Abstract
Background & Aims
The blood group antigen binding adhesin (BabA) has been proposed to play a role in disease pathogenesis. This hypothesis is based on the functional BabA status as determined by polymerase chain reaction (PCR) analysis to distinguish functional babA2 genes from nonfunctional babA1 genes.
Methods
We compared the ability of published PCR-based methods to assess BabA status with BabA immunoblotting and Lewis b (Leb) binding activity assays. We also used immunoblotting to examine the relationship between clinical presentation and levels of BabA expression.
Results
Immunoblotting and Leb binding assays for 80 strains revealed 3 levels of BabA expression: BabA high producers (BabA-H) with Leb binding activity, BabA low producers (BabA-L) without Leb binding activity, and BabA-negative. BabA-negative strains lacked the babA gene. PCR methods to determine BabA status yielded poor results. babA1 sequences were never detected. BabA expression was examined in 250 strains from Western countries and 270 strains from East Asia. The results failed to confirm any relationship between triple-positive status (cagA-positive/vacA s1/BabA-H) and clinical outcome. BabA-negative strains typically were cagA-negative/vacA s2 and were associated with gastritis. BabA-L strains showed a higher level of mucosal injury and were associated more frequently with duodenal ulcer and gastric cancer than the other groups.
Conclusions
Information gained from currently used PCR-based methods must be interpreted with caution. Leb binding activity does not accurately reflect the severity of mucosal damage or the clinical outcome. Quantitation of BabA expression revealed that Leb-nonbinding BabA-L strains are associated with higher levels of mucosal injury and clinical outcome.
Bacterial adherence is believed to play an important role in the colonization of gastric epithelium by Helicobacter pylori. Fucosylated ABO blood group antigens and the related sialyl-Lewis x/a antigens are thought to serve as one group of functional receptors for H pylori adherence.1,2 The ABO antigens are recognized by the blood group antigen binding adhesin (BabA)3,4 and sialyl-Lewis x/a antigens are recognized by the H pylori sialic acid binding adhesin.2
A number of studies have suggested a relation between babA2-positive H pylori and increased cellular mucosal inflammations and an increased risk of developing clinical outcomes (Table 1). However, the role of BabA in the pathogenesis of H pylori–related disease remains unclear. The babA genes initially were cloned from the strain CCUG17875, which contains a silent babA1 gene and an expressed babA2 gene.3 The sequence of these 2 genes differed only by the presence of a 10 – base pair (bp) deletion in the signal peptide sequence of babA1 that eliminates its translational initiation codon.3 Most studies evaluating BabA status have used polymerase chain reaction (PCR) techniques based on detection of the 10-bp deletion to distinguish between the babA2 and babA1 genes (Table 1).5–22 However, the ability of such PCR-based strategies to detect the presence of a functional babA gene is questionable. For example, H pylori strains unable to bind to Lewis b antigen (Leb) have been reported to contain signal peptide sequences that are identical to those of strains with a functional babA2 gene (eg, strain 26695).23 It also has been suggested that babA expression may be regulated transcriptionally24 and that there are chimeric babA– babB genes (chimera babA/B or babB/A).24,25 Finally, not all of these PCR-based studies have shown an association between babA2 and intense cellular mucosal inflammations and/or increased risk of peptic ulcer diseases and gastric cancer (Table 1).10–13,19,20,22 Therefore, in the current study, we have compared results from PCR-based strategies that currently are used to predict BabA functional status with those from strategies that measure actual babA expression (ie, immunoblotting) and Leb binding.
Table 1.
PCR-Based Genotyping for the babA2 Gene
| Prevalence of babA2 gene |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Year | Population | Primer pair |
Total | Gastritis | PUD | Gastric cancer |
MALT | Reflux esophagitis |
Related to diseases |
Related to cagA/vacA |
| Original strategy using primer pair A | |||||||||||
| Western | |||||||||||
| Gerhard et al5a | 1999 | Germany | A | 114 (72%) | 18 (51%) | 23 (100%) | 21 (78%) | 20 (69%) | Yes | Yes | |
| Prinz et al6b,c | 2001 | Germany | A | 145 (39%) | 57 (39%) | Yes | |||||
| Oleastro et al7 | 2003 | Portugal | A | 140 (32%) | 24 (23%) | 21 (58%) | Yes | ||||
| Podzorski et al8 | 2003 | United States | A | 61 (36%) | 22 (36%) | No | |||||
| Oliveira et al9 | 2003 | Brazil | A | 208 (46%) | 24 (32%) | 43 (54%) | 29 (56%) | Yes | Yes | ||
| Lehours et al10 | 2004 | France | A | 82 (49%) | 21 (54%) | 19 (44%) | No | Yes | |||
| Gatti et al11 | 2005 | Brazil | A | 89 (47%) | 37 (53%) | 3 (20%) | 1 (100%) | 1 (33%) | No | Yes | |
| Olfat et al12 | 2005 | Germany | A | 92 (45%) | 19 (28%) | 22 (88%) | Yes | Yes | |||
| Sweden | A | 74 (45%) | 21 (48%) | 12 (40%) | No | Yes | |||||
| Portugal | A | 91 (34%) | 12 (20%) | 19 (63%) | Yes | Yes | |||||
| Finland | A | 57 (60%) | 12 (46%) | 22 (71%) | P = .06 | Yes | |||||
| Western total | 1153 (45%) | 267 (37%) | 165 (61%) | 51 (64%) | 40 (53%) | ||||||
| East Asian | |||||||||||
| Mizushima et al13 | 2001 | Japan | A | 179 (85%) | 34 (81%) | 73 (85%) | 36 (90%) | 9 (82%) | No | No | |
| Yu et al14 | 2002 | China | A | 104 (80%) | 83 (80%) | No | |||||
| Han et al15 | 2004 | China | A | 141 (64%) | 28 (65%) | 50 (65%) | 12 (57%) | Yes (GU vs DU) |
No | ||
| Asian total | 325 (77%) | 145 (77%) | 123 (75%) | 48 (79%) | 9 (82%) | ||||||
| Modified PCR primer pair from primer pair A | |||||||||||
| Western | |||||||||||
| Rad et al16b | 2002 | Germany | B | 141 (38%) | 54 (38%) | Yes | |||||
| Rad et al17d | 2004 | Germany | B | 207 (35%) | 73 (35%) | Yes | |||||
| Zambon et al18 | 2003 | Italy | C | 167 (36%) | 26 (28%) | 20 (49%) | Yes | Yes | |||
| East Asian | |||||||||||
| Lai et al19 | 2002 | Taiwan | D | 101 (100%) | 41 (100%) | 46 (100%) | 14 (100%) | No | No | ||
| PCR primer pair using different strategy | |||||||||||
| Western | |||||||||||
| Mattar et al20 | 2005 | Brazil | E | 104 (69%) | 104 (69%) | No | |||||
| East Asian | |||||||||||
| Sheu et al21 | 2003 | Taiwan | E | 188 (100%) | 98 (100%) | 90 (100%) | No | No | |||
| Lai et al22 | 2005 | Taiwan | E | 143 (100%) | 90 (100%) | 53 (100%) | No | No | |||
PUD, peptic ulcer disease; MALT, mucosa-associated lymphoid tissue.
Reverse-transcription PCR also was performed in this study, and the results were identical to those of PCR.
A total of 88% had German nationality and 12% were from other European countries. The same patients were examined in both studies.
Samples were examined from the antrum and the corpus, and the corpus data are presented (in the antrum, 55 were babA2 positive).
A total of 89% had German nationality and 11% were from other southern European countries.
Materials and Methods
Helicobacter pylori
We examined H pylori isolates cultured from patients in East Asia and North and South America with clinical presentations including simple H pylori gastritis, duodenal ulcer (DU), and noncardiac gastric adenocarcinoma. Ulcers were defined endoscopically, and patients with either ulcer scars or DU and gastric ulcers were excluded. Gastritis was defined as histologic gastritis with no peptic ulcers or gastric cancer. No patients received treatment for their H pylori infection, and those patients who used nonsteroidal anti-inflammatory drugs were excluded. Because of the small number of patients available, gastric cancer patients from the United States were not included.
We obtained gastric biopsy specimens to isolate H pylori. Isolation was performed using standard culture methods according to protocols approved by local ethics committees. To minimize the risk of phase variations, all H pylori samples were obtained from a single bacterial colony that was passaged in vitro less than 4 times.26 The control strain was strain J99,27 which contains a babA gene and possesses Leb binding ability.3 We also used strain 26695,23 which contains a babA gene but does not bind to Leb,3 and strain TN2GF4,28 which contains a babA gene and possesses Leb binding ability. In addition, we constructed isogenic babA mutants from strains J99 and TN2GF4 using methods previously described.29
Histology
Gastric mucosal biopsy specimens were stained with H&E for morphologic observations and either Genta stain30 or El-Zimaity triple stain31 (for US, Colombian, and Korean specimens) or modified Giemsa stain (for Japanese specimens) for detection of H pylori. We examined up to 5 biopsy specimens from the antrum and up to 6 specimens from the corpus. Each specimen was scored for H pylori density, neutrophil infiltration, and atrophy. All of the variables were scored using a visual analogue scale graded from 0 (absent/normal) to 5 (maximal intensity), as described previously.32 Scores for each site were averaged in both the antrum and the corpus specimens.
Detection of BabA Protein and Lewis b Antigen Binding
Whole protein extracts from H pylori isolates were obtained by suspending the bacteria in Laemmli sample buffer and boiling this suspension at 100°C for 10 minutes. Immunoblotting was performed using standard methods. Anti-BabA antiserum (AK277)33 was used as the primary antibody at a 1:5000 dilution. Horseradish-peroxidase– conjugated protein A (1:3000) (Bio-Rad Laboratory, Hercules, CA) was used as the secondary antibody. Detection was performed using enhanced chemiluminescence reagents (Amersham Life Science, Arlington Heights, IL) and radiographic film exposure. The specificity of this antiserum for BabA during immunoblotting was confirmed previously.33 For semiquantitation, 4, 2, and 0.5 μg of J99 strain protein extracts were transferred to each membrane, and the density of the band detected in the 4-μg sample from each strain was expressed numerically using the Image J 1.36 software from the National Institutes of Health (available at: http://rsbweb.nih.gov/ij/). We generated a standard curve using extracts from the J99 strain, and the density of bands detected in the samples from each strain was expressed as the percentage density relative to the J99 strain 4-μg sample. BabA status also was evaluated as the ability to bind to 125I-labeled fucosylated blood group Leb antigens using previously described methods.3
babA Genotypes
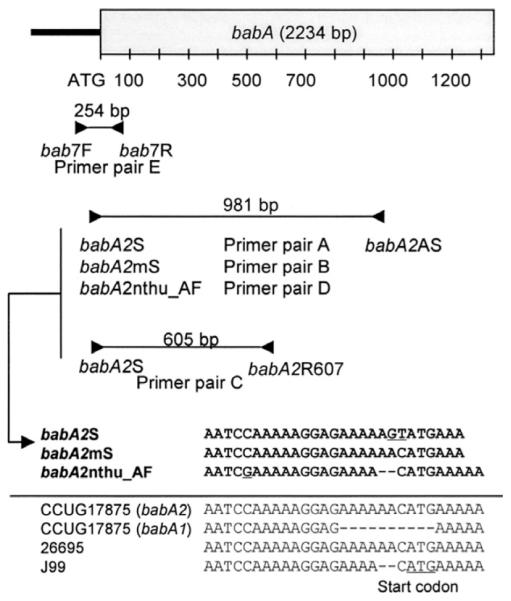
Genomic H pylori DNA was extracted using a commercially available DNA extraction kit (Qiagen Inc., Valencia, CA). babA status (babA2 positive or negative) was determined by PCR using primer pairs and amplification conditions previously described (Figure 1).5–22 Most of these primers were designed to detect the 10-bp signal sequence deletion of babA1, which is absent in babA2. Most previous studies used primer pair A (babA2S and babA2AS) (Table 1) in which 2 nucleotides of the forward primer were changed from the known sequences of the babA2 gene from strain CCUG17875 (ie, AC to GT) (Figure 1). Other published primer pairs (denoted as primer pairs B–D) are modified from this original primer pair (Figure 1). A few studies have used a forward primer (bab7F) that is within the promoter region of the babA gene, a region that is identical to the sequence of babA2 but different from that of babA1 in the strain CCUG17875.20–22 In these studies, they also used a unique reverse primer (bab7R) that is within the 5′ region of the babA gene, a region that is identical in both babA1 and babA2 in the strain CCUG17875 (primer pair E). In some experiments, PCR products were sequenced directly at Macrogen, Ltd. in Seoul, Korea.
Figure 1.
Location of primer pairs used in this study. Most previous studies used primer pair A (babA2S and babA2AS); 2 nucleotides in the forward primer sequence were changed (AC to GT) from the GenBank sequence of the babA2 gene from the strain CCUG17875. The forward primer babA2mS has the same sequence arrangement as the babA2 gene from the strain CCUG17875. This primer was used in combination with the original reverse primer (primer pair B). babA2R607 is a newly designed reverse primer used in combination with the original forward primer (primer pair C). nthu_babA2F is a modified forward primer design based on GenBank sequence data from Taiwanese strains. It has 2 nucleotides deleted near the 3′ end of the original forward primer sequences. This primer was used in combination with the original reverse primer (primer pair D). The forward primer bab7F is located in the promoter region of the babA gene and was used in combination with a unique reverse primer from the 5′ region of the babA gene (bab7R) (primer pair E).
Because babA status has been reported to be related closely to cagA status and vacA genotypes (s region) (Table 1), we also examined cagA status (cagA positive or negative) and vacA s genotypes (s1 or s2) using methods previously described.34
Presence of the babA Gene
By using primers designed by Pride et al,35,36 we tested for the presence of the babA gene (both babA1 and babA2). The presence of the babA gene was confirmed by Southern blotting using 1 μg of genomic DNA digested with either HindIII or SspI and a 542-bp babA-specific probe that was amplified from strain J99 using the following primers: 5′-GCTTACCCGCGCTCAAAG-3′ and 5′-CTCCGTGAAAGGGTTGAAAG-3′. The probe was labeled with horseradish-peroxidase and chemiluminescence was detected using enhanced chemiluminescence reagents (Amersham Pharmacia Biotech) and radiographic film exposure. Samples were scored as babA-gene positive if the PCR and/or Southern blot data yielded a positive result.
babA Messenger RNA Expression
Total H pylori RNA was extracted using a commercially available RNA extraction kit (Qiagen Inc.) and treated with DNase I (Roche Molecular Biochemical, Nutley, NJ). Primers specific for the 16S ribosomal RNA and babA genes were designed by using the Prime3 software (Whitehead Institute for Biomedical Research, Cambridge, MA) (available at: http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi). The sequences of the primers were as follows: 16S ribosomal RNA, 5′-CGTAAGGGCCATGATGACTT-3′ and 5′-CAGCTCGTGTCGTGAGATGT-3′, and babA, 5′-CACGATCAGTTCAAAAG-3′ and 5′-TTRATGAGCGTGCTCGCTTGCG-3′. Reverse transcription, complementary DNA amplification, and real-time PCR were performed using previously described methods,25 with the exception that we used relative quantitative analysis with 16S ribosomal RNA levels as a control for the standardization of babA gene transcriptional activity. In our preliminary experiments, 16S ribosomal RNA levels were similar among the strains we used (data not shown).
Data Analysis
χ2 tests were used for the univariate analysis of the relationship between clinical outcomes and bacterial factors. Mann–Whitney rank sum tests and Kruskal–Wallis tests with the Scheffe test were used for the univariate analysis of the relationship between histology and bacterial factors (cagA status/vacA s genotypes and BabA types, respectively).
A multiple logistic regression analysis was performed to determine which bacterial factor(s) was the most predictive of clinical outcome. Bacterial factors, age, sex, and country were used as explanatory variables. Multiple linear regression analyses were used for analyzing the histologic data. In these analyses, the bacterial factors, sex, age, country, and clinical outcomes were the explanatory variables, and the mutually adjusted associations with the criterion variables were calculated. A P value of less than .05 was accepted as statistically significant. Calculations were performed using the statistical software HALBAU (Gendai-sugaku-sha, Kyoto, Japan).
Results
Lewis b Antigen Binding Assay
We first evaluated the accuracy of assessing BabA status by immunoblotting and PCR analysis as compared with assessing Leb binding activity. Eighty H pylori strains from our Colombian and Japanese H pylori stocks (40 strains from each country) were examined. Leb binding activity was detectable in 68 (85%) of these strains (83% of the Colombian strains and 88% of the Japanese strains).
Comparison Between BabA Immunoblotting and Lewis b Antigen Binding Assay
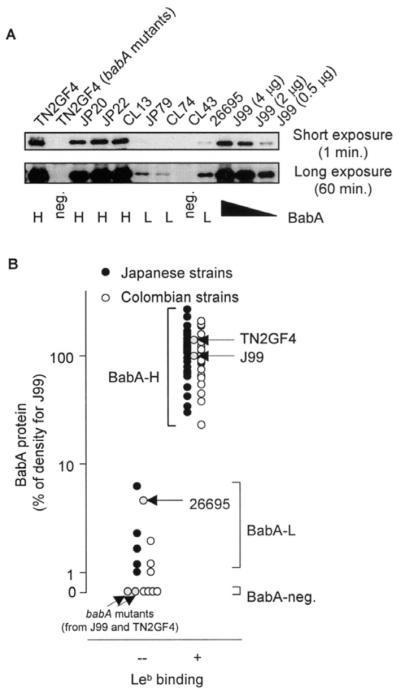
Based on the results of immunoblotting analyses, the H pylori strains were divided into 2 major groups: BabA positive or BabA negative. The BabA-negative strains included 4 Colombian strains with no detectable BabA-specific band even after a long (60-min) exposure (Figure 2). Semiquantitative analyses of the BabA-positive strains allows this group of strains to be classified further into 2 distinct groups: those with high levels of BabA expression (68 strains that express BabA to levels more than 20% of that detected in the J99 strain) or those with low levels of BabA expression (8 strains that express BabA to levels less than 10% of that detected in the J99 strain). All of the strains that showed Leb binding activity were high BabA producers. Both the low- and no-producer strains did not show Leb binding activity. Based on this finding, we classified the strains into 3 distinct groups based on their expression levels of BabA: (1) BabA-high producers (BabA-H), which produce BabA protein to high enough levels to mediate Leb binding, (2) BabA-low producers (BabA-L), which produce a small amount of BabA but not enough to mediate Leb binding, and (3) BabA-negative strains, which do not produce any BabA protein (Figure 2A). Five (10%) Japanese strains and 3 (6%) Colombian strains were classified as BabA-L strains.
Figure 2.
Immunoblot analyses of BabA protein levels. (A) Semiquantitative analyses for BabA protein levels. For semiquantitation, 4, 2, and 0.5 μg of protein extracted from strain J99 was transferred to each membrane along with 4 μg of protein extracted from each experimental strain. The density of the detected band was expressed numerically using Image J 1.36 software (National Institutes of Health). A BabA band was not observed in strain JP79, strain CL74, or strain 26695 (upper panel) after short exposure, whereas the BabA band clearly appeared after longer exposure (lower panel). This result suggests that these strains produced the BabA protein (2.6%, 1.3%, and 6.1% relative to the amount of BabA produced in strain J99 [4 μg]). In contrast, the BabA band did not appear, even after long exposure, in the isogenic babA mutant strains TN2GF4 and strain CL43. Strain names beginning with JP signify strains from Japanese patients; CL signifies strains from Colombian patients. (B) Relationship between the density of BabA band (BabA protein levels) and Leb binding activity. Three groups can be distinguished clearly based on BabA expression levels.
Relation Between the babA Gene and babA Messenger RNA Expression
We tested for the presence of the babA gene using various PCR primer pairs located within the middle regions of the gene.35,36 By using these primers, none of the 4 (5%) clinical isolates classified as BabA-negative contained a babA gene. We also examined the babA gene by PCR using the primer pairs A–E, which can distinguish between babA2 and babA1. As expected, the primer pairs A–D did not yield any PCR products in the BabA-negative strains. Southern blot analyses confirmed that BabA-negative strains lacked the babA gene (data not shown). Interestingly, primer pair E resulted in specific amplification in 2 of the BabA-negative strains, and these products were confirmed by sequence analysis to be the babB gene (data not shown). Analysis of the sequence of primer pair E (bab7F and bab7R) revealed that they are homologous to the babB gene, with 29% of the 72 GenBank-deposited babA sequences being identical to that of the bab7R primer. Because the primer pair E can detect both babA and babB, we did not use this primer pair in subsequent experiments. By definition, babA1 strains do not produce detectable BabA protein and therefore are categorized as BabA-negative strains, although they do possess a babA gene. None of the BabA-negative strains used in the present study contain a babA gene, suggesting that naturally occurring babA1 sequences must be very rare. This hypothesis is supported by the fact that none of the GenBank-deposited babA sequences contain the reported 10-bp deletion. Therefore, assigning BabA status based on PCR techniques to detect the 10-bp deletion is not a reliable method.
We also performed real-time reverse-transcription PCR to analyze babA expression in all BabA-L strains, including the 26695 strain and 10 randomly selected BabA-H strains and the J99 and TN2GF4 strains (Figure 3). babA messenger RNA (mRNA) levels correlated with BabA protein levels as detected by immunoblot, confirming the prediction that BabA expression is regulated at the transcription level.
Figure 3.
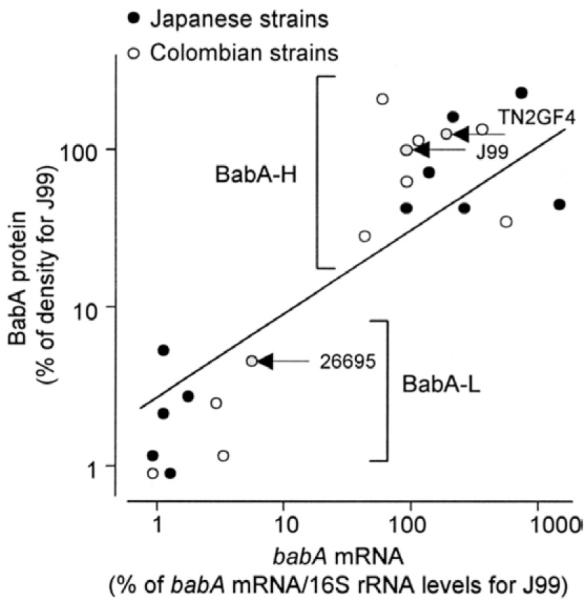
Relationship between the density of BabA band (BabA protein levels) and babA mRNA levels. babA mRNA levels were measured using real-time reverse-transcription PCR, and BabA protein levels were measured using semiquantitative immunoblotting.
Sequencing of the Promoter and Signal Region of the babA Gene
Although all strains containing a babA gene are capable of expressing babA mRNA and producing BabA protein, some strains (BabA-L strains) do not express sufficient babA mRNA/BabA protein levels to mediate Leb binding in vitro. We tested for possible differences between BabA-H and BabA-L strains using direct PCR-based sequencing of the promoter region and signal peptide regions of the babA gene using the following primers: bab7F and babA2AS or babA2R607.5,18,20 We sequenced all 8 of the BabA-L strains and 12 randomly selected BabA-H strains.
BabA status has been reported to be regulated by the number of adenine [poly(A)] residues within the −10 to −35 region of the babA gene promoter.24,25 This region is stable when the number of adenines was 14, but would become nonfunctional when the number was reduced to 10.24 The number of adenine residues was 8 –14 in the strains we analyzed. However, we were unable to confirm any relationship between the number of adenine residues within the −10 to −35 region and BabA status (Figure 4). Although previous reports have suggested that chimeric babA/B or babB/A points were located close to the start codon,24,25 we did not detect any chimeric formation within the region 600 bp from the start codon (data not shown).
Figure 4.
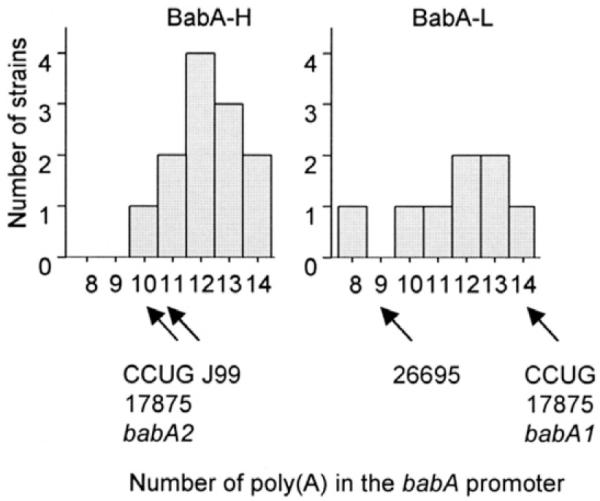
Number of poly(A)s in the babA promoter relative to BabA protein levels. There was no correlation between the number of poly(A)s and BabA expression (eg, between BabA-H and BabA-L strains).
Accuracy of Polymerase Chain Reaction Using Previously Published Primers for babA Gene
The earlier-described results suggest that previously used PCR methods that found the 10-bp difference between the babA1 and babA2 genes are unreliable for determining BabA status. By using Leb binding ability as the gold standard for assigning BabA status, we performed PCR with the previously reported primer pairs and confirmed that these PCR methods had low sensitivity and specificity (Table 2). By definition, BabA-H and BabA-L strains contain babA2 gene sequences within the signal peptide region. However, the primer pairs had very low sensitivity and could not even detect the babA2 gene in the BabA-H and BabA-L strains (eg, only 45% [34 of 76] of the strains yielded a positive result using primer pair A). These data suggested there are sequence variations within these primer regions. Therefore, we examined the 137 GenBank-deposited sequences, including the sequence of the primer babA2AS (from primer pair A, B, and D) and 98 GenBank-deposited sequences, including the sequence of the primer babA2R607 (from primer pair C). Only 14 (10%) and 36 (37%) of the GenBank-deposited sequences were identical to the sequences of the babA2AS and babA2R607 primers, respectively. Importantly, 92 (67%) of the 137 deposited sequences differed from the sequence of primer babA2AS at 1 nucleotide from the 3′ annealing site, a site known to be critical for successful PCR amplification. These results provide further explanation as to why commonly used PCR methods have low sensitivity for assigning BabA status.
Table 2.
Comparison Among PCR, Immunoblot, and Leb Binding for Evaluating BabA Status
| BabA-H | BabA-L | BabA negative | |||
|---|---|---|---|---|---|
| babA gene | + | + | + | ||
| BabA protein | + | weak | − | Lebbinding as gold standard |
|
| Leb binding | + | − | − | Sensitivity, % | Specificity, % |
| Colombia and Japan | n = 68 | n = 8 | n = 4 | ||
| Primer pair A | 29 | 5 | 0 | 43 | 58 |
| Primer pair B | 38 | 8 | 0 | 56 | 33 |
| Primer pair C | 55 | 8 | 0 | 81 | 33 |
| Primer pair D | 46 | 8 | 0 | 68 | 33 |
| Primer pair A–D | 61 | 8 | 0 | 90 | 33 |
| Colombia | n = 33 | n = 3 | n = 4 | ||
| Primer pair A | 12 | 0 | 0 | 36 | 100 |
| Primer pair B | 15 | 3 | 0 | 45 | 57 |
| Primer pair C | 26 | 3 | 0 | 79 | 57 |
| Primer pair D | 26 | 3 | 0 | 79 | 57 |
| Primer pair A–D | 30 | 3 | 0 | 91 | 57 |
| Japan | n = 35 | n = 5 | n = 0 | ||
| Primer pair A | 17 | 5 | — | 49 | 0 |
| Primer pair B | 23 | 5 | — | 66 | 0 |
| Primer pair C | 29 | 5 | — | 83 | 0 |
| Primer pair D | 20 | 5 | — | 57 | 0 |
| Primer pair A–D | 31 | 5 | — | 89 | 0 |
Relationship Between BabA Status and Clinical Presentation
We examined the relationship between the BabA-H, BabA-L, and BabA-negative strain classification, which is based on the presence or absence of detectable BabA protein, and in vitro Leb binding activity, clinical presentation, gastric injury, and the presence of the virulence factors cagA and vacA. We analyzed BabA protein levels in 520 H pylori isolates (including 250 strains from Western countries [150 strains from Colombia, 100 from the United States] and 270 from East Asia [150 from Korea and 120 from Japan]) using immunoblotting.
As noted earlier, the BabA-H strains are defined as those strains that express 20% more BabA protein than the J99 strain, and the BabA-L strains are defined as those strains that express BabA protein to a level that is less than 10% of the level expressed in the J99 strain. BabA-negative strains are defined as those strains with no detectable BabA expression. The 10 strains (1.8%) that expressed BabA protein to levels between 10% and 20% of that in the J99 strain were regarded as strains with a borderline BabA status and were excluded from further analyses.
All strains from East Asia expressed BabA protein. Twenty-four (9.8%) of the Western strains were BabA-negative (Table 3). For these strains, the BabA-negative status was correlated inversely with cagA or vacA s status (ie, only 1 [4.2%] and none [0%] of these BabA-negative strains were cagA- or vacA s1-positive, respectively). Most (91%) Western strains were classified as either cagA-positive/vacA s1-positive/BabA-H (triple positive, 76%), cagA-positive/vacA s1-positive/BabA-L (6.1%), or cagA-negative/vacA s2-positive/BabA-negative strains (9.4%). We were unable to confirm the reported relationship between the triple positive strains and clinical outcome (Table 3).5
Table 3.
Univariate Analysis of Relationship Between H pylori Typing and Clinical Outcomes
| Western countries (n = 244) |
East Asia (n = 266) |
|||||
|---|---|---|---|---|---|---|
| Gastritis n = 98 |
DU n = 97 |
Cancer n = 49 |
Gastritis n = 86 |
DU n = 90 |
Cancer n = 90 |
|
| BabA-H | 75 (77%) | 85 (88%) | 41 (84%) | 81 (94%) | 80 (89%) | 78 (93%) |
| BabA-L | 4 (4.1%) | 9 (9.3%) | 6 (12%) | 5 (5.8%) | 10 (11%) | 12 (13%) |
| BabA negative | 19 (19%) | 3 (3.1%)a | 2 (4.1%)b | 0 | 0 | 0 |
| cagA positive | 77 (79%) | 85 (88%) | 43 (88%) | 82 (95%) | 90 (100%) | 88 (98%) |
| vacA s1 | 76 (78%) | 88 (91%)b | 43 (88%) | 86 (100%) | 90 (100%) | 90 (100%) |
| cagA positive, s1, BabA-H | 72 (73%) | 76 (78%) | 37 (76%) | 77 (90%) | 80 (89%) | 76 (84%) |
| cagA positive, s1, BabA-L | 3 (3.1%) | 8 (8.2%) | 4 (8.2%) | 5 (5.8%) | 10 (11%) | 12 (13%) |
| cagA negative, s2, BabA negative | 18 (18%) | 3 (3.1%)c | 2 (4.1%)b | 0 | 0 | 0 |
| Other types | 5 (5.1%) | 10 (10%) | 6 (12%) | 4 (4.7%) | 0 | 2 (2.2%) |
NOTE. Six samples from Western countries and 4 from East Asia had borderline results for BabA and were excluded. The P value was determined by the χ2 test.
P < .001 compared with gastritis.
P < .05 compared with gastritis.
P < .01 compared with gastritis.
Both BabA-L and BabA-negative strains lacked Leb binding ability in vitro and therefore were either functional or actual BabA-negative strains. However, these strains showed marked differences in their associations with gastric cancer and DU. Patients with BabA-L strains were twice as likely to develop these conditions as gastritis (Table 3). In contrast, BabA-negative strains typically (79%) were associated with gastritis only and their presence was related inversely to the development of DU or gastric cancer (Table 3). Multiple logistic regression analyses confirmed that BabA-negative strains were related inversely to the development of DU or gastric cancer in Western countries (Table 4).
Table 4.
Multiple Logistic Regression Analysis: H pylori Factors Associated With Clinical Presentation
| Factors | P value | Adjusted odds ratio | 95% confidence interval | |
|---|---|---|---|---|
| DU vs gastritis | ||||
| Western countries | BabA | .001 | BabA-L vs -H, 2.6 | 0.8–8.9 |
| BabA-H vs negative, 19.8 | 2.8–137 | |||
| BabA-L vs negative, 54.8 | 6.4–468 | |||
| East Asia | None | — | — | — |
| Gastric cancer vs gastritis | ||||
| Western countries | BabA | .021 | BabA-L vs -H, 1.9 | 0.4–8.3 |
| BabA-H vs negative, 18.2 | 1.7–198 | |||
| BabA-L vs negative, 33.9 | 2.8–411 | |||
| East Asia | None | — | — | — |
Relationship Between BabA Status and Gastric Mucosal Histology
Histologic analyses were performed in gastritis and DU cases. Twenty samples from Korea were not included in this analysis because there were 2 or fewer biopsy specimens available. Independent univariate analysis indicated that BabA status was related to gastric histology both in Western and East Asian countries (Table 5). Gastric mucosal damage and H pylori density was most severe in patients infected with BabA-L strains, less severe in patients infected with BabA-negative strains, and even less severe in patients infected with BabA-H strains. In East Asian countries, no BabA-negative strain samples were found, yet gastric mucosa damage and H pylori density were greater in patients with BabA-L strains than in those with the BabA-H strain. Backward stepwise multiple linear regression analysis confirmed these findings (Table 6).
Table 5.
Univariate Analysis of Relationship Between H pylori Genotyping and Histology
| Antrum |
Corpus |
||||||
|---|---|---|---|---|---|---|---|
| n | H pylori density | Neutrophil infiltration |
Atrophy | H pylori density | Neutrophil infiltration |
Atrophy | |
| Western countries | |||||||
| BabA | |||||||
| BabA-H | 160 | 2.5 (2.7) | 2.6 (2.5) | 1.3 (0.8) | 2.5 (2.7) | 1.8 (1.9) | 0.3 (0) |
| BabA-L | 13 | 3.4 (3.5) | 3.3 (3.0) | 2.1 (2.0) | 3.2 (3.3) | 2.4 (2.0) | 0.8 (0.5) |
| BabA negative | 22 | 2.1 (2.3) | 1.6 (1.4) | 0.8 (0) | 1.8 (2.0) | 1.0 (0.8) | 0.1 (0) |
| P value (L vs H) | <.001 | .083 | .002 | <.001 | .006 | .004 | |
| P value (H vs negative) | NS | .053 | .013 | .029 | .001 | .004 | |
| P value (L vs negative) | <.001 | .003 | <.001 | <.001 | <.001 | <.001 | |
| cagA | |||||||
| Positive | 162 | 2.6 (2.8) | 2.7 (2.5) | 1.4 (1.0) | 2.6 (2.8) | 1.9 (2.0) | 0.4 (0) |
| Negative | 33 | 2.3 (2.5) | 2.0 (2.3) | 0.8 (0) | 2.0 (2.0) | 1.2 (0.9) | 0.1 (0) |
| P value | NS | .043 | .007 | <.001 | .006 | .099 | |
| vacA | |||||||
| s1 | 164 | 2.6 (2.8) | 2.7 (2.5) | 1.3 (1.0) | 2.6 (2.7) | 1.9 (2.0) | 0.3 (0) |
| s2 | 31 | 2.3 (2.5) | 2.0 (2.4) | 0.9 (0.5) | 2.0 (2.0) | 1.2 (1.0) | 0.2 (0) |
| P value | NS | .034 | .062 | .004 | .003 | NS | |
| East Asia | |||||||
| BabA | |||||||
| BabA-H | 141 | 2.6 (2.5) | 2.2 (2.0) | 1.1 (1.0) | 2.2 (2.0) | 1.5 (1.5) | 0.6 (0) |
| BabA-L | 15 | 3.3 (3.0) | 3.1 (3.0) | 2.0 (2.0) | 2.8 (3.0) | 2.1 (2.0) | 1.1 (1.0) |
| P value | .011 | .008 | .015 | .029 | NS | .091 | |
| cagA | |||||||
| Positive | 152 | 2.6 (3.0) | 2.3 (2.0) | 1.1 (1.0) | 2.3 (2.0) | 1.6 (1.5) | 0.6 (1.0) |
| Negative | 4 | 2.0 (1.5) | 1.8 (1.5) | 1.8 (2.0) | 2.3 (1.5) | 1.4 (1.8) | 1.8 (0.5) |
| P value | NS | NS | NS | NS | NS | NS | |
NOTE. Analyses were performed by Mann–Whitney rank sum test (cagA status/vacA s genotypes) and Kruskal-Wallis test with Scheffe test (BabA types). Nine samples (5 from Western countries and 4 from East Asia) had borderline BabA status and were excluded. Twenty samples from Korea also were not included because the number of biopsy specimens was ≤2. For NS, P > .10. Data for vacA s region in Japan were not presented because all strains studied were vacA s1 genotype. For histologic scores (minimum 0, maximum 5), the means (median) are presented.
Table 6.
Final Model Using Multiple Linear Regression Analysis: H pylori Factors Associated With Histology
| Pathology | Sites | Factors | Partial regression coefficient ± SE |
P value | Multiple correlation coefficient |
|---|---|---|---|---|---|
| Western countries | |||||
| H pylori density | Antrum | BabA (L vs H) | 0.77 ± 0.29 | .022 | 0.35 |
| BabA (H vs negative) | 0.12 ± 0.25 | ||||
| BabA (L vs negative) | 0.89 ± 0.36 | ||||
| Corpus | cagA (positive vs negative) | 0.62 ± 0.25 | .013 | 0.43 | |
| BabA (L vs H) | 0.71 ± 0.20 | .019 | |||
| BabA (H vs negative) | −0.01 ± 0.31 | ||||
| BabA (L vs negative) | 0.70 ± 0.37 | ||||
| Neutrophil infiltration | Antrum | BabA (L vs H) | 0.54 ± 0.35 | .012 | 0.41 |
| BabA (H vs negative) | 0.76 ± 0.30 | ||||
| BabA (L vs negative) | 1.30 ± 0.45 | ||||
| Corpus | BabA (L vs H) | 0.62 ± 0.31 | <.001 | 0.27 | |
| BabA (H vs negative) | 0.84 ± 0.26 | ||||
| BabA (L vs negative) | 1.46 ± 0.39 | ||||
| Atrophy | Antrum | BabA (L vs H) | 0.86 ± 0.33 | .022 | 0.40 |
| BabA (H vs negative) | 0.31 ± 0.41 | ||||
| BabA (L vs negative) | 1.17 ± 0.49 | ||||
| cagA (positive vs negative) | 0.69 ± 0.33 | .040 | |||
| Corpus | BabA (L vs H) | 0.44 ± 0.18 | .007 | 0.42 | |
| BabA (H vs negative) | 0.34 ± 0.25 | ||||
| BabA (L vs negative) | 0.78 ± 0.27 | ||||
| cagA (positive vs negative) | 0.52 ± 0.22 | .106 | |||
| East Asia | |||||
| H pylori density | Antrum | BabA (L vs H) | 0.68 ± 0.29 | .023 | 0.32 |
| Corpus | BabA (L vs H) | 0.53 ± 0.29 | .067 | 0.38 | |
| Neutrophil infiltration | Antrum | BabA (L vs H) | 0.85 ± 0.26 | .001 | 0.58 |
| Corpus | BabA (L vs H) | 0.61 ± 0.27 | .029 | 0.40 | |
| Atrophy | Antrum | BabA (L vs H) | 0.88 ± 0.32 | .006 | 0.48 |
| Corpus | BabA (L vs H) | 0.48 ± 0.22 | .027 | 0.58 |
NOTE. In the analyses, the partial regression coefficient 0.77 of BabA (BabA-L vs BabA-H) for H pylori density can be interpreted as showing that the H pylori density score with BabA-L strains would be expected to be 0.77 points greater than with BabA-H strains.
Discussion
We show that PCR-based methods designed to detect a 10-bp deletion in the signal region of the babA gene do not reliably reflect BabA expression as determined by immunoblotting or Leb binding activity. These results call into question the conclusions of previous studies using those techniques to relate BabA functional status and histology or clinical outcome.
BabA-negative status is associated with mild gastric injury and lower H pylori density. BabA-negative strains also are associated infrequently with DU or gastric cancer. However, because BabA-negative status is linked closely to cagA-negative/vacA s2 status, potential interactions between these different putative virulence factors cannot be ruled out. Although we did not measure OipA status in this study, we previously showed that cagA-positive status also is related closely to functional OipA status,37 suggesting that less-virulent strains are BabA negative and CagA negative and contain vacA s2 and a nonfunctional OipA.
We identified a small class of strains (BabA-L) that were BabA-positive but produced low levels of the BabA protein and lacked Leb binding activity. These strains are functionally BabA negative and typically are CagA positive. They are more likely to be associated with DU, gastric cancer, and increased mucosal inflammation and atrophy than BabA-positive strains that show in vitro Leb binding activity (BabA-H strains) and BabA-negative strains. Interestingly, neither BabA-negative nor low-producing BabA strains showed Leb binding activity, yet they differ markedly in their correlation with gastric injury. The finding that BabA-L strains lack Leb binding activity suggests that either gastric mucosal injury is not dependent on Leb binding activity or that in vitro binding activity does not accurately reflect in vivo conditions.
It remains unclear how BabA expression is regulated or if expressing low levels of BabA has a direct role in the pathogenesis of DU or gastric cancer. Although the quantitative analysis of BabA protein levels by immunoblotting has led to the identification of BabA-L strains that infrequently are associated with simple gastritis, the clinical importance of this finding is unclear. It is possible that BabA expression is influenced by the intragastric environment and that the phenotype of the BabA-L strains is a reflection rather than a cause of disease. It also is possible that BabA expression down-regulates the proinflammatory effects of other putative virulence factors, such as the cag pathogenicity island and OipA. Overall, these results suggest that in vitro Leb binding activity does not reflect virulence accurately.
Recent evidence suggests that BabA status may be regulated by transcription24 and/or by the formation of chimeric babA–babB genes.24,25,36 We examined the prediction of Bäckström et al24 that the babA1 gene is transcriptionally silenced by 4 additional adenines between the −10 and −35 sites of the promoter. Such repetitive poly(A) sequences between the −10 and −35 sites also should make this region prone to slippage mutations, thereby possibly altering the transcription levels of downstream genes as well. Our sequence data and the fact that there were 11 adenines in control strain J99, which contains a functional BabA, do not support the hypothesis that differences in the promoter region influence the expression level of BabA. However, because we did not perform primer extension analyses, we are unable to make a definitive statement regarding whether the −10 to −35 spacing plays an important role in BabA expression.
It is possible that strong Leb binding activity is associated with an enhanced immune response resulting in a severely inflamed mucosa. If so, the ability to change BabA status from a high producer to a low producer (ie, Leb binding to Leb nonbinding) would be advantageous for the organism. Solnick et al25 reported the chimeric babA/B formation, which resulted in loss of BabA protein expression during experimental infection of rhesus monkeys with H pylori J166 strains that contained both the babA and babB genes. Those data suggested that, in rhesus monkeys, BabA-producing/Leb binding activity might be disadvantageous and result in a survival advantage for an organism in which binding was switched off. Although we did not find chimeric babA/B or babB/A near the start codon in strains from human beings, it is possible that the mechanism(s) that might regulate BabA status in monkeys is similar to that in human beings. Our finding that a BabA-positive/Leb nonbinding phenotype is associated with severe inflammation, DU, and gastric cancer is consistent with the hypothesis that BabA-L reflects an adaptation of H pylori that enhances survival in inflamed stomachs.
Finally, it should be noted that in a previous study we evaluated the relationship between clinical outcome (gastritis vs DU) and BabA status using immunoblotting with the anti-BabA antiserum AK253, which is directed against amino acid 55-725.38 In the present study, we used the anti-BabA antiserum AK277 that is directed against recombinant BabA containing amino acid 123-432. Our preliminary data show that AK277 more accurately reflects Leb binding activity than does AK253 (data not shown). Possibly, the use of the longer peptide fragments resulted in production of antibodies that are capable of cross-reacting with other H pylori outer-membrane proteins.
Acknowledgments
This report is based on work supported in part by grants from the National Institutes of Health (R01 DK62813 to Y.Y.), the Office of Research and Development, Medical Research Service Department of Veterans Affairs (to D.Y.G.), the Deutsche Forschungsgemeinschaft (project OD 21/1-1) (to S.O.), and a Public Health Service grant (DK56338) that funds the Texas Gulf Coast Digestive Diseases Center. O.O.O. was supported by a UNESCO BAC short-term fellowship and a Macarthur Foundation/University of Ibadan Staff Training Grant.
The authors thank Dr Oscar Gutierrez (Universidad Nacional de Colombia, Bogota, Colombia), the late Dr Jong G. Kim (Guru Hospital, Korea University College of Medicine, Seoul, Korea), Dr Tadashi Kodama and Dr Shoji Mitsufuji (Kyoto Prefectural University of Medicine, Kyoto, Japan), and Dr Masahiro Asaka and Dr Mototsugu Kato (Hokkaido University of Medicine, Sapporo, Japan) for providing clinical samples, and Dr Hala El-Zimaity (Baylor College of Medicine, Houston, TX) for histologic evaluations.
Abbreviations used in this paper
- BabA
blood group antigen binding adhesin
- BabA-H
BabA high producers
- BabA-L
BabA low producers
- bp
base pair
- DU
duodenal ulcer
- Leb
Lewis b antigen
- PCR
polymerase chain reaction
References
- 1.Borén T, Falk P, Roth KA, et al. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892–1895. doi: 10.1126/science.8018146. [DOI] [PubMed] [Google Scholar]
- 2.Mahdavi J, Sonden B, Hurtig M, et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ilver D, Arnqvist A, Ogren J, et al. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 4.Aspholm-Hurtig M, Dailide G, Lahmann M, et al. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science. 2004;305:519–522. doi: 10.1126/science.1098801. [DOI] [PubMed] [Google Scholar]
- 5.Gerhard M, Lehn N, Neumayer N, et al. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci U S A. 1999;96:12778–12783. doi: 10.1073/pnas.96.22.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prinz C, Schoniger M, Rad R, et al. Key importance of the Helicobacter pylori adherence factor blood group antigen binding adhesin during chronic gastric inflammation. Cancer Res. 2001;61:1903–1909. [PubMed] [Google Scholar]
- 7.Oleastro M, Gerhard M, Lopes AI, et al. Helicobacter pylori virulence genotypes in Portuguese children and adults with gastroduodenal pathology. Eur J Clin Microbiol Infect Dis. 2003;22:85–91. doi: 10.1007/s10096-002-0865-3. [DOI] [PubMed] [Google Scholar]
- 8.Podzorski RP, Podzorski DS, Wuerth A, et al. Analysis of the vacA, cagA, cagE, iceA, and babA2 genes in Helicobacter pylori from sixty-one pediatric patients from the Midwestern United States. Diagn Microbiol Infect Dis. 2003;46:83–88. doi: 10.1016/s0732-8893(03)00034-8. [DOI] [PubMed] [Google Scholar]
- 9.Oliveira AG, Santos A, Guerra JB, et al. babA2- and cagA-positive Helicobacter pylori strains are associated with duodenal ulcer and gastric carcinoma in Brazil. J Clin Microbiol. 2003;41:3964–3966. doi: 10.1128/JCM.41.8.3964-3966.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehours P, Menard A, Dupouy S, et al. Evaluation of the association of nine Helicobacter pylori virulence factors with strains involved in low-grade gastric mucosa-associated lymphoid tissue lymphoma. Infect Immun. 2004;72:880–888. doi: 10.1128/IAI.72.2.880-888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatti LL, Fagundes e Souza EK, Leite KR, et al. cagA vacA alleles and babA2 genotypes of Helicobacter pylori associated with gastric disease in Brazilian adult patients. Diagn Microbiol Infect Dis. 2005;51:231–235. doi: 10.1016/j.diagmicrobio.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Olfat FO, Zheng Q, Oleastro M, et al. Correlation of the Helicobacter pylori adherence factor BabA with duodenal ulcer disease in four European countries. FEMS Immunol Med Microbiol. 2005;44:151–156. doi: 10.1016/j.femsim.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Mizushima T, Sugiyama T, Komatsu Y, et al. Clinical relevance of the babA2 genotype of Helicobacter pylori in Japanese clinical isolates. J Clin Microbiol. 2001;39:2463–2465. doi: 10.1128/JCM.39.7.2463-2465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J, Leung WK, Go MY, et al. Relationship between Helicobacter pylori babA2 status with gastric epithelial cell turnover and premalignant gastric lesions. Gut. 2002;51:480–484. doi: 10.1136/gut.51.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han YH, Liu WZ, Zhu HY, et al. Clinical relevance of iceA and babA2 genotypes of Helicobacter pylori in a Shanghai population. Chin J Dig Dis. 2004;5:181–185. doi: 10.1111/j.1443-9573.2004.00175.x. [DOI] [PubMed] [Google Scholar]
- 16.Rad R, Gerhard M, Lang R, et al. The Helicobacter pylori blood group antigen-binding adhesin facilitates bacterial colonization and augments a nonspecific immune response. J Immunol. 2002;168:3033–3341. doi: 10.4049/jimmunol.168.6.3033. [DOI] [PubMed] [Google Scholar]
- 17.Rad R, Dossumbekova A, Neu B, et al. Cytokine gene polymorphisms influence mucosal cytokine expression, gastric inflammation, and host specific colonisation during Helicobacter pylori infection. Gut. 2004;53:1082–1089. doi: 10.1136/gut.2003.029736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zambon CF, Navaglia F, Basso D, et al. Helicobacter pylori babA2, cagA, and s1 vacA genes work synergistically in causing intestinal metaplasia. J Clin Pathol. 2003;56:287–291. doi: 10.1136/jcp.56.4.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai CH, Kuo CH, Chen YC, et al. High prevalence of cagA- and babA2-positive Helicobacter pylori clinical isolates in Taiwan. J Clin Microbiol. 2002;40:3860–3862. doi: 10.1128/JCM.40.10.3860-3862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattar R, dos Santos AF, Eisig JN, et al. No correlation of babA2 with vacA and cagA genotypes of Helicobacter pylori and grading of gastritis from peptic ulcer disease patients in Brazil. Helicobacter. 2005;10:601–608. doi: 10.1111/j.1523-5378.2005.00360.x. [DOI] [PubMed] [Google Scholar]
- 21.Sheu BS, Sheu SM, Yang HB, et al. Host gastric Lewis expression determines the bacterial density of Helicobacter pylori in babA2 genopositive infection [erratum: 2005;54:442] Gut. 2003;52:927–932. doi: 10.1136/gut.52.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai CH, Poon SK, Chen YC, et al. Lower prevalence of Helicobacter pylori infection with vacAs1a, cagA-positive, and babA2-positive genotype in erosive reflux esophagitis disease. Helicobacter. 2005;10:577–585. doi: 10.1111/j.1523-5378.2005.00363.x. [DOI] [PubMed] [Google Scholar]
- 23.Tomb JF, White O, Kerlavage AR, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 24.Bäckström A, Lundberg C, Kersulyte D, et al. Metastability of Helicobacter pylori bab adhesin genes and dynamics in Lewis b antigen binding. Proc Natl Acad Sci U S A. 2004;101:16923–16928. doi: 10.1073/pnas.0404817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solnick JV, Hansen LM, Salama NR, et al. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc Natl Acad Sci U S A. 2004;101:2106–2111. doi: 10.1073/pnas.0308573100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaoka Y, Ojo O, Fujimoto S, et al. Helicobacter pylori outer membrane proteins and gastroduodenal disease. Gut. 2006;55:775–781. doi: 10.1136/gut.2005.083014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alm RA, Ling LS, Moir DT, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori [erratum: 1999;397:719] Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe T, Tada M, Nagai H, et al. Helicobacter pylori infection induces gastric cancer in Mongolian gerbils. Gastroenterology. 1998;115:642–648. doi: 10.1016/s0016-5085(98)70143-x. [DOI] [PubMed] [Google Scholar]
- 29.Yamaoka Y, Kwon DH, Graham DY. A Mr 34,000 proinflammatory outer membrane protein (oipA)of Helicobacter pylori. Proc Natl Acad Sci U S A. 2000;97:7533–7538. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genta RM, Robason GO, Graham DY. Simultaneous visualization of Helicobacter pylori and gastric morphology: a new stain. Hum Pathol. 1994;25:221–226. doi: 10.1016/0046-8177(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 31.El-Zimaity HM, Ota H, Scott S, et al. A new triple stain for Helicobacter pylori suitable for the autostainer: carbol fuchsin/Alcian blue/hematoxylin-eosin. Arch Pathol Lab Med. 1998;122:732–736. [PubMed] [Google Scholar]
- 32.El-Zimaity HMT, Graham DY, Al-Assi MT, et al. Interobserver variation in the histopathological assessment of Helicobacter pylori gastritis. Hum Pathol. 1996;27:35–41. doi: 10.1016/s0046-8177(96)90135-5. [DOI] [PubMed] [Google Scholar]
- 33.Odenbreit S, Kavermann H, Puls J, et al. CagA tyrosine phosphorylation and interleukin-8 induction by Helicobacter pylori are independent from alpAB, HopZ and bab group outer membrane proteins. Int J Med Microbiol. 2002;292:257–266. doi: 10.1078/1438-4221-00205. [DOI] [PubMed] [Google Scholar]
- 34.Yamaoka Y, Kodama T, Gutierrez O, et al. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol. 1999;37:2274–2279. doi: 10.1128/jcm.37.7.2274-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pride DT, Meinersmann RJ, Blaser MJ. Allelic variation within Helicobacter pylori babA and babB. Infect Immun. 2001;69:1160–1171. doi: 10.1128/IAI.69.2.1160-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pride DT, Blaser MJ. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J Mol Biol. 2002;316:629–642. doi: 10.1006/jmbi.2001.5311. [DOI] [PubMed] [Google Scholar]
- 37.Yamaoka Y, Kikuchi S, el-Zimaity HM, et al. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology. 2002;123:414–424. doi: 10.1053/gast.2002.34781. [DOI] [PubMed] [Google Scholar]
- 38.Yamaoka Y, Souchek J, Odenbreit S, et al. Discrimination between cases of duodenal ulcer and gastritis on the basis of putative virulence factors of Helicobacter pylori. J Clin Microbiol. 2002;40:2244–2246. doi: 10.1128/JCM.40.6.2244-2246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]