP, TRA2A, UBXN1, WSX1-gp130 | 42 | 32 | RNA Post-Transcriptional Modification, Cancer, Cellular Growth and Proliferation |
| 4 | 3 BETA HSD, AKR7A2, CCNL1, CNN1, CRIM1, CSRP2, DDX5, DUSP1, GLIS2, HELZ, HOXB8, HSD3B7, MAT2A, MEIS1, MIR21 (includes EG:406991), NAMPT, NBL1, NKIRAS2, PDGF BB, RNA polymerase II, RPL13, SERPINA3, SLC1A1, SP2, TAGLN, TEAD2, TEAD4, TIA1, TNNC1, TOB1, UNC5B, XAB2, XDH, YAP1, ZNF83 | 42 | 32 | Cellular Assembly and Organization, Drug Metabolism, Genetic Disorder |
| 5 | ABCG1, ADAMTS9, AEBP1, ATF5, CARD10, CLIP2, CRISP3 (includes EG:10321), CXCL16, DBP, GFER, HDL, HES1, HEY2, LCN2, NCOA7, NFkB (complex), NFKBIZ, Notch, NOTCH2, NOTCH3, PLTP, PRDX2, PYCARD, RBCK1, RIOK3, RTKN, RTN4R, S100P, Secretase gamma, SLC3A1, SLC7A1, TAX1BP3, TCEA2, WTAP, ZMYND11 | 40 | 31 | Amino Acid Metabolism, Molecular Transport, Small Molecule Biochemistry |
Gene Ontology Categories in Severe Versus Mild Endometriosis Throughout the Menstrual Cycle
The most common gene ontology (GO) biological process groups in all comparisons were transcription, transport, cell adhesion, nuclear messenger RNA (mRNA) splicing, proteolysis, translation, and cell cycle, with angiogenesis and apoptosis processes having significant representation in the secretory (ESE and MSE) phase (Table 2). The main cellular components involved were nucleus, cytoplasm, extracellular and intracellular regions, and membranes (Table 2), demonstrating the ubiquitous participation of all cellular components in molecular differences between severe and mild endometriosis. The main GO molecular function categories included nucleotide binding, protein and DNA binding, receptor activity, actin binding, signal transducer activity, and others.
Microarray Validation by Real-Time RT-PCR
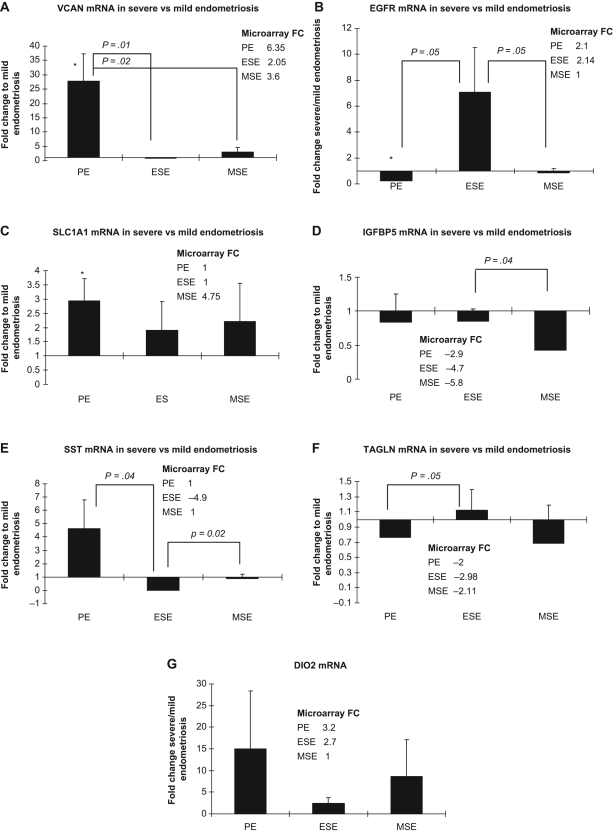
Some of the highly up- or downregulated genes, as well as genes dysregulated in all cycle phases between severe and mild endometriosis groups were selected randomly for validation using real-time RT-PCR: DIO2, IGFBP5), VCAN, SLC1A1, SST, TAGLN, and EGFR (Figure 2 ; Supplemental Tables 1-3). Most of the validated genes (VCAN, IGFBP5, SST, DIO2) follow the trend differences of the microarray results.
Figure 2.
Quantitative real-time reverse transcriptase−polymerase chain reaction (QPCR) validation of microarray data. Panels A, VCAN; B, EGFR; C, SLC1A1; D, IGFBP5; E, SST; F, TAGLN; G, DIO2 gene expression in severe endometriosis expressed as fold change compared to the expression in mild endometriosis, throughout the menstrual cycle. Microarray data are presented in the insert. *Statistically significant differences (P < .05) between the same cycle phases in severe vs mild endometriosis determined by the (Mann-Whitney test). Error bars represent ± SEM. PE indicates proliferative phase endometrium; ESE, early secretory phase endometrium; MSE, mid-secretory phase endometrium; SEM, standard error of the mean; VCAN, versican; EGFR, epidermal growth factor receptor; SLC1A1, solute carrier family 1, member 1; IGFBP5, insulin-like growth factor binding protein 5; SST, somatostatin; TAGLN, transgelin; DIO2, thyroxine deiodinase 2.
Analysis of Networks and Canonical Pathways Regulated in Severe Versus Mild Endometriosis
Ingenuity pathway analysis of gene expression profiles revealed several associated network functions identified as different between severe versus mild endometriosis in proliferative, early secretory, and mid-secretory phase samples (Table 3). As expected, several genes are involved in more than 1 network/pathway. Comparison of canonical pathways regulated in severe versus mild endometriosis revealed large differences in eutopic endometrium in these 2 disease stages, as shown by the high number of regulated genes in the pathways. The major canonical pathways regulated are presented in Table 4 .
Table 4.
Top 50 Canonical Pathways
| Ingenuity Canonical Pathways | −log(P Value) | P Value | Ratio | Molecules |
|---|---|---|---|---|
| Pathways regulated in proliferative endometrium (PE) from severe endometriosis vs mild PE | ||||
| Neuregulin signaling | 6.25E00 | .000001 | 0.11 | ITGB1, BAD, PTPN11, HSP90AB1, RRAS, PIK3R1, DCN, HSP90AA1, PDPK1, PTEN, EGFR |
| Prostate cancer signaling | 4.83E00 | .000015 | 0.09 | BAD, TFDP1, PIK3C2A, HSP90AB1, RRAS, PIK3R1, HSP90AA1, PDPK1, PTEN |
| Non-small cell lung cancer signaling | 4.64E00 | .000023 | 0.10 | CDKN2A, BAD, TFDP1, PIK3C2A, RRAS, PIK3R1, PDPK1, EGFR |
| PI3K/AKT signaling | 4.35E00 | .000045 | 0.07 | ITGB1, JAK1, BAD, HSP90AB1, RRAS, PIK3R1, YWHAZ, HSP90AA1, PDPK1, PTEN |
| Chronic myeloid leukemia signaling | 4.27E00 | .000054 | 0.09 | CDKN2A, CTBP1, BAD, TFDP1, PTPN11, PIK3C2A, RRAS, PIK3R1, CBLC |
| Myc-mediated apoptosis signaling | 4.17E00 | .000068 | 0.12 | CDKN2A, BAD, PIK3C2A, RRAS, PIK3R1, YWHAZ, BAX |
| Melanoma signaling | 3.98E00 | .000105 | 0.13 | CDKN2A, BAD, PIK3C2A, RRAS, PIK3R1, PTEN |
| IGF-1 signaling | 3.69E00 | .000204 | 0.08 | BAD, PTPN11, PIK3C2A, RRAS, PIK3R1, YWHAZ, PDPK1, IGFBP5 |
| P70S6K signaling | 3.51E00 | .000309 | 0.07 | GNAI2, JAK1, BAD, PIK3C2A, RRAS, PIK3R1, YWHAZ, PDPK1, EGFR |
| Endometrial cancer signaling | 3.45E00 | .000355 | 0.11 | BAD, PIK3C2A, RRAS, PIK3R1, PDPK1, PTEN |
| Docosahexaenoic acid (DHA) signaling | 3.39E00 | .000407 | 0.11 | BAD, PIK3C2A, PIK3R1, PDPK1, BAX |
| FAK signaling | 3.1E00 | .000794 | 0.07 | ITGB1, PIK3C2A, RRAS, PIK3R1, PDPK1, PTEN, EGFR |
| PTEN signaling | 2.89E00 | .001288 | 0.07 | ITGB1, BAD, RRAS, PIK3R1, PDPK1, PTEN, EGFR |
| FLT3 signaling in hematopoietic progenitor cells | 2.83E00 | .001479 | 0.08 | BAD, PTPN11, PIK3C2A, RRAS, PIK3R1, PDPK1 |
| Glioma signaling | 2.75E00 | .001778 | 0.06 | CDKN2A, TFDP1, PIK3C2A, RRAS, PIK3R1, PTEN, EGFR |
| CNTF signaling | 2.75E00 | .001778 | 0.09 | JAK1, PTPN11, PIK3C2A, RRAS, PIK3R1 |
| Insulin receptor signaling | 2.68E00 | .002089 | 0.06 | JAK1, BAD, PTPN11, PIK3C2A, RRAS, PIK3R1, PDPK1, PTEN |
| Pancreatic adenocarcinoma signaling | 2.6E00 | .002512 | 0.06 | CDKN2A, JAK1, BAD, TFDP1, PIK3C2A, PIK3R1, EGFR |
| IL-2 signaling | 2.56E00 | .002754 | 0.09 | JAK1, PTPN11, PIK3C2A, RRAS, PIK3R1 |
| Molecular mechanisms of cancer | 2.49E00 | .003236 | 0.04 | CDKN2A, JAK1, PIK3C2A, BAD, TFDP1, RRAS, PIK3R1, MAP3K7IP2, GNA11, BAX, APH1A (includes EG:51107), GNAI2, PTPN11, FZD2 |
| FcγRIIB signaling in B lymphocytes | 2.31E00 | .004898 | 0.07 | PIK3C2A, RRAS, PIK3R1, PDPK1 |
| JAK/Stat signaling | 2.3E00 | .005012 | 0.08 | JAK1, PTPN11, PIK3C2A, RRAS, PIK3R1 |
| p53 signaling | 2.29E00 | .005129 | 0.07 | CDKN2A, SCO2 (includes EG:9997), PIK3C2A, PIK3R1, BAX, PTEN |
| Glucocorticoid receptor signaling | 2.28E00 | .005248 | 0.04 | HSPA8, JAK1, PIK3C2A, HSP90AB1, RRAS, GTF2H4, PIK3R1, ANXA1, HSP90AA1, CCL5, UBE2I |
| Angiopoietin signaling | 2.27E00 | .005370 | 0.07 | BAD, PTPN11, PIK3C2A, RRAS, PIK3R1 |
| ERK5 signaling | 2.24E00 | .005754 | 0.07 | BAD, PTPN11, RRAS, YWHAZ, EGFR |
| Neurotrophin/TRK signaling | 2.18E00 | .006607 | 0.06 | PTPN11, PIK3C2A, RRAS, PIK3R1, PDPK1 |
| IL-3 signaling | 2.04E00 | .009120 | 0.07 | JAK1, BAD, PIK3C2A, RRAS, PIK3R1 |
| Agrin interactions at neuromuscular junction | 2.04E00 | .009120 | 0.07 | ITGB1, RRAS, LAMB1, CTTN, EGFR |
| EGF signaling | 2.01E00 | .009772 | 0.08 | JAK1, PIK3C2A, PIK3R1, EGFR |
| Aryl hydrocarbon receptor signaling | 1.89E00 | .012882 | 0.04 | CDKN2A, NFIC, TFDP1, HSP90AB1, HSP90AA1, GSTT2, BAX |
| 14-3-3-mediated signaling | 1.86E00 | .013804 | 0.05 | BAD, PIK3C2A, RRAS, PIK3R1, YWHAZ, BAX |
| Aldosterone signaling in epithelial cells | 1.83E00 | .014791 | 0.05 | HSPA8, PIK3C2A, PIK3R1, HSP90AA1, PDPK1 |
| EIF2 signaling | 1.79E00 | .016218 | 0.05 | PIK3C2A, RRAS, PIK3R1, EIF4G3, PDPK1 |
| Thrombopoietin signaling | 1.77E00 | .016982 | 0.06 | PTPN11, PIK3C2A, RRAS, PIK3R1 |
| Mitotic roles of polo-like kinase | 1.77E00 | .016982 | 0.06 | ANAPC4, HSP90AB1, HSP90AA1, CDC16 |
| IL-9 signaling | 1.73E00 | .018621 | 0.08 | JAK1, PIK3C2A, PIK3R1 |
| Bladder cancer signaling | 1.69E00 | .020417 | 0.05 | CDKN2A, FGFR3, TFDP1, RRAS, EGFR |
| Colorectal cancer metastasis signaling | 1.68E00 | .020893 | 0.04 | JAK1, BAD, PIK3C2A, RRAS, PIK3R1, IFNGR1, BAX, FZD2, EGFR |
| G beta gamma signaling | 1.63E00 | .023442 | 0.04 | GNAI2, RRAS, GNA11, PDPK1, EGFR |
| IL-15 signaling | 1.6E00 | .025119 | 0.06 | JAK1, PIK3C2A, RRAS, PIK3R1 |
| Hypoxia signaling in the Cardiovascular system | 1.58E00 | .026303 | 0.06 | HSP90AB1, HSP90AA1, PTEN, UBE2I |
| GM-CSF signaling | 1.58E00 | .026303 | 0.06 | PTPN11, PIK3C2A, RRAS, PIK3R1 |
| IL-4 signaling | 1.53E00 | .029512 | 0.06 | JAK1, PIK3C2A, RRAS, PIK3R1 |
| Macropinocytosis signaling | 1.53E00 | .029512 | 0.06 | ITGB1, PIK3C2A, RRAS, PIK3R1 |
| Erythropoietin signaling | 1.51E00 | .030903 | 0.05 | PIK3C2A, RRAS, PIK3R1, PDPK1 |
| Fc Epsilon RI signaling | 1.51E00 | .030903 | 0.05 | PTPN11, PIK3C2A, RRAS, PIK3R1, PDPK1 |
| Huntington disease signaling | 1.48E00 | .033113 | 0.03 | HSPA8, PIK3C2A, PIK3R1, GNA11, STX16, PDPK1, BAX, EGFR |
| HGF signaling | 1.47E00 | .033884 | 0.05 | CDKN2A, PTPN11, PIK3C2A, RRAS, PIK3R1 |
| iCOS-iCOSL signaling in T Helper Cells | 1.46E00 | .034674 | 0.04 | BAD, PIK3C2A, PIK3R1, PDPK1, PTEN |
| Pathways regulated in early secretory endometrium (ESE) from severe endometriosis vs mild ESE | ||||
| Complement system | 3.54E00 | .00029 | 0.19 | CFD, SERPING1, C1S, CD55, C1QA, CD46, C1QB |
| Hepatic fibrosis/hepatic stellate cell activation | 2.92E00 | .00120 | 0.10 | VCAM1, FN1, PDGFA, IFNGR1, IGFBP5, BAX, CCL5, PGF, MYL9 (includes EG:10398), IGF1, TIMP1, IGFBP3, EGFR |
| IL-8 signaling | 2.72E00 | .00191 | 0.08 | NAPEPLD, VCAM1, PIK3C2A, RRAS, PIK3R1, BAX, IRAK1, PGF, EIF4EBP1, ROCK2, HMOX1, CCND2, RHOD, RHOF, EGFR |
| Clathrin-mediated endocytosis signaling | 2.71E00 | .00195 | 0.08 | ACTR2, PIK3C2A, PDGFA, PIK3R1, ITGB8, PGF, HSPA8, SNX9 (includes EG:51429), IGF1, FGF18, RAB5C, DAB2, CTTN, ITGB5 |
| Docosahexaenoic acid (DHA) signaling | 2.67E00 | .00214 | 0.13 | BAD, FOXO1, PIK3C2A, PIK3R1, BIK, BAX |
| IGF-1 signaling | 2.65E00 | .00224 | 0.10 | IGFBP6, IGF1, BAD, FOXO1, PIK3C2A, RRAS, PIK3R1, IGFBP3, IGFBP5, SFN |
| Prostate cancer signaling | 2.41E00 | .00389 | 0.09 | BAD, TFDP1, FOXO1, PIK3C2A, HSP90AB1, RRAS, SRD5A1, PIK3R1, CREB3L4 |
| Myc-mediated apoptosis signaling | 2.23E00 | .00589 | 0.12 | IGF1, BAD, PIK3C2A, RRAS, PIK3R1, BAX, SFN |
| NRF2-mediated oxidative stress response | 2.1E00 | .00794 | 0.08 | AKR7A2, MGST1, PIK3C2A, RRAS, PIK3R1, MAF, CLPP, MAFF, HMOX1, DNAJC4, GSTT2, GSTO2, FKBP5, PTPLAD1 |
| Ubiquinone biosynthesis | 2.07E00 | .00851 | 0.06 | NDUFB11, NDUFS8, NDUFS7, MGMT, NDUFB7, NDUFA3, ALDH6A1 |
| IL-4 signaling | 1.92E00 | .01202 | 0.10 | HLA-DQB1, SOCS1, HLA-DMA, JAK1, PIK3C2A, RRAS, PIK3R1 |
| VEGF signaling | 1.81E00 | .01549 | 0.08 | ROCK2, BAD, FOXO1, PIK3C2A, RRAS, PIK3R1, SFN, PGF |
| Pancreatic adenocarcinoma signaling | 1.77E00 | .01698 | 0.08 | HMOX1, NAPEPLD, JAK1, BAD, TFDP1, PIK3C2A, PIK3R1, PGF, EGFR |
| Virus entry via endocytic pathways | 1.75E00 | .01778 | 0.08 | PIK3C2A, RRAS, PIK3R1, CD55, CAV1, ITGB8, ITGB5, FOLR1 |
| Caveolar-mediated endocytosis signaling | 1.72E00 | .01905 | 0.09 | CD55, RAB5C, CAV1, COPE, ITGB8, ITGB5, EGFR |
| Axonal guidance signaling | 1.65E00 | .02239 | 0.06 | ACTR2, FZD10, PIK3C2A, UNC5A, RRAS, PDGFA, PIK3R1, GNA11, EFNA4, PGF, MYL9 (includes EG:10398), ROCK2, FZD8, IGF1, GLIS2, RHOD, NTRK3, WNT4, BMP7, EFNB3, RTN4R, FZD2, GLIS1 |
| Role of NANOG in mammalian embryonic stem cell pluripotency | 1.62E00 | .02399 | 0.09 | FZD8, FZD10, JAK1, PIK3C2A, RRAS, PIK3R1, WNT4, BMP7, TCF7L1, FZD2 |
| Fructose and mannose metabolism | 1.56E00 | .02754 | 0.03 | AKR7A2, GMDS, GALK1, FBP1, ALDOA |
| Colorectal cancer metastasis signaling | 1.55E00 | .02818 | 0.06 | FZD10, MMP7, JAK1, PIK3C2A, BAD, RRAS, PIK3R1, IFNGR1, BAX, PGF, FZD8, RHOD, WNT4, RHOF, FZD2, EGFR |
| Interferon signaling | 1.55E00 | .02818 | 0.13 | SOCS1, JAK1, IFITM1, IFNGR1 |
| Glucocorticoid receptor signaling | 1.52E00 | .03020 | 0.06 | VCAM1, JAK1, PIK3C2A, RRAS, PIK3R1, CCL5, SLPI, HSPA8, HSP90AB1, DUSP1, GTF2H4, CDKN1C, FKBP5, POLR2I, UBE2I, ADRB2 |
| VDR/RXR activation | 1.52E00 | .03020 | 0.09 | IGFBP6, FOXO1, PDGFA, IGFBP3, IGFBP5, HES1, CCL5 |
| ILK signaling | 1.51E00 | .03090 | 0.06 | MYL9 (includes EG:10398), PARVB, FN1, PIK3C2A, RHOD, PIK3R1, CREB3L4, ITGB8, RHOF, PPP1R14B, ITGB5, PGF |
| Basal cell carcinoma signaling | 1.47E00 | .03388 | 0.10 | FZD8, FZD10, GLIS2, WNT4, BMP7, FZD2, GLIS1 |
| PI3K/AKT signaling | 1.46E00 | .03467 | 0.07 | JAK1, BAD, FOXO1, HSP90AB1, RRAS, PIK3R1, SFN, EIF4EBP1, THEM4 |
| Human embryonic stem cell pluripotency | 1.44E00 | .03631 | 0.07 | FZD8, FZD10, PIK3C2A, PDGFA, NTRK3, PIK3R1, SMAD6, WNT4, BMP7, FZD2 |
| Wnt/β-catenin signaling | 1.44E00 | .03631 | 0.07 | SOX4, FZD8, SFRP4, FZD10, MMP7, FRZB, CDH3, WNT4, TCF7L1, PIN1, DKK1, FZD2 |
| Macropinocytosis signaling | 1.42E00 | .03802 | 0.08 | PIK3C2A, RRAS, PDGFA, PIK3R1, ITGB8, ITGB5 |
| Non-small cell lung cancer signaling | 1.39E00 | .04074 | 0.08 | BAD, TFDP1, PIK3C2A, RRAS, PIK3R1, EGFR |
| Riboflavin metabolism | 1.39E00 | .04074 | 0.05 | ACP5, ENPP3, ENPP1 |
| FLT3 signaling in hematopoietic progenitor cells | 1.31E00 | .04898 | 0.08 | BAD, PIK3C2A, RRAS, PIK3R1, CREB3L4, EIF4EBP1 |
| Glutathione metabolism | 1.29E00 | .05129 | 0.05 | GPX3, MGST1, GPX1, GSTT2, GSTO2 |
| Selenoamino acid metabolism | 1.28E00 | .05248 | 0.04 | MGMT, PAPSS1 (includes EG:9061), MAT2A |
| p53 signaling | 1.27E00 | .05370 | 0.08 | SCO2 (includes EG:9997), CCND2, PIK3C2A, PIK3R1, RPRM, BAX, SFN |
| IL-2 signaling | 1.26E00 | .05495 | 0.09 | SOCS1, JAK1, PIK3C2A, RRAS, PIK3R1 |
| IL-3 signaling | 1.26E00 | .05495 | 0.08 | JAK1, BAD, FOXO1, PIK3C2A, RRAS, PIK3R1 |
| Amyotrophic lateral sclerosis signaling | 1.25E00 | .05623 | 0.06 | IGF1, PIK3C2A, PIK3R1, RAB5C, GPX1, BAX, PGF |
| 14-3-3-mediated signaling | 1.24E00 | .05754 | 0.07 | BAD, FOXO1, PIK3C2A, RRAS, PIK3R1, BAX, SFN, PLCL1 |
| PDGF signaling | 1.23E00 | .05888 | 0.08 | JAK1, PIK3C2A, RRAS, PDGFA, PIK3R1, CAV1 |
| Acute phase response signaling | 1.22E00 | .06026 | 0.06 | HMOX1, SOCS1, SERPING1, FN1, RBP7, RRAS, C1S, PIK3R1, SERPINA3, SERPINA1, IRAK1 |
| Integrin signaling | 1.21E00 | .06166 | 0.06 | ACTR2, PARVB, PIK3C2A, RHOD, RRAS, PIK3R1, CAV1, ITGB8, RHOF, TSPAN4, ITGB5, RAP2A |
| Nitric oxide signaling in the cardiovascular system | 1.21E00 | .06166 | 0.07 | PIK3C2A, HSP90AB1, PIK3R1, CAV1, SLC7A1, PGF |
| Molecular mechanisms of cancer | 1.21E00 | .06166 | 0.05 | FZD10, JAK1, PIK3C2A, BAD, TFDP1, RRAS, PIK3R1, GNA11, SMAD6, BAX, APH1A (includes EG:51107), FZD8, CCND2, FOXO1, RHOD, IHH, BMP7, RHOF, FZD2, RAP2A |
| HMGB1 signaling | 1.19E00 | .06457 | 0.07 | VCAM1, PIK3C2A, RHOD, RRAS, PIK3R1, IFNGR1, RHOF |
| Nicotinate and nicotinamide metabolism | 1.17E00 | .06761 | 0.05 | ENPP3, PRKX, ENPP1, QPRT, NAMPT, HIPK1, IRAK1 |
| Leukocyte extravasation signaling | 1.14E00 | .07244 | 0.06 | ROCK2, CLDN10, MMP7, VCAM1, PIK3C2A, ICAM3, TIMP1, PIK3R1, RDX, MLLT4, CTTN |
| Glioma signaling | 1.11E00 | .07762 | 0.06 | IGF1, TFDP1, PIK3C2A, RRAS, PDGFA, PIK3R1, EGFR |
| Methionine metabolism | 1.1E00 | .07943 | 0.04 | DNMT3A, IL4I1, MAT2A |
| mTOR signaling | 1.1E00 | .07943 | 0.06 | HMOX1, NAPEPLD, PIK3C2A, RHOD, RRAS, PIK3R1, RHOF, PGF, EIF4EBP1 |
| Melanoma signaling | 1.1E00 | .07943 | 0.09 | BAD, PIK3C2A, RRAS, PIK3R1 |
| Pathways regulated in mid-secretory endometrium (MSE) from severe endometriosis vs mild MSE | ||||
| Neuregulin signaling | 3.13E00 | .00074 | 0.14 | ITGB1, BAD, PIK3R1, DCN, PDPK1, PTEN, ERBB2IP (includes EG:55914), PRKCI, HSP90AB1, PTPN11, CDK5, HSP90AA1, PSEN1, EGFR |
| Acute phase response signaling | 2.63E00 | .00234 | 0.11 | IL6ST, SOCS1, MAP2K7, C1S, PIK3R1, PDPK1, CP, SERPINA3, MAP3K5 (includes EG:4217), IL1R1, TCF3, IRAK1, HMOX1, SOD2, RIPK1, PTPN11, C4BPA, MAP3K7, CRABP2, SERPINA1 |
| Docosahexaenoic acid (DHA) SIGNALING | 2.37E00 | .00427 | 0.16 | BAD, FOXO1, PIK3C2A, PIK3R1, BIK, PDPK1, BAX |
| Wnt/β-catenin signaling | 2.34E00 | .00457 | 0.12 | SOX4, SOX7, FZD10, FRAT1, TCF7L1, TCF3, SOX17, CSNK1E, FZD8, TGFB1, MAP3K7, DKK3, CD44, PIN1, SOX18, SFRP1, DKK1, FZD2, FZD7 |
| Germ cell-sertoli cell junction signaling | 2.14E00 | .00724 | 0.11 | EPN3, ITGB1, MAP2K7, PIK3C2A, CDC42, TJP1, PIK3R1, TUBG1, TUBB2A, PDPK1, MAP3K5 (includes EG:4217), IQGAP1, GSN, RHOQ, TGFB1, MAP3K7, PPAP2B, ACTN1 |
| Clathrin-mediated endocytosis signaling | 2.11E00 | .00776 | 0.10 | ITGB1, ACTR2, PIK3C2A, RAB5A, CDC42, PDGFA, PIK3R1, CLTB, HSPA8, MET, ARRB1, FGF18, RAB5C, DAB2, TFRC, CTTN, ITGB5 |
| Prostate cancer signaling | 2.09E00 | .00813 | 0.12 | BAD, TFDP1, FOXO1, PIK3C2A, HSP90AB1, SRD5A1, PIK3R1, HSP90AA1, PDPK1, CREB3L4, PTEN |
| Mitochondrial dysfunction | 1.94E00 | .01148 | 0.09 | NDUFS7, XDH, NDUFA13, APH1A (includes EG:51107), MAOB, NDUFB11, SOD2, NDUFS8, TXN2, NDUFB7, NDUFA3, CYC1, UQCRC1, PSEN1, MAOA |
| Virus entry via endocytic pathways | 1.89E00 | .01288 | 0.12 | ITGB1, PRKCI, CDC42, PIK3C2A, PIK3R1, CLTB, CD55, CAV1, TFRC, ITGB5, FOLR1 |
| Macropinocytosis signaling | 1.86E00 | .01380 | 0.13 | ITGB1, MET, PRKCI, RAB5A, CDC42, PIK3C2A, PDGFA, PIK3R1, ITGB5 |
| Leukocyte extravasation signaling | 1.83E00 | .01479 | 0.09 | ITGB1, PIK3C2A, CDC42, PIK3R1, RDX, THY1, ROCK2, GNAI2, CLDN23, PRKCI, PTPN11, ICAM3, EZR, CD44, MMP11, CTTN, ACTN1, TIMP2 |
| VDR/RXR activation | 1.78E00 | .01660 | 0.13 | IGFBP6, SPP1, PRKCI, FOXO1, PDGFA, IGFBP5, NCOR2, HES1, CCL5, KLF4 |
| Notch signaling | 1.77E00 | .01698 | 0.16 | RFNG, NOTCH2, NOTCH3, HEY2, HES1, APH1A (includes EG:51107), PSEN1 |
| NRF2-mediated oxidative stress response | 1.5E00 | .03162 | 0.09 | AKR7A2, MAP2K7, PIK3C2A, PIK3R1, SLC35A2, DNAJC13, DNAJC3, MAP3K5 (includes EG:4217), DNAJA1, CLPP, MAFF, MAFG, HMOX1, PRKCI, SOD2, DNAJC4, MAP3K7 |
| Huntington disease signaling | 1.43E00 | .03715 | 0.08 | MAP2K7, CAPN6, PIK3C2A, PIK3R1, GNA11, HSPA9, PDPK1, CREB3L4, BAX, GNG7, HDAC5, HSPA8, PRKCI, GNG11, CDK5, STX16, NCOR2, POLR2I, EGFR |
| Hepatic fibrosis/hepatic stellate cell activation | 1.42E00 | .03802 | 0.10 | PDGFA, FGFR1, FGFR2, IGFBP5, IL1R1, BAX, CCL5, MET, COL1A2, TGFB1, COL3A1, TIMP2, EGFR |
| Insulin receptor signaling | 1.42E00 | .03802 | 0.09 | JAK1, BAD, PIK3C2A, PIK3R1, PDPK1, VAMP2, PTEN, EIF4EBP1, PRKCI, RHOQ, FOXO1, PTPN11, PPP1R12A |
| IGF-1 signaling | 1.42E00 | .03802 | 0.10 | IGFBP6, PRKCI, BAD, FOXO1, PTPN11, PIK3C2A, PIK3R1, YWHAZ, PDPK1, IGFBP5 |
| Neurotrophin/TRK signaling | 1.39E00 | .04074 | 0.10 | MAP2K7, CDC42, PTPN11, PIK3C2A, PIK3R1, PDPK1, CREB3L4, MAP3K5 (includes EG:4217) |
| G beta gamma signaling | 1.39E00 | .04074 | 0.09 | GNAI2, GNAS, GNG11, PRKCI, CDC42, GNA11, CAV1, PDPK1, GNG7, EGFR |
| PI3K/AKT signaling | 1.38E00 | .04169 | 0.09 | ITGB1, JAK1, BAD, FOXO1, HSP90AB1, PIK3R1, YWHAZ, HSP90AA1, PDPK1, MAP3K5 (includes EG:4217), PTEN, EIF4EBP1 |
| Semaphorin signaling in neurons | 1.38E00 | .04169 | 0.13 | ROCK2, ITGB1, MET, RHOQ, CDK5, DPYSL4, NRP1 |
| Mitotic roles of polo-like kinase | 1.38E00 | .04169 | 0.11 | ANAPC4, HSP90AB1, TGFB1, CDC7, HSP90AA1, CDC16, STAG2 |
| Histidine metabolism | 1.37E00 | .04266 | 0.05 | PRPS2, ALDH3B2 (includes EG:222), MAOB, MGMT, ABP1, MAOA |
| SAPK/JNK signaling | 1.36E00 | .04365 | 0.10 | MAP2K7, GNG11, RIPK1, TRD@, CDC42, PIK3C2A, MAP3K7, PIK3R1, MAP3K5 (includes EG:4217), GNG7 |
| Type I diabetes mellitus signaling | 1.34E00 | .04571 | 0.10 | SOCS1, HLA-DMA, MAP2K7, JAK1, RIPK1, TRD@, MAP3K7, IL1R1, MAP3K5 (includes EG:4217), HSPD1, IRAK1 |
| FGF signaling | 1.3E00 | .05012 | 0.10 | MET, PTPN11, PIK3C2A, FGF18, PIK3R1, FGFR1, FGFR2, CREB3L4, MAP3K5 (includes EG:4217) |
| Chronic myeloid leukemia signaling | 1.25E00 | .05623 | 0.10 | CTBP1, RBL2, BAD, TFDP1, PTPN11, PIK3C2A, TGFB1, PIK3R1, CBLC, HDAC5 |
| RAR activation | 1.25E00 | .05623 | 0.08 | CYP26A1, PIK3R1, NR2F2, PDPK1, MAP3K5 (includes EG:4217), PTEN, PRKCI, DUSP1, GTF2H4, TGFB1, CRABP2, RDH5, NCOR2, CARM1, SCAND1 |
| Phenylalanine metabolism | 1.25E00 | .05623 | 0.05 | ALDH3B2 (includes EG:222), MAOB, ABP1, MAOA, PRDX2 |
| 14-3-3-mediated signaling | 1.24E00 | .05754 | 0.10 | PRKCI, BAD, FOXO1, PIK3C2A, PIK3R1, TUBB2A, TUBG1, YAP1, YWHAZ, BAX, MAP3K5 (includes EG:4217) |
| Caveolar-mediated endocytosis signaling | 1.23E00 | .05888 | 0.10 | ITGB1, RAB5A, CD55, RAB5C, CAV1, COPE, ITGB5, EGFR |
| Ubiquinone biosynthesis | 1.17E00 | .06761 | 0.06 | NDUFB11, NDUFS8, NDUFS7, MGMT, NDUFB7, NDUFA3, NDUFA13 |
| RAN signaling | 1.17E00 | .06761 | 0.13 | KPNB1, RANBP2, RAN |
| ILK signaling | 1.11E00 | .07762 | 0.08 | MUC1, ITGB1, PIK3C2A, CDC42, PIK3R1, FERMT2, PDPK1, CREB3L4, PTEN, PARVB, RHOQ, PPAP2B, CHD1 (includes EG:1105), ACTN1, ITGB5 |
| Human embryonic stem cell pluripotency | 1.07E00 | .08511 | 0.08 | FZD8, GNAS, FZD10, PIK3C2A, PDGFA, TGFB1, PIK3R1, FGFR1, FGFR2, PDPK1, FZD2, FZD7 |
| B Cell receptor signaling | 1.06E00 | .08710 | 0.08 | MAP2K7, BAD, CDC42, PIK3C2A, PIK3R1, CREB3L4, MAP3K5 (includes EG:4217), PTEN, PTPRC, PTPN11, CARD10, MAP3K7, PAG1 |
| Ephrin receptor signaling | 1.04E00 | .09120 | 0.08 | ITGB1, ACTR2, CDC42, PDGFA, GNA11, CREB3L4, GNG7, EFNA4, ROCK2, GNAI2, EPHB6, GNAS, GNG11, PTPN11, EFNB3 |
| Pentose phosphate pathway | 9.86E-01 | .10328 | 0.04 | PRPS2, PGLS, FBP1, ALDOA |
| Melanoma signaling | 9.75E-01 | .10593 | 0.11 | BAD, PIK3C2A, PIK3R1, CHD1 (includes EG:1105), PTEN |
| CCR5 signaling in macrophages | 9.7E-01 | .10715 | 0.08 | GNAI2, GNAS, GNG11, PRKCI, TRD@, CCL5, GNG7 |
| FLT3 signaling in hematopoietic progenitor cells | 9.45E-01 | .11350 | 0.10 | BAD, PTPN11, PIK3C2A, PIK3R1, PDPK1, CREB3L4, EIF4EBP1 |
| Oxidative phosphorylation | 9.28E-01 | .11803 | 0.08 | ATP6V0E2, ATP6V0B, ATP5D, NDUFS7, TCIRG1, ATP6V1A, NDUFA13, NDUFB11, NDUFS8, NDUFB7, NDUFA3, CYC1, UQCRC1 |
| IL-8 signaling | 9.27E-01 | .11830 | 0.07 | PIK3C2A, PIK3R1, BAX, GNG7, EIF4EBP1, IRAK1, ROCK2, GNAI2, HMOX1, GNAS, GNG11, PRKCI, RHOQ, EGFR |
| Fructose and mannose metabolism | 9.1E-01 | .12303 | 0.04 | AKR7A2, TSTA3, GMPPA, FBP1, ALDOA, FUK |
| Cdc42 signaling | 9E-01 | .12589 | 0.08 | ITGB1, ANAPC4, ACTR2, PRKCI, TRD@, CDC42, CDC42EP5, PPP1R12A, CDC16, IQGAP1 |
| Arginine and proline metabolism | 8.96E-01 | .12706 | 0.04 | CKB, MAOB, VNN1, GAMT, LOXL1, ABP1, MAOA |
| HGF signaling | 8.69E-01 | .13521 | 0.09 | MET, MAP2K7, PRKCI, CDC42, PTPN11, PIK3C2A, MAP3K7, PIK3R1, MAP3K5 (includes EG:4217) |
| EGF signaling | 8.5E-01 | .14125 | 0.10 | MAP2K7, JAK1, PIK3C2A, PIK3R1, EGFR |
| Nitric oxide signaling in the cardiovascular system | 8.49E-01 | .14158 | 0.08 | PIK3C2A, HSP90AB1, PIK3R1, CAV1, HSP90AA1, SLC7A1, CACNA1A |
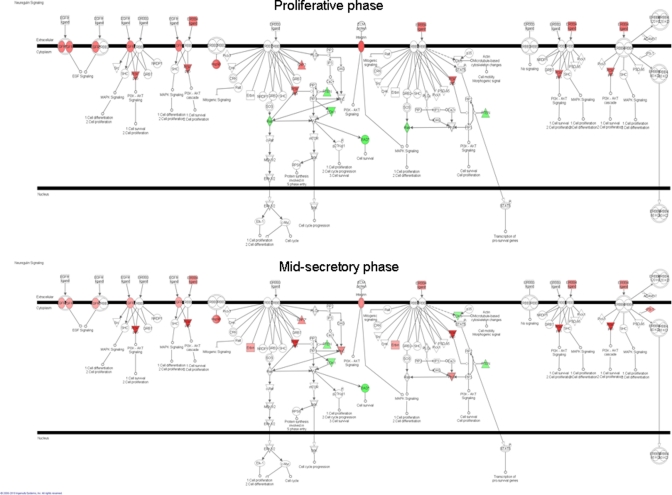
Major differences in neuregulin signaling, which involves members of the EGFR family, were observed in the proliferative and mid-secretory phases between severe versus mild endometriosis (Figure 3 ). Epidermal growth factor receptor mRNA was upregulated in severe versus mild endometriosis in PE and ESE (Figure 2B), indicating its involvement in severe disease, and confirming our earlier reports of the involvement of EGF family in the pathophysiology of severe endometriosis.13,24
Figure 3.
Neuregulin pathway regulation in proliferative endometrium (PE) and mid-secretory endometrium (MSE) from participants with severe vs mild endometriosis analyzed by ingenuity pathway analysis (IPA). Red color indicates upregulation of a gene; green color, down-regulation.
Epidermal Growth Factor Receptor Protein Immunoreactivity
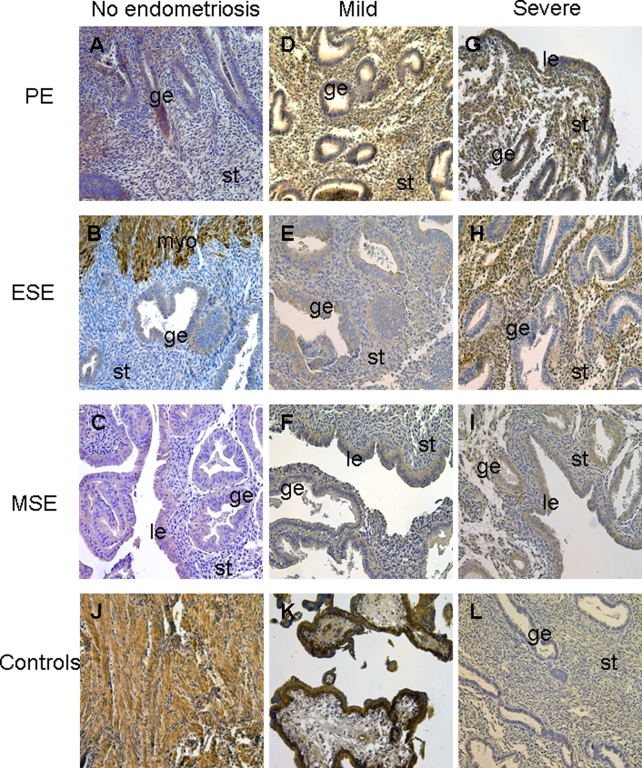
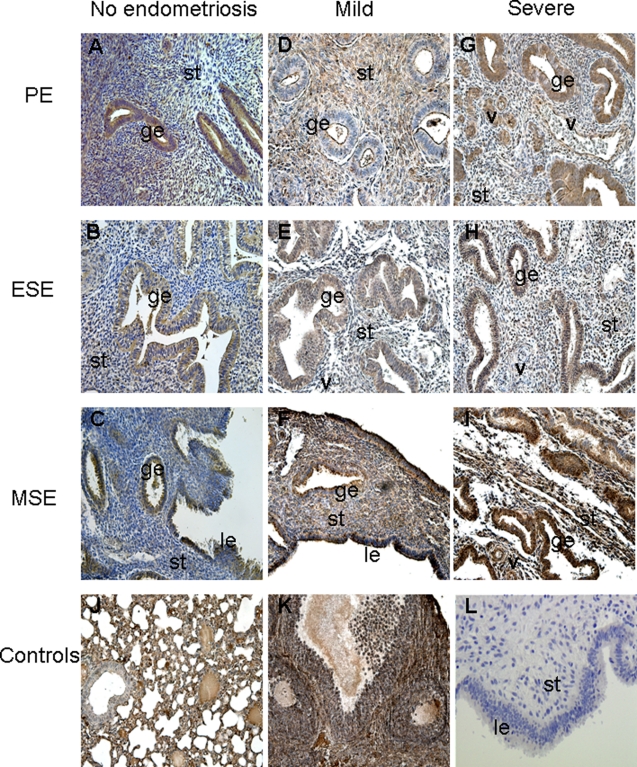
As presented in Figure 4 and Table 5 , EGFR protein was expressed throughout the menstrual cycle in women with mild as well as severe endometriosis. Interestingly, the most dramatic difference in EGFR protein immunoreactivity was observed in the early secretory phase, where strong stromal expression was observed in severe compared to mild endometriosis, consistent with the real-time RT-PCR data (Figure 2B, Figure 4E,H, Table 5). There was a slight increase in epithelial EGFR immunostaining in the proliferative phase of severe endometriosis samples (Figure 4D,G, Table 5), whereas immunostaining was similar in MSE samples, regardless of the endometriosis stage (Figure 4F, I, Table 5). Immunohistochemical analysis of EGFR in endometrial samples from women without endometriosis throughout the menstrual cycle revealed weak expression of this protein in epithelial and/or stromal compartments (in particular, in ESE stroma), compared to the endometrium from women with endometriosis (Figure 4A-C, Table 5).
Figure 4.
Epidermal growth factor receptor (EGFR) immunoreactivity in endometrial tissue from women without endometriosis (A-C), women with mild (D-F), and severe (G-I) endometriosis in the proliferative phase ([PE] n = 4, n = 4, and n = 5, respectively; A, D, G), early secretory phase ([ESE] n = 4, n = 3, and n = 7, respectively; B, E, H), and mid-secretory phase ([MSE] n = 4, n = 3, and n = 5, respectively; C, F, I) of the cycle. Human myometrial tissue was used as an internal positive control (on the same slide as endometrium, adjacent to the basalis endometrium, J). Human 12-week gestation placental tissue (K) was used as an additional positive control. L indicates negative control, nonimmune IgG-treated human endometrium; Le, luminal epithelium; ge, glandular epithelium; st, stroma; myo, myometrium; IgG, immunoglobulin G. Magnification ×200.
Table 5.
Semiquantitative Evaluation of VCAN and EGFR Immunostaining in Human Endometrial Tissue Sections From Women With Mild And Severe Endometriosis, As Well As Without Endometriosis
| Protein | PE Mild Endometriosis |
ESE Mild Endometriosis |
MSE Mild Endometriosis |
PE Severe Endometriosis |
ESE Severe Endometriosis |
MSE Severe Endometriosis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Epithelium | Stroma | Epithelium | Stroma | Epithelium | Stroma | Epithelium | Stroma | Epithelium | stroma | Epithelium | Stroma | |
| EGFR | + | ++ | + | +/++ | ++ | ++ | ++ | ++ | + | +++ | ++ | ++ |
| VCAN | −/+ | +++ | ++ | + | ++ | ++ | +++ | + | ++ | + | +++ | ++ |
| PE no endometriosis |
ESE no endometriosis |
MSE no endometriosis |
||||||||||
| Epithelium |
Stroma |
Epithelium |
Stroma |
Epithelium |
Stroma |
|||||||
| EGFR | ++ | ++ | + | + | ++ | + | ||||||
| VCAN | ++ | + | +/++ | + | ++ | + | ||||||
Abbreviations: PE, proliferative phase endometrium; ESE, early secretory phase endometrium; MSE, mid-secretory endometrium; VACN, versican; EGFR, epidermal growth factor receptor; −, no staining, −/+, a few stained cells; +, faint staining; ++, moderate staining; +++, strong staining.
Versican Protein Immunoreactivity
In the proliferative phase, there was a strong stromal and epithelial VCAN immunostaining in severe and mild endometriosis, respectively (Figure 5D,G , Table 5). In ESE, similar VCAN immunostaining was observed in the stroma and epithelial compartments (Figure 5E,H, Table 5), and epithelial VCAN immunostaining in MSE tended to be stronger in the severe endometriosis samples (Figure 5F,I, Table 5). Of note, diffuse stromal reactivity of VCAN in both cellular and extracellular compartments was observed. Remarkable was the VCAN immunoreactivity in the vasculature—in the smooth muscle layer and endothelial cells, regardless of disease status, stage, or cycle phase. Versican immunostaining in endometrial tissue from women without endometriosis throughout the menstrual cycle demonstrated overall weaker expression compared to samples from women with mild or severe endometriosis. This was particularly evident in PE epithelium and stroma in patients with severe and mild endometriosis, respectively, as well as MSE epithelium in severe endometriosis samples (Figure 5A-C, Table 5). Positive and negative controls demonstrated specificity of the observed results (Figure 5J-L).
Figure 5.
Versican (VCAN) immunoreactivity in endometrial tissue from women without endometriosis (A-C), and women with mild (D-F), and severe (G-I) endometriosis in the proliferative ([PE] n = 4, n = 4, and n = 5, respectively; A, D, G), early secretory ([ESE]; n = 4, n = 3, and n = 7, respectively; B, E, H), and mid-secretory ([MSE]; n = 4, n = 3, and n = 5, respectively; C, F, I) phases of the menstrual cycle. Mouse lung tissue (J) and mouse ovary (K) were used as positive controls. L indicates negative control, nonimmune IgG-treated human endometrium; Le, luminal epithelium; ge, glandular epithelium; st, stroma; v, blood vessel. Magnification ×200.
Discussion
General Comments
The main finding of this study is the demonstrated difference in global gene expression in eutopic endometrium from participants with severe versus mild endometriosis, throughout the menstrual cycle. These 2 endometriosis stages are distinct in their clinical presentation, as well as therapeutic and surgical management, although the corresponding scientific literature is limited. The data herein underscore significant molecular and signaling pathway differences between these 2 stages of endometriosis in distinct hormonal milieu, suggesting that eutopic endometrium in severe versus mild endometriosis has different functional capacities.
Comparison of severe versus mild endometriosis samples by cycle phase revealed the dysregulation of several cyclic adenosine monophosphate (cAMP) and/or progesterone regulated gene, such as downregulation of IHH, SST, and TAGLN in ESE and upregulation of DKK1, MAO, IL15, and IL1R1 in MSE. Upregulation of DIO2 and downregulation of TRH transcripts between severe and mild endometriosis samples indicate potential involvement of thyroid hormone homeostasis and metabolism in the pathophysiology of this endometrial disorder.
Women with severe endometriosis experience higher rates of implantation failure during IVF treatment cycles. Of the 25 human receptivity-related genes identified by analysis of endometrial tissue from healthy fertile women,25 TAGLN and calponin 1 transcripts were dysregulated in MSE from women with severe versus mild endometriosis, suggesting their potential role in the impaired implantation process in women with severe disease.
Dysregulation of Neuregulin Signaling and EGFR in Severe Versus Mild Endometriosis
Of interest is the association of neuregulin signaling with endometriosis. Neuregulin signaling involves ligands for the transmembrane tyrosine kinase receptors ERBB1 (EGFR), ERBB2, ERBB3, and ERBB4—members of the EGFR family.26 Ligand binding activates intracellular signaling cascades and the induction of cellular responses including proliferation, migration, differentiation, and survival or apoptosis in different organs and systems.27 Neuregulin genes, though not regulated themselves in the current study, influence proliferation, migration, and differentiation of epithelial, neuronal, glial, cardiac, and other types of cells.28–30 This canonical pathway was highly regulated herein in PE and MSE between severe and mild endometriosis samples. Neuregulin (also known as heregulin) signals through HER3 and HER4 receptors; although no changes were observed herein between disease stages or in different cycle phases.
Epidermal growth factor receptor is the major player of neuregulin-signaling pathway. Epidermal growth factor receptor (ERBB1) expression in normal eutopic endometrium on the mRNA and protein levels during different menstrual cycle phases was demonstrated herein and confirms earlier reports.31–33 We have observed that EGFR gene expression is increased in eutopic endometrium of women with severe endometriosis compared to women without disease in ESE and MSE, but not PE, and is not regulated in mild endometriosis versus nonendometriosis samples throughout the cycle (Aghajanova et al, unpublished data). Herein, we have found that EGFR is dysregulated in severe versus mild endometriosis throughout the menstrual cycle on both mRNA and protein levels, with the most dramatic difference (upregulation) in ESE. Furthermore, transducer of ERBB2 was upregulated in ESE. Thus, the present study demonstrates differences in EGFR expression between different stages of endometriosis and also supports earlier studies noting involvement of EGF family members in the pathophysiology of endometriosis.13,25,34 Interestingly, EGFR is a tumor marker, particularly for epithelial tumors such as colon cancer, lung cancer, prostate cancer, breast cancer, or other solid tumors.26,35 Whether it is a marker of a severity of endometriosis remains to be determined.
Dysregulation of ECM Molecules in Severe Versus Mild Endometriosis
This is the first study to demonstrate mRNA expression and immunoreactivity of the ECM proteoglycan VCAN in human endometrium. In participants without endometriosis, VCAN immunoreactivity was weak, especially in PE; whereas, strong immunoreactivity was observed in endometrial stroma from women with severe endometriosis and in epithelium of samples from women with mild disease. Versican can bind to integrins on the cell surface,36 stimulating cell proliferation and inhibiting apoptosis.37 Versican has multiple functions and interactions in different model systems. For example, overexpression of VCAN in a pheochromocytoma cell line upregulates EGFR.38 Also, VCAN expression is increased in endothelial cells with increased migrating capacity.39 Thus, high levels of VCAN may promote an invasive phenotype of endometrial cells in endometriosis by affecting their proliferation, apoptosis, adhesion, and migration, and also may participate in or be causative of the upregulation of EGFR in endometriosis. These functions in endometriosis await further investigation.
Dysregulation of MicroRNAs in Severe Versus Mild Endometriosis
MicroRNA (miRNA) 21 (MIR21) was found to be upregulated on the array, herein, in eutopic endometrium throughout the menstrual cycle in severe versus mild endometriosis. It has recently been shown to be upregulated in eutopic endometrium of women with versus without endometriosis.40 Some of the predicted target genes for this miRNA are the tumor-suppressor gene PTEN (downregulated 2.18- and 2.3-fold in severe vs mild endometriosis in PE and MSE, respectively), PDCD4, E2F1, and TGFBRII.41–43 Interestingly, downregulation of MIR21 inhibits expression of EGFR in human glioblastoma cells.44 Whether there is such a mechanism operating in human endometrial stromal fibroblasts (hESF) remains to be determined.
Herein, we observed the upregulation of DICER1 in ESE and MSE from severe versus mild endometriosis (Supplement Tables 2 and 3). The transcript for DICER1 (dicer1, ribonuclease type III), which is a repressor of gene expression due to its involvement in the biogenesis of microRNAs and small interfering RNAs, demonstrates cyclic variation throughout the normal human menstrual cycle.45 Female mice with a conditional knockout of Dicer1 in mesenchyme-derived cells of the oviducts and uterus are sterile, in part, due to uterine defects.46 Although endometrial stromal Dicer1 expression was absent, the decidualization process was not compromised,46 consistent with the recent finding that DICER1 knockdown in hESF does not affect decidualization.45 Increased expression of DICER1 in secretory endometrium from women with severe versus mild endometriosis may lead to downregulation of apoptosis-associated genes and dysregulation of adhesion molecules, leading to resistance to apoptosis and increased migratory functions in endometrial cells, as observed with endothelial cells.47
Dysregulation of Canonical Pathways in Severe Versus Mild Endometriosis
Several pathways regulated between severe and mild endometriosis are of interest. Severe endometriosis samples exhibited dysregulation of second-messenger signaling pathways, including PI3K/AKT, JAK/STAT, SPK/JNK, and MAPK, confirming recent reports.48 Regulation of neurotrophin/TRK (neurotrophic tyrosine kinase) signaling in PE and MSE and axonal signaling in ESE are consistent with the presence of nerve fibers in eutopic endometrium and perhaps their role in the pathogenesis of endometriosis-associated pain.49,50 However, other functions of these pathways (and their members) may be operating in endometrium, not related to pain. The current study demonstrates differences in these pathways between severe and mild stages of endometriosis, although pain and stage are not necessarily correlated.51,52
Of note is the involvement of cancer-associated pathways, such as prostate, endometrial, bladder, colorectal, pancreatic cancer, and basal cell carcinoma signaling, suggesting commonalities in the pathophysiology between severe endometriosis and epithelial cancers.
Wnt signaling, NRF2-mediated oxidative stress response signaling (nuclear factor [erythroid-derived 2]-like 2, involved in apoptosis and the oxidative stress response), and retinoid X receptor (RXR) signaling were significantly regulated in secretory endometrium (ESE and MSE; Supplememtal Table 2), probably indicating the differences in the endometrial response to progesterone between severe and mild endometriosis.
Summary
Taken together, these data demonstrate the complexity of the processes and gene interactions and pathways involved in the endometrium of women with endometriosis and the molecular differences in the setting of severe versus mild disease. Whether these differences account for the observed differences in clinical presentations of women with severe versus mild endometriosis, that is lower implantation and pregnancy rates in women with severe disease, remain to be determined. The signaling pathways identified may serve for development of targeted therapies to correct the phenotype at the endometrial level.
Footnotes
The authors declared no potential conflicts of interests with respect to the authorship and/or publication of this article.
The authors disclosed receipt of the following financial support for the research and/or authorship of this article: the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)/NIH through cooperative agreement U54HD055764-04 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research.
References
- 1. Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364(9447):1789–1799 [DOI] [PubMed] [Google Scholar]
- 2. Taylor RN, Yu J, Torres PB, et al. Mechanistic and therapeutic implications of angiogenesis in endometriosis. Reprod Sci. 2009;16(2):140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. The American Fertility Society Revised American Fertility Society classification of endometriosis. Fertil Steril. 1985;43(3):351–352 [DOI] [PubMed] [Google Scholar]
- 4. Rock JA. The revised American Fertility Society classification of endometriosis: reproducibility of scoring. ZOLADEX Endometriosis Study Group. Fertil Steril. 1995;63(5):1108–1110 [DOI] [PubMed] [Google Scholar]
- 5. Nisolle M, Donnez J. Peritoneal endometriosis, ovarian endometriosis, and adenomyotic nodules of the rectovaginal septum are three different entities. Fertil Steril. 1997;68(4):585–596 [DOI] [PubMed] [Google Scholar]
- 6. Donnez J, Nisolle M, Smoes P, Gillet N, Beguin S, Casanas-Roux F. Peritoneal endometriosis and “endometriotic” nodules of the rectovaginal septum are two different entities. Fertil Steril. 1996;66(3):362–368 [PubMed] [Google Scholar]
- 7. Matsuzaki S, Maleysson E, Darcha C. Analysis of matrix metalloproteinase-7 expression in eutopic and ectopic endometrium samples from patients with different forms of endometriosis. Hum Reprod. 2010;25(3):742–750 [DOI] [PubMed] [Google Scholar]
- 8. D’Hooghe TM, Debrock S, Hill JA, Meuleman C. Endometriosis and subfertility: is the relationship resolved?. Semin Reprod Med. 2003;21(2):243–254 [DOI] [PubMed] [Google Scholar]
- 9. Kuivasaari P, Hippelainen M, Anttila M, Heinonen S. Effect of endometriosis on IVF/ICSI outcome: stage III/IV endometriosis worsens cumulative pregnancy and live-born rates. Hum Reprod. 2005;20(11):3130–3135 [DOI] [PubMed] [Google Scholar]
- 10. Barnhart K, Dunsmoor-Su R, Coutifaris C. Effect of endometriosis on in vitro fertilization. Fertil Steril. 2002;77(5):1148–1155 [DOI] [PubMed] [Google Scholar]
- 11. Matalliotakis IM, Cakmak H, Mahutte N, Fragouli Y, Arici A, Sakkas D. Women with advanced-stage endometriosis and previous surgery respond less well to gonadotropin stimulation, but have similar IVF implantation and delivery rates compared with women with tubal factor infertility. Fertil Steril. 2007;88(6):1568–1572 [DOI] [PubMed] [Google Scholar]
- 12. Kao LC, Germeyer A, Tulac S, et al. Expression profiling of endometrium from women with endometriosis reveals candidate genes for disease-based implantation failure and infertility. Endocrinology. 2003;144(7):2870–2881 [DOI] [PubMed] [Google Scholar]
- 13. Burney RO, Talbi S, Hamilton AE, et al. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology. 2007;148(8):3814–3826 [DOI] [PubMed] [Google Scholar]
- 14. Bulun SE. Endometriosis. N Engl J Med. 2009;360(3):268–279 [DOI] [PubMed] [Google Scholar]
- 15. Aghajanova L, Horcajadas JA, Weeks JL, et al. The protein kinase A pathway-regulated transcriptome of endometrial stromal fibroblasts reveals compromised differentiation and persistent proliferative potential in endometriosis. Endocrinology. 2010;151(3):1341–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Talbi S, Hamilton AE, Vo KC, et al. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology. 2006;147(3):1097–1121 [DOI] [PubMed] [Google Scholar]
- 17. Noyes RW, Hertig AT, Rock J. Dating the endometrial biopsy. Fertil Steril. 1950;1:3–25 [DOI] [PubMed] [Google Scholar]
- 18. Aghajanova L, Hamilton A, Kwintkiewicz J, Vo KC, Giudice LC. Steroidogenic enzyme and key decidualization marker dysregulation in endometrial stromal cells from women with versus without endometriosis. Biol Reprod. 2009;80(1):105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Russell DL, Ochsner SA, Hsieh M, Mulders S, Richards JS. Hormone-regulated expression and localization of versican in the rodent ovary. Endocrinology. 2003;144(3):1020–1031 [DOI] [PubMed] [Google Scholar]
- 20. Kaplan F, Comber J, Sladek R, et al. The growth factor midkine is modulated by both glucocorticoid and retinoid in fetal lung development. Am J Respir Cell Mol Biol. 2003;28(1):33–41 [DOI] [PubMed] [Google Scholar]
- 21. Bulmer JN, Thrower S, Wells M. Expression of epidermal growth factor receptor and transferrin receptor by human trophoblast populations. Am J Reprod Immunol. 1989;21(3-4):87–93 [DOI] [PubMed] [Google Scholar]
- 22. Smith K, LeJeune S, Harris AH, Rees MC. Epidermal growth factor receptor in human uterine tissues. Hum Reprod. 1991;6(9):619–622 [DOI] [PubMed] [Google Scholar]
- 23. Heiner JS, Cai L, Ding H, Rutgers JK. Myometrial expression of mRNA encoding epidermal growth factor receptor (EGFR) throughout the menstrual cycle. Am J Reprod Immunol. 1994;32(3):152–156 [DOI] [PubMed] [Google Scholar]
- 24. Velarde MC, Aghajanova L, Nezhat CR, Giudice LC. Increased mitogen-activated protein kinase kinase/extracellularly regulated kinase activity in human endometrial stromal fibroblasts of women with endometriosis reduces 3,’5’-cyclic adenosine 5’-monophosphate inhibition of cyclin D1. Endocrinology. 2009;150(10):4701–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horcajadas JA, Pellicer A, Simón C. Wide genomic analysis of human endometrial receptivity: new times, new opportunities. Hum Reprod Update. 2007;13(1):77–86 [DOI] [PubMed] [Google Scholar]
- 26. Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5(5):341–354 [DOI] [PubMed] [Google Scholar]
- 27. Earp HS, 3rd, Calvo BF, Sartor CI. The EGF receptor family—multiple roles in proliferation, differentiation, and neoplasia with an emphasis on HER4. Trans Am Clin Climatol Assoc. 2003;114:315–333 [PMC free article] [PubMed] [Google Scholar]
- 28. Birchmeier C, Nave KA. Neuregulin-1, a key axonal signal that drives Schwann cell growth and differentiation. Glia. 2008;56(4):1491–1497 [DOI] [PubMed] [Google Scholar]
- 29. Esper RM, Pankonin MS, Loeb JA. Neuregulins: versatile growth and differentiation factors in nervous system development and human disease. Brain Res Rev. 2006;51(2):161–175 [DOI] [PubMed] [Google Scholar]
- 30. Xu Y, Li X, Zhou M. Neuregulin-1/ErbB signaling: a druggable target for treating heart failure. Curr Opin Pharmacol. 2009;9(2):214–219 [DOI] [PubMed] [Google Scholar]
- 31. Imai T, Kurachi H, Adachi K, et al. Changes in epidermal growth factor receptor and the levels of its ligands during menstrual cycle in human endometrium. Biol Reprod. 1995;52(4):928–938 [DOI] [PubMed] [Google Scholar]
- 32. Ejskjaer K, Sorensen BS, Poulsen SS, Mogensen O, Forman A, Nexo E. Expression of the epidermal growth factor system in human endometrium during the menstrual cycle. Mol Hum Reprod. 2005;11(8):543–551 [DOI] [PubMed] [Google Scholar]
- 33. Aghajanova L, Bjuresten K, Altmae S, Landgren BM, Stavreus-Evers A. HB-EGF but not amphiregulin or their receptors HER1 and HER4 is altered in endometrium of women with unexplained infertility. Reprod Sci. 2008;15(5):484–492 [DOI] [PubMed] [Google Scholar]
- 34. Ejskjaer K, Sorensen BS, Poulsen SS, Mogensen O, Forman A, Nexo E. Expression of the epidermal growth factor system in eutopic endometrium from women with endometriosis differs from that in endometrium from healthy women. Gynecol Obstet Invest. 2009;67(2):118–126 [DOI] [PubMed] [Google Scholar]
- 35. Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009;21(2):177–184 [DOI] [PubMed] [Google Scholar]
- 36. Wu YJ, La Pierre DP, Wu J, Yee AJ, Yang BB. The interaction of versican with its binding partners. Cell Res. 2005;15(7):483–494 [DOI] [PubMed] [Google Scholar]
- 37. Sheng W, Wang G, Wang Y, et al. The roles of versican V1 and V2 isoforms in cell proliferation and apoptosis. Mol Biol Cell. 2005;16(3):1330–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu Y, Chen L, Cao L, Sheng W, Yang BB. Overexpression of the C-terminal PG-M/versican domain impairs growth of tumor cells by intervening in the interaction between epidermal growth factor receptor and beta1-integrin. J Cell Sci. 2004;117(11):2227–2237 [DOI] [PubMed] [Google Scholar]
- 39. Cattaruzza S, Schiappacassi M, Ljungberg-Rose A, et al. Distribution of PG-M/versican variants in human tissues and de novo expression of isoform V3 upon endothelial cell activation, migration, and neoangiogenesis in vitro. J Biol Chem. 2002;277(49):47626–47635 [DOI] [PubMed] [Google Scholar]
- 40. Luo X, Prucha MS, Chegini N. The expression, regulation and function of miR-21 in the endometrium and endometriosis. Reprod Sci. 2010;17(3):349A [Google Scholar]
- 41. Pan Q, Luo X, Chegini N. MicroRNA 21: response to hormonal therapies and regulatory function in leiomyoma, transformed leiomyoma and leiomyosarcoma cells. Mol Hum Reprod. 2010;16(3):215–227 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42. Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta. 2010;411(11-12):846–852 [DOI] [PubMed] [Google Scholar]
- 43. Qi L, Bart J, Tan LP, et al. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer. 2009;9:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou X, Ren Y, Moore L, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90(2):144–155 [DOI] [PubMed] [Google Scholar]
- 45. Ran L, Arias P, Han D, Andreu-Vieyra C, Matzuk M, Hawkins S. Characterization of DICER gene expression in the human reproductive tract. Reprod Sci. 2010;17(2):137A [Google Scholar]
- 46. Nagaraja AK, Andreu-Vieyra C, Franco HL, et al. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol. 2008;22(10):2336–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Asada S, Takahashi T, Isodono K, et al. Downregulation of Dicer expression by serum withdrawal sensitizes human endothelial cells to apoptosis. Am J Physiol Heart Circ Physiol. 2008;295(6):2512–2521 [DOI] [PubMed] [Google Scholar]
- 48. Zhang H, Zhao X, Liu S, Li J, Wen Z, Li M. 17betaE2 promotes cell proliferation in endometriosis by decreasing PTEN via NFkappaB-dependent pathway. Mol Cell Endocrinol. 2010;317(1-2):31–43 [DOI] [PubMed] [Google Scholar]
- 49. Tokushige N, Markham R, Russell P, Fraser IS. High density of small nerve fibres in the functional layer of the endometrium in women with endometriosis. Hum Reprod. 2006;21(3):782–787 [DOI] [PubMed] [Google Scholar]
- 50. Tokushige N, Markham R, Russell P, Fraser IS. Different types of small nerve fibers in eutopic endometrium and myometrium in women with endometriosis. Fertil Steril. 2007;88(4):795–803 [DOI] [PubMed] [Google Scholar]
- 51. Porpora MG, Koninckx PR, Piazze J, Natili M, Colagrande S, Cosmi EV. Correlation between endometriosis and pelvic pain. J Am Assoc Gynecol Laparosc. 1999;6(4):429–434 [DOI] [PubMed] [Google Scholar]
- 52. Vercellini P, Fedele L, Aimi G, Pietropaolo G, Consonni D, Crosignani PG. Association between endometriosis stage, lesion type, patient characteristics and severity of pelvic pain symptoms: a multivariate analysis of over 1000 patients. Hum Reprod. 2007;22(1):266–271 [DOI] [PubMed] [Google Scholar]