Abstract
Agmatine, an amine formed by decarboxylation of L-arginine by arginine decarboxylase (ADC), has been recently discovered in mammalian brain and other tissues. While the cloning and sequencing of ADC from plant and bacteria have been reported extensively, the structure of mammalian enzyme is not known. Using homology screening approach, we have identified a human cDNA clone that exhibits ADC activity when expressed in COS-7 cells. The cDNA and deduced amino acid sequence of this human ADC clone is distinct from ADC of other forms. Human ADC is a 460-amino acid protein that shows about 48% identity to mammalian ornithine decarboxylase (ODC) but has no ODC activity. While naive COS-7 cells do not make agmatine, these cells are able to produce agmatine, as measured by HPLC, when transfected with ADC cDNA. Northern blot analysis using the cDNA probe indicated the expression of ADC message in selective human brain regions and other human tissues.
Keywords: Agmatine, Arginine, Polyamine, Arginine decarboxylase, Ornithine decarboxylase
1. Introduction
In plants, bacteria and invertebrates, agmatine, an amine and organic cation, is formed by the decarboxylation of L-arginine by the enzyme arginine decarboxylase (ADC) (EC 4.1.1.19). While agmatine and ADC have been recognized since early in this century as products of non-mammalian tissues/cells, they were not believed to be expressed in mammals [1]. In 1994, we discovered agmatine and ADC activity in rat and bovine brain [2] and subsequently both have been identified in many other organs and cell types [2–8]. In all species, agmatine is metabolized by hydrolysis to putrescine by agmatinase (agmatine ureo hydrolase) (E.C.3.5.3.11). It was originally believed that agmatine merely served as a metabolic intermediate in a pathway for polyamine biosynthesis paralleling another pathway by which putrescine is synthesized from ornithine by ornithine decarboxylase (ODC) (EC 4.1.1.17). The recent findings that agmatine is synthesized, stored, and released in brain and other cells and is present in serum [9], suggest that the amine may have other physiological functions including a role as a neurotransmitter [10], and as a regulator of cellular proliferation [7,11] and inflammation [12]. In this light, it is evident that ADC plays a crucial role in determining the availability of agmatine in tissues/cells and in the circulation.
However, little is known about the molecular structure of mammalian ADC. It may differ from ADC of bacteria, plants, and the nematode C. elegans suggested by the reaction properties of mammalian ADC [13]. Based on biochemical properties, the ADC of C. elegans appears to be closer to the mammalian enzyme as both are membrane-associated and can decarboxylate ornithine and arginine [14]. Indeed, it has been proposed that ODC and ADC may represent isoforms of the same enzyme [15]. While the mammalian form of agmatinase has been recently cloned by two laboratories [16,17], the structure of mammalian ADC has not been reported. In this study, we report the cloning and expression of mammalian ADC that is distinct from ADC of bacteria and plants but similar to mammalian ODC. Our findings provide the first molecular evidence that mammalian ADC is an unique enzyme, distinct but related to ODC and different from mammalian basic amino acid decarboxylases and ADC of bacteria and plants.
2. Materials and methods
2.1. cDNA of human ADC
We initially screened rat tissue cDNA library using PCR with oligo primers designed from homology screening of cDNA sequences of mammalian ODC and plant ADC. We obtained several PCR products from rat kidney and brain that were sequenced. Among these PCR products, one product of 700 bases was identical to a previously described ODC-like protein with no ODC activity [18]. We obtained this cDNA clone from Invitrogen gene collection and subcloned into the pCMV.SPORT6 vector (Invitrogen, NY). Once we confirmed that this cDNA codes for a protein with ADC activity in COS-7 cells, it was then sequenced with both vector and insert-derived primers.
2.2. Molecular phylogenetic analyses
Phylogenetic analyses were carried out using the PHY-LIP package version 3.57 [19]. Multiple-sequence alignments of the human ADC with sequences of ODC and ADC from other species were performed using the CLUSTAL W program, version 1.82 [20]. The unrooted phylogenetic tree was constructed by the neighbor-joining method [21], followed by the creation of a majority-rules, strict consensus phylogram with confidence intervals using CONSENSE [22]. Relative measures of internal support were obtained using 100 bootstrap replications [22,23].
2.3. Transient transfections in COS-7 cells
All cell culture reagents were obtained from Gibco BRL (Gaithersburg, MD). COS-7 cells were grown in Dulbecco’s medium (DMEM) supplemented with the heat-inactivated fetal bovine serum (10%), streptomycin (100 μg/ml) and penicillin (100 units/ml) at 37 °C in humidified air containing 5% CO2. Transient transfection was performed with the Lipofectamine 2000 kit (Invitrogen) in COS-7 cells (passage number below 10) according to the manufacturers’ instructions. Transfected cells were harvested at 24–72 h after transfection for measurements of ADC and ODC activity and cellular agmatine levels.
2.4. Assay of ADC and ODC
The activity of ADC and ODC was measured in cytosolic and membrane fractions prepared from control and cDNA-transfected COS-7 cells. Briefly, harvested cells were homogenized in ADC assay buffer (5 mM Tris–HCl, pH 8.7, containing the protease inhibitors 0.5 mM PMSF, 100 μM benzamide, and 10 μM pepstatin A) and centrifuged at 30,000 × g for 15 min to obtain cytosolic and membrane fractions. The membrane pellet was resuspended in the ADC assay buffer for enzyme measurements. ADC activity was measured as described earlier using the standard enzyme reaction [24], carried out in 250 μl of ADC assay buffer consisting of 10 mM Tris–HCl (pH 8.25), 0.1 mM pyridoxal phosphate, 1 mM DTT, 0.5 mM PMSF, 0.2 mM EDTA, 1 mM MgSO4, 0.2 mM arginine and 0.4 μCi of (1-14C)-arginine (specific activity 55 mCi/mmol) at 30 °C for 30 min. The reaction was performed in glass tubes with a center well inserted into a tightly closed rubber stopper. The center wells contain strips of filter paper moistened with 1 M KOH to trap the 14CO2 generated. The reaction was performed in a shaking water bath, except when indicated otherwise, for 1 h at 30 °C. The reaction was terminated by the addition of 100 μl of 40% trichloroacetic acid injected through the rubber stopper. After further incubation for 20 min at 37 °C, the filter paper strips were transferred to scintillation vials and counted by liquid scintillation spectroscopy. The ODC activity was measured by the method of Metcalf et al. [25] using 114C-ornithine (specific activity 55 mCi/mmol) as substrate and measuring the 14CO2 formed. Briefly, the tissue preparation was incubated at 37 °C for 60 min in an assay buffer consisting of 10 mM sodium phosphate buffer (pH 7.0), 0.1 mM pyridoxal phosphate, 5 mM dithiothreitol, 0.1 mM 114C-ornithine (0.1 μCi). The 14CO2 is trapped in filter papers and the radioactivity counted as described above for ADC assay.
2.5. Measurement of agmatine by HPLC
The amount of agmatine in control and ADC-transfected COS-7 cells was measured by HPLC as described earlier for tissue agmatine measurements [9]. COS-7 cells were harvested 3 days after transfection and suspended in phosphate buffer (pH 5.7) containing amino picollinate (APP) as internal standard. The cell suspension was homogenized in 10% TCA, centrifuged to remove proteins and the supernatant was used for HPLC analysis. Briefly, the samples were derivatized with o-pthalaldehyde (OPA) and injected into a HPLC system comprising of a reverse phase column (5 μm) and fluorescence detector. The recovery of agmatine was calculated from the added external standard and the results are expressed as ng/106 cells.
2.6. Northern blot analysis
The expression of ADC message in transfected COS-7 cells as well as in human tissues was determined by Northern blot analysis using labeled cDNA probe. The distribution of mRNA in human brain and other tissues was assessed using Multiple Tissue Northern blots, purchased from Clontech. The Northern blot analysis was performed using a 32P-dCTP (3000 Ci/mmol; Amersham, Piscataway, NJ)-labeled human ADC cDNA probe that was labeled by Rediprime II Random Prime Labeling System (Amersham). Hybridization was carried out overnight at 42 °C in 10 ml of ULTRAhyb hybridization solution (Ambion, Austin, TX). After washing with equal volumes of 2 × SSC/0.1% SDS to a final stringency of 0.1 × SSC/0/1% SDS at 42 °C, the blots were exposed to Kodak Biomax MR films with intensifying screen at −80 °C until the required signal was achieved.
2.7. Protein assay
Proteins were measured by Bio-Rad protein assay kit (Bio-Rad) using bovine serum albumin (BSA) as a standard.
3. Results
3.1. cDNA and amino acid sequence of human ADC
We report the cDNA sequence of a previously characterized ODC-like protein (GenBank accession no. AY325129) that encodes for a protein with ADC activity. This cDNA sequence contained a 5′ untranslated region of about 360 bp and a coding region of 2.6 kb. The ORF predicted a protein of 460 residues. The alignment with human genome database revealed that the gene is located in chromosome 1 and stretches to a total size of 39 kb containing 11 exons.
3.2. Phylogenetic analyses
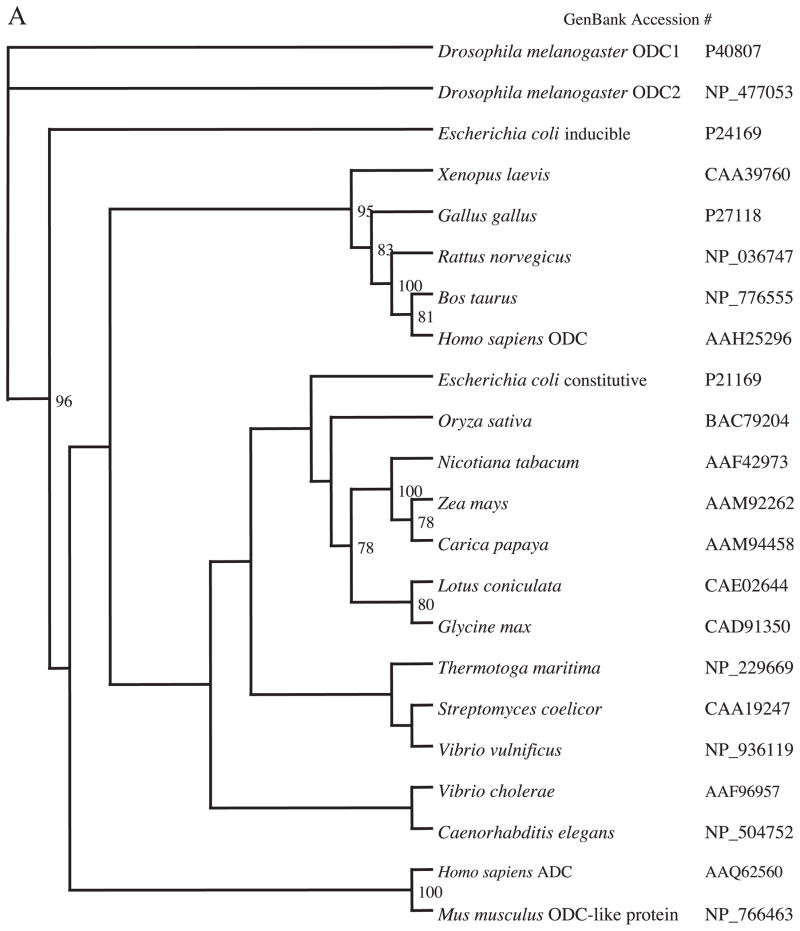
Using CLUSTAL W, a multi-sequence alignment of ADC protein with ODC and ADC sequences from different species was carried out. These sequences were input into the PHYLIP distance algorithm (PROTDIST) to calculate unrooted phylograms using the neighbor-joining method, which was generated by the program TREEVIEW (Fig. 1). Only bootstrap values above 50% are shown at the nodal branches. The phylogram separates all the similar groups into different clusters inferring distinct phylogenies. It confirmed that even though human ADC and ODC may be related, they clearly fall into distinct clades or clusters. Because ADC and ODC show the highest amino acid identity (48%) with functionally conserved and semi-conserved amino acid substitutions amongst the entire group, CLUSTAL W was used to show a direct comparison (alignment) of the two amino acid sequences, also including ODCp (supposed ADC homolog of mouse), which shows an 86% homology with human ADC (Fig. 2). While some key amino acid residues involved in decarboxylation reaction and pyridoxal phosphate binding domain are conserved between these proteins, the 360 cysteine residue in ODC is substituted by valine in human ADC is reported here (Fig. 2) as well as in an earlier paper [18]. This substitution results in the altered selectivity of the substrate binding, namely arginine instead of ornithine, making this protein an ADC. The sequences of ADC from bacteria and plants showed the least sequence identity if any.
Fig. 1.
Phylogenetic tree (unrooted) based on amino acid sequences showing the relationship of the Homo sapiens ADC compared to ODC (A) and ADC from mammals, bacteria and plants (B). The numbers at the forks indicate the number of times the group consisting of species to the right of that fork occurred among 100 trees calculated using PROTDIST from the PHYLIP package.
Fig. 2.
Comparison of the amino acid sequences of ODC-like protein (mouse), ADC (human), and ODC (human) using CLUSTAL W. Shaded areas show identical amino acids, colons show functionally conserved amino acid substitutions, and periods show semi-conserved amino acid substitutions between the three proteins. Numbers on the right refer to the last amino acid on each line.
3.3. Expression of ADC activity
To investigate the functional expression of ADC activity, we expressed this cDNA clone in COS-7 cells and measured enzyme activity as well as the production of agmatine. In initial experiments, ADC and ODC activities were measured in COS-7 cells transiently transfected with ADC cDNA and control vector. The activity of ADC in COS-7 cells, transfected with ADC cDNA, ranged from 524 to 1805 nmol/h/mg protein depending on the transfection efficiency with the mean activity of 945 nmol/h/mg protein from five different transfection experiments (Table 1). Most of the ADC activity is associated with the membrane fraction with very little activity in the soluble fraction. No significant ADC activity was expressed in naive cells or cells transfected with only vector. In contrast, ODC activity was similar in the cytosol fraction of wild-type, vector-transfected and cDNA-transfected cells with no detectable activity in membrane fraction (Table 1). We also tested whether ADC in transfected cells could be inhibited by DFMO, a known inhibitor of ODC, and calcium, a known inhibitor of mammalian ADC. As shown in Table 2, membrane ADC activity in transfected cells was not inhibited by DFMO (1 mM) but significantly inhibited by calcium (1 mM). In contrast, soluble ODC activity was markedly inhibited by DFMO but not by Ca+ +.
Table 1.
The activity of ADC and ODC in transfected COS-7cells
| ADC
|
ODC
|
|||
|---|---|---|---|---|
| Cytosol | Membrane | Cytosol | Membrane | |
| Wild-type | none | none | 0.17 ± 0.02 | none |
| Vector-transfected | 0.04 ± 0.01 | 0.14 ± 0.08 | 0.25 ± 0.03 | 0.016 ± 0.08 |
| cDNA-transfected | 14.7 ± 0.58 | 945 ± 378 | 0.31 ± 0.07 | 0.21 ± 0.05 |
Cytosol and membrane fractions were prepared from transfected and control COS-7 cells. ADC and ODC activities measured using 114C-arginine and 114C-ornithine, respectively. Activity is expressed as nmol/h/mg protein and values are mean ± S.E. from five transfection experiments and each assay was carried out in triplicates.
Table 2.
Effect of inhibitors on ADC activity in transfected COS-7 cells
| ADC activity (nmol/h/mg protein) | ODC activity (nmol/h/mg protein) | |
|---|---|---|
| cDNA-transfected cells | 582 ± 21 | 0.41 ± 0.09 |
| cDNA-transfected cells + DFMO (1 mM) | 514 ± 31 | 0.11 ± 0.06* |
| cDNA-transfected cells + Ca+ (1 mM) | 243 ± 45* | 0.43 ± 0.07 |
Membrane and cytosol fractions were prepared from transfected COS-7 cells. ADC (membrane) and ODC (cytosol) activities were measured in the presence of DFMO or calcium. Values are mean ± S.E. from two transfection experiments and each assay was carried out in triplicates.
P < 0.001 compared to cDNA-transfected cells.
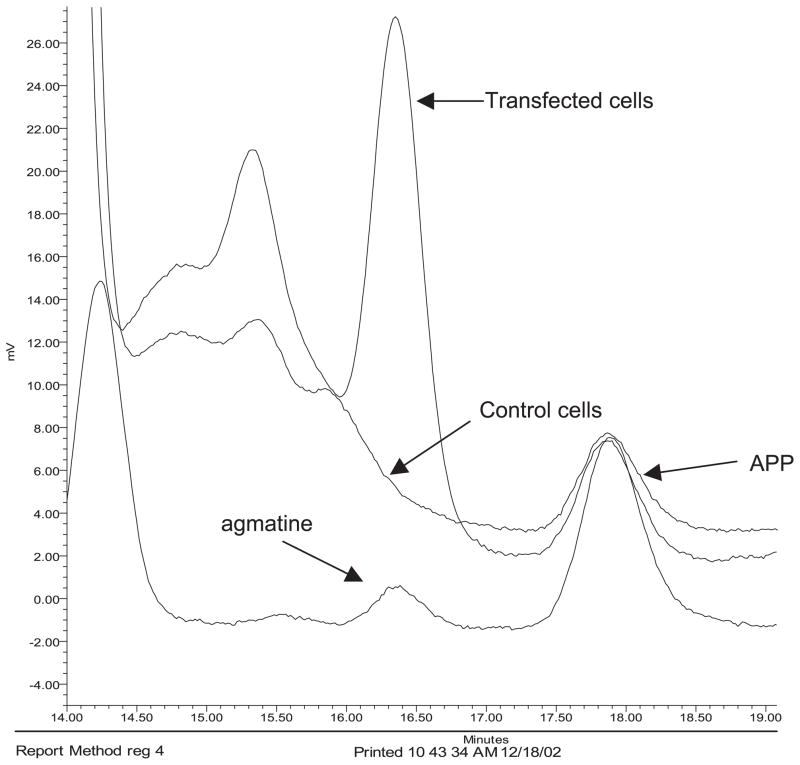
The amount of agmatine in COS-7 cells was measured by HPLC to determine whether the transfected cells were able to make agmatine. A typical HPLC tracing shows a large agmatine peak in cytosolic sample prepared from ADC cDNA-transfected COS-7 cells compared to vector-transfected cells (Fig. 3). The amount of agmatine ranged from 10.5 to 20.0 ng/106 cells in ADC-transfected cells while control cells had no agmatine.
Fig. 3.
HPLC tracing of the measurement of agmatine in control and ADC cDNA-transfected COS-7 cells. The bottom tracing is from the agmatine standard.
3.4. Tissue distribution of ADC message
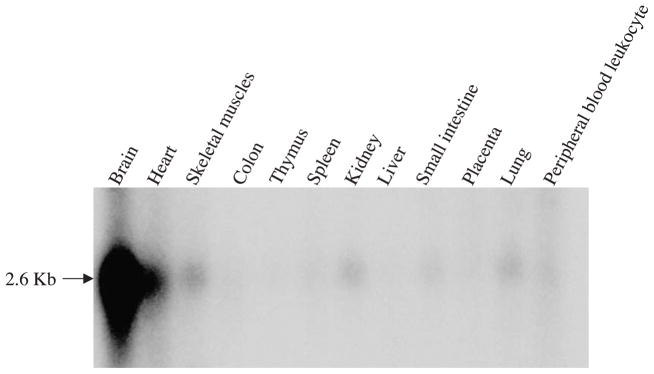
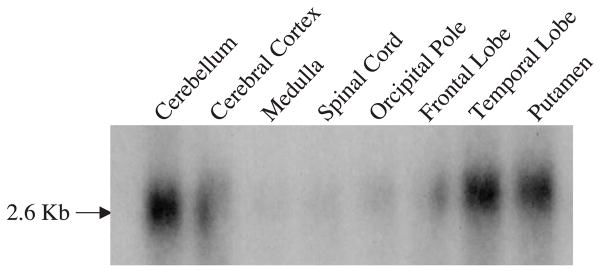
The expression of ADC message in human tissues and brain was determined by Northern blot analysis using 32P labeled ADC cDNA, probed with human Multiple Tissue Northern blots obtained from Clontech. The tissue distribution of ADC message is shown in Fig. 4. The highest level of the expression was observed in brain, followed by heart, kidney, lung, skeletal muscle and peripheral leukocytes. In the human brain, the ADC message was expressed in an uneven manner with higher expression levels in the cerebellum, cerebral cortex, putamen, and lower levels in the medulla and spinal cord (Fig. 5). Among the cortical hemispheres, the temporal lobe has the highest level of expression.
Fig. 4.
Analysis of the expression of ADC message in human tissues using human multiple tissue Northern blot. The RNA blot was probed using the labeled ADC cDNA.
Fig. 5.
Analysis of the expression of ADC message in human brain regions using human multiple tissue Northern blot. The RNA blot was probed using the labeled ADC cDNA.
4. Discussion
Since our discovery of agmatine and ADC in rat brain in 1994 [2], we have been working to identify the structure of mammalian ADC. While the sequences of ADC from several plants and bacteria has been reported, the mammalian sequence is not known. While one study reported a partial ADC sequence from rat kidney, no subsequent study on the full sequence or expression of ADC activity was reported. Using homology screening and PCR approach, we have identified a human cDNA clone that expresses ADC activity and produces agmatine when transfected in COS-7 cells. This clone is identical to a previously cloned protein that was described as an ODC-like protein without ODC activity [18]. In this earlier study, the expressed protein was tested only for ODC activity because of its similarity to ODC sequence. Our sequence data indicate that this gene has the ORF that encodes for a protein of 460 amino acids similar to the previously published ODC-like protein sequence. However, we have shown that this protein is actually able to decarboxylate arginine but not ornithine and, therefore, should be considered as human ADC. Human ADC shows about 48% identity to human ODC (Fig. 1B) but largely distinct from ADC of other species. Although this was not expected, it has been reported that ADC is a diversified enzyme with distinct regulatory sequences. For example, ADC is expressed in constitutive (biosynthetic) and inducible (biodegradative) forms in E. coli that share very little sequence homology and have different reaction properties [13,26–28]. In contrast, oat ADC is a pyruvyl enzyme and activated by proteolytic cleavage [29] while ADC of C. elegans is a membrane-bound ornithine/arginine decarboxylase [14].
The differences in terms of phylogeny observed in the ODC and ADC group of enzymes from different species is not surprising because the pyridoxal-5-phosphate-dependent enzymes (B6 enzymes) that act on amino acid substrates are of multiple evolutionary origins. The numerous common mechanistic features of these enzymes are not historical traits passed on from a common ancestor enzyme but rather reflect evolutionary or chemical necessities [30]. According to Mehta and Christen [30], functional specialization of the enzymes already occurred in the universal ancestor making it very difficult to study the evolution of this group. This is shown by the fact that decarboxylases are divided into four apparently unrelated groups numbered in the order of the Enzyme Commission (EC). The prokaryotic ornithine, lysine and ADCs (biodegradative type) belong to group III while the eukaryotic ODC and ADCs as well as prokaryotic ADCs (biosynthetic type) constitute group IV. These evidences lend credence to our results and indicate that human ADC is different from ODC even though they share a 48% identity at the amino acid level. It is interesting to see that the mouse homolog of ADC which is also classified as an ODC-like protein shares an 86% amino acid identity with ADC does not cluster with the mammalian ODC’s but with ADC with 100% bootstrap value, clearly indicative of a separate group compared to the mammalian ODC. When compared to the other ADC’s from plants and prokaryotes (Fig. 1B), the evidence is less convincing due to the extremely low bootstrap values and this reinforces the fact that the group is classified according to mechanistic features and not to evolutionary relationship.
Based on reaction properties of mammalian ADC, we predicted that mammalian enzyme may be largely distinct from the enzyme of other species [24]. For example, unlike the bacterial and plant isoforms, but like that of C. elegans, mammalian ADC is membrane-associated. However, like the bacterial enzyme, mammalian ADC has a very basic pH optimum (8.23). Unlike other isoforms of ADC, the rat tissue ADC [24] as well as expressed human ADC (Table 2) are significantly inhibited by Ca+ +. Thus, the present findings on the reaction properties of human ADC clearly indicates that this activity and gene is distinct from the non-mammalian ADC in its nucleotide and amino acid sequences. In fact, these differences in the sequences of mammalian and non-mammalian ADC (Fig. 1A) would also explain the failure of DFMA, a well-known inhibitor of non-mammalian ADC, to inhibit rat ADC activity [18].
Although human ADC shows about 48% similarity to human ODC (Fig. 1B), most of the conserved residues are in the middle part of the protein. A large portion of the N-terminal (amino acids 1 through 40) residues shows very little homology between ADC and ODC. These differences could contribute to the observed dissimilarities in reaction/biochemical properties of ADC and ODC [24]. For example, the failure of DFMO, a strong inhibitor of ODC by binding at the substrate site, to inhibit ADC activity. The amino acid sequences of ODC and ADC indicated that the C360 residue, crucial for ornithine binding in ODC, is replaced by valine in ADC [31]. Since mammalian ODC can also use arginine as substrate, it has been proposed that the decarboxylation of arginine in brain can be attributed to ODC [15]. However, based on our present data and the reaction/biochemical properties of ADC and ODC [24], it is evident that these two enzymes are homologous but separate proteins.
The traditional view holds that an important physiological role of ODC is to provide polyamines from ornithine which in turn are essential for cellular growth and proliferation [32]. Although the functional role of agmatine in cells is not clear, it appears in those systems, in which it has been examined, agmatine does not promote, but rather inhibits, proliferation and cell growth [7,11]. The differential expression of ADC and ODC in rat tissues and cultured cells indicates such distinct physiological functions. ODC, an enzyme with a very high turnover, is induced during cellular proliferation to provide putrescine, the precursor for the polyamines, spermine and spermidine. The fact that ADC is not induced during proliferating cells and its activity is highest in confluent cells, suggests that agmatine is not a precursor for the production of polyamines. In fact, it has been shown that agmatine induces antizyme to ODC thereby suppressing the activity of ODC and the production of polyamines in kidney cells [11].
In conclusion, mammalian ADC is a novel enzyme of arginine metabolism and is distinct from ODC. As the enzyme responsible for the biosynthesis of agmatine, ADC’s biological importance is closely linked to the function of this amine. There are increasing evidences that agmatine may have important biological functions including actions as a novel neurotransmitter [10,33,34], as a modulator of cellular proliferation [7,11] and inflammation [12], and in regulating renal and gastric function [35,36]. Further characterization of ADC gene and its regulation will allow us to explore the functions of agmatine in mammalian systems.
Acknowledgments
This work was supported by NIH grant NS 39445.
References
- 1.Tabor CW, Tabor H. Polyamines. Ann Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 2.Li G, Regunathan S, Barrow CJ, Eshraghi J, Cooper R, Reis DJ. Agmatine: an endogenous clonidine-displacing substance in the brain. Science. 1994;263:966–969. doi: 10.1126/science.7906055. [DOI] [PubMed] [Google Scholar]
- 3.Raasch W, Regunathan S, Li G, Reis DJ. Agmatine, the bacterial amine, is widely distributed in mammalian tissues. Life Sci. 1995;56:2319–2330. doi: 10.1016/0024-3205(95)00226-v. [DOI] [PubMed] [Google Scholar]
- 4.Li G, Regunathan S, Reis DJ. Agmatine is synthesized by a mitochondrial arginine decarboxylase in rat brain. Ann N Y Acad Sci. 1995;763:325–329. doi: 10.1111/j.1749-6632.1995.tb32418.x. [DOI] [PubMed] [Google Scholar]
- 5.Lortie MJ, Novotny WF, Peterson OW, Vallon V, Malvey K, Mendonca M, Satriano J, Insel P, Thomson SC, Blantz RC. Agmatine, a bioactive metabolite of arginine. J Clin Invest. 1996;97:413–420. doi: 10.1172/JCI118430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sastre M, Galea E, Reis DJ, Regunathan S. Metabolism of agmatine in macrophages: modulation by lipopolysacharides and inhibitory cytokines. Biochem J. 1998;330:1405–1409. doi: 10.1042/bj3301405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Regunathan S, Youngson C, Raasch W, Wang H, Reis DJ. Imidazoline receptors and agmatine in blood vessels: a novel system inhibiting vascular smooth muscle proliferation. J Pharmacol Exp Ther. 1996;276:1272–1282. [PubMed] [Google Scholar]
- 8.Regunathan S, Feinstein DL, Raasch W, Reis DJ. Agmatine, the decarboxylated arginine is localized and synthesized in glial cells. NeuroReport. 1995;6:1897–1900. doi: 10.1097/00001756-199510020-00018. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, Halaris AE, Piletz JE. Determination of agmatine in brain and plasma using high-performance liquid chromatography with flourescence detection. J Chromatogr. 1997;691:277–286. doi: 10.1016/s0378-4347(96)00458-6. [DOI] [PubMed] [Google Scholar]
- 10.Reis DJ, Regunathan S. Agmatine: a novel neurotransmitter? Adv Pharmacol. 1998;42:645–649. doi: 10.1016/s1054-3589(08)60834-0. [DOI] [PubMed] [Google Scholar]
- 11.Satriano J, Matsufuji S, Murakami Y, Lortie MJ, Schwartz D, Kelly CJ, Hayashi S, Blantz RC. Agmatine suppresses proliferation by frameshift induction of antienzyme and attenuation of cellular poly-amine levels. J Biol Chem. 1998;273:15313–15316. doi: 10.1074/jbc.273.25.15313. [DOI] [PubMed] [Google Scholar]
- 12.Regunathan S, Feinstein DL, Reis DJ. Anti-proliferative and anti-inflammatory actions of imidazoline agents: are imidazoline receptors involved? Ann NY Acad Sci. 1999;881:410–419. doi: 10.1111/j.1749-6632.1999.tb09389.x. [DOI] [PubMed] [Google Scholar]
- 13.Wu WH, Morris DR. Biosynthetic arginine decarboxylase from Escherichia coli: purification and properties. J Biol Chem. 1973;248:1687–1695. [PubMed] [Google Scholar]
- 14.Schaeffer JM, Donatelli MR. Characterization of a high-affinity membrane-associated ornithine decarboxylase from the free-living nematode Caenorhabditis elegans. Biochem J. 1990;270:599–604. doi: 10.1042/bj2700599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilad GM, Gilad VH, Rabey JM. Arginine and ornithine decarboxylation in rodent brain—coincidental changes during development and after ischemia. Neurosci Lett. 1996;216:33–36. doi: 10.1016/0304-3940(96)12996-7. [DOI] [PubMed] [Google Scholar]
- 16.Iyer RK, Kim HK, Tsoa RW, Grody WW, Cederbaum SD. Cloning and characterization of human agmatinase. Mol Genet Metab. 2002;75:209–218. doi: 10.1006/mgme.2001.3277. [DOI] [PubMed] [Google Scholar]
- 17.Mistry SK, Burwell TJ, Chambers RM, Rudolph-Owen L, Spaltman F, Cook WJ, Morris SM. Cloning of human agmatinase. An alternate path for polyamine synthesis induced in liver by hepatitis B virus. Am J Physiol: Gasterointest Liver Physiol. 2002;282:375–381. doi: 10.1152/ajpgi.00386.2001. [DOI] [PubMed] [Google Scholar]
- 18.Pitkanen L, Heiskala M, Andersson L. Expression of a novel human ornithine decarboxylase-like protein in the central nervous system and testes. Biochem Biophys Res Commun. 2001;287:1051–1057. doi: 10.1006/bbrc.2001.5703. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. PHYLIP (Phylogeny Inference Package), version 3.57. Department of Genetics, University of Washington; Seattle, USA: 1995. [Google Scholar]
- 20.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 23.Sanderson MJ. Confidence limits on phylogenies: the bootstrap revisited. Cladistics. 1989;5:113–129. doi: 10.1111/j.1096-0031.1989.tb00559.x. [DOI] [PubMed] [Google Scholar]
- 24.Regunathan S, Reis DJ. Characterization of arginine decarboxylase in rat brain and liver: distinction from ornithine decarboxylase. J Neurochem. 2000;74:2201–2208. doi: 10.1046/j.1471-4159.2000.0742201.x. [DOI] [PubMed] [Google Scholar]
- 25.Metcalf BW, Bey P, Danzin C. Catalytic irreversible inhibition of mammalian ornithine decarboxylase by substrate and product analogues. J Am Chem Soc. 1978;100:2551–2563. [Google Scholar]
- 26.Stim-Herndon KP, Flores TM, Bennett GN. Molecular characterization of adiY, a regulatory gene which affect expression of the biodegradative, acid-induced arginine decarboxylase gene (adiA) of Escherichia coli. Microbiology. 1996;142:1311–1320. doi: 10.1099/13500872-142-5-1311. [DOI] [PubMed] [Google Scholar]
- 27.Stim KP, Bennett GN. Nucleotide sequence of the adi gene which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli. J Bacteriol. 1993;175:1221–1234. doi: 10.1128/jb.175.5.1221-1234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buch JK, Boyle SM. Biosynthetic arginine decarboxylase in Escherichia coli is synthesized as a precursor and located in the cell envelope. J Bacteriol. 1985;163:522–527. doi: 10.1128/jb.163.2.522-527.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malmberg RL, Cellino ML. Arginine decarboxylase of oats is activated by enzymatic cleavage into two polypeptides. J Biol Chem. 1994;269:2703–2706. [PubMed] [Google Scholar]
- 30.Mehta PK, Christen P. The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Adv Enzymol Relat Areas Mol Biol. 2000;74:129–184. doi: 10.1002/9780470123201.ch4. [DOI] [PubMed] [Google Scholar]
- 31.Jackson LK, Brooks HB, Osterman AL, Goldsmith EJ, Phillips MA. Altering the reaction specificity of eukaryotic ornithine decarboxylase. 2000;39:11247–11257. doi: 10.1021/bi001209s. [DOI] [PubMed] [Google Scholar]
- 32.Pegg AE, Shantz LM, Coleman CS. Ornithine decarboxylase: structure, function and translational regulation. Biochem Soc Trans. 1994;22:846–852. doi: 10.1042/bst0220846. [DOI] [PubMed] [Google Scholar]
- 33.Yang XC, Reis DJ. Agmatine selectively blocks the NMDA subclass of glutamate receptor channels in cultured mouse hippocampal neurons. J Pharmacol Exp Ther. 1999;288:544–549. [PubMed] [Google Scholar]
- 34.Reis DJ, Yang XC, Milner TA. Agmatine containing axon terminals in rat hippocampus form synapses on pyramidal cells. Neurosci Lett. 1998;250:185–188. doi: 10.1016/s0304-3940(98)00466-2. [DOI] [PubMed] [Google Scholar]
- 35.Glavin GB, Carlisle MA, Smyth DD. Agmatine, an endogenous imidazoline receptor agonist, increases gastric secretion and worsens experimental gastric mucosal injury in rats. J Pharmacol Exp Ther. 1995;274:741–744. [PubMed] [Google Scholar]
- 36.Schwartz D, Peterson OW, Mendonca M, Satriano J, Lortie MJ, Blantz RC. Agmatine effects glomerular filtration via a nitric oxide synthase-dependent mechanism. Am J Physiol. 1997;272:F597–F601. doi: 10.1152/ajprenal.1997.272.5.F597. [DOI] [PubMed] [Google Scholar]