Abstract
Although the critical role for epigenetic mechanisms in development and cell differentiation has long been appreciated, recent evidence reveals that these mechanisms are also employed in post-mitotic neurons as a means of consolidating, and stabilizing cognitive-behavioral memories. In this review, we discuss evidence for an “epigenetic code” in the central nervous system that mediates synaptic plasticity, learning, and memory. We consider how specific epigenetic changes are regulated and may interact with each other during memory formation, and how these changes manifest functionally at the cellular and circuit levels. We also describe a central role for mitogen-activated protein kinases in controlling chromatin signaling in plasticity and memory. Finally, we consider how aberrant epigenetic modifications may lead to cognitive disorders that affect learning and memory, and review the therapeutic potential of epigenetic treatments for the amelioration of these conditions.
Keywords: Mitogen-activated Protein Kinase, MAPK, ERK, MSK, CREB, learning, memory, LTP, transcription factor, gene expression, chromatin, DNA, methylation, histone, acetylation, phosphorylation, epigenesis, epigenetic, demethylation, HDAC, drug addiction, Rett syndrome, imprinting
Introduction
Biologists have long recognized the conceptual parallels between cellular development and cognitive-behavioral memory formation (Marcus et al., 1994). Both cellular development and memory formation rely on transient environmental signals to trigger lasting, even lifelong, cellular changes. There is a clear analogy between developmental “memory” where cell phenotypes and properties are triggered during development and stored and manifest for a lifetime, and cognitive-behavioral memory where information is acquired through experience and is subsequently available for long-term recollection.
Investigation of the precise molecular mechanisms in both cellular development and memory has increased over the past two decades, and an interesting new understanding has emerged: developmental regulation of cell division and cell terminal differentiation involves many of the same molecular signaling cascades that are employed in learning and memory storage. Therefore, cellular development and cognitive memory processes are not just analogous but homologous at the molecular level.
There are several specific known examples in mammalian systems that substantiate this generalization. One example is the role of developmental growth factors such as BDNF and reelin in triggering plasticity and long term behavioral memories in the adult CNS (Bekinschtein et al., 2007; Herz and Chen, 2006; Patterson et al., 1996; Rattiner et al., 2004; Weeber et al., 2002). Also, the prototypic signal transduction cascades that regulate cell division and differentiation developmentally, the Mitogen-activated Protein Kinases (the MAPKs), are a central and conserved signaling pathway subserving adult synaptic plasticity and memory (Sharma and Carew, 2004; Sweatt, 2001; Thomas and Huganir, 2004). Finally and perhaps most strikingly, a series of studies over the last decade has demonstrated a role for epigenetic molecular mechanisms, specifically DNA methylation, chromatin modification, and prion-like mechanisms in generating and maintaining experience-driven behavioral change in young and old animals (Levenson and Sweatt, 2006).
Here, we provide an overview of recent findings that suggest epigenetic mechanisms, comprising an epigenetic code, are utilized in long-term memory formation in the adult CNS. We also briefly illustrate the parallel utilization of cellular signal transduction cascades in both development and memory formation, focusing on MAPK signaling and its role in controlling learning and memory-associated gene expression. We also discuss the emerging role of the MAPK cascade in regulating memory-associated epigenetic modifications in the CNS . We then present several possibilities as to how an epigenetic code might manifest itself to drive functional changes in neurons within a memory-encoding neural circuit, describing results implicating gene targets such as BDNF in this process. Finally, we discuss the potential relevance of these studies to the human condition, describing examples of what might be considered “epigenetic” disorders of cognitive function, and the idea that epigenetic mechanisms represent a new therapeutic target for disorders of learning, memory, and drug abuse.
Cracking the epigenetic code
The histone code and its role in learning and memory
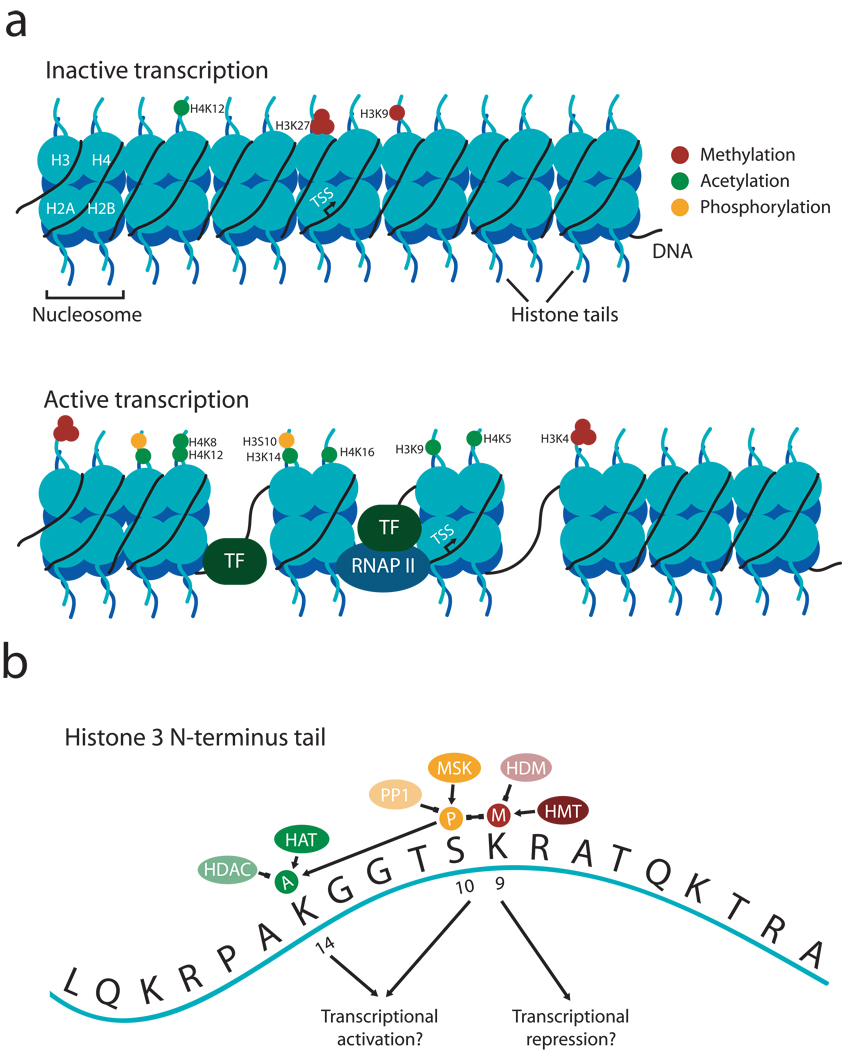
Within a cell nucleus, 147 base pairs of DNA is wrapped tightly around an octamer of histone proteins (two each of H2A, H2B, H3, and H4) to form the basic unit of chromatin called the nucleosome. Each histone protein is composed of a central globular domain and an N-terminal tail that contains multiple sites for potential modifications, including acetylation, phosphorylation, methylation, ubiquitination, and ADP-ribosylation (see Figure 1). Each of these marks is bidirectionally catalyzed or removed by a specific set of enzymes (Strahl and Allis, 2000). Thus, histone acetyltransferases (HATs) catalyze the transfer of acetyl groups to histone proteins, whereas histone deacetylases (HDACs) cause the removal of acetyl groups. Likewise, histone methylation is initiated by histone methyltransferases (HMTs) such as G9a whereas histone demethylases (HDMs) such as LSD1 remove methylation marks (Shi et al., 2004; Tachibana et al., 2001). Interestingly, a number of histone sites can undergo dimethylation or even tri-methylation (Scharf and Imhof, 2010; Shi and Whetstine, 2007). Finally, phosphorylation of serine or threonine residues on histone tails can be accomplished by a broad range of nuclear kinases, such as MSK-1, and dephosphorylated by protein phosphatases such as protein phosphatase 1 (PP1) (Brami-Cherrier et al., 2009; Koshibu et al., 2009).
Figure 1. Dynamic regulation of histone modifications directs transcriptional activity.
A, Individual residues on histone tails undergo of a number of unique modifications, including acetylation, phosphorylation, and mono-, di-, and tri-methylation surround the transcription start site (TSS) for a given gene. These modifications in turn correlate with transcriptional repression (top), in which DNA is tightly condensed on the nucleosome and therefore inaccessible, or transcriptional activation (bottom), in which transcription factors (TF) or RNA polymerase II (RNAP II) can access the underlying DNA to promote gene expression. The specific epigenetic marks listed correlate with transcriptional activation or repression, although this list is by no means exhaustive. B, Expanded view of individual modifications on the tail of histone H3. See text for details and acronyms. The concept of a histone “code” suggests that individual marks interact with each other to form a combinatorial outcome. In this case, methylation at lysine 9 on H3 (a mark of transcriptional repression) and phosphorylation at serine 10 on H3 repress each other, whereas phosphorylation at serine 10 enhances acetylation on lysine 14 (a mark of transcriptional activation.
Importantly, histone modifications are capable of being both gene-specific within the genome and site-specific within a given chromatin particle, meaning that they are in an ideal position to selectively influence gene expression. Site-specific modifications are known to directly alter chromatin state and transcription through a number of mechanisms. For example, acetylation of histone proteins is thought to activate transcription by relaxing the charged attraction between a histone tail and DNA, thereby increasing access of transcription factors or RNA polymerase to DNA sites. Additionally, site-specific acetylation of a histone tail enables transcription factors that contain a bromodomain to bind to the histone and initiate chromatin remodeling (Dyson et al., 2001). Likewise, methylated lysines are bound by proteins with a chromodomain, although the affinity of these proteins for their respective modification is highly dependent on the overall context and presence of other modifications (Scharf and Imhof, 2010). Moreover, while some modifications such as histone acetylation or phosphorylation are generally associated with transcriptional activation, others are more closely correlated with transcriptional repression (Barski et al., 2007; Wang et al., 2008).
Given that histone proteins can be modified at a number of sites, this raises the possibility that specific modifications could work together as a sort of “code”, which would ultimately dictate whether a specific gene was transcribed. This hypothesis, first formalized nearly a decade ago (Jenuwein and Allis, 2001; Strahl and Allis, 2000; Turner, 2000) and more recently supported experimentally (Campos and Reinberg, 2009), suggests that certain combinations of modifications will lead to transcriptional activation whereas others would lead to transcriptional repression. Indeed, analysis of histone modifications across the human genome using ChIP-Seq (chromatin immunoprecipitation sequencing) has demonstrated that a specific combination of 17 modifications tended to co-occur at the level of the individual nucleosome and was associated with increased gene expression (Wang et al., 2008). Importantly, this group of modifications was observed at thousands of gene promoters, indicating that it is a relatively general mechanism by which histone modifications may alter gene transcription (Wang et al., 2008). Although small groups of histone modifications tend to occur together, these modifications are only correlated with (rather than explicitly predictive of) increased gene expression. Moreover, exact combinations of modifications across a nucleosome are seldom repeated at different genes, indicating complex and gene-specific regulation of histone modifications. Thus, the histone code hypothesis has since been modified to consider both the context of a specific modification as well as the final outcome (Lee et al., 2010; Turner, 2007), where the histone code is considered the “language” that controls gene expression rather than an explicit combination of modifications that always generate an identical response.
Theoretically, the incorporation of multiple histone modifications into a code could occur in a number of ways. For instance, a specific modification may recruit other histone modifying enzymes that either repress or facilitate nearby marks (Figure 1B). This appears to be the case with phosphorylation at Ser10 on H3, which both represses methylation at lysine 9 and encourages acetylation at lysine 14 (Cheung et al., 2000; Fischle et al., 2005). Interestingly, this type of interaction may occur between different histone tails as well as on the same tail (Zippo et al., 2009). Another possibility is that although certain marks may act as transcriptional repressors under some cases, they may facilitate transcription in the presence of another mark on the same histone tail. This would explain why a number of histone modifications have been associated with both transcriptional activation and transcriptional repression, and sets of marks that are both independently correlated with transcriptional activation do not necessarily always occur together (Barski et al., 2007). Yet another means by which specific histone modifications could combine to produce a unique epigenetic signature is via the inherent kinetics underlying each reaction. Histone acetylation and phosphorylation are likely reversed very rapidly, whereas histone methylation may persist for longer periods of time. This would allow these mechanisms to synergistically control gene expression across unique time courses despite having no direct interactions.
Overwhelming evidence indicates that histone modifications in the CNS are essential components of memory formation and consolidation. Indeed, multiple types of behavioral experiences are capable of inducing histone modifications in several brain regions (Bredy et al., 2007; Chwang et al., 2007; Fischer et al., 2007; Gupta et al., 2010; Koshibu et al., 2009; Levenson et al., 2004; Lubin and Sweatt, 2007; Peleg et al., 2010; Swank and Sweatt, 2001). For example, contextual fear conditioning, a hippocampus-dependent form of memory, coincides with increases in H3K9 dimethylation, H3K4 trimethylation, H3S10 phosphorylation, and H3S10/H3K14 phospho-acetylation in the CA1 region of the hippocampus (Chwang et al., 2006; Gupta et al., 2010). Moreover, contextual fear conditioning coincides with enhanced acetylation at multiple sites on the tails of H3 and H4, including H3K9, H3K14, H4K5, H4K8, and H4K12 in the hippocampus (Peleg et al., 2010). None of these changes occur in control animals that are exposed to the same context but receive no fear conditioning, indicating that these modifications are specific to associative learning. Importantly, interference with the molecular machinery that regulates histone acetylation, phosphorylation, and methylation disrupts associative learning and long term potentiation (LTP; a cellular correlate of memory) (Alarcon et al., 2004; Chwang et al., 2007; Fischer et al., 2007; Gupta et al., 2010; Korzus et al., 2004; Koshibu et al., 2009; Levenson et al., 2004; Vecsey et al., 2007). Specifically, upregulating histone acetylation using HDAC inhibitors enhances memory formation and LTP (Levenson et al., 2004), whereas genetic mutations in CREB Binding Protein (CBP), a known HAT, disrupts memory formation and LTP (Alarcon et al., 2004). Likewise, mice with deletion of a specific HDAC (HDAC2) display enhanced fear conditioning and hippocampal LTP, whereas overexpression of HDAC2 in the hippocampus impairs memory and blunts LTP (Guan et al., 2009). Similarly for histone phosphorylation, inhibition of nuclear PP1, which is implicated in the removal of histone phosphorylation marks, results in improved long term memory (Koshibu et al., 2009), whereas genetic deletion of specific histone methyltransferases impairs memory formation (Gupta et al., 2010).
Overall, these modifications are consistent with the involvement of a “histone code” in learning and memory, where specific sets of changes are produced in response to specific types of behavioral experiences and these modifications are necessary for memory formation and/or consolidation. However, in the context of learning and memory, it appears that it is the combination of histone modifications, rather than the sum of individual modifications, that produces unique changes in gene expression required for memory formation. Specifically, the co-occurrence of acetylation at H3K9, H3K14, H4K5, H4K8, and H4K12 in the hippocampus following fear conditioning is associated with changes in the transcription of hundreds of genes in young mice (Peleg et al., 2010). In contrast, elderly mice that lack acetylation ony at H4K12 following fear conditioning manifest learning deficits and show almost no conditioning-induced changes in gene expression. This suggests that a specific combination of histone modifications is necessary to initiate learning-related gene expression programs. Consistent with this hypothesis, treatment with an HDAC inhibitor selectively restored H4K12 acetylation, enabled the conditioning-induced changes in gene expression, and improved fear memory formation (Peleg et al., 2010).
The DNA methylation code and its role in learning and memory
DNA methylation, or the addition of a methyl group to the 5’ position on a cytosine pyrimidine ring, can also occur at multiple sites within a gene. However, methylation is generally limited to cytosine nucleotides followed by guanine nucleotides, or so-called CpG sites. These sites, though under-represented throughout the genome, are occasionally clustered in CpG “islands”. Interestingly, CpG sites tend to exist in the promoter regions of active genes, suggesting the ability to control transcription. DNA methylation is catalyzed by two groups of enzymes, known as DNA methyltransferases (DNMTs). The first group, de novo DNMTs, methylates “naked” or non-methylated cytosines on either DNA strand. The second group, maintenance DNMTs, recognizes hemi-methylated DNA and attaches a methyl group to the complementary cytosine base. DNMTs insure self-perpetuating DNA methylation in the face of ongoing passive demethylation, allowing for persitent chemical modification throughout the lifetime of a single cell (Day and Sweatt, 2010).
Like histone modifications, DNA methylation may constitute an epigenetic code (Turner, 2007), although this idea is more recent and has been less fully explored. Clearly, methylation at promoter regions is capable of altering transcription due to the affinity of certain proteins for methylated cytosine (methyl binding domain proteins, or MBDs). The prototypical example of an MBD is MeCP2, which is mutated in the neurodevelopmental disorder Rett Syndrome and dramatically affects synaptic plasticity in the hippocampus and memory formation (Amir et al., 1999; Chao et al., 2007; Moretti et al., 2006). Mechanistically, MeCP2 is capable of recruiting both repressive and activating transcription factors or chromatin remodeling complexes such as HDACs (Chahrour et al., 2008). Importantly, MBDs like MeCP2 have different affinities for fully methylated and hemi-methylated DNA, meaning that the difference between these two states may actually be a critical component of the methylation code (Valinluck et al., 2004). In the adult CNS, hydroxymethylation of cytosines that tags methyl groups for removal can affect MBD protein binding to DNA (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). It is less clear, however, if hydroxymethylation represents a distinct epigenetic marker, or an intermediate stage of an existing methylation marker.
What might a DNA methylation code look like? Given that methylation/demethylation machinery can produce at least three different outcomes for each CpG in question (no methylation, hemi-methylation, and full methylation of both DNA strands) and that the promoter and intragenic regions of a plasticity gene may contain hundreds of CpG sites, the potential combinatorial complexity of a DNA methylation code is astounding. Indeed, it is conceivable that even within a small stretch of DNA, CpG sites could exhibit any of the three possibilities, thereby leading to site-specific outcomes (as illustrated in Figure 2). Therefore, understanding how DNA methylation contributes to transcriptional efficacy will require examination of DNA methylation changes at the single nucleotide level. It is also important to note that the context of the DNA methylation – i.e., where methylation occurs relative to transcription factor binding site or transcription start site – may dramatically influence its potential effect on gene transcription (Klose et al., 2005; Weber et al., 2007). To date, existing studies have typically only examined CpG methylation in relatively small stretches of DNA near gene transcription start sites.
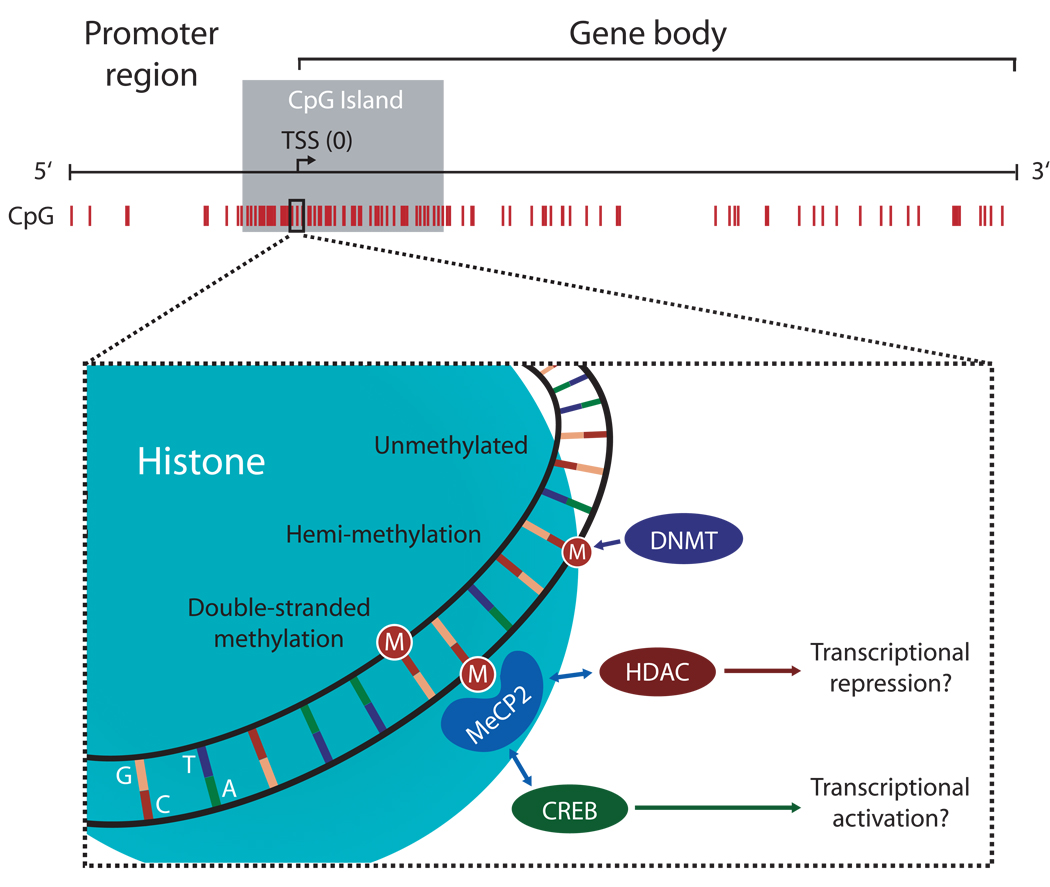
Figure 2. DNA methylation status affects gene transcription.
A number of plasticity-related genes in the brain possess large CpG islands within the gene promoter region. Each CpG dinucleotide in the DNA sequence can undergo methylation by DNA methyltransferases (DNMTs), resulting in hemimethylation and/or double-stranded DNA methylation. Proteins with methyl binding domains, bind to methylated DNA and associate with other co-factors, such as HDACs or transcription factors like CREB, to alter gene expression. It is presently unclear is the specific combination of CpG methylation marks constitutes a “code” for unique outcomes, or if the overall or average density of methylation is a larger determinant of transcriptional efficacy.
Recent evidence indicates that, like histone modifications, changes in DNA methylation represent a critical molecular component of both the formation and maintenance of long term memories (Feng et al., 2010; Lubin et al., 2008; Miller et al., 2008; Miller et al., 2010; Miller and Sweatt, 2007). Interestingly, contextual fear conditioning consequently increases and decreases methylation of memory-related genes expressed in the hippocampus, implicating methylation and demethylation as a molecular mechanism underlying learning and memory (Day and Sweatt, 2010; Miller et al., 2010; Miller and Sweatt, 2007). Consistent with the idea that these changes are necessary for memory formation, inhibition of DNMTs within the hippocampus, which produces a hypomethylated state in naïve animals, results in impaired expression of contextual fear memories (Lubin et al., 2008; Miller and Sweatt, 2007). Likewise, DNMT inhibitors impair the induction of LTP at hippocampal synapses, providing an important cellular correlate of learning deficits induced by blocking DNA methylation (Levenson et al., 2006). Interestingly, DNMT inhibition in the prefrontal cortex impairs the recall of existing memories, but not the formation of new memories, indicating circuit-specific roles for DNA methylation in memory formation and maintenance (Miller et al., 2010). One challenge in interpreting the results of these studies is that the nucleoside analogs conventionally used to inhibit DNMT activity, such as zebularine and 5-aza-deoxycytidine, are believed to require DNA replication to incorporate into DNA and function as DNMT inhibitors (Szyf, 2009). Therefore, in the largely post-mitotic brain, the mechanism by which these compounds enter DNA is less clear, leading to speculation as to whether these drugs are capable of inhibiting DNA methylation in the adult CNS (Day and Sweatt, 2010). To circumvent this problem, recent studies have employed a distinct DNMT inhibitor, RG108, which acts at DNMTs active site and therefore does not require DNA replication. Studies have shown that RG108 produces the same deleterious effects on learning and memory as nucleoside DNMT inhibitors (Lubin et al., 2008; Miller et al., 2010). Likewise, conditional forebrain- and neuron-specific deletion of DNMT1 and DNMT3a impairs performance on the Morris Water Maze and fear learning (Feng et al., 2010), providing genetic confirmation of a role for DNMTs in cognition.
Methodological considerations in testing the epigenetic hypothesis of memory
As discussed above, changes in histone modifications and DNA methylation in the CNS occur in association with memory formation, while experimental manipulation of DNA and histone methylation/acetylation can alter memory formation. These findings strongly support the involvement of an epigenetic code in processes of learning and memory. However, the vast majority of the experiments undertaken thus far have not attempted to directly test the idea that specific patterns of histone and DNA chemical modifications are translated in a combinatorial fashion to subserve specific aspects of memory. No doubt, addressing this defining feature of the epigenetic code is a large undertaking that requires multiple independent lines of experimentation. In this section we will briefly comment on a few of the methodological challenges in testing the epigenetic code hypothesis, keeping in mind that defining some of these challenges may help conceptualize advances designed to overcome them.
To illustrate the critical involvement of an epigenetic code in memory formation and storage, it will be necessary to experimentally demonstrate that neurons of memory-encoding circuits generate a combinatorial set of epigenetic marks in response to a memory-evoking experience. To further substantiate the “epigenetic code” theory, more refined experiments would be required to show that disrupting this specific combinatorial pattern, without altering the overall sum of modifications across the epigenome, suppresses memory function. Moreover, it will be necessary to illustrate that this combinatorial code occurs at the level(s) of a single gene or allele, perhaps at a single CpG island, at an individual chromatin particle, or even at a single histone amino-terminal tail. Finally, all contemporary models of memory storage posit sparse encoding of memories within a memory circuit, meaning that measuring changes at the level of individual neurons is a necessary and relevant parameter. Taken in sum, these considerations present an immense set of technical hurdles to overcome in order to test the epigenetic code hypothesis.
Nevertheless, several recent technical advances will likely aid in more directly testing the epigenetic theory of memory formation. In particular, modern genetic engineering approaches now allow single nucleotide mutations to be introduced into the genome of a mouse that can manifest in single cell types, restricted to one or a few brain subregions, and temporally restricted to post-developmental time points. A reasonable number of memory-associated genes are epigenetically modified in response to experience, including bdnf, reelin, zif268, PP1, arc, and calcineurin, providing a set of candidates for the assessment of combinatorial epigenetic changes at the single-gene or single-exon/intron level using precise genetic engineering approaches. Moreover, the application of genome-wide tools to this problem will enable examination of DNA methylation patterns in a much wider pool of genes, which is currently lacking. Finally, chromatin immunoprecipitation (ChIP) procedures also allow detection of methyl-DNA binding proteins and specific histone modifications at the level of these and other specific gene loci. Additionally, new molecular biological methods have emerged for identifying changes in DNA methylation at the single-cell and single-allele level. Bisulfite sequencing, considered the “gold standard” for assaying DNA methylation, provides single-nucleotide information about a cytosine’s methylation state. Global analysis of all DNA from a given brain region cannot distinguish between DNA methylation changes in different cell types (e.g., neurons vs. glial cells; glutamatergic vs. GABAergic cells, etc), which is a current limitation. However, bacterial subcloning of single pieces of DNA, which originate from single alleles within a single cell, allows isolation of DNA from single CNS cells. Thus, direct bisulfite sequencing combined with DNA subcloning enables quantitative interrogation of single-allele changes in methylation, at the single nucleotide level, in single cells from brain tissue (Miller et al., 2010). Such an approach may be especially powerful for interrogating the sparsely encoded, environmentally-induced neuronal changes that occur during learning and memory. Overall these recent and emerging techniques pave the way for substantive experimental interrogation of experience-driven epigenetic changes, potentially aiding in the identification of an epigenetic code, that underlie memory formation. The ultimate challenge for future studies will be to determine in a comprehensive fashion how DNA methylation and chromatin remodeling at the single-cell level is regulated and translated into changes in neural circuit function and behavior in the context of learning and memory.
The role of MAPK signaling in regulating epigenetic changes
The MAPK cascade was first established as the prototypic regulator of cell division and differentiation in non-neuronal cells (Bading and Greenberg, 1991; English and Sweatt, 1996; Fiore et al., 1993; Murphy et al., 1994). The prominent expression and activation of MAPKs in the mature nervous system, particularly in the hippocampus, prompted researchers to question the role of the MAPK cascade in terminally differentiated, non-dividing neurons in the brain (Bading and Greenberg, 1991; English and Sweatt, 1996). It was speculated that the cascade might have been co-opted in the mature nervous system to subserve synaptic plasticity and memory formation, thereby proposing a mechanism of molecular homology between cellular development and learning and memory (Atkins et al., 1998; English and Sweatt, 1996; English and Sweatt, 1997; Sweatt, 2001).
Since then, there has been a rich literature detailing the importance of the MAPK in neuronal functions, including plasticity (Thomas and Huganir, 2004). As a brief example, the first experiments to begin to test the idea that the MAPK cascade is critical in neuronal processes demonstrated that the Extracellular-Signal Regulated Kinase (ERK) isoforms of MAPK are activated with LTP induction in hippocampal slices, where ERK activation is necessary for NMDA receptor-dependent LTP in area CA1 (English and Sweatt, 1996; English and Sweatt, 1997). Subsequent studies showed that ERK is activated in the hippocampus with associative learning, and is necessary for contextual fear conditioning and spatial learning (Atkins et al., 1998). Studies from a wide variety of laboratories have now shown that MAPK signaling cascades are involved in many forms of synaptic plasticity and learning across many species (Reissner et al., 2006). Moreover, recent studies from Alcino Silva’s group have directly implicated mis-regulation of the ras/ERK pathway in a human learning disorder, Neurofibromatosis-associated Mental Retardation (Ehninger et al., 2008). As the ERK cascade plays a fundamental role in regulating synaptic function, elucidating the targets and regulation of ERK is critical to understanding basic biochemical mechanisms of hippocampal synaptic plasticity and memory formation (Ehninger et al., 2008; Weeber and Sweatt, 2002).
ERK is a pluripotent signaling mechanism as it impinges upon targets in the neuronal membrane, in the cytoplasm, and within the nucleus in order to effect changes in synaptic function and connectivity (Figure 3). ERK regulation is especially complex in the hippocampus – the cascade is downstream of a multitude of cell surface receptors and upstream regulators. The prevailing model is that ERK serves as a biochemical signal integrator that allows the neuron to decide whether or not to trigger lasting changes in synaptic strength (Sweatt, 2001). The canonical role of the ERK pathway in all cells is regulation of gene expression, and studies of the role of ERK signaling in synaptic plasticity, memory formation, drug addiction, and circadian rhythms have borne this out in the adult CNS as well (Girault et al., 2007; Sweatt, 2001; Valjent et al., 2001). There are several mechanisms through which ERK has been shown to regulate gene transcription in the CNS (Figure 3). One regulatory mechanism is transcription factor phosphorylation, and we and others have shown that ERK is required for CREB phosphorylation in hippocampal pyramidal neurons (Eckel-Mahan et al., 2008; Impey et al., 1998; Roberson et al., 1999; Sindreu et al., 2007). The efficacy of phospho-CREB in modulation of transcription also depends upon the recruitment and activation of a number of transcriptional coactivators, including CBP (Vecsey et al., 2007). Thus, regulation of transcription by CREB depends upon the activity of HATs (McManus and Hendzel, 2001; Ogryzko et al., 1996; Perissi et al., 1999; Yuan and Gambee, 2001). In addition, histone phosphorylation contributes to regulating gene transcription, in particular through Serine 10 phosphorylation of histone H3, which is associated with transcriptional activation.
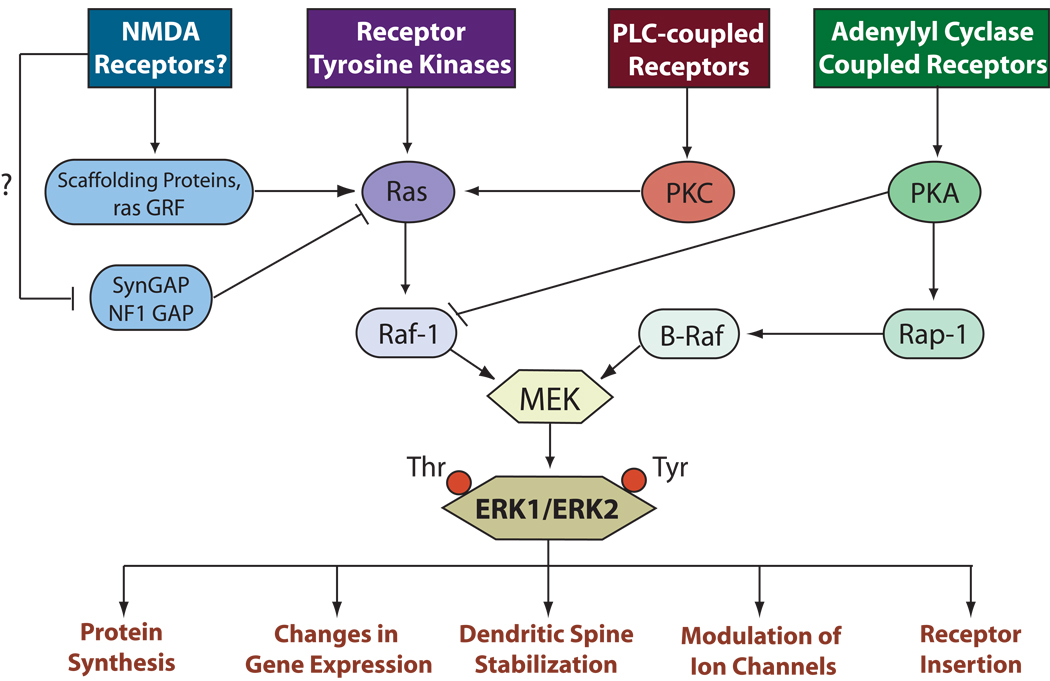
Figure 3. The ERK/MAP kinase cascade in the hippocampus.
The ERK/MAPK cascade can integrate a wide variety of signals and result in a final common output. The ERK cascade is initiated by the activation of Raf kinase via the small GTP-binding protein, ras, or the ras-related protein, rap-1. Activated Raf then phosphorylates MEK, a dual specific kinase. MEK phosphorylates ERK 1 and 2 on a tyrosine and threonine residue. Once activated, ERK exerts many downstream effects, including the regulation of cellular excitability and the activation of transcription factors leading to altered gene expression. Each MAP kinase cascade (ERK, JNK, and p38 MAPK) is composed of three distinct kinases activated in sequence, and despite the fact that many separate MAP kinase families exist, there is limited crosstalk between these highly homologous cascades. While many of the steps of the ERK cascade have been elucidated, the mechanisms by which the components of the MAP kinase cascade come into physical contact have not been investigated. In this context it is interesting to note that there are multiple upstream regulators of ERK in the hippocampus: NE, DA, nicotinic ACh, muscarinic ACh, histamine, estrogen, serotonin, BDNF, NMDA receptors, metabotropic glutamate receptors, AMPA receptors, voltage-gated calcium channels, reactive oxygen species, various PKC isoforms, PKA, NO, NF1, and multiple ras isoforms and homologs.
ERK MAPKs are also central to controlling histone post-translational modifications in synaptic plasticity and experience-driven behavioral changes (Borrelli et al., 2008; Brami-Cherrier et al., 2009; Levenson et al., 2004; Reul et al., 2009; Swank and Sweatt, 2001). Acetylation of histone H3 in the hippocampus, which is accociated with long-term memory consolidation (Fischer et al., 2007; Korzus et al., 2004; Levenson et al., 2004; Wood et al., 2006a), is dependent on the activation of NMDA receptors and of ERK MAPK (Levenson et al., 2004). Activation of NMDA receptors and other memory- and plasticity- associated cell surface receptors also increases acetylation of histone H3, and these effects are blocked by inhibition of ERK signaling (Brami-Cherrier et al., 2007; Brami-Cherrier et al., 2009; Levenson et al., 2004; Reul et al., 2009). Moreover, activation of ERK through either the PKC or PKA pathways, biochemical events known to be involved in long term memory formation, also increase histone H3 acetylation (Brami-Cherrier et al., 2007; Brami-Cherrier et al., 2009; Levenson et al., 2004; Reul et al., 2009). Moreover, ERK/MAPK signaling also regulates histone phosphorylation, and changes in hippocampal histone phosphorylation following fear conditioning are ERK/MAPK dependent (Chwang et al., 2006; Wood et al., 2006b). Overall, a large body of results indicate that histone-associated heterochromatin undergoes ERK-dependent regulation, and that these histone modifications and changes in heterochromatin are necessary for hippocampal LTP and memory formation (Alarcon et al., 2004; Korzus et al., 2004; Levenson et al., 2004; Wood et al., 2006a).
Typically, ERK does not directly affect nuclear targets, but rather acts through intermediary kinases. In a series of experiments, Chwang et al investigated the role of Mitogen-and Stress-activated protein Kinase 1 (MSK1), a nuclear kinase downstream of ERK, in chromatin remodeling during hippocampal-dependent memory formation. Mice lacking MSK1 showed impaired Pavlovian fear conditioning and spatial learning, as well as a deficiency in histone phosphorylation and acetylation in the hippocampus after fear training. This study identified MSK1 as an important regulator of chromatin remodeling in long-term memory, identifying a central signal transduction pathway in plasticity and memory – the ERK-MSK1-histone phospho-acetylation pathway (Figure 4).
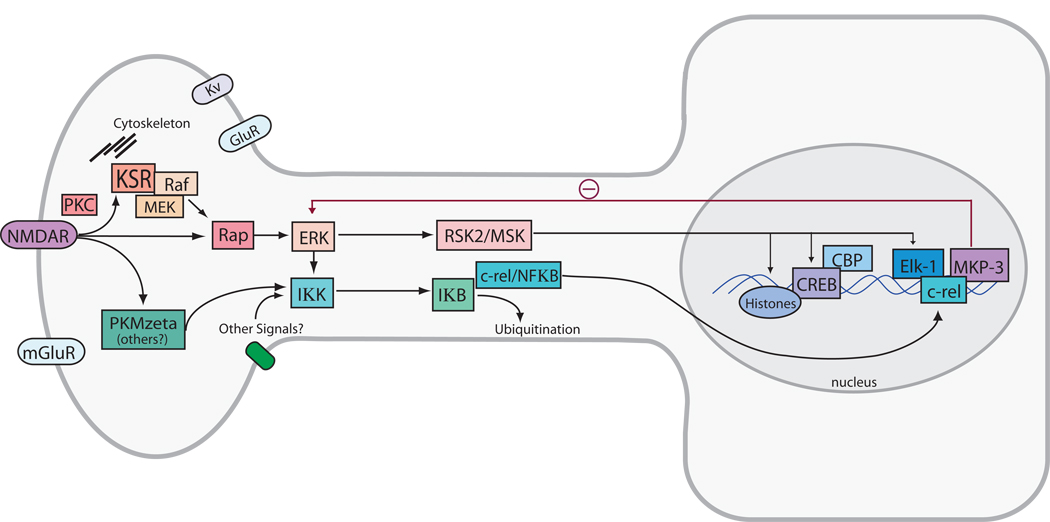
Figure 4. Model for ERK-mediated regulation of histone acetylation and gene transcription.
Activation of the NMDA subtype of glutamate receptors (NMDARs) and voltage-gated Ca++ channels leads to influx of Ca++ and activation of the ras-MEK-ERK signaling cascade in adult neurons. This leads to activation of CREB-mediated transcription via intermediary actions of RSK2 or the Mitogen Stimulated Kinase, MSK. A downstream target of these kinases, CREB, is postulated to facilitate transcription through interaction with CREB-binding protein (CBP) and acetylation of histones. Additional pathways for regulating chromatin structure in memory include metabotropic glutamate receptor (mGluR) and NMDAR activation of Protein Kinase M zeta (PKMzeta) and downstream targeting of NFkappaB signaling in the nucleus. ERK MAPK signaling can also activate this pathway as an ancillary mechanism for chromatin regulation. Targets of this pathway include the transcription factors c-rel and Elk-1, which can regulate the expression of the MAPK Phosphatase MKP-3, which represents a likely site of negative feedback control of the pathway. See text and (Lubin and Sweatt, 2007) for additional discussion.
Overall, studies demonstrating a role for MAPK regulation in memory formation and in triggering lasting behavioral change are interesting in two contexts. First, these observations are consistent with one of the broad themes we are developing in this review, which is that molecular mechanisms that operate in cell differentiation and development have been co-opted in the mature nervous system to subserve lasting functional changes related to memory formation. In this vein, then, MAPK signaling and a role for epigenetic mechanisms in memory provide two prominent examples of molecular homologies between memory and development. Second, the role of MAPKs in regulating histone post-translational modifications in memory and plasticity provides a direct mechanistic link between epigenetic modifications and learning and memory, indeed illustrating a striking conservation of a pluripotent cell-surface-to-epigenome signal transduction pathway in cellular development and cognitive memory.
How does the epigenetic code manifest functional change?
Integration of multiple epigenetic modifications
As discussed above, it is well understood that certain histone modifications interact with each other by preventing access to or recruiting histone modifying enzymes. However, it is less clear how DNA methylation affects histone modifications and vice-versa. One possibility is that DNA methylation patterns are established and maintained by specific combinations of chromatin modifications. For example, HDACs are known to interact with DNMTs, whereas transcription factors that recruit HAT enzymes can trigger demethylation of DNA (D'Alessio and Szyf, 2006; D'Alessio et al., 2007). Likewise, HDAC inhibitors are capable of inducing DNA demethylation (Cervoni and Szyf, 2001; Szyf, 2009).
Conversely, it is also possible that DNA methylation regulates important aspects of chromatin state, indicating a bi-directional relationship between histone and DNA modifications. Consistent with this hypothesis, MeCP2, which binds preferentially to fully methylated DNA, can associate with both HDAC machinery as well as histone methyltransferases to alter specific histone modifications (Bird, 2002; Fuks et al., 2003a; Fuks et al., 2003b). Additionally, DNMT inhibitors block changes in H3 acetylation associated with memory formation (Miller et al., 2008). Furthermore, deficits in memory and hippocampal synaptic plasticity induced by DNMT inhibitors can be reversed by pre-treatment with an HDAC inhibitor (Miller et al., 2008). Taken together, these results reveal a complex relationship between histone modifications and DNA methylation, and suggest that simple considerations of a “histone code” or “DNA methylation code” will each be inadequate in terms of predicting transcriptional output. The interactions between the two mechanisms need to be fully understood in order to formulate a more comprehensive epigenetic code hypothesis for transcriptional regulation in memory.
Regulation of gene expression
To produce a diverse array of cell classes despite working with identical underlying genetic material, cells must be capable expressing or repressing a given set of genes to generate a neuron, a hepatocyte, or a hematocyte. These complex gene transcription programs initiated during cellular differentiation and division appear to be epigenetically regulated (Ng and Gurdon, 2008), and ultimately insure a given cell lineage can be maintained through multiple rounds of cell division or prolonged life in the case of non-dividing cells.
Similarly, many types of long-lasting synaptic plasticity such as LTP, required for memory consolidation, initiate complex gene transcription programs (Alberini, 2008; Davis and Squire, 1984; Frey et al., 1988). In fact, activity-dependent changes in gene expression have long been implicated in learning and memory processes in the CNS (Flavell and Greenberg, 2008; Loebrich and Nedivi, 2009). Therefore, epigenetic modifications may play a similar role in the CNS, initiating functional consequences within a cell or a circuit by modulating gene expression. Accumulating evidence already supports the hypothesis that gene expression programs are a functional readout of epigenetic marking in the CNS in memory formation. As reviewed above, these gene programs are largely dependent on intracellular signaling cascades (such as the MAPK pathway) and activation of critical transcription factors that bind to specific sequences in gene promoter regions. Indeed, it may be this specificity in transcription factor binding sites that leads certain signal transduction cascades to target specific genes and induce specific epigenetic changes. For example, when phosphorylated, CREB binds to cAMP responsive element sites in gene promoters and interacts with CBP, which possesses HAT activity (Gonzalez et al., 1989; Montminy et al., 1990a; Montminy et al., 1990b; Silva et al., 1998). Interestingly, stimuli that produce long lasting LTP also increase CREB phosphorylation in the hippocampus (Deisseroth et al., 1996), and CREB manipulaitons impair memory formation in multiple tasks (Silva et al., 1998). Likewise, blocking cAMP-dependent transcription alone is sufficient to impair LTP maintenance (Frey et al., 1993; Impey et al., 1996). Thus, given that transcriptional machinery such as CREB has long been established as a regulator of cellular and behavioral memory (Frank and Greenberg, 1994; Shaywitz and Greenberg, 1999; Silva et al., 1998), it is perhaps not surprising that epigenetic modifications have been found to interact with these systems (Chahrour et al., 2008; Renthal and Nestler, 2008).
Other epigenetic targets have also been identified in regulating overall transcription rates of specific genes in the establishment, consolidation, and maintenance of behavioral memories (Guan et al., 2009; Lubin et al., 2008; Miller et al., 2008; Miller et al., 2010; Peleg et al., 2010). Specifically, contextual fear conditioning induces a rapid but reversible methylation of the memory suppressor gene PP1 within the hippocampus, and demethylation of reelin, a gene involved in cellular plasticity and memory (Miller and Sweatt, 2007). Importantly, each of these DNA methylation changes are functionally relevant, leading to decreased expression of PP1 and increased expression of reelin (Miller and Sweatt, 2007). Moreover, consistent with the finding that blocking DNA methylation in the anterior cingulate cortex prevents remote memory maintenance, another study reported long-lasting changes in methylation of the memory suppressor gene calcineurin within this brain area following contextual fear conditioning (Miller et al., 2010). These changes in calcineurin methylation persisted at least 30 days following conditioning, suggesting the change is stable enough to maintain a memory over time despite ongoing cellular activity and molecular turnover. Thus, calcineurin is an excellent candidate for a molecular storage device. Likewise, although they are too numerous to name here, histone modifications have been repeatedly associated with changes in gene transcription and expression in multiple organisms, systems, and brain subregions (Brami-Cherrier et al., 2005; Dulac, 2010; Guan et al., 2002; Gupta et al., 2010; Koshibu et al., 2009; Renthal and Nestler, 2008). Thus, these results reveal that even within non-dividing neurons in the adult CNS, epigenetic mechanisms regulate patterns of gene expression in a functionally relevant manner. Indeed, when viewed through this lens, epigenetic changes can simply be viewed as one of the final steps (or perhaps the final step) in a long cascade of events that leads to learning-related gene transcription (Kornhauser et al., 2002; Shaywitz and Greenberg, 1999; Sweatt, 2001).
Alternative splicing
A related means for epigenetic control of gene expression involves the unique regulation of specific protein isoforms, or differently spliced versions of the same protein. This can occur in multiple ways, such as increased expression of one exon over another competing exon or silencing of an entire exon. By regulating the expression of splice variants with different cellular functions or different affinities for effector proteins, the potential uses of the same gene locus can be expanded in a multiplicative fashion (Nilsen and Graveley, 2010).
The mechanisms that regulate alternative splicing are currently unclear. However, histone modifications appear to modulate this process by recruiting different splicing regulators that determine splicing outcome (Luco et al., 2010). DNA methylation is also likely involved in the differential expression of BDNF exons following fear learning (Lubin et al., 2008). Contextual fear conditioning produces a rapid increase in mRNA for BDNF exon IV, thereby decreasing methylation at this locus in area CA1 of the hippocampus. Interestingly, context exposure alone (no conditioning) produced increases in BDNF exon I and VI mRNA, which also corresponded to decreased CpG methylation at these sites. Moreover, intra-hippocampal infusions of the DNMT inhibitors zebularine or RG108 impaired fear memory expression, despite the fact that they increase expression of all BDNF exons in naïve animals. Importantly, the same study reported that in animals that underwent contextual fear conditioning, zebularine blocked the learning-related decreases in BDNF exon IV methylation. Together, these results reveal that DNA methylation regulates expression of BDNF splice variants in a complex, experience-dependent manner, and that the effects of DNMT inhibitors likely depend on the overall behavioral and cellular context. Experience-dependent regulation of BDNF isoforms by DNA methylation represents the clearest evidence of a CpG methylation “code” in the formation and consolidation of behavioral memories.
Imprinting and allelic tagging
Adult fully differentiated cells in placental mammals can manifest differential handling of paternal and maternal copies of somatic genes, a phenomenon referred to as imprinting. Thus specific genes expressed in non-germline cells including neurons, which are not on the X or Y chromosome, can be “imprinted” with DNA methylation. These imprinting marks cross the generations through the germline, and designate a particular copy (allele) of a gene as having originated with the mother versus the father. In traditional cases of genetic imprinting, one copy of the gene is fully silenced, leaving one parent’s copy of the gene the exclusive source of cellular mRNA product..
One prominent example of an imprinted gene involved in cognition is ube3a, which encodes ubiquitin E3 ligase. Imprinted (i.e.methylated) alleles of the ube3a gene are preferentially expressed in a brain subregion-specific fashion: for example, the maternal copy is selectively expressed in neurons in the cerebellum and forebrain, including the hippocampus (Jiang et al, 1998). Mutations in the maternal copy of the ube3a gene result in Angelman Syndrome, a disability characterized by autism-like symptoms accompanied with severe learning and memory deficits and a near complete absence of speech learning. Studies of Angelman Syndrome were the first to implicate the epigenetic mechanism of imprinting in learning, memory, and synaptic plasticity (Jiang et al., 1998). Notably, mice with a maternal deficiency in UBE3A function display deficits in hippocampal-dependent learning and memory and a loss of hippocampal long-term potentiation at Schaffer/collateral synapses (Jiang et al., 1998).
For many years, imprinting of genes in the adult CNS was assumed to be restricted to a few genes, 30–50 or so being a common assumption. However, gene imprinting has recently been found to occur at much higher levels than this: a recent pair of exciting papers from Catherine Dulac’s laboratory have greatly expanded our view of the importance of gene imprinting in CNS function in the adult nervous system (Gregg et al., 2010a; Gregg et al., 2010b). This work from Dulac and colleagues demonstrated that over 1300 gene loci in the adult CNS manifest differential read-out of the paternal versus maternal allele. Many of these differentially regulated genes also exhibited brain subregion-selective expression as well. These findings identify parental expression bias as a major mode of epigenetic regulation in the adult CNS, and one important implication of these studies is that epigenetic control of the expression of parent-specific alleles is a driving factor for regulating gene transcription broadly in the brain. The control of the specific expression of one parental allele over another through imprinting of genes in the mature CNS may greatly increase the complexity and subtlety of transcriptional control that operates in cognition. The traditional view of imprinting assumes all-or-none silencing of one allele, rather than a partial expression bias. The work of Dulac and colleagues may necessitate a re-definition of imprinting to incorporate the concept of widespread partial attenuation of one allele, where paternal and maternal alleles are differentially handled and expressed. The function of these genetic parent-of-origin effects may be “allelic tagging” of specific copies of a gene within a neuron (Day and Sweatt, In Press). By this mechanism one allele of a gene (e.g. the paternal copy) could be modified separately from the other allele, providing two templates of the same gene in the same cell that can be differentially regulated by plasticity-related epigenetic mechanisms. Differential epigenetic modification of the two available copies of a given gene within a cell would allow each allele to be handled and expressed differently across the lifespan. As a speculative example for illustrative purposes, a tagged paternal allele of the BDNF gene in a single neuron might be used exclusively during development, and epigenetically regulated as appropriate for its role during early life. The maternal BDNF allele might then be reserved for use in the adult, wherein memory-associated epigenetic mechanisms might operate upon a fresh template of the gene as necessary for triggering short- or long-term activity-dependent changes in BDNF transcription. Epigenetic imprinting of the parental versus maternal alleles would be a prerequisite for this sort of differential epigenetic handling.
Epigenetically based disorders of cognition and novel therapeutic targets
Epigenetic mechanisms of pathogenesis have been implicated in several CNS diseases, including neurodevelopmental disorders of cognition where disruptions in learning and memory are the primary clinical sequelae. Disorders in this category are Angelman Syndrome and Rubinstein-Taybi Syndrome, Fragile X Mental retardation (FMR), and Rett Syndrome. In addition, recent work has implicated derangement of epigenetic mechanisms in post-developmental neurodegenerative disorders of aging such as Alzheimer’s Disease and neuropsychiatric conditions such as drug addiction. Given the protracted and often devastating nature of these disorders, drugs that target the underlying epigenetic defect could provide potentially groundbreaking therapeutic avenues.
In this section we discuss recent exciting findings that explore the manipulation of epigenetic modifications as a therapeutic avenue for the treatment of cognitive dysfunction. We then describe Rett Syndrome as one of the best-established epigenetic disorders specifically dependent on DNA methylation. Finally, we address the emerging role of epigenetic mechanisms in substance abuse and drug addiction.
Histone acetylation and HDAC inhibitors
A promising avenue for therapeutic intervention involves the use of drugs that target HDAC inhibitors to prevent the removal of acetyl groups on histone tails (Kazantsev and Thompson, 2008; Szyf, 2009). This class of drug, such as trichostatin A (TSA), suberoylanilide hydroxamic acid (SAHA), and sodium butyrate, inhibit several isoforms of HDAC enzymes and result in global histone hyperacetylation. A number of these drugs have already been approved for clinical use in patients or a currently in clinical trials in the cancer arena (Szyf, 2009). As discussed above, histone acetylation is robustly associated with “activated” gene transcription, and the formation of new memories produces increases in histone acetylation in the hippocampus (Peleg et al., 2010). In this context, treatment with HDAC inhibitors has been shown to improve memory formation in hippocampal-dependent tasks, and enhance hippocampal LTP (Levenson et al., 2004). Moreover, HDAC inhibitors have been shown to selectively reverse deficits in histone acetylation in aged animals, effectively restoring the ability to learn new associations (Peleg et al., 2010). Finally, even after the induction of severe neuronal atrophy, HDAC inhibitors restore memory formation and even enable access to previously formed long-term memories (Fischer et al., 2007). This result is especially exciting given that a number of patients who present with dementia or Alzheimer’s Disease have difficulty retrieving previously formed memories (American Psychological Association, 2000).
Importantly, the memory-enhancing effects of HDAC inhibitors may be mediated by specific HDAC isoforms. Selective overexpression of HDAC2 in neurons produces a decrease in spine density and impairs synaptic plasticity and memory formation, whereas overexpression of HDAC1 had little effect (Guan et al., 2009). Likewise, deficiency in HDAC2 or chronic treatment with HDAC inhibitors resulted in increased spine density and improved memory function (Guan et al., 2009). In contrast, another study indicated that systemic inhibition of HDACs (and specifically class 1 HDACs) dramatically improved contextual memory function in a mouse model of Alzheimer’s Disease (Kilgore et al., 2010). Thus, future research will be required to parse the effects of HDAC inhibitors on memory function in normal, aged, and diseased mouse models. Nevertheless, the use of HDAC inhibitors in the treatment of learning and memory disorders or neurodegenerative diseases possesses clear therapeutic potential.
Histone methylation
Histone methylation and demethylation represent a second set of modifications that may possess therapeutic interest in relation to disorders of learning and memory. However, unlike histone acetylation, histone methylation is not universally associated with either transcriptional repression or transcriptional activation (Ng et al., 2003; Scharf and Imhof, 2010). Instead, certain modifications, such as dimethylation at H3K9, are associated with transcriptional repression whereas other modifications, such as dimethylation or trimethylation of H3K4, are associated with transcriptional activation (Scharf and Imhof, 2010; Wang et al., 2008). However, there are a number of relatively selective compounds capable of modifying specific methylation marks (Allis et al., 2007; Greiner et al., 2005; Scharf and Imhof, 2010; Shi and Whetstine, 2007; Szyf, 2009), such as the small-molecule inhibitor of the G9a methyltransferase, which reverses H3K9 dimethylation (Kubicek et al., 2007). In rodents, forebrain specific deletion of the GLP/G9a histone methyltransferase complex results in a number of learning-related behavioral deficits, in part by enabling the expression on non-neuronal genes (Schaefer et al., 2009). Similarly, mice with a heterozygous deletion of Mll, a H3K4 specific methyltransferase, exhibited significant impairment in the formation of long-term contextual (but not cued) fear memories (Gupta et al., 2010). Thus, although the therapeutic potential of histone methylation modifying enzymes is relatively unexplored at the present time, these results indicate that selective antagonists of H3K4 demethylating enzymes may be interesting candidates for treating learning and memory disorders (Shi et al., 2004).
DNA methylation and cognitive dysfunction: Insights from Rett Syndrome
Rett Syndrome is a disorder that affects around 1 in 10,000 to 15,000 females. Typically, females with Rett Syndrome appear developmentally normal until between 6 and 18 months of age, at which time development stagnates and subsequently regresses. Classic Rett Syndrome is characterized by profound cognitive impairment, communication dysfunction, stereotypic movements, and pervasive growth failure (Wan et al., 1999). In a breakthrough discovery, mutations in the gene encoding MeCP2 were found to be responsible for at least 95% of classic Rett Syndrome cases (Amir et al., 1999). This seminal finding provided a link between DNA methylation, specifically involving the methyl-DNA binding protein MeCP2, and intellectual dysfunction.
The identification of mecp2 as the mutated gene in Rett Syndrome led to the creation of several transgenic mouse models of Rett Syndrome. Initial attempts to create MeCP2-null mice resulted in embryonic lethality (Tate et al., 1996). To circumvent this problem, two groups independently used the Cre/LoxP recombination system to delete portions of the MeCP2 gene. The Jaenisch lab used a targeted construct that deleted exon 3, which encodes for most of the MBD, while the Bird lab deleted exons 3 and 4, which encode for all but the first 8 amino acids of the protein (Chen et al., 2001; Guy et al., 2001). MeCP2-null mice from the Jaenisch and Bird labs have impairments in hippocampal physiology and behavior, as well as a number of more general physical deficits including early post-natal lethality. Symptomatic male mice have altered hippocampal NMDA receptor expression and impairments in LTP and LTD (Asaka et al., 2006). Male mutant mice also display deficits in cued fear conditioning, while mutant mice of both sexes display deficits in object recognition and altered anxiety (Stearns et al., 2007).
The MeCP2-null mice generated by the Bird and Jaenisch labs display an early onset of symptoms and short lifespan that differentiates them from classic Rett syndrome and limits the analysis of symptoms. Two groups have developed models that attempted to address these limitations. The Zoghbi group generated the mutant mouse model MeCP2308/Y, possessing a premature stop after codon 308, where mutations have been frequently indentified in humans with Rett Syndrome. These mice exhibit a milder phenotype, presumably because the truncated protein retains partial function, characterized by impaired motor function, reduced activity, stereotypic forelimb-clasping movement, and abnormal social interactions (Moretti et al., 2005). MeCP2308/Y also display impaired LTP, increased basal synaptic transmission, and deficits in the induction of LTD, as well as corresponding disruptions in spatial memory, contextual fear conditioning, and long-term social memory (Moretti et al., 2006). Importantly, these mice possess hyperacetylation of H3 (Shahbazian et al., 2002). The Tam group generated another line of MeCP2-null mice (Mecp2tm1Tam) with a deletion of the methyl-binding domain. Behavioral testing of these mice revealed deficits in cerebellar learning and impairments in both cued and contextual fear conditioning and contextual association (Pelka et al., 2006). In a collaborative effort the Zoghbi and Sweatt laboratories showed that MeCP2-deficient animals have deficits in spatial learning, contextual fear conditioning, and LTP deficits (Moretti et al., 2006). Moreover, they also showed that overexpression of MeCP2 resulted in enhanced fear conditioning, and enhanced LTP (Collins et al., 2004). Since Rett syndrome is caused by mutations in MeCP2, enhancing MeCP2 levels could therefore be a therapeutic option. Overall these findings strongly support the idea that MeCP2 might be involved in regulation of LTP and hippocampal-dependent memory formation.
Rett Syndrome has classically been viewed as a neurodevelopmental disorder, the underlying genetic basis of which is mutation/deletion of the MeCP2 gene and resultant disruption of normal MeCP2 function during prenatal and early postnatal development. This model is consistent with the fact that the mutated gene product is present throughout development. However, the mutant gene product is also present in the fully developed adult CNS. Thus, it is unclear if Rett Syndrome is caused exclusively by disruption of MeCP2 function during development, or whether loss of MeCP2 in the mature CNS might also contribute to neurobehavioral and cognitive dysfunction in Rett patients. Recent data from Adrian Bird’s group has suggested that loss of normal MeCP2 function in the adult nervous system contributes to neurobehavioral dysfunction in Rett Syndrome. Specifically, inducible expression of MeCP2 in adult animals extensively rescued the neurological phenotypes in MeCP2-deficient animals. Moreover, exciting work from Greenberg and colleagues has reveal activity-dependent acute regulation of MeCP2 function in neurons, specifically through phosphorylation of specific serine residues (Chen et al., 2003; Tao et al., 2009; Zhou et al., 2006). These and other recent findings (Deng et al., 2010) strongly suggest a dynamic role for MeCP2 in the adult CNS in the regulation of activity-dependent gene transcription during learning and memory. Therefore, MeCP2 function may be necessary in an ongoing fashion for normal learning and memory and synaptic plasticity in the mature CNS. A new understanding of the role of MeCP2 in the adult CNS might allow the development of new therapeutic approaches to Rett treatment based on restoration or augmentation of MeCP2 function after CNS development is largely completed. Findings from studies of Rett syndrome patients and genetically engineered mouse models implicate DNA methylation as a central regulator of adult memory formation. That animals deficient in methyl-DNA binding proteins have deficits in memory and long-term synaptic plasticity is in line with this conceptual framework. Finally, these observations are consistent with the overall theme we are developing in this review, which is the co-opting of developmental molecular mechanisms to subserve long-lasting functional changes in the adult CNS.
Epigenetic modifications and maladaptive behaviors: Insights from drug addiction
Drug addiction is a chronic, relapsing disorder in which drug-related associations (e.g., discrete drug cues, locations in which drugs were consumed, and drug paraphernalia) are capable of exerting tremendous control over behavior long after drug taking has ceased. On this basis, drug addiction has long been considered and interpreted as a disorder of learning and memory (Berke and Hyman, 2000; Hyman, 2005; Hyman et al., 2006; Kelley, 2004). A hallmark feature of drugs of abuse is that they result in persistent functional and structural alterations in brain reward circuits such as the nucleus accumbens (LaPlant et al., 2010; Nestler, 2001; Robinson and Kolb, 1997). These changes occur alongside equally long-lasting changes in expression of genes such as ΔFosB, BDNF, and creb (Kumar et al., 2005; McClung and Nestler, 2003; Nestler, 2001), leading to the suggestion that epigenetic mechanisms may be critical components of drug-related responses (Nestler, 2001). A number of pioneering reports by Eric Nestler and colleagues have largely confirmed this hypothesis, revealing that epigenetic mechanisms are involved in both biochemical and behavioral responses to drugs of abuse. The first of these studies employed a chromatin immunoprecipitation approach to identify histone modifications at individual gene-targets in the nucleus accumbens following cocaine treatment (Kumar et al., 2005). This technique revealed that acute cocaine administration produced a dynamic increase in phospho-acetylation at H3 (S10/K14) and increased acetylation on H4, both surrounding the promoter region of c-fos, an immediate early gene. In contrast, prolonged cocaine exposure produced an increase in acetylation at H3K9 and H3K14 at the promoter for FosB, BDNF, and Cdk5 genes, while leaving c-fos unchanged. This is critical given that FosB and BDNF have been implicated in the transition from casual to chronic drug use and cocaine craving during withdrawal, respectively (Grimm et al., 2003; McClung and Nestler, 2003). Interestingly, the increase in H3 acetylation at the BDNF gene persists for at least a week following cessation of cocaine, which overlaps with the withdrawal-related increases in BDNF levels across multiple brain regions (Grimm et al., 2003).
Further experiments have demonstrated that these modifications are important regulators of the rewarding properties of cocaine. Treatment with an HDAC inhibitor prior to cocaine or morphine exposure enhances behavioral preferences for places associated with drug delivery (so-called conditioned place preference, or CPP) (Kumar et al., 2005; Renthal et al., 2007; Sanchis-Segura et al., 2009). Additionally, antagonism of sirtuins (Sirt1 and Sirt2, a unique class of HDACs) in the nucleus accumbens reduces CPP and operant responding for cocaine reward (Renthal et al., 2009). In contrast, overexpression of HDAC4 in the nucleus accumbens impairs the development of a conditioned place preference for cocaine and decreases the break point for cocaine self-administration, indicative of blunted motivation to consume the drug (Kumar et al., 2005; Wang et al., 2010). Similarly, viral overexpression of HDAC5 in the nucleus accumbens blunts the development of cocaine CPP, whereas global deletion of the HDAC5 gene enhances CPP (Renthal et al., 2007). Conversely, a recent report found that HDAC inhibitors delivered during extinction sessions facilitate the extinction of cocaine CPP in mice, indicating that histone acetylation may also play a critical role in the reversal of drug-related memories (Malvaez et al., 2010). Together, these findings suggest that HDAC inhibitors facilitate learning and memory, whether it is during associative conditioning or extinction. Therefore, HDACs may be promising candidates for drug abuse treatments, especially when combined with behavioral therapy.
Although the majority of experiments have focused on histone acetylation, it is now abundantly clear that other histone modifications, including phosphorylation and methylation, are critical components of the epigenetic response to drugs of abuse (Maze et al., 2010; Stipanovich et al., 2008). Indeed, cocaine induces a robust phosphorylation of H3S10 within the nucleus accumbens at the promoters of c-fos and c-jun (Brami-Cherrier et al., 2009). Importantly, this response is regulated by two distinct signal transduction cascades, both of which are downstream of a major target of drug-induced increases in striatal dopamine concentration: the activation of dopamine D1 receptors in the striatonigral (direct) pathway. H3S10 phosphorylation is positively regulated by the same MAPK pathways reviewed above, including phosphorylation of ERK and MSK-1-induced phosphorylation of H3 (Bertran-Gonzalez et al., 2008; Brami-Cherrier et al., 2005). Likewise, nuclear accumulation of 32-kDa dopamine-regulated and cyclic-AMP-regulated phosphoprotein (DARPP-32), which also occurs following D1 receptor activation, acts to inhibit PP1, thereby preventing histone dephosphorylation (Stipanovich et al., 2008). Critically, these pathways are instrumental in controlling behavioral responses to cocaine and morphine, as inhibition of D1 receptors, ERK, DARPP-32, MSK-1, all diminish drug-induced locomotor responses or drug CPP (Brami-Cherrier et al., 2009; Brami-Cherrier et al., 2005; Stipanovich et al., 2008).
Much like the emergent evidence that DNA methylation regulates hippocampal-dependent memory formation, recent reports have revealed that DNA methylation in the striatum is associated with drug-related behaviors. For example, acute cocaine administration produces rapid changes in expression of DNMT isoforms within the nucleus accumbens (Anier et al., 2010; LaPlant et al., 2010), suggesting dynamic control of DNA methylation by drugs of abuse. Consistent with this observation, cocaine produces a hypermethylation at the promoter region of PP1c (the catalytic subunit of PP1) in the nucleus accumbens, resulting in enhanced MeCP2 binding to the PP1c promoter (Anier et al., 2010). Conversely, cocaine decreases methylation at the FosB promoter, which coincides with the transcriptional upregulation of FosB and is consistent with the observed decrease in MeCP2 binding to FosB (Anier et al., 2010). Importantly, systemic inhibition of DNA methyltransferase activity significantly impairs the development of locomotor sensitization induced by repeated cocaine administration (Anier et al., 2010), and site-specific DNMT inhibition in the nucleus accumbens boosts the development of cocaine CPP (LaPlant et al., 2010). In contrast, overexpression of the DNMT3a isoform within the nucleus accumbens disrupts cocaine CPP (LaPlant et al., 2010), whereas MeCP2 knockdown in the dorsal striatum prevents escalation of cocaine self-administration during extended access (Im et al., 2010). Additionally, DNA methylation within the hippocampus and prelimbic cortex is also necessary for the establishment and maintenance of cocaine CPP, respectively, indicating that epigenetic changes in brain regions outside of the striatum are also key regulators of drug memories (Han et al., 2010). These results reveal that DNA methylation within the striatum is an important biochemical step in the short- and long-term behavioral response to drugs of abuse, and suggest that interfering with methylation machinery may constitute a possible avenue for therapeutic treatment. However, it is important to note here that epigenetic mechanisms likely do not exist to solely support the formation and persistence of drug-related memories. Indeed, the same biochemical pathways that regulate epigenetic modifications are involved in unlearned and learned responses to natural rewards like food, mating, and social interaction (Aragona et al., 2003; Aragona et al., 2006; Aragona and Wang, 2007; Bureau et al., 2010; Day, 2008; Kelley and Berridge, 2002; Kelley et al., 1997; Shiflett et al., 2008; Shiflett et al., 2009; Stipanovich et al., 2008). Therefore, future studies will be required to determine whether these events also induce epigenetic changes, and in what ways these changes differ from those induced by drug exposure.
Although drug-taking is remarkably conserved across species, it is clear that not all members of a population will exhibit signs of addiction (e.g., inability to cease drug taking, high motivation to take the drug, and continued drug use in spite of harmful consequences) despite equivalent drug availability or drug history (Deroche-Gamonet et al., 2004; Kreek et al., 2005). Therefore, a critical component in the development of drug addiction is individual variability. While genetic polymorphisms resulting in differences in risk-taking and drug effects may help to account for this difference, only 30–60% of addiction vulnerability is thought to be heritable in the strict genomic sense (Kreek et al., 2005). Another potential explanatory factor for vulnerability to addictive disease are the long-lasting epigenetic effects of early life experiences or even transgenerational epigenetic inheritance (Champagne and Curley, 2009; Roth et al., 2009; Weaver et al., 2004; Weaver et al., 2005), which is capable of stochastic variation at a much higher rate than mutation of DNA bases (Petronis, 2010). Thus, in addition to potentially explaining how drugs of abuse produce long-lasting changes in neuronal plasticity, epigenetic mechanisms hold tremendous potential to reveal why some individuals are more prone to take drugs and/or develop full-blown addiction.
Conclusions
In writing this review, we have endeavored to provide an overview of an emerging topic at the cross-section of developmental biology and cognitive neuroscience. We have attempted to provide a novel synthesis of ideas across modalities of epigenetic modification and cellular and behavioral processes of learning and memory. There are interesting and compelling new avenues of inquiry, such as potential novel therapeutics, that arise from recent work implicating both DNA methylation and histone regulation as critical molecular mechanisms underlying memory consolidation and memory storage in the adult CNS. In a broader sense, these findings have established behavioral epigenetics as a subfield in its own right.
Finally, the main overarching theme of this review is that cell “developmental” molecular mechanisms, e.g. growth factor regulation, MAPK signaling and epigenetic mechanisms, are conserved in the adult CNS to subserve long-term plasticity and memory formation (Ehninger et al., 2008; Marcus et al., 1994; Weeber and Sweatt, 2002). That cellular development and adult memory are molecular homologs, i.e., share identical molecular and biochemical mechanisms, provides an explanation for one of the long-standing questions in neuroscience: why can’t neurons divide? One of the critical roles for most adult neurons is to be plastic; to be able to modulate their function over time. Moreover, in many instances the cellular changes need to be either long-lasting or permanent in order for the neuron to serve the appropriate function in a given neural circuit. The terminally differentiated adult neuron has adapted many of the molecular mechanisms used to regulate cell division and perpetuate cell phenotype in order to perform one of its primary functions, long-term plasticity. These processes can therefore no longer be utilized to trigger cell division or alter cell phenotype.
Acknowledgements
The authors wish to thank Tom Carew, Huda Zoghbi, Art Beaudet, and Eric Kandel for many helpful discussions. We apologize to the many authors whose primary work was not directly cited, owing to limitations of space. Research in the authors’ laboratory is supported by funds from the NINDS, NIMH, NIA, NIDA, the Rett Syndrome Foundation, the Ellison Medical Foundation, and the Evelyn F. McKnight Brain Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Alberini CM. The role of protein synthesis during the labile phases of memory: revisiting the skepticism. Neurobiol Learn Mem. 2008;89:234–246. doi: 10.1016/j.nlm.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- American Psychological Association. Diagnostic and statistical manual of mental disorders. Revised 4th Edition. Washington, D.C.: Author; 2000. [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Anier K, Malinovskaja K, Aonurm-Helm A, Zharkovsky A, Kalda A. DNA Methylation Regulates Cocaine-Induced Behavioral Sensitization in Mice. Neuropsychopharmacology. 2010 doi: 10.1038/npp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona BJ, Liu Y, Curtis JT, Stephan FK, Wang Z. A critical role for nucleus accumbens dopamine in partner-preference formation in male prairie voles. J Neurosci. 2003;23:3483–3490. doi: 10.1523/JNEUROSCI.23-08-03483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona BJ, Liu Y, Yu YJ, Curtis JT, Detwiler JM, Insel TR, Wang Z. Nucleus accumbens dopamine differentially mediates the formation and maintenance of monogamous pair bonds. Nat Neurosci. 2006;9:133–139. doi: 10.1038/nn1613. [DOI] [PubMed] [Google Scholar]
- Aragona BJ, Wang Z. Opposing regulation of pair bond formation by cAMP signaling within the nucleus accumbens shell. J Neurosci. 2007;27:13352–13356. doi: 10.1523/JNEUROSCI.3216-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaka Y, Jugloff DG, Zhang L, Eubanks JH, Fitzsimonds RM. Hippocampal synaptic plasticity is impaired in the Mecp2-null mouse model of Rett syndrome. Neurobiol Dis. 2006;21:217–227. doi: 10.1016/j.nbd.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Atkins CM, Selcher JC, Petraitis JJ, Trzaskos JM, Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nat Neurosci. 1998;1:602–609. doi: 10.1038/2836. [DOI] [PubMed] [Google Scholar]
- Bading H, Greenberg ME. Stimulation of protein tyrosine phosphorylation by NMDA receptor activation. Science. 1991;253:912–914. doi: 10.1126/science.1715095. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Igaz LM, Bevilaqua LR, Izquierdo I, Medina JH. Persistence of long-term memory storage requires a late protein synthesis- and BDNF- dependent phase in the hippocampus. Neuron. 2007;53:261–277. doi: 10.1016/j.neuron.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Berke JD, Hyman SE. Addiction, dopamine, and the molecular mechanisms of memory. Neuron. 2000;25:515–532. doi: 10.1016/s0896-6273(00)81056-9. [DOI] [PubMed] [Google Scholar]
- Bertran-Gonzalez J, Bosch C, Maroteaux M, Matamales M, Herve D, Valjent E, Girault JA. Opposing patterns of signaling activation in dopamine D1 and D2 receptor-expressing striatal neurons in response to cocaine and haloperidol. J Neurosci. 2008;28:5671–5685. doi: 10.1523/JNEUROSCI.1039-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brami-Cherrier K, Lavaur J, Pages C, Arthur JS, Caboche J. Glutamate induces histone H3 phosphorylation but not acetylation in striatal neurons: role of mitogen- and stress-activated kinase-1. J Neurochem. 2007;101:697–708. doi: 10.1111/j.1471-4159.2006.04352.x. [DOI] [PubMed] [Google Scholar]
- Brami-Cherrier K, Roze E, Girault JA, Betuing S, Caboche J. Role of the ERK/MSK1 signalling pathway in chromatin remodelling and brain responses to drugs of abuse. J Neurochem. 2009;108:1323–1335. doi: 10.1111/j.1471-4159.2009.05879.x. [DOI] [PubMed] [Google Scholar]
- Brami-Cherrier K, Valjent E, Herve D, Darragh J, Corvol JC, Pages C, Arthur SJ, Girault JA, Caboche J. Parsing molecular and behavioral effects of cocaine in mitogen- and stress-activated protein kinase-1-deficient mice. J Neurosci. 2005;25:11444–11454. doi: 10.1523/JNEUROSCI.1711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14:268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau G, Carrier M, Lebel M, Cyr M. Intrastriatal inhibition of extracellular signal-regulated kinases impaired the consolidation phase of motor skill learning. Neurobiol Learn Mem. 2010;94:107–115. doi: 10.1016/j.nlm.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Cervoni N, Szyf M. Demethylase activity is directed by histone acetylation. J Biol Chem. 2001;276:40778–40787. doi: 10.1074/jbc.M103921200. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA, Curley JP. Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neurosci Biobehav Rev. 2009;33:593–600. doi: 10.1016/j.neubiorev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Chao HT, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Chwang WB, Arthur JS, Schumacher A, Sweatt JD. The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J Neurosci. 2007;27:12732–12742. doi: 10.1523/JNEUROSCI.2522-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwang WB, O'Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13:322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, Sweatt JD, Zoghbi H. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet. 2004;13:2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- D'Alessio AC, Szyf M. Epigenetic tete-a-tete: the bilateral relationship between chromatin modifications and DNA methylation. Biochem Cell Biol. 2006;84:463–476. doi: 10.1139/o06-090. [DOI] [PubMed] [Google Scholar]
- D'Alessio AC, Weaver IC, Szyf M. Acetylation-induced transcription is required for active DNA demethylation in methylation-silenced genes. Mol Cell Biol. 2007;27:7462–7474. doi: 10.1128/MCB.01120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis HP, Squire LR. Protein synthesis and memory: a review. Psychol Bull. 1984;96:518–559. [PubMed] [Google Scholar]
- Day JJ. Extracellular signal-related kinase activation during natural reward learning: a physiological role for phasic nucleus accumbens dopamine? J Neurosci. 2008;28:4295–4297. doi: 10.1523/JNEUROSCI.0776-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Cognitive neuroepigenetics: A role for epigenetic mechanisms in learning and memory. Neurobiol Learn Mem. doi: 10.1016/j.nlm.2010.12.008. (In Press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Bito H, Tsien RW. Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron. 1996;16:89–101. doi: 10.1016/s0896-6273(00)80026-4. [DOI] [PubMed] [Google Scholar]
- Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci. 2010;13:1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroche-Gamonet V, Belin D, Piazza PV. Evidence for addiction-like behavior in the rat. Science. 2004;305:1014–1017. doi: 10.1126/science.1099020. [DOI] [PubMed] [Google Scholar]
- Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson MH, Rose S, Mahadevan LC. Acetyllysine-binding and function of bromodomain-containing proteins in chromatin. Front Biosci. 2001;6:D853–D865. doi: 10.2741/dyson. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Phan T, Han S, Wang H, Chan GC, Scheiner ZS, Storm DR. Circadian oscillation of hippocampal MAPK activity and cAmp: implications for memory persistence. Nat Neurosci. 2008;11:1074–1082. doi: 10.1038/nn.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Li W, Fox K, Stryker MP, Silva AJ. Reversing neurodevelopmental disorders in adults. Neuron. 2008;60:950–960. doi: 10.1016/j.neuron.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English JD, Sweatt JD. Activation of p42 mitogen-activated protein kinase in hippocampal long term potentiation. J Biol Chem. 1996;271:24329–24332. doi: 10.1074/jbc.271.40.24329. [DOI] [PubMed] [Google Scholar]
- English JD, Sweatt JD. A requirement for the mitogen-activated protein kinase cascade in hippocampal long term potentiation. J Biol Chem. 1997;272:19103–19106. doi: 10.1074/jbc.272.31.19103. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore RS, Murphy TH, Sanghera JS, Pelech SL, Baraban JM. Activation of p42 mitogen-activated protein kinase by glutamate receptor stimulation in rat primary cortical cultures. J Neurochem. 1993;61:1626–1633. doi: 10.1111/j.1471-4159.1993.tb09796.x. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DA, Greenberg ME. CREB: a mediator of long-term memory from mollusks to mammals. Cell. 1994;79:5–8. doi: 10.1016/0092-8674(94)90394-8. [DOI] [PubMed] [Google Scholar]
- Frey U, Huang YY, Kandel ER. Effects of cAMP simulate a late stage of LTP in hippocampal CA1 neurons. Science. 1993;260:1661–1664. doi: 10.1126/science.8389057. [DOI] [PubMed] [Google Scholar]
- Frey U, Krug M, Reymann KG, Matthies H. Anisomycin, an inhibitor of protein synthesis, blocks late phases of LTP phenomena in the hippocampal CA1 region in vitro. Brain Res. 1988;452:57–65. doi: 10.1016/0006-8993(88)90008-x. [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003a;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003b;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Girault JA, Valjent E, Caboche J, Herve D. ERK2: a logical AND gate critical for drug-induced plasticity? Curr Opin Pharmacol. 2007;7:77–85. doi: 10.1016/j.coph.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Yamamoto KK, Fischer WH, Karr D, Menzel P, Biggs W, 3rd, Vale WW, Montminy MR. A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature. 1989;337:749–752. doi: 10.1038/337749a0. [DOI] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010a;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010b;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat Chem Biol. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- Grimm JW, Lu L, Hayashi T, Hope BT, Su TP, Shaham Y. Time-dependent increases in brain-derived neurotrophic factor protein levels within the mesolimbic dopamine system after withdrawal from cocaine: implications for incubation of cocaine craving. J Neurosci. 2003;23:742–747. doi: 10.1523/JNEUROSCI.23-03-00742.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D, Kandel ER. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Han J, Li Y, Wang D, Wei C, Yang X, Sui N. Effect of 5-aza-2-deoxycytidine microinjecting into hippocampus and prelimbic cortex on acquisition and retrieval of cocaine-induced place preference in C57BL/6 mice. Eur J Pharmacol. 2010;642:93–98. doi: 10.1016/j.ejphar.2010.05.050. [DOI] [PubMed] [Google Scholar]
- Herz J, Chen Y. Reelin, lipoprotein receptors and synaptic plasticity. Nat Rev Neurosci. 2006;7:850–859. doi: 10.1038/nrn2009. [DOI] [PubMed] [Google Scholar]
- Hyman SE. Addiction: a disease of learning and memory. Am J Psychiatry. 2005;162:1414–1422. doi: 10.1176/appi.ajp.162.8.1414. [DOI] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ. Neural Mechanisms of Addiction: The Role of Reward-Related Learning and Memory. Annu Rev Neurosci. 2006 doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Mark M, Villacres EC, Poser S, Chavkin C, Storm DR. Induction of CRE-mediated gene expression by stimuli that generate long-lasting LTP in area CA1 of the hippocampus. Neuron. 1996;16:973–982. doi: 10.1016/s0896-6273(00)80120-8. [DOI] [PubMed] [Google Scholar]
- Impey S, Obrietan K, Wong ST, Poser S, Yano S, Wayman G, Deloulme JC, Chan G, Storm DR. Cross talk between ERK and PKA is required for Ca2+ stimulation of CREB-dependent transcription and ERK nuclear translocation. Neuron. 1998;21:869–883. doi: 10.1016/s0896-6273(00)80602-9. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- Kelley AE. Ventral striatal control of appetitive motivation: role in ingestive behavior and reward-related learning. Neurosci Biobehav Rev. 2004;27:765–776. doi: 10.1016/j.neubiorev.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Kelley AE, Berridge KC. The neuroscience of natural rewards: relevance to addictive drugs. J Neurosci. 2002;22:3306–3311. doi: 10.1523/JNEUROSCI.22-09-03306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley AE, Smith-Roe SL, Holahan MR. Response-reinforcement learning is dependent on N-methyl-D-aspartate receptor activation in the nucleus accumbens core. Proc Natl Acad Sci U S A. 1997;94:12174–12179. doi: 10.1073/pnas.94.22.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Kornhauser JM, Cowan CW, Shaywitz AJ, Dolmetsch RE, Griffith EC, Hu LS, Haddad C, Xia Z, Greenberg ME. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron. 2002;34:221–233. doi: 10.1016/s0896-6273(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshibu K, Graff J, Beullens M, Heitz FD, Berchtold D, Russig H, Farinelli M, Bollen M, Mansuy IM. Protein phosphatase 1 regulates the histone code for long-term memory. J Neurosci. 2009;29:13079–13089. doi: 10.1523/JNEUROSCI.3610-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreek MJ, Nielsen DA, Butelman ER, LaForge KS. Genetic influences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nat Neurosci. 2005;8:1450–1457. doi: 10.1038/nn1583. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iniguez SD, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms: a common theme in vertebrate and invertebrate memory formation. Cell Mol Life Sci. 2006;63:1009–1016. doi: 10.1007/s00018-006-6026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebrich S, Nedivi E. The function of activity-regulated genes in the nervous system. Physiol Rev. 2009;89:1079–1103. doi: 10.1152/physrev.00013.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Sweatt JD. The IkappaB kinase regulates chromatin structure during reconsolidation of conditioned fear memories. Neuron. 2007;55:942–957. doi: 10.1016/j.neuron.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, Sanchis-Segura C, Vo D, Lattal KM, Wood MA. Modulation of chromatin modification facilitates extinction of cocaine-induced conditioned place preference. Biol Psychiatry. 2010;67:36–43. doi: 10.1016/j.biopsych.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus EA, Emptage NJ, Marois R, Carew TJ. A comparison of the mechanistic relationships between development and learning in Aplysia. Prog Brain Res. 1994;100:179–188. doi: 10.1016/s0079-6123(08)60784-0. [DOI] [PubMed] [Google Scholar]
- Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- McManus KJ, Hendzel MJ. CBP, a transcriptional coactivator and acetyltransferase. Biochem Cell Biol. 2001;79:253–266. [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Montminy MR, Gonzalez GA, Yamamoto KK. Characteristics of the cAMP response unit. Metabolism. 1990a;39:6–12. doi: 10.1016/0026-0495(90)90198-l. [DOI] [PubMed] [Google Scholar]
- Montminy MR, Gonzalez GA, Yamamoto KK. Regulation of cAMP-inducible genes by CREB. Trends Neurosci. 1990b;13:184–188. doi: 10.1016/0166-2236(90)90045-c. [DOI] [PubMed] [Google Scholar]
- Moretti P, Bouwknecht JA, Teague R, Paylor R, Zoghbi HY. Abnormalities of social interactions and home-cage behavior in a mouse model of Rett syndrome. Hum Mol Genet. 2005;14:205–220. doi: 10.1093/hmg/ddi016. [DOI] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TH, Blatter LA, Bhat RV, Fiore RS, Wier WG, Baraban JM. Differential regulation of calcium/calmodulin-dependent protein kinase II and p42 MAP kinase activity by synaptic transmission. J Neurosci. 1994;14:1320–1331. doi: 10.1523/JNEUROSCI.14-03-01320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Patterson SL, Abel T, Deuel TA, Martin KC, Rose JC, Kandel ER. Recombinant BDNF rescues deficits in basal synaptic transmission and hippocampal LTP in BDNF knockout mice. Neuron. 1996;16:1137–1145. doi: 10.1016/s0896-6273(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Pelka GJ, Watson CM, Radziewic T, Hayward M, Lahooti H, Christodoulou J, Tam PP. Mecp2 deficiency is associated with learning and cognitive deficits and altered gene activity in the hippocampal region of mice. Brain. 2006;129:887–898. doi: 10.1093/brain/awl022. [DOI] [PubMed] [Google Scholar]
- Perissi V, Dasen JS, Kurokawa R, Wang Z, Korzus E, Rose DW, Glass CK, Rosenfeld MG. Factor-specific modulation of CREB-binding protein acetyltransferase activity. Proc Natl Acad Sci U S A. 1999;96:3652–3657. doi: 10.1073/pnas.96.7.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- Rattiner LM, Davis M, French CT, Ressler KJ. Brain-derived neurotrophic factor and tyrosine kinase receptor B involvement in amygdala-dependent fear conditioning. J Neurosci. 2004;24:4796–4806. doi: 10.1523/JNEUROSCI.5654-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissner KJ, Shobe JL, Carew TJ. Molecular nodes in memory processing: insights from Aplysia. Cell Mol Life Sci. 2006;63:963–974. doi: 10.1007/s00018-006-6022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, 3rd, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ. Epigenetic mechanisms in drug addiction. Trends Mol Med. 2008;14:341–350. doi: 10.1016/j.molmed.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reul JM, Hesketh SA, Collins A, Mecinas MG. Epigenetic mechanisms in the dentate gyrus act as a molecular switch in hippocampus-associated memory formation. Epigenetics. 2009;4:434–439. doi: 10.4161/epi.4.7.9806. [DOI] [PubMed] [Google Scholar]
- Roberson ED, English JD, Adams JP, Selcher JC, Kondratick C, Sweatt JD. The mitogen-activated protein kinase cascade couples PKA and PKC to cAMP response element binding protein phosphorylation in area CA1 of hippocampus. J Neurosci. 1999;19:4337–4348. doi: 10.1523/JNEUROSCI.19-11-04337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson TE, Kolb B. Persistent structural modifications in nucleus accumbens and prefrontal cortex neurons produced by previous experience with amphetamine. J Neurosci. 1997;17:8491–8497. doi: 10.1523/JNEUROSCI.17-21-08491.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchis-Segura C, Lopez-Atalaya JP, Barco A. Selective boosting of transcriptional and behavioral responses to drugs of abuse by histone deacetylase inhibition. Neuropsychopharmacology. 2009;34:2642–2654. doi: 10.1038/npp.2009.125. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, Tarakhovsky A, Greengard P. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64:678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf AN, Imhof A. Every methyl counts - Epigenetic calculus. FEBS Lett. 2010 doi: 10.1016/j.febslet.2010.11.029. [DOI] [PubMed] [Google Scholar]
- Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, Armstrong D, Paylor R, Zoghbi H. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- Sharma SK, Carew TJ. The roles of MAPK cascades in synaptic plasticity and memory in Aplysia: facilitatory effects and inhibitory constraints. Learn Mem. 2004;11:373–378. doi: 10.1101/lm.81104. [DOI] [PubMed] [Google Scholar]
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Shiflett MW, Martini RP, Mauna JC, Foster RL, Peet E, Thiels E. Cue-elicited reward-seeking requires extracellular signal-regulated kinase activation in the nucleus accumbens. J Neurosci. 2008;28:1434–1443. doi: 10.1523/JNEUROSCI.2383-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett MW, Mauna JC, Chipman AM, Peet E, Thiels E. Appetitive Pavlovian conditioned stimuli increase CREB phosphorylation in the nucleus accumbens. Neurobiol Learn Mem. 2009 doi: 10.1016/j.nlm.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- Sindreu CB, Scheiner ZS, Storm DR. Ca2+ -stimulated adenylyl cyclases regulate ERK-dependent activation of MSK1 during fear conditioning. Neuron. 2007;53:79–89. doi: 10.1016/j.neuron.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns NA, Schaevitz LR, Bowling H, Nag N, Berger UV, Berger-Sweeney J. Behavioral and anatomical abnormalities in Mecp2 mutant mice: a model for Rett syndrome. Neuroscience. 2007;146:907–921. doi: 10.1016/j.neuroscience.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Stipanovich A, Valjent E, Matamales M, Nishi A, Ahn JH, Maroteaux M, Bertran-Gonzalez J, Brami-Cherrier K, Enslen H, Corbille AG, et al. A phosphatase cascade by which rewarding stimuli control nucleosomal response. Nature. 2008;453:879–884. doi: 10.1038/nature06994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Swank MW, Sweatt JD. Increased histone acetyltransferase and lysine acetyltransferase activity and biphasic activation of the ERK/RSK cascade in insular cortex during novel taste learning. J Neurosci. 2001;21:3383–3391. doi: 10.1523/JNEUROSCI.21-10-03383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. The neuronal MAP kinase cascade: a biochemical signal integration system subserving synaptic plasticity and memory. J Neurochem. 2001;76:1–10. doi: 10.1046/j.1471-4159.2001.00054.x. [DOI] [PubMed] [Google Scholar]
- Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Hu K, Chang Q, Wu H, Sherman NE, Martinowich K, Klose RJ, Schanen C, Jaenisch R, Wang W, Sun YE. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc Natl Acad Sci U S A. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate P, Skarnes W, Bird A. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat Genet. 1996;12:205–208. doi: 10.1038/ng0296-205. [DOI] [PubMed] [Google Scholar]
- Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valjent E, Caboche J, Vanhoutte P. Mitogen-activated protein kinase/extracellular signal-regulated kinase induced gene regulation in brain: a molecular substrate for learning and memory? Mol Neurobiol. 2001;23:83–99. doi: 10.1385/MN:23:2-3:083. [DOI] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Lee SS, Zhang X, Houwink-Manville I, Song HR, Amir RE, Budden S, Naidu S, Pereira JL, Lo IF, et al. Rett syndrome and beyond: recurrent spontaneous and familial MECP2 mutations at CpG hotspots. Am J Hum Genet. 1999;65:1520–1529. doi: 10.1086/302690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lv Z, Hu Z, Sheng J, Hui B, Sun J, Ma L. Chronic cocaine-induced H3 acetylation and transcriptional activation of CaMKIIalpha in the nucleus accumbens is critical for motivation for drug reinforcement. Neuropsychopharmacology. 2010;35:913–928. doi: 10.1038/npp.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Beffert U, Jones C, Christian JM, Forster E, Sweatt JD, Herz J. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J Biol Chem. 2002;277:39944–39952. doi: 10.1074/jbc.M205147200. [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Sweatt JD. Molecular neurobiology of human cognition. Neuron. 2002;33:845–848. doi: 10.1016/s0896-6273(02)00634-7. [DOI] [PubMed] [Google Scholar]
- Wood MA, Attner MA, Oliveira AM, Brindle PK, Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn Mem. 2006a;13:609–617. doi: 10.1101/lm.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Hawk JD, Abel T. Combinatorial chromatin modifications and memory storage: a code for memory? Learn Mem. 2006b;13:241–244. doi: 10.1101/lm.278206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan LW, Gambee JE. Histone acetylation by p300 is involved in CREB-mediated transcription on chromatin. Biochim Biophys Acta. 2001;1541:161–169. doi: 10.1016/s0167-4889(01)00141-0. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]