Abstract
We present evidence that a bacterial signal transduction cascade that couples morphogenesis with cell cycle progression is regulated by dynamic localization of its components. Previous studies have implicated two histidine kinases, DivJ and PleC, and the response regulator, DivK, in the regulation of morphogenesis in the dimorphic bacterium Caulobacter crescentus. Here, we show that the cytoplasmic response regulator, DivK, exhibits a dynamic, cyclical localization that culminates in asymmetric distribution of DivK within the two cell types that are characteristic of the Caulobacter cell cycle; DivK is dispersed throughout the cytoplasm of the progeny swarmer cell and is localized to the pole of the stalked cell. The membrane-bound DivJ and PleC histidine kinases, which are asymmetrically localized at the opposite poles of the predivisional cell, control the temporal and spatial localization of DivK. DivJ mediates DivK targeting to the poles whereas PleC controls its release from one of the poles at times and places that are consistent with the activities and location of DivJ and PleC in the late predivisional cell. Thus, dynamic changes in subcellular location of multiple components of a signal transduction cascade may constitute a novel mode of prokaryotic regulation to generate and maintain cellular asymmetry.
The physical location of proteins that carry and process messages inside the cell is widely used in eukaryotic cell signaling. It is well established that the location, or perhaps more importantly, the relocation of a signaling protein can directly affect its target response (1). Thus, the physical organization of the cell provides an additional layer of control upon chemical reaction networks that govern signal reception. The recent discovery in bacteria that several signal transduction histidine kinases exhibit dynamic localization has raised the possibility that this mode of regulation is not unique to eukaryotic systems and may have a more primitive existence than previously thought (2, 3).
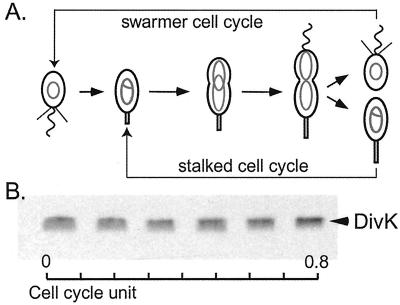
Caulobacter crescentus provides a valuable system to ask questions regarding bacterial polarity because its “spatial awareness” is readily discernible by the presence of polar organelles (4). Invariably, wild-type Caulobacter divides asymmetrically to give rise to two dissimilar daughter cells (Fig. 1A). The smaller swarmer cell has a single polar flagellum and several polar pili and is unable to initiate DNA replication, whereas the stalked cell is able to immediately replicate its chromosome. The nascent swarmer cell must differentiate into a stalked cell before initiating DNA replication. Stalked cells, produced either by differentiation from the swarmer cell or by cell division, elongate into a polarized predivisional cell with the existing stalk at one pole and a newly built flagellum at the other pole. Thus, growth of these bacteria is actually described by two cell cycles (Fig. 1A); one in which the stalked cell behaves much like a stem cell that repeatedly divides to yield a stalked cell and a new swarmer cell, and the other, longer cycle in which the swarmer cell first differentiates into a stalked cell before forming the asymmetric dividing cell.
Figure 1.
DivK protein is present throughout the cell cycle. (A) Schematic diagram of C. crescentus cell cycle. Nonreplicating chromosomes are indicated as gray ovals and replicating chromosomes as gray theta structures. (B) Cell extracts from a synchronous population of cells were analyzed for the presence of DivK by immunoblot. Samples were taken every 20 min of a 120-min cell cycle, as indicated as cell cycle units.
Two-component signal transduction proteins play a crucial role in coordinating cell differentiation with cell cycle progression in Caulobacter (2, 5–9). Members of the two-component signaling family typically contain a sensor kinase that is autophosporylated on a conserved histidine residue upon receiving a signal and a cognate response regulator that is activated by transfer of the phosphoryl group to a conserved aspartate residue. If the response regulator has an attached DNA-binding domain, phosphorylation results in the modulation of target gene transcription. In Caulobacter, the essential response regulator CtrA controls multiple cell cycle and differentiation events (8). It is a negative regulator of the initiation of DNA replication (10) and controls the transcription of many cell cycle-regulated promoters, including those driving genes governing DNA methylation, cell division, and flagellar biogenesis (8, 11). An essential histidine kinase, CckA, is responsible, directly or indirectly, for the phosphorylation of CtrA, to create the active form of CtrA (CtrA∼P) (2). It was recently shown that the CckA histidine kinase exhibits dynamic localization, alternating between dispersed distribution around the cell membrane and tight accumulation at the poles during the course of the cell cycle (2).
Additional two-component signal transduction proteins that have been shown to play a role in Caulobacter cell cycle regulation include the membrane-bound histidine kinases PleC and DivJ and the response regulator DivK (6, 7, 12, 13). A null pleC mutant produces seemingly symmetric predivisional cells and is defective in multiple aspects of polar development, including stalk biosynthesis and flagellar rotation. The DivJ histidine kinase plays a role in controlling the location of the stalk and its length and, to some extent, cell division. PleC and DivJ are differentially localized during the cell cycle and their location in part reflects the time and site of their action (3).
The divK gene encodes a single-domain response regulator, suggesting that it may be part of a multicomponent signal transduction pathway (7). Genetic and biochemical studies support the conclusion that PleC, DivJ, and DivK act on the same phosphorelay pathway (3, 5, 14). However, unlike PleC and DivJ, DivK is essential for cell viability (6, 7, 13). Temperature-sensitive mutants of divK become highly filamentous when shifted to the restrictive temperature, suggesting that DivK may function in two different signal transduction pathways that control both polar differentiation and cell division (7).
We show here that the DivK response regulator is a soluble protein that exhibits a differential cellular distribution as a function of the cell cycle. This cyclical migration of DivK culminates in its polar localization in the progeny stalked cell and cytoplasmic distribution in the progeny swarmer cell. The PleC and DivJ histidine kinases that have been genetically and biochemically linked to DivK function have opposite effects on DivK localization. A divJ null mutation completely disrupts the cell cycle-regulated localization of DivK and results in its cytoplasmic distribution throughout the cell cycle. In the morphologically symmetrical pleC∷tn5 mutant, the polar localization of DivK in the two daughter cells is identical, in striking contrast to wild-type daughter cells. These findings suggest that the mechanism of morphological asymmetry may rely on the asymmetric distribution of regulatory proteins within the cell and that dynamic spatial sequestering of a cytoplasmic response regulator to a localized complex is a feature of the establishment and maintenance of cellular asymmetry.
Materials and Methods
Bacterial Strains, Plasmids, and Media.
Wild-type C. crescentus CB15N and derivative strains were grown in peptone-yeast extract (PYE complex media) or M2G or M5G supplemented with glutamate (1 mM) (minimal and low-phosphate media, respectively) (15, 16). Plasmids were mobilized from Escherichia coli strain S17–1 into C. crescentus by bacterial conjugation (15). The ΔdivJ mutant (LS3196), pleC∷tn5 mutant (LS3194), and ΔdivJ;pleC∷tn5 double mutant (LS3197) have been described (3). The plasmid-encoded divK-gfp was generated as follows: The divK gene was PCR amplified by using a T7 primer and another primer that contained a built-in EcoRI site at the 3′ end. The PCR product was cloned into the SacI and EcoRI sites of pEGFP-N2 (CLONTECH) to create pdivK-EGFP. A 1.4-kb SacI/NotI fragment from pdivK-EGFP was cloned into pBluescriptII KS+. The divK-gfp fragment was further cloned into the SacI and HindIII sites of low-copy plasmids, pMR10 and pMR20, to make pMR10divK-EGFP and pMR20divK-EGFP, respectively. The chomosomal divK null allele was constructed by using a two-step knockout technique. A PCR product that resulted in an in-frame deletion of divK was ligated between the upstream and dowstream DNA sequences of the divK gene. The disrupted gene was moved to an integration plasmid, pNPTS138, carrying the sacB gene. The resulting plasmid was introduced into CB15N expressing divK from a pMR10 plasmid. Kanamycin- resistant integrants were selected that contained tandem copies of the wild-type divK and the mutant ΔdivK DNA separated by plasmid sequence. Excision of the integrated plasmid was selected for by growth on 3% sucrose, inducing the toxic activity of the sacB-encoded levansucrase. The cells were further tested for kanamycin sensitivity to avoid false positives due to sacB inactivation. The structure of the divK locus in kanamycin-sensitive and sucrose-resistant isolates was verified by PCR analysis. The Caulobacter strains expressing divJ-gfp (LS3200) and pleC-gfp (LS3338) have been described (3).
Purification of DivK Protein and Production of Anti-DivK Antibodies.
The divK gene was PCR-amplified by using the T3 primer and a primer with a built-in NdeI site and cloned into the pET21a vector (Novagen). The DivK protein was overexpressed in E. coli strain BL21(DE3) (Novagen). Cells were disrupted by using a French pressure cell, and inclusion bodies-enriched with DivK proteins were isolated by centrifugation. Solubilization of the inclusion bodies was achieved by mixing with 6 M urea. After dialysis against 50 mM Tris buffer, pH 8.0, the protein preparation was subjected to ion-exchange chromatography on a Mono-Q column (Amersham Pharmacia), and DivK was eluted with a gradient of 1 M NaCl. The purified preparation of DivK protein was separated by SDS/PAGE; the DivK band was excised and used to immunize two rabbits (Covance, Berkeley, CA).
Synchronization, Immunoblots, Immunoprecipitations, and Biochemical Fractionation.
Pure populations of swarmer cells were isolated, resuspended to an optical density at 660 nm of 0.2 and allowed to proceed synchronously through the cell cycle (16). Samples were taken at 20-min intervals with an average cell cycle length of 120 min. Immunoblot analysis was carried out as described (16) except that samples were normalized for protein content and equal amounts of total protein were loaded in each gel lane. Cytoplasmic and membrane protein fractions were prepared from log-phase cultures as described (2).
Cell Cycle in Vivo Phosphorylation Assay.
In vivo phosphorylation experiments were performed as described (16) with the following modifications. The M5G culture medium was supplemented with glutamate (1 mM). Importantly, the culture used for the synchrony must be at an optical density at 660 nm of no more than 0.15. The synchronized population of swarmer cells derived from this culture was resuspended in its filtered culture media at the same OD660. This ensured that the synchronized cells would divide before the culture reached an OD660 of 0.35 when cells tend to accumulate one chromosome (similar to a G1 stall), presumably due to phosphate starvation. The cells were then labeled for 3 min with either 30 μCi [γ-32P] ATP or 100 μCi [32P]H3PO4, with comparable results. After labeling, the samples were immunoprecipitated by using anti-DivK sera. Radiolabeled precipitates were resolved on a 15% SDS polyacrylamide gel and visualized on a Molecular Dynamics PhosphoImager.
Fluorescence Microscopy.
Images of cells were taken with a ×100 differential interference contrast (DIC) objective by using a 5-MHz Micromax 5600 digital camera (Princeton Instruments, Trenton, NJ) controlled through Metamorph (Universal Imaging, Media, PA). Images were processed with metamorph software. Time-lapse experiments were performed as described (2) with the exception that bright-field images were taken with DIC microscopy.
Flow Cytometry.
Samples for flow cytometry were prepared as described (17) and analyzed in a Beckton Dickinson FACStar Plus machine. Data were collected and analyzed by using facs/desk software (Stanford University).
Results
The Response Regulator DivK Is Present and Phosphorylated Throughout the Cell Cycle.
The essential single domain response regulator, DivK, is involved in polar differentiation and cell division in Caulobacter (5, 7). To approach an analysis of the regulation of DivK during the cell cycle, we first addressed when the DivK protein is present during the cell cycle and when it is phosphorylated. The transcription of divK was previously shown to be cell cycle-regulated, with a sharp peak of expression in the late predivisional cell (7). To determine whether the protein levels of DivK followed the temporal pattern of divK transcription, antibodies to DivK were generated and immunoblot assays to detect DivK protein were performed on cell extracts of samples from a synchronized culture of wild-type CB15N (Fig. 1B). To our surprise, DivK was present throughout the cell cycle, showing only a slight increase in the late predivisional cell. Overall, cell cycle-regulated transcription of divK does not directly correlate with the levels of DivK protein during the cell cycle. If DivK is a stable protein relative to the length of the cell cycle, one should expect little influence from the transcriptional regulation, especially if, as is the case, the divK promoter is not strong (7). The stability of DivK therefore was determined by performing a pulse–chase experiment with [35S]methionine followed by immunoprecipitation using anti-DivK antibodies. No significant degradation of DivK was observed for over a period of 4 h (data not shown). Under the experimental conditions used to assay DivK turnover, the cell cycle takes about 2 h. Thus, DivK remains stable during the entire cell cycle, which could explain the small influence of transcriptional regulation on the overall protein abundance during the cell cycle.
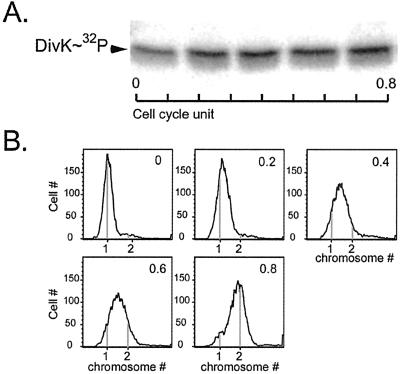
The activity of response regulators usually depends on the phosphorylation state of the protein (18). Therefore, synchronous populations of wild-type CB15N cells were labeled with [γ-32P] ATP and DivK∼32P was immunoprecipitated with anti-DivK antibodies (Fig. 2A). The incorporation of 32P into DivK was detected by autoradiography. To ensure the quality of the synchrony, an aliquot of each time point was analyzed for DNA content by flow cytometry (Fig. 2B). At 0 cell cycle units, all cells had one chromosome, which is the characteristic of swarmer cells. By 0.8 cell cycle units, most cells had two chromosomes representing predivisional cells that have completed DNA replication. As shown in Fig. 2A, DivK did not appear to undergo dramatic cell cycle-regulated changes in phosphorylation state. The most significant difference was observed in swarmer cells (cells with one chromosome) where DivK phosphorylation was lower (50 ± 20%) than later in the cell cycle.
Figure 2.
DivK is phosphorylated throughout the cell cycle. (A) Cells from a synchronous population of CB15N were labeled with [γ-32P] ATP every 20 min of a 100-min cell cycle, indicated as cell cycle units. DivK∼32P was immunoprecipitated with anti-DivK sera. (B) The quality of the synchrony was ensured by flow cytometry analysis. DNA content was measured by flow cytometry of chromomycin-stained cells that were collected at each time point shown as cell cycle units of the in vivo phosphorylation experiment.
DivK Fused to Green Fluorescent Protein (DivK-GFP) Dynamically Changes its Cellular Location During the Cell Cycle.
Because DivK is implicated in the coordination of differentiation events during the cell cycle (7), it is reasonable that its activity would change during the cell cycle. Yet, the protein is present and phosphorylated throughout the cell cycle. Three transmembrane histidine kinases, CckA, PleC, and DivJ, which also are involved in polar differentiation and cell division, have been shown to change their cellular location during the cell cycle (2, 3). We investigated the subcellular location of DivK by generating a translational divK-gfp fusion under the control of the divK promoter on a low copy plasmid. This construct fully complemented a divK null mutant (which is not viable without a functional copy of divK). The divK gene resides in an operon with a gene encoding another response regulator, pleD (7). The null mutation in divK was engineered to prevent polar effects on pleD expression (see Materials and Methods). The existence of the full-length GFP fusion protein was verified by immunoblot using anti-DivK and anti-GFP sera (data not shown).
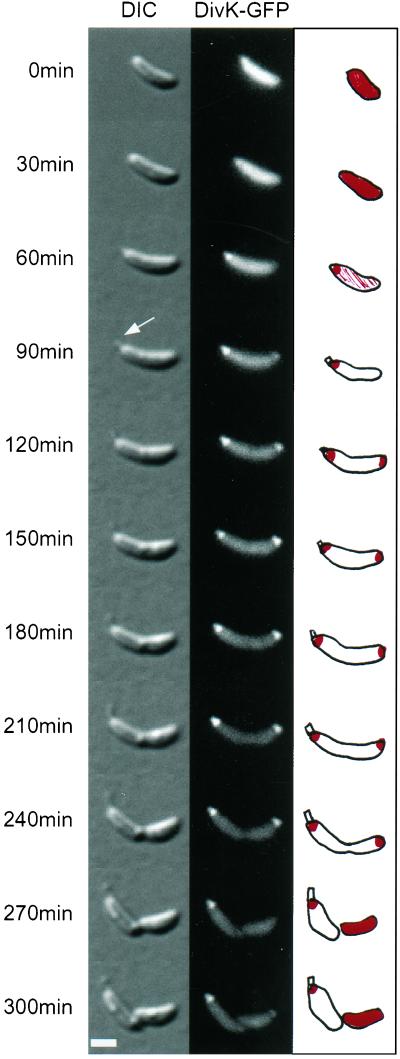
Time-lapse fluorescence microscopy was performed to assess DivK-GFP localization during the course of the cell cycle. Fig. 3 illustrates a time-lapse experiment with DIC and fluorescent images. The fluorescent signal was evenly distributed within the swarmer cell. As the swarmer cell differentiated into a stalked cell, DivK-GFP accumulated at the pole where the nascent stalk formed (shown by the arrow). Thus, DivK-GFP first recognized the “stalked” pole. Later, it formed another focus at the opposite pole. DivK-GFP maintained this bipolar localization until the cell divided when the fusion protein was specifically released from the “flagellated” pole (pole opposite the stalk) but remained anchored at the stalked pole. Thus, division gives rise to two daughter cells with a differential distribution of the DivK-GFP protein: a daughter swarmer cell with a uniform distribution of DivK-GFP, and a daughter stalked cell with DivK-GFP localized at the stalked pole. The daughter stalked cell could easily be distinguished from the daughter swarmer cell by the presence of the stalk and its larger size. A sequence of time-lapse images illustrating DivK-GFP localization for a period close to two cell cycle lengths can be viewed in quicktime Movie 1, which is published as supplemental data on the PNAS web site, www.pnas.org.
Figure 3.
DivK-GFP exhibits dynamic changes in cellular localization. Swarmer cells isolated from a culture of a ΔdivK deletion strain expressing divK-gfp were immobilized on an agarose pad and allowed to progress synchronously through a 250-min cell cycle. DIC and fluorescence images of growing and differentiating cells were taken every 30 min. The white arrow shows a new elongating stalk. A schematic representation of DivK-GFP (in red) is illustrated. The white horizontal bar represents 1 μm.
Because DivK is a stable protein that is present throughout the course of the cell cycle, this change in DivK distribution does not appear to be due to targeted mRNA translation nor localized proteolysis. Instead, the DivK protein itself has a dynamic localization that is rigorously controlled during the cell cycle.
The Two Histidine Kinases, DivJ and PleC, Have Opposite Effects on DivK Localization.
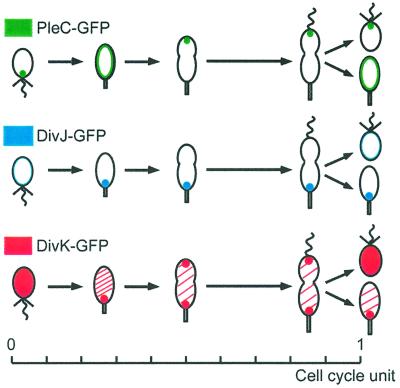
Genetic and biochemical data suggest that the two histidine kinases, PleC and DivJ, control polar differentiation by modulating DivK activity (3, 5, 14). These two histidine kinases are also dynamically localized during the course of the cell cycle (3). Because PleC, DivJ, and DivK may interact with each other at different times during the cell cycle, we examined the relative localization of these three signaling proteins as a function of the cell cycle. We performed independent time-lapse microscopy experiments on synchronous, living wild-type cells expressing pleC, divJ, and divK fused to GFP (Fig. 4). As described (3), PleC-GFP accumulated at the flagellated pole of the swarmer cells. As the swarmer cells differentiated into stalked cells, PleC-GFP was released from the pole and distributed around the membrane; it then reaccumulated at the pole opposite the stalk in the early predivisional cell. At cell division, PleC-GFP was maintained at the pole of the progeny swarmer. In a completely different manner, the DivJ-GFP fluorescent signal was below detection in swarmer cells, but, as the cells differentiated into stalked cells, the signal concentrated at the stalked pole. DivJ-GFP kept this polar localization during the rest of the cell cycle, confirming previous observations (3). The DivK-GFP was evenly distributed in swarmer cells, but coalesced at the stalked pole of the newly differentiated stalked cell. Soon after, the cells acquired a second focus of DivK-GFP at the pole opposite the stalk. Thus, the presence of DivK at the stalked pole is coincident with DivJ localization for most of the cell cycle, and DivK and PleC colocalize at the pole opposite the stalk of the predivisional cell, but do not colocalize in the progeny swarmer cells. These results suggest that PleC and DivJ may physically interact with DivK at the poles to control its activity at different times during the cell cycle.
Figure 4.
Relative localization of PleC, DivJ, and DivK during the cell cycle. Schematics summarizing the localization results from time-lapse experiments performed on synchronous, living cells expressing pleC-gfp (in green), divJ-gfp (in blue), or divK-gfp (in red), in strains with the same doubling time and differentiation patterns.
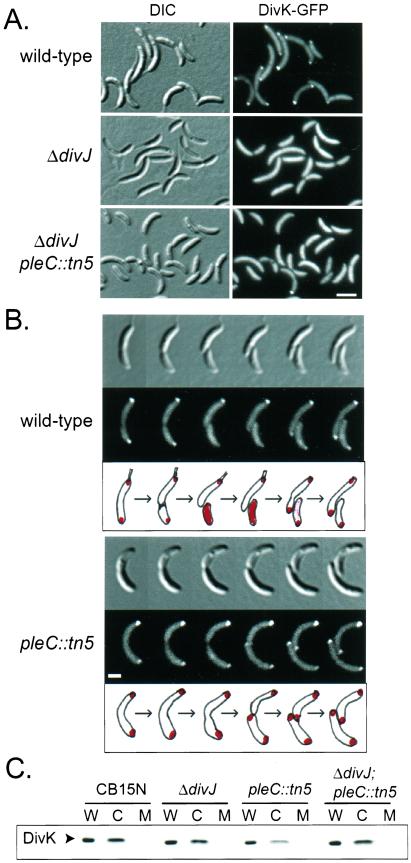
To determine whether the physical presence of the PleC and DivJ histidine kinases affect DivK localization, a divK-GFP fusion carried on a low copy plasmid was expressed in wild-type CB15N, a ΔdivJ deletion mutant, a pleC∷tn5 insertion mutant, and a ΔdivJ;pleC∷tn5 double mutant (Fig. 5 A and B). Fig. 5A shows the typical distribution of DivK-GFP in the wild-type, ΔdivJ, and ΔdivJ;pleC∷tn5 backgrounds. We observed a mixture of uniform, polar, and bipolar patterns of DivK-GFP localization as expected for a wild-type asynchronous culture. In the ΔdivJ mutant background, no apparent foci were localized. Thus, the DivJ histidine kinase is required for DivK polar localization. In the ΔdivJ;pleC∷tn5 double mutant cells, the fluorescent signal from DivK-GFP was mostly uniform, which suggested that the divJ mutation is dominant over the pleC mutation confirming earlier work (3, 5). However, a small percentage (4%) of ΔdivJ;pleC∷tn5 mutant cells exhibited a fluorescent signal that was more intense at the pole(s) (Fig. 5A), a phenomenon that was not observed in the ΔdivJ mutant.
Figure 5.
DivJ and PleC histidine kinases exert opposite effects on localization of the soluble DivK response regulator. (A) DivJ is required for polar localization of DivK. Fluorescence microcopy of asynchronous wild-type, ΔdivJ mutant, and ΔdivJ;pleC∷tn5 double mutant cells expressing a plasmid encoded divK-gfp is shown. The corresponding DIC are on the left. The white horizontal bar represents 2 μm. (B) PleC controls DivK release from the flagellated pole after cell division. Time-lapse fluorescence microscopy experiments on synchronized wild-type and pleC∷tn5 predivisional cells expressing a plasmid-encoded divK-gfp. Fluorescent and DIC images were taken every 30 min; below is a schematic representation of DivK-GFP localization. The white horizontal bar represents 1 μm. (C) DivK is a soluble, cytoplasmic protein. Cytoplasmic (C) and membrane (M) fractions were prepared from log-phase cultures of CB15N (wild type), ΔdivJ, pleC∷tn5, and ΔdivJ;pleC∷tn5 cells. Proteins from all fractions, including a whole-cell control (W), were resolved on 15% SDS-polyacrylamide gel and immunoblotted by using anti-DivK sera.
On the other hand, the pleC∷tn5 mutant had no problem localizing DivK-GFP at the poles but was unable to release DivK-GFP from either poles after cell division. This was observed in time-lapse experiments, starting with predivisional cells (Fig. 5B). In the wild-type background, DivK-GFP was bipolarly localized in the predivisional cell and was released into the cytoplasm of the swarmer daughter cell at or immediately after cell division. This pattern is identical to that observed in the divK null background (Fig. 3), indicating that the presence of the chromosomal wild-type copy of divK had no effect on the localization pattern of the plasmid-encoded DivK-GFP. In the pleC∷tn5 mutant background, however, the localization pattern was different. As the predivisional cell divided, DivK-GFP remained localized at both poles, resulting in the generation of two daughter cells with polarly localized DivK-GFP.
Thus, it appears that DivJ is essential for the localization of DivK to the poles whereas PleC is specifically needed for the release of DivK from the flagellated pole when the cell divides. The fact that both PleC and DivJ are required for coordinating polar morphogenesis with cell cycle progression, for establishing the morphological asymmetries between the two daughter cells, and for mediating phosphorylation of DivK (3, 5, 14), strongly suggests that the temporal and spatial control of DivK localization plays a key role in regulating these events.
Three regulatory proteins known to dynamically change their cellular location during the Caulobacter cell cycle, the CckA, DivJ, and PleC histidine kinases, are all membrane-bound proteins (2, 3). In contrast, the DivK response regulator segregated in the soluble fraction after biochemical fractionation of the wild-type CB15N strain (Fig. 5C), indicating that DivK is a cytoplasmic protein. This was also observed in ΔdivJ, pleC∷tn5, and ΔdivJ;pleC∷tn5 double mutant backgrounds. Thus, DivK must be joining a complex at the cell poles and both its localization and release from that complex are subject to temporal control, probably mediated by DivJ and PleC.
Discussion
Two-component signal transduction proteins regulate many Caulobacter cell cycle events, and the regulation of their activity is therefore critically important in controlling cell cycle progression. Temporally regulated phosphorylation and selective proteolysis of the CtrA response regulator are central to the regulation of its activity, which controls cell cycle progression and determines the fate of the two daughter cells (8, 16). DivK is yet another essential response regulator that controls both cell differentiation and cell division in Caulobacter (7). However, unlike CtrA, DivK is present and phosphorylated throughout the cell cycle. The most distinct cell cycle regulation of the DivK response regulator is its dynamic spatial distribution within the cell (Fig. 3). We showed that a functional DivK-GFP is dispersed throughout the cytoplasm of swarmer cells. As these cells differentiate into stalked cells, DivK-GFP rapidly coalesces at the pole where a new stalk will elongate. Shortly thereafter, DivK-GFP accumulates at both poles. At division, DivK-GFP is specifically released from the flagellated pole of the daughter swarmer cell but remains localized at the stalked pole of the daughter stalked cell. Together with the genetic evidence that implicates DivK in morphogenesis (7), these results suggest that the dynamic localization of divK plays a role in coupling morphogenesis to cell cycle progression. In addition, both genetic and biochemical evidence suggests that the two histidine kinases, DivJ and PleC, mediate the phosphorylation reactions that activate the DivK response regulator (3, 5, 14). It may be that DivK is phosphorylated at different times in the cell cycle by these two histidine kinases, which themselves show dynamic patterns of polar localization. Thus, where DivK is activated may be as important as when it is activated.
How is DivK localization controlled? Our data indicate that the DivJ histidine kinase is required for DivK localization at the poles; in ΔdivJ mutants, DivK is dispersed in the cytoplasm throughout the cell cycle. In contrast, the PleC histidine kinase is essential for the release of DivK from the pole of the daughter swarmer cell; pleC∷tn5 mutant cells maintain bipolar localization of DivK after division, yielding two daughter cells with identical polar localization of DivK. In wild-type cells, DivK is distributed in the cytoplasm of swarmer cells and is localized at the pole of stalked cells. This is accomplished by the PleC-mediated release of DivK from the flagellated pole at division, and through DivJ-mediated localization of DivK at the stalked pole during the swarmer-to-stalked cell transition. Thus, both DivJ and PleC are necessary for the dynamic localization of DivK during the cell cycle. Both histidine kinases have been shown to regulate morphogenic events. pleC∷tn5 mutants produce two seemingly identical daughter cells; they are the same size, they lack polar pili and stalks and possess one or more inactive flagella, sometimes at both poles (6, 13, 19). ΔdivJ mutants have long stalks with a high frequency of stalks at both ends or in the middle of the cell (3). These data are consistent with the idea that the defects in the pleC∷tn5 and ΔdivJ mutants are due to mislocalization of DivK during the cell cycle.
Biochemical fractionation identified DivK as a soluble, cytoplasmic protein. The fact that DivK is localized to one or both poles at specific times in the cell cycle suggests that this cytoplasmic protein interacts with a membrane-bound complex. The PleC and DivJ sensor kinases are obvious candidate components of the membrane-bound complexes at each pole. They are integral membrane proteins that are required for the cell cycle-dependent localization of DivK, they are asymmetrically localized to the poles of predivisional cells, and each colocalizes separately with DivK at the stalked and flagellated poles of predivisional cells. However, the cell cycle pattern of localization of these signaling proteins (Fig. 4) also clearly indicates that the physical presence, alone, of PleC and DivJ at the poles is not sufficient to control DivK localization. It is likely that the action of another player, yet to be identified, and/or the activation of the kinases through phosphorylation also are involved in controlling DivK localization. Previous studies suggest that PleC and DivJ control DivK phosphorylation (3, 7). In the ΔdivJ mutant where DivK is uniformly dispersed in the cytoplasm, DivK phosphorylation is considerably reduced. In contrast, DivK phosphorylation is slightly increased in the pleC∷tn5 mutant (3) that is defective for DivK release from the flagellated pole after cell division. Additionally, we showed that the DivK∼P level was lower in swarmer cells (where the protein is dispersed) relative to later stages in the cell cycle. Thus, low phosphorylation level of DivK protein tends to correlate with dispersed distribution of the protein in the cytoplasm, suggesting that the level of DivK phosphorylation may play a role in DivK targeting to the poles. Our results argue that the asymmetric localization of the two sensor kinases, PleC and DivJ, contributes, at least partially, to the specificity of the cell type by controlling DivK localization, possibly by regulating its level of phosphorylation.
The cell cycle localization of DivK is clearly not essential for viability because pleC∷tn5 and ΔdivJ mutants in which the timed localization of DivK is disrupted are viable. This indicates that the essential function of DivK is regulated at another level, supporting the contention that DivK is a pleiotropic signaling protein with multiple functions (7). DivK and, as we propose, localization of DivK, controls the coordination of morphogenesis and cell cycle progression, which is not essential for viability (3, 5, 7). The essential function of DivK is likely to be its role in regulating cell division (7), for which DivK localization at the poles is probably not essential.
Changes in cellular location represent a mechanism that frequently is used by eukaryotic cells to control the activity of regulatory proteins (1). In bacteria, the regulatory function of protein localization is less obvious. The localization of DivK at the poles must affect the local concentration of DivK (or DivK∼P). If DivK interacts with upstream partners also localized at the pole, as we think it might with the DivJ and PleC histidine kinases, it would greatly enhance their local concentrations and therefore their local activities. This is achieved by concentrating the proteins in a small volume in the pole area. Such localized signaling systems would be advantageous for the cell because localization increases the number of complexes between cognate signal transduction proteins and hence amplifies the activation of downstream events.
Polar location of signaling transduction proteins is not unique to intrinsically polar bacteria like Caulobacter. The chemotaxis signal transduction proteins have been shown to preferentially accumulate at the cell poles in a wide variety of bacteria and an archeon (20–23). The function of the polar location of these signaling proteins is not well understood, but its conservation among distantly related microorganisms underlines its biological importance. The broad range of Caulobacter events controlled by DivK requires regulation of its interactions with multiple upstream partners, including the DivJ and PleC sensor kinases, which, as our results suggest, is mostly controlled by protein localization. This underscores the importance of spatial organization of a bacterial signal transduction cascade to cell specificity. We suspect that many prokaryotic processes are regulated by the subcellular localization of signal transduction cascades, which if true, will significantly expand our views of prokaryotic regulatory processes.
Supplementary Material
Acknowledgments
We thank Harley MacAdams for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM32506/5120 M2 and Office of Naval Research Grant N00014–99-1–0563-00002.
Abbreviations
- DIC
differential interference contrast
- GFP
green fluorescent protein
References
- 1.Stein G S, van Wijnen A J, Stein J L, Lian J B, Montecino M, Choi J, Zaidi K, Javed A. J Cell Sci. 2000;113:2527–2533. doi: 10.1242/jcs.113.14.2527. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs C, Domian I J, Maddock J R, Shapiro L. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 3.Wheeler R T, Shapiro L. Mol Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- 4.Brun Y V, Marczynski G, Shapiro L. Annu Rev Biochem. 1994;63:419–450. doi: 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- 5.Sommer J M, Newton A. Genetics. 1991;129:623–630. doi: 10.1093/genetics/129.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S P, Sharma P L, Schoenlein P V, Ely B. Proc Natl Acad Sci USA. 1993;90:630–634. doi: 10.1073/pnas.90.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hecht G B, Lane T, Ohta N, Sommer J M, Newton A. EMBO J. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Ohta N, Zhao J L, Newton A. Proc Natl Acad Sci USA. 1999;96:13068–13073. doi: 10.1073/pnas.96.23.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burton G J, Hecht G B, Newton A. J Bacteriol. 1997;179:5849–5853. doi: 10.1128/jb.179.18.5849-5853.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta N, Lane T, Ninfa E G, Sommer J M, Newton A. Proc Natl Acad Sci USA. 1992;89:10297–10301. doi: 10.1073/pnas.89.21.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, Ohta N, Newton A. Proc Natl Acad Sci USA. 1998;95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ely B. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 16.Domian I J, Quon K C, Shapiro L. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 17.Winzeler E, Shapiro L. J Mol Biol. 1995;251:346–365. doi: 10.1006/jmbi.1995.0439. [DOI] [PubMed] [Google Scholar]
- 18.Hoch J A. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 19.Sommer J M, Newton A. J Bacteriol. 1989;171:392–401. doi: 10.1128/jb.171.1.392-401.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maddock J R, Shapiro L. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 21.Alley M R, Maddock J R, Shapiro L. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- 22.Sourjik V, Berg H C. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 23.Guestwicki J E, Lamanna A C, Harshey R M, McCarter L L, Kiessling L L, Adler J. J Bacteriol. 2000;182:6499–6502. doi: 10.1128/jb.182.22.6499-6502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.