Abstract
Cholesterol is a critical component of neuronal membranes, required for normal signal transduction. We showed previously that adult hippocampal neurons co-express high levels of cholesterogenic enzymes, and that their expression is under the control of the p75 neurotrophin receptor (p75NTR). Most of the cellular effects of p75NTR are mediated via interacting proteins, including neurotrophin receptor interacting factor (NRIF). In this study, we tested the hypothesis that p75NTR-dependent regulation of cholesterol and lipid biosynthesis genes is mediated by NRIF. We found that in vitro down regulation of NRIF expression decreased the mRNA for two main cholesterogenic enzymes, 3-hydroxy-3-methylglutaryl-coenzyme A reductase (Hmgcr; EC 2.3.3.10) and 7-dehydrocholesterol reductase (Dhcr7; EC 1.3.1.21). Further analyses revealed that NRIF-dependent and Dhcr7-dependent transcriptional changes show a high degree of overlap, and that NRIF reduction resulted in reduced expression of sterol-sensing domain protein SCAP, followed by a decrease in mRNA levels of SRE-motif containing genes (HMGCR, FASN, SREBP2, S1P, and SQS1). Finally, a reduction in cholesterol biosynthesis-related gene expression was also observed in hippocampal tissue of mice with NRIF deletion. Our combined in vitro and in vivo studies suggest that hippocampal neuronal cholesterol biosynthesis is regulated through the p75NTR interacting factor NRIF.
Keywords: Cholesterol, Neuroblastoma, NRIF KO mice, SREBP, Gene expression, p75NTR, Neurotrophin
Introduction
Cholesterol is critical for the normal function of the mammalian brain (Dietschy and Turley 2001). Recent studies have demonstrated that disturbances in cholesterol homeostasis result in altered CNS structure and function, leading to a wide variety of disorders (Maxfield and Tabas 2005). For example, deficient cholesterol biosynthesis in oligodendrocytes delays myelination (Saher et al. 2005) and various mutations in enzymes of the cholesterol biosynthesis pathway lead to pathologies that include a neurological phenotype (Nwokoro et al. 2001). Furthermore, defects in cholesterol trafficking lead to Niemann–Pick Type C disease (Paul et al. 2004), while altered cholesterol biosynthesis is seen in Huntington’s disease (Valenza and Cattaneo 2006). In addition, partial or complete lack of 7-dehydrocholesterol reductase (Dhcr7) enzymatic activity leads to severe developmental malformations and mental retardation, as seen in patients with Smith–Lemli–Opitz syndrome (Smith et al. 1964; Tierney et al. 2000).
Studies by Suzuki et al. 2007 showed that BDNF can regulate the expression level of cholesterol and cholesterogenic enzymes in primary cortical neurons (Suzuki et al. 2007). In addition, our recent studies revealed that cholesterogenic enzymes are co-expressed with p75 neurotrophin receptor (p75NTR) at a high level in adult hippocampal and cholinergic neurons (Korade et al. 2007), and that genes critically involved in cholesterol biosynthesis are regulated through p75NTR. This was observed in both neuroblastoma cell lines and primary cerebellar neurons (Korade et al. 2007).
Most of the cellular effects of p75NTR are mediated via interacting proteins, including neurotrophin receptor interacting factor (NRIF, zfp110) (Casademunt et al. 1999). NRIF is a zinc finger, DNA-binding protein that associates with the intracellular domain (ICD) of p75NTR (Casademunt et al. 1999). Upon ligand binding, p75NTR undergoes proteolysis by the metalloprotease TACE followed by the gamma-secretase complex (Kenchappa et al. 2006). This processing releases the ICD of the receptor and NRIF, which then undergoes ubiquitination by Traf6, an E3 ubiquitin ligase that associates with p75NTR, and nuclear translocation (Geetha et al. 2005; Kenchappa et al. 2006). Presumably, once in the nucleus NRIF binds specific DNA sequence and acts as transcriptional repressor. However, the specific downstream target genes have yet to be determined. Recent evidence suggests that NRIF is involved in p75NTR-mediated apoptosis in sympathetic neurons (Kenchappa et al. 2006). However, its spatial and temporal distribution is much broader than that of p75NTR, suggesting that it has other roles beyond p75NTR signaling (Kendall et al. 2003). Moreover, activation of p75NTR can lead to a wide variety of responses from regulation of neurite outgrowth to survival (Chao and Bothwell 2002; Schor 2005; Yano and Chao 2004). Which of these signaling pathways other than cell death involve NRIF is not known (Gentry et al. 2004).
The current study hypothesized that the p75NTR effect on the expression of neuronal cholesterol biosynthesis genes is mediated through NRIF. Based on this overall hypothesis, we put forth several testable predictions: (1) NRIF down regulation will lead to altered expression of Hmgcr and Dhcr7, two critical enzymes of the cholesterol biosynthesis pathway, (2) NRIF- and Dhcr7-dependent transcript changes will overlap, (3) NRIF reduction will affect the expression of genes that contain sterol-regulatory element sequences, and (4) the most critical changes in the expression of cholesterol biosynthesis genes produced by absence of NRIF will be present under both in vitro and in vivo conditions.
Materials and Methods
Cell Culture and Reagents
Neuroblastoma cell lines were purchased from American Type Culture Collection (Rockville, MD, USA). Neuro2a cells were maintained in alpha modification of minimal essential medium (Eagle) with Earle’s salt, and supplemented with L-glutamine, sodium bicarbonate, non-essential amino acids, sodium pyruvate, 10% fetal bovine serum (FBS) (Thermo Scientific HyClone), penicillin/streptomycin at 37°C and 5% CO2. To evaluate the role of lipoproteins on gene expression, cells were cultured with medium containing (a) 10% FBS serum or (b) 10% lipid reduced FBS containing no detectable cholesterol levels (Thermo Scientific HyClone lipid reduced FBS). The Neuro2a cells were subcultured once a week, and the culture medium was changed every 2 days. shRNA (short hairpin RNA) plasmids (OligoID V2LMM_27337 pGIPZ and Non-silencing-GIPZ controls) were purchased from Open Biosystems through Vanderbilt Microarray Shared Resource. These plasmids contain short hairpin RNA expressed as human microRNA-30 primary transcripts. They also contain turbo green florescent protein sequence and the shRNAmir allows visualization of construct-expressing cells. Furthermore, the transfected cells express a puromycin drug resistance marker for selecting stable cell lines.
Stable shRNA Transfections
Neuro2a cells were cultured for 2 days before transfections. Cells were transfected with either NRIF shRNA or Non-silencing shRNA plasmids (Open Biosystems) using a Nucleofector instrument and Nucleofector kit V (Amaxa GmbH, Cologne, Germany) optimized for use with Neuro2a cells. Briefly, 2×106 cells were resuspended in 100 μl of transfection buffer (Amaxa) and shRNA plasmid was added to cells that were transferred to the cuvettes and electroporated using program T-24. To establish stable down regulation of NRIF, after transfection of cells with pGIPZ plasmids, cells were grown in presence of puromycin. The expression of NRIF was monitored by immunoprecipitation and Western blotting (Fig. 1) and quantitative reverse transcription polymerase chain reaction (RT-PCR) using NRIF-specific primers (Fig. 2).
Figure 1.
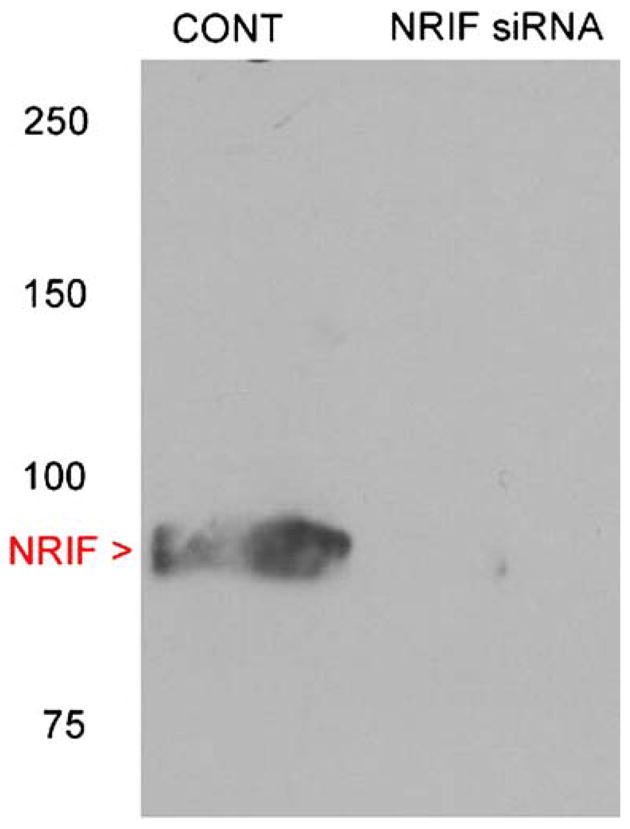
Western blotting of NRIF protein expression in shRNA-transfected Neuro2a cells. Control non-silencing shRNA- and NRIF-shRNA-transfected Neuro2a cells were cultured for 5 days, lysed, immunoprecipitated with NRIF antibody and analyzed by Western blotting. The blot was probed with anti-NRIF antibody. While control cells express NRIF protein, NRIF-shRNA-transfected cells do not show any NRIF protein expression
Figure 2.
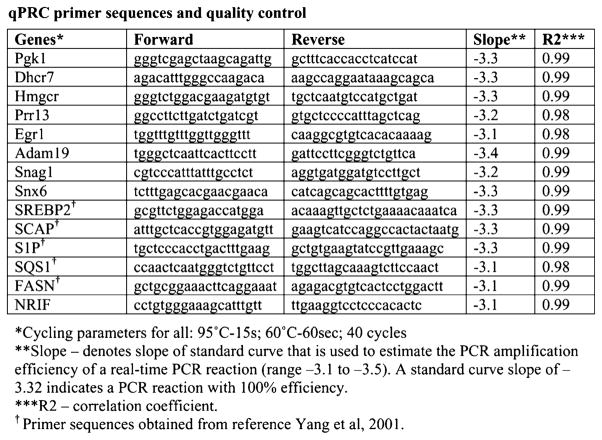
qPCR primer sequences and quality control All mouse gene-specific primers used in our experiments showed a slope between −3.10 and −3.58 with R2>0.98. Lipid gene specific qPCR primers were described in (Yang et al. 2001)
RNA Preparation and Quantitative PCR
Total RNA was isolated from the cells using Trizol (Life Technologies, Rockville, MD, USA). Purification of total RNA was performed using RNeasy Mini Kit (Qiagen) and it included on-column digestion of DNA during RNA purification step using RNase-Free DNase Set (Qiagen). The concentration of total RNA was measured on a NanoDrop instrument (Thermo Scientific, Wilmington, DE) and the integrity of the RNA was established by electrophoresis using the Agilent Bioanalyzer 2100 instrument (Agilent Technologies, Santa Clara, CA, USA).
Total RNA (100 ng) from each sample was reverse transcribed to cDNA using High Capacity cDNA Archive Kit (Applied Biosystems, Foster, City, CA, USA). Real-time PCR was performed with an ABI Prism 7300 System (Applied Biosystems, Foster, City, CA, USA) using 1 ng cDNA per 50-μl reaction volume, 2X SYBR green master mix and gene-specific primers. All samples were run in triplicate. Data from the PCR reactions were analyzed using the comparative cycle number determined as threshold (Ct) method (Kurrasch et al. 2004). Differential expression was calculated as ΔΔCt against expression of Pgk1 as normalizer. We designed gene-specific primers (~20 bp) to yield 85–110 bp PCR amplicons using Primer3 software (http://frodo.wi.mit.edu/) for different genes. For each gene, we designed four sets of primers. Each primer set was tested to determine the efficiency of PCR using no template and serial dilution of a specific template. From this PCR, we calculated the efficiency of PCR primers and R2 value (coefficient of correlation). All mouse gene-specific primers are listed in Fig. 2 and (Yang et al. 2001). The validated primer sets showed slopes between −3.10 and −3.58 with R2> 0.99. Dissociation curve analysis was performed using Dissociation Curve 1.0 software (ABI) for each PCR reaction.
NRIF1 KO Mice
Mice with NRIF deletion were described previously (Casademunt et al. 1999). Sv129NRIF1−/− mice were kept and bred in Division of Animal Care facilities at Vanderbilt University. Whole brain was removed from the skull and placed into Hank’s Balanced Salt Solution. The hippocampi were dissected using a Leica microscope following a procedure described by Banker and Goslin (Goslin et al. 1998). All procedures were performed in accordance with the Guide for the Humane Use and Care of Laboratory Animals and the use of animals in this study was approved by the IACUC of the Vanderbilt University.
Cell Lysis, Immunoprecipitation, and Immunoblotting
Western blotting was performed as described previously (Mirnics et al. 2005). Briefly, the cultured cells were lysed in lysis buffer (150 mM NaCl, 50 mM Tris-HCL pH 7.4, 5 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate with PMSF, protease inhibitor cocktail [AEBSF, Aprotinin, Bestatin hydrochloride, E-64, Leupeptin hemisulfate salt, Pepstatin A] and phosphatase inhibitor cocktail [Sodium orthovana-date, Sodium molybdate, Sodium tartrate, Imidazole] by Sigma), sonicated and subjected to immunoprecipitation. Protein concentration was determined by modified Lowry assay (Bio-Rad DC protein assay kit). NRIF was immunoprecipitated using affinity purified NRIF antibody overnight at +4°C as described previously (Kenchappa et al. 2006), then incubated with protein A sepharose for 2 h at +4°C and subjected to SDS-PAGE. After transfer of proteins to a nitrocellulose membrane and blotting with 5% milk in TBST, the blot was probed with primary (rabbit polyclonal anti-NRIF) and then secondary (anti-rabbit or HRP coupled IgG—Sigma) antibody. The expression level of the proteins against which the primary antibodies were directed was determined using enhanced chemiluminescence reagent (Western Blotting Luminol Reagent, Pierce) and exposing membranes to X-ray film (Kodak MR film).
Results
NRIF Regulates Transcription of Hmgcr and Dhcr7
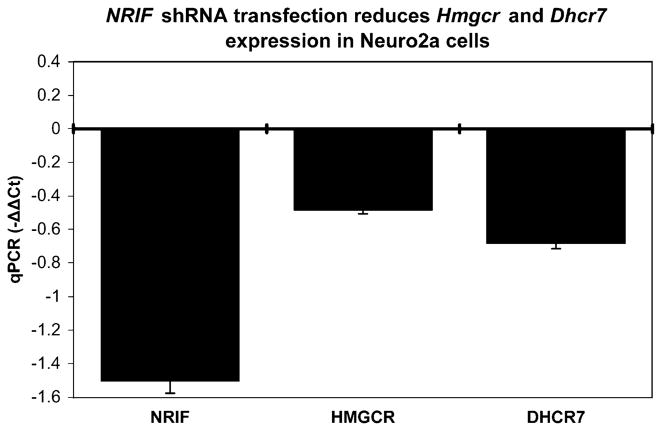
The Neuro2a neuroblastoma cell line expresses the p75NTR and its intracellular interacting protein, Neurotrophin receptor interacting factor, which is believed to function as a transcriptional repressor. After p75NTR cleavage by gamma-secretase, NRIF binds the p75ICD and is translocated into the nucleus (Kenchappa et al. 2006). Since our previous studies showed that p75NTR regulates expression of the cholesterogenic enzymes Hmgcr and Dhcr7, the first and last enzymes in the cholesterol biosynthesis pathway, we hypothesized that this regulation is mediated through NRIF. To test our hypothesis, we developed stably transfected Neuro2a cells with down regulated NRIF expression. Neuro2a cells were transfected with either pGIPZ NRIF shRNA or pGIPZ non-silencing-shRNA plasmid. Both constructs contained a puromycin selection cassette and GFP, which enabled monitoring the expression of the shRNA. We used qPCR and immunoprecipitation and Western blotting to confirm the shRNA-induced reduction of NRIF mRNA and protein expression (Figs. 1, 2, and 3). QPCR analysis of NRIF-shRNA-transfected and control cells demonstrated that both Hmgcr and Dhcr7 were down regulated in NRIF-shRNA expressing Neuro2a cells (−ΔΔCt=−0.48, p<0.01 and −ΔΔCt= −0.68, p<0.01, respectively) (Fig. 3), suggesting that NRIF is involved in regulation of cholesterogenic transcripts.
Figure 3.
NRIF regulates transcription of Hmgcr and Dhcr7, the first and last enzymes in cholesterol biosynthesis pathway. Neuro2a cells were transfected with either NRIF shRNA or non-silencing shRNA and stable cell lines were established using puromycin selection. qPCR analysis shows that in addition to NRIF down regulation, both Hmgcr and Dhcr7 transcripts were significantly down regulated (-ΔΔCt=−0.48, p<0.01 and -ΔΔCt=−0.68, p<0.01, respectively)
NRIF-dependent and Dhcr7-dependent Transcript Changes Overlap in Neuro2a Cells
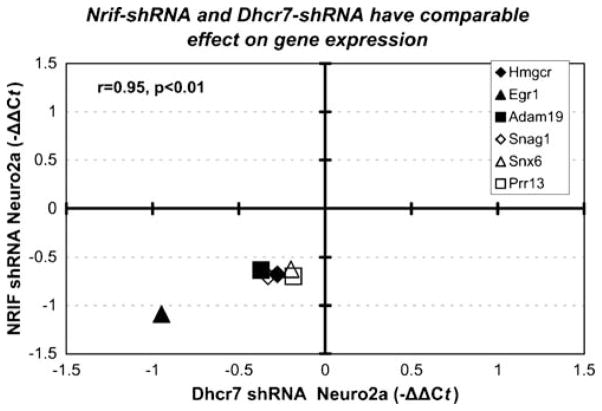
The NRIF-dependent regulation of Dhcr7 expression suggested that Dhcr7-dependent changes will be also observed after NRIF silencing. Our recent microarray study of Dhcr7-shRNA-transfected Neuro2a cells identified multiple Dhcr7-dependent transcripts, several of which were validated by qPCR. Therefore, we analyzed the expression of these genes in the NRIF-shRNA-silenced Neuro2a cells. As predicted, we observed a strong overlap between Dhcr7-dependent and NRIF-dependent transcripts (Fig. 4), which included comparable and highly correlated (Pearson correlation r=0.95, p<0.01) expression changes in Hmgcr, Egr1 (early growth response 1), Adam19 (a disintegrin and metallopeptidase domain 19), Prr13 (proline rich 13), Snag1 (sorting nexin associated golgi protein 1) and Snx6 (sorting nexin 6).
Figure 4.
NRIF-dependent transcript changes are similar to those observed in Dhcr7-deficient Neuro2a cells. Both axes denote the average −ΔΔCt from three independent experiments, two independent reverse transcriptions for each experiment and four replicates for each RT. −ΔΔCt was calculated against Pgk1 as a normalizer. X-axis values are derived form qPCR analysis of Dhcr7-shRNA-transfected Neuro2a cells (Korade et al. 2008; minor revisions); Y-axis values are derived from qPCR analysis of stably transfected NRIF shRNA Neuro2a cells. Each symbol represents a single gene. Note that the expression changes were virtually identical across both systems (r= 0.95, p<0.01)
NRIF Silencing Leads to Reduced Expression of Genes Involved in Sterol and Fatty Acid Biosynthesis
Since both Dchr7 and Hmgcr were regulated by NRIF, and NRIF-dependent and Dhcr7-dependent cholesterol biosynthesis transcripts significantly overlapped, we further hypothesized that NRIF is critical for the regulation of cholesterol-dependent transcripts. We focused our attention on the sterol-regulatory element binding proteins (SREBPs), which have a central role in control of lipid homeostasis (Bengoechea-Alonso and Ericsson 2007; Brown and Goldstein 1999). SREBPs are transcription factors that bind to DNA sterol-regulatory element sequences found in the promoter regions of many genes involved in cholesterol and fatty acid biosynthesis (Yokoyama et al. 1993) and regulate their expression and cholesterol homeostasis (Eberle et al. 2004).
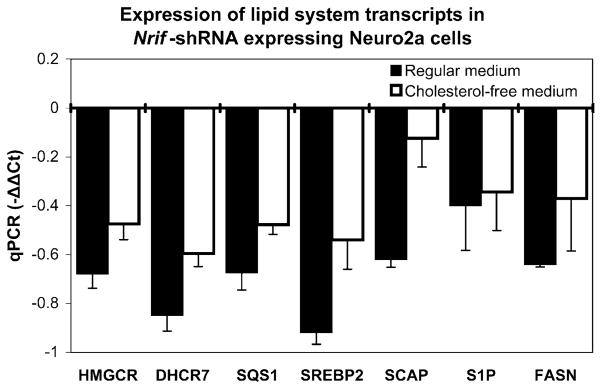
Using qPCR, we compared the expression level of lipid metabolic genes between the cells transfected with NRIF shRNA and non-silencing control shRNA. All tested lipid-related genes, which included fatty acid synthase (FASN), sterol-regulatory element binding protein 2 (SREBP2), SREBF chaperone (SCAP), site-1 protease (S1P, MBTPS1), and squalene synthase (SQS1), showed significantly reduced expression in the NRIF-shRNA-transfected Neuro2a cells (Fig. 5). This suggests that NRIF is a strong regulator of the SREBP system in cells of neuronal origin, further confirming the critical role of NRIF in the regulation of neuronal cholesterol homeostasis.
Figure 5.
NRIF-shRNA expressing cells show altered expression of lipid transcripts in both cholesterol-deficient and regular medium. The experiment was performed with the same parameters as described in Fig. 4. Note that all tested lipid system-related transcripts (HMGCR, DHCR7, FASN, SREBP2, SCAP, S1P, and SQS1) showed significantly reduced expression in NRIF-shRNA-transfected cultures compared to cells transfected with non-silencing shRNA, regardless if the cells were grown in regular or cholesterol-deficient cell culture media
Exogenous Cholesterol has no Effect on NRIF-shRNA-mediated Expression Changes
In the absence of intrinsic cholesterol production, cells can obtain cholesterol from the extracellular environment. To test if the observed expression changes can be modulated by addition of external cholesterol, we cultured cells in either regular medium (that contains 10% FBS) or medium with cholesterol-deficient serum (lipid-reduced 10% FBS; LRS medium). Stably transfected control and NRIF-deficient cells showed similar transcript changes in either the presence of regular or cholesterol-deficient serum (Fig. 5), suggesting that addition of exogenous cholesterol cannot supplement for endogenous cholesterol synthesis in cells of neuronal origin.
NRIF-dependent Transcripts are Similar in Neuro2a and NRIF KO Mice
While Neuro2a cells are of neuronal origin and represent an easy to use in vitro system, we wanted to test if our findings would be reproduced in an in vivo system, where neuronal cholesterol homeostasis may be also regulated by additional factors. Therefore, we analyzed the expression of the genes whose levels were changed by NRIF depletion in Neuro2a cells in the hippocampus of NRIF KO mice (Casademunt et al. 1999). Postnatal day 7 hippocampus (P7) was isolated from wild type NRIF +/+ and NRIF−/− mice and the expression levels of the previously discussed genes analyzed by qPCR (Adam19, Prr13, Snag1, Snx6, Hmgcr, Fasn, Srebp2, S1p, and Sqs1). We found a high concordance (r=0.62, p<0.05) between the effects of NRIF ablation in vivo (NRIF KO mice) and in vitro (NRIF-shRNA expressing Neuro2a cells) in the expression of the cholesterol biosynthesis genes (Fig. 6).
Figure 6.
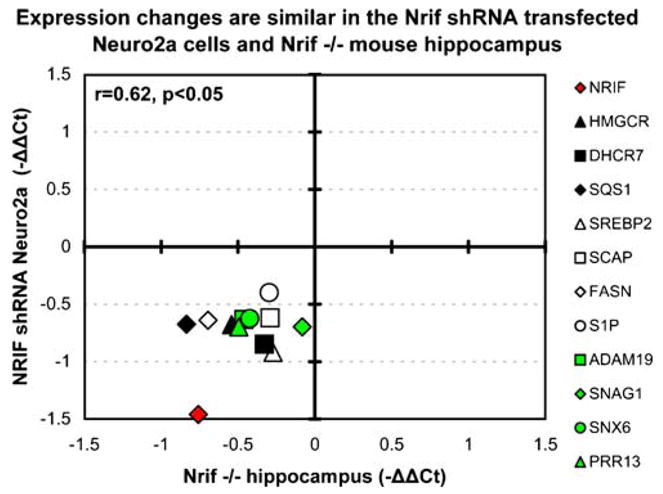
Transcript changes are similar in NRIF-shRNA expressing Neuro2a cells and NRIF−/− mice. The figure layout is similar to that in Fig. 4 Black symbols denote cholesterol biosynthesis genes, white symbols correspond to genes with SRE binding domain, while green symbols denote Dhcr7-regulated genes. Note that all of these gene expression changes in the NRIF-shRNA expressing Neuro2a cells are highly correlated to those observed in hippocampus of 7-day old NRIF KO mice (r=0.62, p<0.05)
Discussion
Our previous study demonstrated that p75NTR regulates the expression of cholesterogenic enzymes in both Neuro2a cells and primary neuronal cultures (Korade et al. 2007). However, p75NTR regulates a myriad of intracellular signals (Gentry et al. 2004) and which of these would contribute to the control of genes for cholesterol biosynthesis is not clear. NRIF fits the profile of a potential effector for this action very well. First, NRIF and p75ICD directly interact and after gamma-secretase cleavage of the receptor NRIF translocates into the nucleus (Kenchappa et al. 2006). Second, NRIF is a DNA-binding protein and as such it is potentially able to regulate the transcription of cholesterol biosynthesis genes. Finally, the anatomical distribution of the p75NTR, Dhcr7, and NRIF transcripts showed a great degree of overlap in the mouse brain (Kendall et al. 2003), opening the possibility that this anatomical co-expression may be a basis for a functional interaction. These converging lines of evidence, in addition to the correlation between p75NTR and NRIF regulation of Dhcr7 (Korade et al. 2007), suggested that NRIF itself would be a very potent transcriptional regulator of the cholesterogenic biosynthesis transcripts in the nervous system.
The results of our study confirmed our hypotheses, as we found that: (1) NRIF regulates transcription of the critical cholesterol biosynthesis genes, Hmgcr and Dhcr7 in Neuro2a neuroblastoma cell line; (2) Dhcr7 and NRIF down regulation have similar consequences on the expression of several signaling genes; (3) NRIF silencing itself leads to reduced expression of genes involved in sterol-regulated biosynthesis; (4) NRIF-dependent transcriptional regulation is present both in the absence and presence of exogenous cholesterol, suggesting that preservation of endogenous control over cholesterol biosynthesis in neurons cannot be replaced by exogenous sources; and (5) the regulatory role of NRIF on gene transcription is conserved across in vitro and in vivo experimental systems (Neuro2a and NRIF KO mice, respectively).
In addition to confirming our initial hypotheses, our findings have several noteworthy implications. Importantly, transcriptional regulation of the cholesterol biosynthesis genes is a novel role for NRIF. NRIF, being a nuclear transcription factor, may directly bind to the promoter of lipid genes. Based on the overlap between the expression profile of NRIF- and Dhcr7-deficient cells, this putative mechanism would suggest that Dhcr7, or a direct regulator of Dhcr7 is a target of NRIF. However, we acknowledge that NRIF regulation of lipid gene biosynthesis may involve a more complex set of cellular events, where NRIF would bind to a transcription complex that is several steps removed from regulation of cholesterol biosynthesis.
What could be the consequences of such transcriptional dysregulation of lipid genes? We speculate that the transcriptional disturbance of lipid biosynthesis genes by altered NRIF expression leads to a broader dysregulation of cellular homeostasis, and this is potentially a result of altered cholesterol biosynthesis and availability. Cholesterol is an essential building block of cell membranes and lipid rafts (Korade and Kenworthy 2008), which are a critical place of receptor insertion, neurotrophin signaling, neurotransmitter release, and regulated intramembrane proteolysis of transmembrane proteins (Pike 2005). Reduction in NRIF leads to reduced Dhcr7 levels, which in turn results in accumulation of 7-dehydrocholesterol (7DHC), which is inserted into the membranes instead of cholesterol (Keller et al. 2004). However, although 7-dehydrocholesterol and cholesterol can both incorporate into membranes, they are structurally different, and this is likely to affect the structure and composition of the lipid rafts: it is known that the presence of 7-dehydrocholesterol in hippocampal membranes impairs ligand-binding activity of the serotonin 1A receptor (Singh et al. 2007). Thus, the hypothesized p75NTR-NRIF-Dhcr7-lipid raft cascade is likely to be very important for neuronal homeostasis, and this hypothesis is supported by numerous literature reports: cholesterol content of rafts affects membrane structure, ion conductance/excitability, trafficking to and from the cell surface, size and number of postsynaptic receptor clusters, and neurotransmitter signaling through G-protein coupled receptors (Korade and Kenworthy 2008). Furthermore, cholesterol is a substrate for biosynthesis of neurosteroids and oxysterols. Reduced “mature” cholesterol levels may result in the use of 7-DHC for neurosteroid synthesis, and it is not clear if the activity of the 7-DHC-generated neurosteroids are similar to those of “normal” neurosteroids.
Finally, as the NRIF-dependent expression changes we observed were present in both cholesterol-containing and cholesterol-free cell culture media, our findings suggest that the NRIF-regulated endogenous cholesterol biosynthesis is critical for neuronal homeostasis, and that external supplementation cannot counteract the detrimental effects of altered intrinsic neuronal cholesterol biosynthesis. This further underscores that through regulation of intrinsic neuronal cholesterol biosynthesis, NRIF may have an important role in mediating many signals impinging on neurons. This role of NRIF should be further investigated in the context of human disorders of cholesterol metabolism, such as Smith–Lemli–Opitz syndrome and Niemann–Pick Type C disease.
Acknowledgments
The current research was supported by K02 MH070786 and R01 MH079299 (KM), R01 NS038220 (BDC), and startup funds from VU Kennedy Center for Research on Human Development (ZK).
Contributor Information
Zeljka Korade, Email: zeljka.korade@vanderbilt.edu, Department of Biochemistry, Vanderbilt University School of Medicine, 8124A MRB III, Nashville, TN 37232, USA. Vanderbilt Kennedy Center for Research on Human Development, Nashville, TN 37232, USA.
Rajappa S. Kenchappa, Department of Biochemistry, Vanderbilt University School of Medicine, 8124A MRB III, Nashville, TN 37232, USA
Karoly Mirnics, Vanderbilt Kennedy Center for Research on Human Development, Nashville, TN 37232, USA. Department of Psychiatry, Vanderbilt University, Nashville, TN 37232, USA.
Bruce D. Carter, Department of Biochemistry, Vanderbilt University School of Medicine, 8124A MRB III, Nashville, TN 37232, USA. Vanderbilt Kennedy Center for Research on Human Development, Nashville, TN 37232, USA
References
- Bengoechea-Alonso MT, Ericsson J. SREBP in signal transduction: cholesterol metabolism and beyond. Current Opinion in Cell Biology. 2007;19:215–222. doi: 10.1016/j.ceb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casademunt E, Carter BD, Benzel I, Frade JM, Dechant G, Barde YA. The zinc finger protein NRIF interacts with the neurotrophin receptor p75(NTR) and participates in programmed cell death. The EMBO Journal. 1999;18:6050–6061. doi: 10.1093/emboj/18.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MV, Bothwell M. Neurotrophins: to cleave or not to cleave. Neuron. 2002;33:9–12. doi: 10.1016/S0896-6273(01)00573-6. [DOI] [PubMed] [Google Scholar]
- Dietschy JM, Turley SD. Cholesterol metabolism in the brain. Current Opinion in Lipidology. 2001;12:105–112. doi: 10.1097/00041433-200104000-00003. [DOI] [PubMed] [Google Scholar]
- Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Geetha T, Kenchappa RS, Wooten MW, Carter BD. TRAF6-mediated ubiquitination regulates nuclear translocation of NRIF, the p75 receptor interactor. The EMBO Journal. 2005;24:3859–3868. doi: 10.1038/sj.emboj.7600845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry JJ, Barker PA, Carter BD. The p75 neurotrophin receptor: multiple interactors and numerous functions. Progress in Brain Research. 2004;146:25–39. doi: 10.1016/S0079-6123(03)46002-0. [DOI] [PubMed] [Google Scholar]
- Goslin K, Asmussen H, Banker G. Rat hippocampal neurons in low-density culture. In: Banker G, Goslin K, editors. Culturing Nerve Cells. 2. Cambridge: The MIT Press; 1998. [Google Scholar]
- Keller RK, Arnold TP, Fliesler SJ. Formation of 7-dehydrocholesterol-containing membrane rafts in vitro and in vivo, with relevance to the Smith-Lemli-Opitz syndrome. Journal of Lipid Research. 2004;45:347–355. doi: 10.1194/jlr.M300232-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenchappa RS, Zampieri N, Chao MV, Barker PA, Teng HK, Hempstead BL, et al. Ligand-dependent cleavage of the P75 neurotrophin receptor is necessary for NRIF nuclear translocation and apoptosis in sympathetic neurons. Neuron. 2006;50:219–232. doi: 10.1016/j.neuron.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Kendall SE, Ryczko MC, Mehan M, Verdi JM. Characterization of NADE, NRIF and SC-1 gene expression during mouse neurogenesis. Brain Research. Developmental Brain Research. 2003;144:151–158. doi: 10.1016/S0165-3806(03)00166-4. [DOI] [PubMed] [Google Scholar]
- Korade Z, Kenworthy A. Lipid rafts, cholesterol, and the brain. Neuropharmacology. 2008 doi: 10.1016/j.neuro pharm.2008.02.019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z, Mi Z, Portugal C, Schor NF. Expression and p75 neurotrophin receptor dependence of cholesterol synthetic enzymes in adult mouse brain. Neurobiology of Aging. 2007;28:1522–1531. doi: 10.1016/j.neurobiolaging.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kurrasch DM, Huang J, Wilkie TM, Repa JJ. Quantitative real-time polymerase chain reaction measurement of regulators of G-protein signaling mRNA levels in mouse tissues. Methods in Enzymology. 2004;389:3–15. doi: 10.1016/S0076-6879(04)89001-3. [DOI] [PubMed] [Google Scholar]
- Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- Mirnics ZK, Yan C, Portugal C, Kim TW, Saragovi HU, Sisodia SS, et al. P75 neurotrophin receptor regulates expression of neural cell adhesion molecule 1. Neurobiology of Disease. 2005;20:969–985. doi: 10.1016/j.nbd.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Nwokoro NA, Wassif CA, Porter FD. Genetic disorders of cholesterol biosynthesis in mice and humans. Molecular Genetics and Metabolism. 2001;74:105–119. doi: 10.1006/mgme.2001.3226. [DOI] [PubMed] [Google Scholar]
- Paul CA, Boegle AK, Maue RA. Before the loss: neuronal dysfunction in Niemann-Pick Type C disease. Biochimica et Biophysica Acta. 2004;1685:63–76. doi: 10.1016/j.bbalip.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Pike LJ. Growth factor receptors, lipid rafts and caveolae: an evolving story. Biochimica et Biophysica Acta. 2005;1746:260–273. doi: 10.1016/j.bbamcr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Saher G, Brugger B, Lappe-Siefke C, Mobius W, Tozawa R, Wehr MC, et al. High cholesterol level is essential for myelin membrane growth. Nature Neuroscience. 2005;8:468–475. doi: 10.1038/nn1426. [DOI] [PubMed] [Google Scholar]
- Schor NF. The p75 neurotrophin receptor in human development and disease. Progress in Neurobiology. 2005;77:201–214. doi: 10.1016/j.pneurobio.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Singh P, Paila YD, Chattopadhyay A. Differential effects of cholesterol and 7-dehydrocholesterol on the ligand binding activity of the hippocampal serotonin(1A) receptor: implications in SLOS. Biochemical and Biophysical Research Communications. 2007;358:495–499. doi: 10.1016/j.bbrc.2007.04.135. [DOI] [PubMed] [Google Scholar]
- Smith DW, Lemli L, Opitz JM. A newly recognized syndrome of multiple congenital anomalies. The Journal of Pediatrics. 1964;64:210–217. doi: 10.1016/S0022-3476(64)80264-X. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Kiyosue K, Hazama S, Ogura A, Kashihara M, Hara T, et al. Brain-derived neurotrophic factor regulates cholesterol metabolism for synapse development. The Journal of Neuroscience. 2007;27:6417–6427. doi: 10.1523/JNEUROSCI.0690-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney E, Nwokoro NA, Kelley RI. Behavioral phenotype of RSH/Smith-Lemli-Opitz syndrome. Mental Retardation and Developmental Disabilities Research Reviews. 2000;6:131–134. doi: 10.1002/1098-2779(2000)6:2<131::AID-MRDD7>3.0. CO;2-R. [DOI] [PubMed] [Google Scholar]
- Valenza M, Cattaneo E. Cholesterol dysfunction in neurodegenerative diseases: is Huntington’s disease in the list? Progress in Neurobiology. 2006;80:165–176. doi: 10.1016/j.pneuro bio.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Yang J, Goldstein JL, Hammer RE, Moon YA, Brown MS, Horton JD. Decreased lipid synthesis in livers of mice with disrupted Site-1 protease gene. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13607–13612. doi: 10.1073/pnas.201524598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano H, Chao MV. Mechanisms of neurotrophin receptor vesicular transport. Journal of Neurobiology. 2004;58:244–257. doi: 10.1002/neu.10321. [DOI] [PubMed] [Google Scholar]
- Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, et al. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]