Abstract
We report a comprehensive genetic, metabolomic, and biochemical study on the catalytic properties of Streptomyces coelicolor cytochrome P450 (P450) 154A1, known to have a unique heme orientation in its crystal structure. Deletion of the P450 154A1 gene compromised the long-term stability of the bacterial spores. A novel dipentaenone (1) with a high degree of conjugation was identified as an endogenous substrate of P450 154A1 using a metabolomics approach. The biotransformation of 1 by P450 154A1 was shown to be an unexpected intramolecular cyclization to a Paternò–Büchi-like product, without oxidation/reduction.
Cytochrome P450 (P450) constitutes a superfamily of enzymes containing a prosthetic heme iron. P450s are arguably the most functionally versatile enzymes and catalyze a great variety of NADPH- and O2-dependent reactions.1 These processes normally involve the insertion of one oxygen atom from molecular dioxgen into the substrate, often in a stereospecific and regioselective manner. For almost all catalytic activities, P450s must be complemented with their electron donor partner proteins, either NADPH-cytochrome P450 reductase (for microsomal eukaryotic P450s) or a ferredoxin/ferredoxin reductase system (for most prokaryotic and mitochondrial P450s).2
Streptomycetes produce a vast array of antibiotics and other pharmacologically active natural products. Streptomyces coelicolor A3(2) is the most studied member of the genus as a model system for genetic investigations. In 2002, 18 P450 sequences from S. coelicolor A3(2) were revealed from the genome-sequencing project,3 presenting a new source for P450 enzymology.4–6 Of these enzymes, the crystal structure of P450 154A1 shows a unique heme orientation that is opposite that of almost all other P450s.7 Whether or not this structural feature influences the catalytic function of P450 154A1 is not understood because its endogenous substrates and physiologic functions remain unknown. Accordingly, we initiated a combined genetic, metabolomic, and biochemical investigation on this enzyme.
The gene encoding P450 154A1 was deleted by a double crossover method to give the mutant strain S. coelicolor K-154A1. The mutant did not show any phenotypic difference from the wild type (WT) strain if fresh spores were used for inoculation. However, the viability of the spores from the mutant was greatly reduced after a long time in storage (>6 months) in 20% glycerol (v/v) at −80 °C. Germination of these aged spores on agar plates was very slow, and only sporadic tiny colonies were formed after 7 days of incubation (Figure S1, Supporting Information). WT spores stored under the same conditions grew into confluence. The K-154A1 spores also demonstrated growth deficiency in a high osmotic pressure liquid medium, with no growth observed in the YEME medium8 (34% sucrose, w/v) unless the sugar was excluded from the medium. This complex phenotype indicates that P450 154A1 may have a physiological function in the spore stability or the overall fitness of the organism. Further studies will be required to fully understand the underlying mechanism. Nevertheless, this result strongly suggested the existence of an endogenous substrate of P450 154A1.
A metabolomic approach was employed to search for substrates from the metabolome of S. coelicolor K-154A1. An organic extract of the mutant was prepared as a chemical library that might contain enriched substrates. The extract was then incubated with recombinant P450 154A1 in the presence of NADPH and spinach ferredoxin and ferredoxin reductase, with subsequent metabolite profiling by LC-MS. The metabolic profile obtained after the reaction was compared with that prior to the reaction using specialized metabolomics software, XCMS9 and mzMine.10 Molecules that were depleted in the enzymatic reaction were considered as potential substrates. A substrate (1) was identified using this method (Figure 1a). Metabolic profiling of WT S. coelicolor did not reveal the presence of 1, providing further evidence that 1 is an authentic substrate of P450 154A1. The molecular formula was determined as C16H18O3 based on the positive ion HRMS of 1 (Figure 1b).
Figure 1.
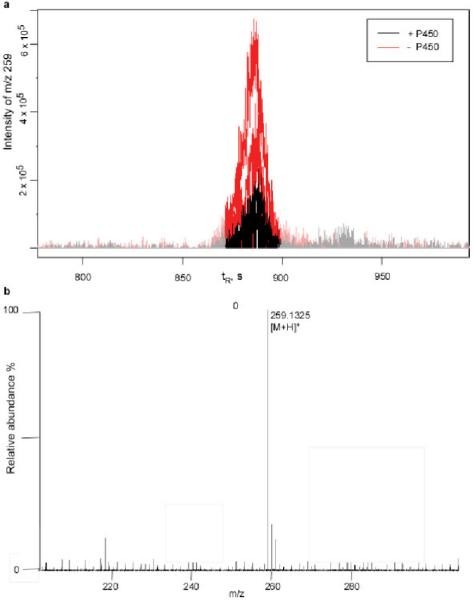
Metabolomics-approach identification of a putative endogenous substrate, 1 (m/z [M + H]+ 259). (a) Data analysis by XCMS showing the depletion of 1 in the presence of P450 154A1. (b) The molecular formula of 1 was determined as C16H18O3 from HRMS (Table S1, Supporting Information).
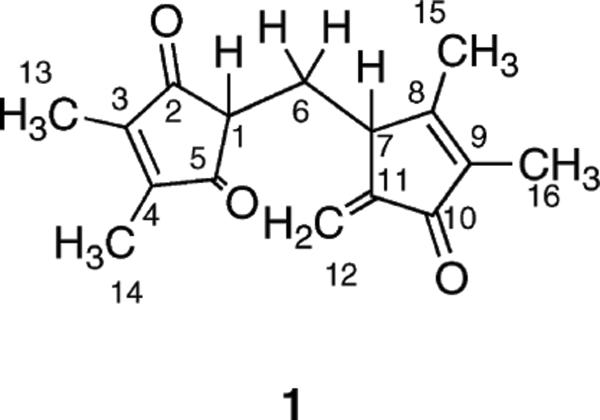
In order to elucidate the structure of 1, about 0.5 mg of the material was prepared from the cell pellet collected from a 5-L culture of S. coelicolor K-154A1. The dipentaenone structure of 1 was elucidated by various types of NMR spectroscopy (Figure 2), including 1H, 13C, COSY, HSQC, and HMBC (Figures S2–S6, Supporting Information). HSQC analysis revealed the presence of four methyl groups (C15, C16, and two equivalent methyl units, C13/C14), one aliphatic methylene group (C6), one olefinic methylene group (C12), and two methine groups (C1 and C7). Additionally, eight quaternary carbons, including five olefinic carbons (C8–C9, C11, and two equivalent olefinic carbons, C3–C4) and three keto groups (C10 and two equivalent keto groups, C2/C5), were identified using HMBC and 13C NMR. The presence of two 1,2-dimethyl olefin units (C13–C3–C4–C14 and C15–C8–C9–C16) was confirmed by the unusual 5J coupling between nonequivalent methyl groups in the COSY experiment and 2J/3J coupling between olefinic carbons and protons from the attached methyl groups (HMBC). From combining all of the above information, the structure of 1 was established except for the absolute configuration at C7. In addition to the two 1,2-dimethyl olefin units, a notable structural feature of 1 is its high degree of conjugation (λmax 246 nm; ε 16 430 M−1 cm−1). To the best of our knowledge, this structure represents a new chemical entity not reported previously. The biosynthetic origin of 1 is unknown, but the pentaenone ring of 1 resembles the scaffold of methylenomycin C, an antibiotic produced by S. coelicolor.11–13
Figure 2.
Structure of 1.
Incubation of 1 with P450 154A1 resulted in one major product, 2a, and one minor product, 2b, as monitored by HPLC-UV spectroscopy (Figure 3a). Comparison of the UV spectra of these compounds showed that the absorption maxima of both products were identical and the wavelengths were 9 nm shorter than that of substrate, indicating decreased conjugation (Figure 3b). Surprisingly, the conversion did not require NADPH/NADH or any redox partners (ferredoxin or ferredoxin reductase). No conversion of 1 was observed in the absence of P450 154A1 or when P450 154A1 was replaced with family member P450 154C1. HRMS of 2a and 2b indicated that the products have the same molecular formula as the substrate, 1. Therefore P450 154A1 must catalyze an unusual rearrangement of 1 independent of oxidation and reduction. Because 2a and 2b were not found in WT S. coelicolor, only small quantities of 2a could be prepared by in vitro enzymatic conversion, making the structural elucidation challenging. Various NMR experiments with 2a showed that three structural features of 1 were conserved (Figures S8–S11, Supporting Information): (1) the presence of two 1,2-dimethyl olefin units, supported by the characteristic long-range coupling between nonequivalent methyl groups in COSY; (2) the methane–methylene–methine (C1–C6–C7) spin system; and (3) two keto groups (C10 and C2). No olefinic protons were detected in 1H NMR, indicating that the double bond C11–C12 in 1 is broken during the reaction. One of the 1,2-dimethyl olefin methyl groups derived from C13–C3–C4–C14 in 1 is no longer symmetric in 2a, indicating that one of the adjacent keto groups (C5 or C2) is directly involved in the rearrangement. Therefore, 2a must be derived from an intramolecular cyclization to a Paternò–Büchilike product14 with a reaction between the C5 keto group and the C11–C12 double bond. Only two oxetane-containing products can be derived from this cyclization (excluding consideration of stereochemistry) (Figure 4). The 13C chemical shift of the C12 methylene is 34 ppm, as established by HSQC, strongly suggesting it is not connected to an oxygen atom. Therefore the indicated structure of 2a can be established as the best fit to all of the spectral data, except for the absolute confirmation at the chiral centers (see Figure S7 for detailed discussion of structure elucidation, Supporting Information). The disappearance of one alkene bond and one keto group in the products is also consistent with a less conjugated structure (UV, Figure 3b). The turnover rate of 1 by P450 154A1 was 1.1 min−1 with the enzyme saturated with excess substrate (54 μM).
Figure 3.
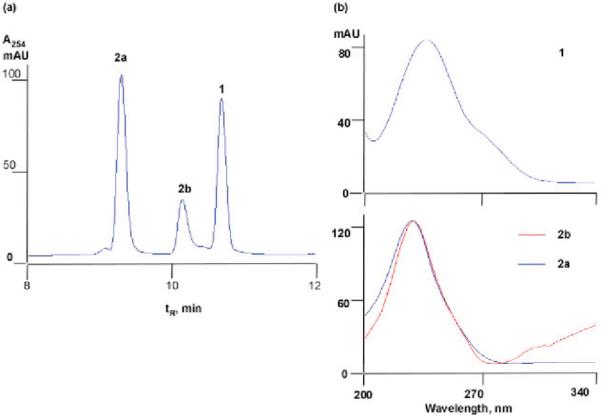
(a) HPLC profile of conversion of 1 by P450 154A1. (b) UV spectra of 1 and its products 2a/2b.
Figure 4.
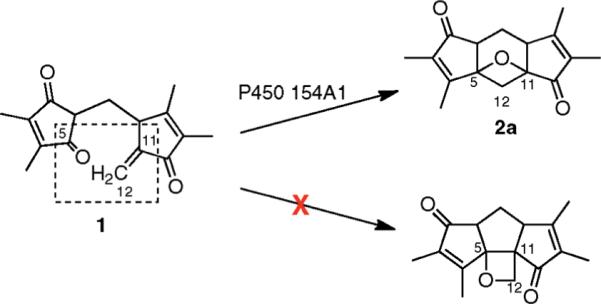
Conversion of 1 to 2a by P450 154A1. The functional groups that directly participate in the rearrangement of 1 are circled in the dotted rectangle.
In conclusion, we demonstrate that S. coelicolor P450 154A1 may play a role in the long term stability of the organism. A novel endogenous substrate of P450 154A1 was identified using a metabolomics approach. Structure elucidation of the substrate 1 and its metabolite 2a revealed an unusual cyclization to a Paternò–Büchilike product catalyzed by P450 154A1 without net oxidation–reduction. Although P450s are well-known to catalyze a great variety of reactions, only a few examples of P450-catalyzed rearrangement without the involvement of external reducing equivalents have been reported,1 e.g. the rearrangement of prostaglandin H215 and fatty acid hydroperoxides.16 The substrates involved in the above-mentioned reactions are all peroxides, and the only other nonredox reactions reported for P450s are hydrolyses.17,18 Therefore, the P450 154A1 catalyzed rearrangement of 1 proposed here is unprecedented and appears to be novel in the field of P450 enzymology. This special catalytic property may be associated with the unique heme orientation in P450 154A1.6
Supplementary Material
Acknowledgment
We thank D. Stec (Vanderbilt University) for assistance in NMR spectroscopy. This work was supported by U.S. Public Health Service Grants R01 GM069970 and P30 ES000267.
Footnotes
Supporting Information Available: Full experimental details and spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Guengerich FP. Chem. Res. Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- (2).Ortiz de Montellano PR. Cytochrome P450: Structure, Mechanism, and Biochemistry. 3rd ed. Kluwer Academic/Plenum Publishers; New York: 2005. [Google Scholar]
- (3).Bentley SD, et al. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- (4).Lamb DC, Waterman MR, Kelly SL, Guengerich FP. Curr. Opin. Biotechnol. 2007;18:504–512. doi: 10.1016/j.copbio.2007.09.010. [DOI] [PubMed] [Google Scholar]
- (5).Lamb DC, Guengerich FP, Kelly SL, Waterman MR. Expert Opin. Drug Metab. Toxicol. 2006;2:27–40. doi: 10.1517/17425255.2.1.27. [DOI] [PubMed] [Google Scholar]
- (6).Lamb DC, Skaug T, Song HL, Jackson CJ, Podust LM, Waterman MR, Kell DB, Kelly DE, Kelly SL. J. Biol. Chem. 2002;277:24000–24005. doi: 10.1074/jbc.M111109200. [DOI] [PubMed] [Google Scholar]
- (7).Podust LM, Bach H, Kim Y, Lamb DC, Arase M, Sherman DH, Kelly SL, Waterman MR. Protein Sci. 2004;13:255–268. doi: 10.1110/ps.03384804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kieser T, Bibb MJ, Buttner MJ, Chater F, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Norwich U.K.: 2000. [Google Scholar]
- (9).Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- (10).Katajamaa M, Miettinen J, Oresic M. Bioinformatics. 2006;22:634–636. doi: 10.1093/bioinformatics/btk039. [DOI] [PubMed] [Google Scholar]
- (11).Corre C, Challis GL. ChemBioChem. 2005;6:2166–2170. doi: 10.1002/cbic.200500243. [DOI] [PubMed] [Google Scholar]
- (12).Aguilar A, Hopwood DA. J. Gen. Microbiol. 1982;128:1893–1901. doi: 10.1099/00221287-128-8-1893. [DOI] [PubMed] [Google Scholar]
- (13).Tius MA, Patterson GM, Astrab DP. J. Antibiot. (Tokyo) 1985;38:1061–1067. doi: 10.7164/antibiotics.38.1061. [DOI] [PubMed] [Google Scholar]
- (14).Büchi G, Inman CG, Lipinsky ES. J. Am. Chem. Soc. 1954;76:4327–4331. [Google Scholar]
- (15).Hecker M, Ullrich V. J. Biol. Chem. 1989;264:141–150. [PubMed] [Google Scholar]
- (16).Song WC, Brash AR. Science. 1991;253:781–784. doi: 10.1126/science.1876834. [DOI] [PubMed] [Google Scholar]
- (17).Zhao B, Lei L, Vassylyev DG, Lin X, Cane DE, Kelly SL, Yuan H, Lamb DC, Waterman MR. J. Biol. Chem. 2009;284:36711–36719. doi: 10.1074/jbc.M109.064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Yun CH, Ahn T, Guengerich FP, Yamazaki H, Shimada T. Arch. Biochem. Biophys. 1999;367:81–88. doi: 10.1006/abbi.1999.1254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.