Abstract
Glioblastoma (GB) is currently characterized by low survival rates and therapies with insufficient efficacy. Here, we describe biodegradable polymers that can deliver genes to primary GB cells as well as GB tumor stem cells in vitro with low non-specific toxicity and transfection efficiencies of up to 60.6±5 % in normal (10%) serum conditions. We developed polymer-DNA nanoparticles that remained more stable in normal serum and could also be stored for at least 3 months in ready-to-use form with no measurable decrease in efficacy, expanding their potential in a practical or clinical setting. A subset of polymers was identified that shows a high degree of specificity to tumor cells compared with healthy astrocytes and human neural stem cells when cultured (separately or in co-culture), yielding higher transfection in GB cells while having little to no apparent effect on healthy cells.
Keywords: Drug delivery, gene therapy, nanoparticle, stem cell, DNA, cancer
1. Introduction
Approximately 184,300 new central nervous system (CNS) tumors are diagnosed annually, with 50,000 of those originating in the CNS and the rest being metastases from other tumors [1–5]. Malignant gliomas are the most common primary brain tumor and are refractory to combinatorial surgical resection, radiation, and chemotherapy [6–8]. These tumors possess the ability to extensively invade surrounding tissue, making curative resection impossible [9–11]. As a result, experimental treatments like gene therapy are attractive for GB patients. Despite the vast potential of gene therapy, viral methods are plagued with safety concerns [12], while non-viral delivery vehicles often lack efficiency [13]. A high-throughput screening approach based on a synthetic library of cationic poly(β-amino esters) (PBAEs) has led to high transfection efficiency and low toxicity in many cell types [14, 15], though they have not previously been tested in brain tumors. Hundreds of these polymers can be synthesized by combinatorial chemistry, and many cause superior transfection compared to leading commercial agents like Lipofectamine 2000 (Invitrogen), while also being less toxic to cells [16].
Also important for eventual clinical use is the stability of the polymer-DNA nanoparticles, as well as their ease of preparation. Because PBAEs are electrostatically complexed with DNA in slightly acidic, aqueous suspension, the particles have rapid release properties due to hydrolytic degradation of the polymers. While these release properties are advantageous for intracellular gene delivery, they are not conducive for practical use by clinicians or large-scale production. It is therefore necessary to develop a way of preparing a dried form of the polymer-DNA nanoparticles that can be stored for long periods of time and easily reconstituted just before use. However, increased aggregation is a complication for colloidal suspensions that are dried without a lyoprotectant [17], and an effective way of lyophilizing and storing these particles is essential for eventual translation of this technology.
We sought to apply and improve upon previously validated methods of non-viral gene delivery for the treatment of primary human brain tumor cells, including the identification of efficacious reagents and the development of more practical and easily-used formulations. Aside from the malignant astrocytes that comprise the bulk of a GB tumor, we are also interested in brain tumor stem cells (BTSCs). BTSCs are hypothesized to be the source of GB tumors and to play a role in the inevitable tumor recurrence following surgery, chemotherapy, and radiotherapy [18–21]. Aside from being resistant to more types of therapy than astrocytes [22], BTSCs can cause regrowth of an entire tumor at low cell concentrations [23], which the malignant astrocytes alone cannot do [20, 21]. The persistence of BTSCs despite therapy, as well as their tumorigenic capacity, make them a particularly good target of experimental therapies like gene delivery. A comparison of the transfection efficiencies in GB astrocytes, BTSCs, and healthy (non-cancerous) versions of both is of interest, since we have recently found that cell-type specificity can be determined simply by the polymer structure itself [15].
2. Materials and methods
2.1. Materials
Lipofectamine 2000 (Invitrogen, Carlsbad, CA), FuGENE® HD (Roche), Opti-MEM I (Invitrogen), pEGFP-N1 DNA (Elim Biopharmaceuticals, Hayward, CA), pDsRed-Max-N1 [24] and FUGW [25] (Addgene DNA plasmids 21718 and 14883, respectively, Cambridge, MA), and cell culture media components were used as received. Label IT® Tracker Cy3 Kit was purchased from Mirus Bio. 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) was purchased from Sigma (Saint Louis, MO) and used as a 750 nM solution in PBS.
2.2. Cell Culture
All intraoperative samples were obtained as described previously under institutionally approved protocols. Human BTSCs (BTSC line 551) was isolated from a GB tumor sample of an adult patient (69 years old) diagnosed with GB, as previously described [26]. Briefly, necrotic tissue and blood vessels were removed from the tumor, which was dissociated mechanically and with trypsin/EDTA. After inhibition of trypsin with 1:1 DMEM/F-12 (Invitrogen) with 10% heat-inactivated FBS and 1% antibiotic-antimycotic (anti-anti, final concentration 1X, Invitrogen), the cells were collected by centrifugation, and the serum-containing medium was replaced with BTSC neurosphere medium [1:1 DMEM/F-12, 2% B-27 serum-free supplement (B-27, final concentration 1X, Gibco, Bethesda, MD), 1% anti-anti, 20 ng/mL basic fibroblast growth factor (bFGF, Invitrogen), and 20 ng/mL epidermal growth factor (EGF, Sigma, Saint Louis, MO)]. A GB astrocyte cell line (GB 319) was derived from BTSCs isolated as above (from a 79-year-old adult GB patient). After isolation, they were maintained in astrocyte medium with serum (1:1 DMEM/F-12, 10% FBS, 1% anti-anti) as adherent cultures.
Human fetal neural stem cells (NSCs) (F34) were derived after 17 weeks of gestation, obtained from elective abortion. Brain cortical tissue was mechanically dissociated and cells were maintained in neurosphere medium consisting of 2:1 high-glucose DMEM with l-glutamine (Invitrogen)/Ham’s F-12 (Cellgro), 1X B-27, 1% anti-anti, 20 ng/mL bFGF, 20 ng/mL EGF, 20 ng/mL leukemia inhibitory factor (LIF, Millipore, Billerica, MA), and 5 μg/mL heparin (Sigma)]. BTSC 551 was stably transduced with EGFP using the lentiviral vector FUGW. Cells were incubated with the virus overnight in the presence of 8 μg/mL polybrene, then sorted by FACS. The GFP+ 551 BTSCs were maintained in neurosphere medium in nonadherent flasks and passaged approximately every 10 days by mechanical dissociation. For transfection experiments, 551 BTSCs were plated on laminin-coated plates, either as neurospheres or dissociated into single cells, and maintained in neurosphere medium following the protocol described by Pollard et al. [27]. F34 fetal neurospheres were maintained either as NSCs or in astrocyte medium with serum for differentiation and were used as non-cancerous controls for BTSC 551 BTSCs and GB 319 astrocytes. All cells were incubated at 37°C in a humid atmosphere of 5% CO2.
2.3. Synthesis of Poly(β-amino esters)
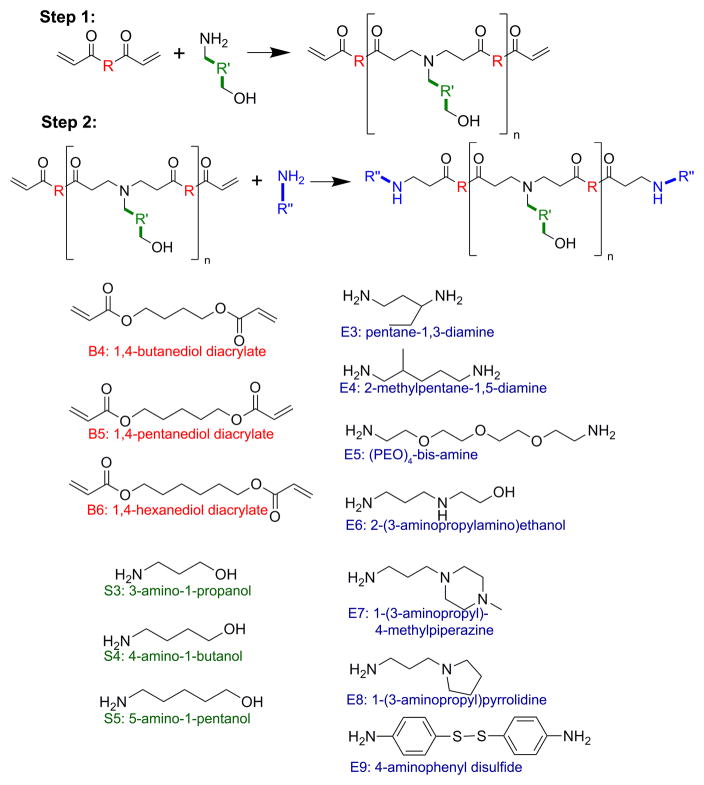
PBAEs were synthesized in a two-step reaction as described previously [14] using small commercially-available molecules (Figure 1). These small molecules were purchased from Acros Organics (E8), Alfa Aesar (B4, B6, S3, S4, S5, E7), Fluka (E6), Monomer-Polymer and Dajac Labs (B5), Sigma-Aldrich (E9), and TCI America (E3, E4, E5), and used as received. Briefly, for each polymer, one backbone monomer (labeled as B4, B5, or B6) was mixed with a sidechain monomer (S3, S4, or S5), either neat or as a 500 mg/mL solution in DMSO, at a 1.1:1 or 1.2:1 ratio of B:S. The mixture was stirred at 90°C for 24 hr to yield acrylate-terminated base polymers by Michael addition. Base polymers were endcapped with a small molecule (E3, E4, E5, E6, E7, E8, or E9) by dissolving endcapping molecules in DMSO (167 mg/mL) and adding 480 μL endcapping solution to 320 μL of 0.5 M base polymer in DMSO, then shaking the mixture at room temperature overnight. Final polymers were stored with dessicant at 4°C as 100 mg/mL solutions in DMSO. Polymers used for optimization experiments were stored in small aliquots at −20°C to minimize freeze-thaw of samples.
Figure 1.
Monomer structures used to synthesize PBAEs. Backbone (B) monomers were polymerized with sidechain (S) monomers. The B-S base polymers were then end-capped with small molecules (E).
2.4. DNA Delivery to GB 319 Astrocytes
2.4.1. Nanoparticle Uptake
Plasmid DNA encoding EGFP was labeled with Cy3 using a Mirus Bio kit according to manufacturer instructions. A solution of 150 μg/mL DNA and 100 μL/mL Label IT Tracker reagent was prepared in buffer, then incubated at 37°C for 3 hr on an orbital shaker. The labeled DNA was precipitated with ethanol in 300 mM sodium acetate buffer and cooled at −20°C for 30 min. The DNA was pelleted by centrifugation at 15,000 g at 4°C for 15 min, then washed with 70% ethanol and centrifuged again. The dried pellet was reconstituted in water at a concentration of 0.5 mg/mL, and concentration was verified using by absorbance at 260 nm using a Biotek Synergy 2 Gen5 Multiplate reader. The absence of protein from the solution was measured by absorbance at 280 nm (A260/A280 = 1.78).
2.4.2. Initial Screenings of Polymer Formulations
GB 319 astrocytes were plated in 24-well plates at a density of 75,000 cells/well 24 hr before transfection. Just before transfection, all culture media were replaced with serum-free DMEM/F-12 (1:1). EGFP plasmid DNA was delivered to GB astrocytes as previously described [28]. Briefly, DNA and polymer were each diluted in 25 mM sodium acetate (pH 5), then combined at a 30:1, 60:1, 90:1, or 120:1 weight/weight ratio (w/w) of polymer to DNA (volume ratio was 1:1 in each case). The mixture was incubated for 10 min to allow complexation to occur, then added to the cell medium at a volumetric ratio of 1:5 complexes to culture medium. Lipofectamine 2000 and FuGENE HD were prepared according to the manufacturers’ instructions and used as positive controls, with FuGENE HD used at a ratio of 4 μL reagent to 1 μg DNA and Lipofectamine at 2.5 μL reagent to 1 μg DNA. The dose used was 1.5 μg DNA per well unless stated otherwise. After 4 hr incubation time at 37°C, the particles and transfection medium were aspirated and replaced with fresh complete astrocyte medium.
After 48 hr, cells were fixed with 10% buffered formalin for 20 min, stained with DAPI for 5 min, and stored in PBS at 4°C for up to 1 week. Using a Zeiss Observer, fluorescent images were taken (total of 6 images per experimental group). The number of DAPI+ cells were counted in ImageJ to calculate viability (expressed as a percentage of untreated controls, mean±standard error). The number of GFP+ cells were counted in ImageJ and normalized to the number of cells in the same image to calculate transfection efficiency (expressed as a percentage, mean±standard error).
Top formulations, along with FuGENE HD and Lipofectamine 2000, were used in repeat experiments with complete culture medium as the transfection medium to assess the efficacy of the reagents in 10% FBS. Doses between 0.15 μg to 1.5 μg DNA per well were tested to find a dose that would allow at least 80% viability. Viability and transfection efficiency were quantified after 48 hr as described above.
2.4.3. Optimization of Stable Nanoparticle Formulations
The top polymer from the initial screenings was used to form nanoparticles with DNA that could be stored in dry form and used at later time points with little or no loss of efficacy. Nanoparticles were formed as above, at twice the normal concentration in 25 mM sodium acetate. Following complexation, the particles were mixed in a 1:1 (v/v) ratio with a solution of D-sucrose in 25 mM sodium acetate at varying sugar concentrations (0, 30, 60, and 90 mg/mL sucrose in sodium acetate added to particles, for a final concentration of 0–45 mg/mL sucrose). The suspension of particles in sucrose and sodium acetate was frozen at −80°C for 2 hr, then lyophilized for 48 hr and stored at room temperature, 4°C, or −20°C with dessicant. Particles were tested immediately after lyophilization (t=0) and after 1, 2, and 3 months of storage.
Before use, particles were reconstituted in the same starting volume of distilled water. Transfection experiments were carried out as described above. For quantification, the total fluorescence from each well was also measured using the plate reader before fixation and staining with DAPI. Transfection efficiency and viability were quantified as above. The number of cells in each well was also used to normalize fluorescent signal from the plate reader (RFU/cell) and normalized to untreated controls (mean± standard error). The direct number-averaged size of the particles and effective concentration was also tested at the same time points using nanoparticle tracking analysis (NTA) with a NanoSight LM10-HS (NanoSight Ltd., Wiltshire, UK). Each sample was diluted in PBS 1:90 from the standard transfection concentration for a total of 3 measurements. The mean diameter, mode diameter, and standard deviation of the size distribution were recorded. Results were pooled and expressed as mean±standard error for each parameter.
2.5. DNA Delivery to GB 551 Brain Tumor Stem Cells
2.5.1. DNA Delivery to Brain Tumor Stem Cells in Monolayer
Before transfections, 96-well plates were coated with 20 μg/mL laminin (Sigma) overnight at 4°C and washed once with PBS. BTSC neurospheres were collected by centrifugation and the supernatant discarded. Neurospheres were mechanically dissociated by trituration with a 200 μL micropipette and plated on laminin at 15,000 cells/well in complete medium. Cells were incubated 24 hr to allow them to adhere. Transfections were carried out as above using plasmid DsRed-Max instead of EGFP, scaled down by a factor of 5 from 24- to 96-well plates. Complete BTSC medium was used, with all growth factors but no serum added. Quantification was carried out with ImageJ using the area of red fluorescence as a marker of successful transfection and the area of GFP signal as a marker of all cells in the image.
2.5.2. DNA Delivery to Brain Tumor Stem Cells as 3-D Neurospheres
Before transfections, 96-well plates were coated with laminin and neurospheres were collected. Using a 5-mL glass pipette, neurospheres were gently resuspended in fresh medium with all growth factors and supplements. Separately, 100 mL of the suspension were triturated with a 200 μL micropipette to mechanically dissociate neurospheres for counting with a hemacytometer. All intact neurospheres were then diluted to a final seeding density of 15,000 cells/well in the laminin-coated plates. Neurospheres were incubated for 24 hr to allow them to adhere before transfections. Transfections were carried out as above, scaled down by a factor of 5 from 24- to 96-well plates.
To quantify 3-D cultures, neurospheres were removed from the wells with Accutase®. Complete medium was added and neurospheres were mechanically dissociated. The cells were collected by centrifugation, Accutase and media were removed, and the pellet was gently triturated to a single-cell suspension in FACS buffer (2% FBS in PBS) with or without 5000 ppm 7-aminoactinomycin D (7-AAD). Cells were kept on ice for no more than 1 hr before analysis using a C6 Flow Cytometer® (Accuri Cytometers, Inc., Ann Arbor, MI). Events with high FL3 and low FL1 signal were counted as non-viable (7AAD+) cells. Events with high FL1 and high FL2 signal were counted as healthy, successfully transfected (DsRed+) cells.
2.6. DNA Delivery to Human Fetal Cells
2.6. Fetal Neural Stem Cells
Before transfections, 96-well plates were coated with laminin and F34 neurospheres were collected, dissociated, and plated as an adherent monolayer following the same 96-well plate procedure described above. Transfections were carried out as using DsRed-Max as was done for BTSCs. Transfection and viability were quantified using DsRed signal and DAPI signal, respectively.
2.6. Fetal Astrocytes
F34 neurospheres were plated in complete astrocyte medium with 10% FBS in a culture flask. After one passage, cells were trypsinized and seeded in 96-well plates for transfections with DsRed-Max. Transfection and quantification were carried out the same as with the other cell types.
2.7. Statistics
All experiments were performed with n=4 replicates unless otherwise noted. Bar graphs indicate mean±standard error of the mean. GraphPad Prism was used for all statistical analysis. One-way ANOVA with a post hoc Dunnett test was used to assess statistical significance.
3. Results and Discussion
Using fluorescent protein-coding plasmid vectors as an indicator of successful gene delivery, we sought to compare the efficacy and nonspecific cytotoxicity of PBAEs in primary GB astrocytes and BTSCs, as well as in healthy primary astrocytes and NSCs. We also explored methods of expanding the practical usage of PBAE-DNA nanoparticles via lyophilization and long-term storage. We identified several PBAE structures that could be used as efficient gene delivery agents to brain cancer cells. Although gene therapy is emerging as an attractive option with great potential to treat GB, most groups have thus far investigated viral or cellular vectors, which may carry additional safety risks. Furthermore, most synthetic gene vectors must be prepared shortly before use and are not stable for long periods of time. Here, we show that synthetic PBAEs can transfect GB astrocytes and BTSCs with high efficiency superior to their efficiency in non-cancerous astrocytes and NSCs, and we show the development of a stable form of the PBAE-DNA nanoparticles that can be easily prepared, stored, and reconstituted with little to no loss of function over three months.
3.1 DNA Nanoparticle Uptake by Glioblastoma Astrocytes
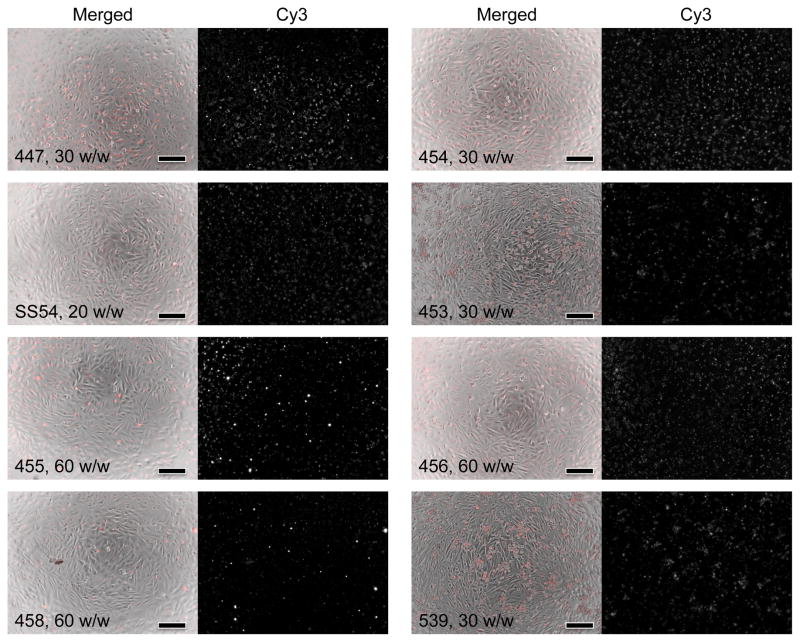
The ability of PBAEs to facilitate DNA entry into the cell was assessed by fluorescence imaging. Using peak absorbance values, one Cy3 molecule was calculated to have been conjugated approximately once per 83 nucleotides. DNA enters GB cells efficiently using a wide range of polymers with varying structures (Figure 2). Within hours after transfection, most if not all cells have taken up nanoparticles. Of note, however, is that the Cy3 signal indicates only the presence of DNA in the cell and does not provide information on the plasmid’s integrity or exact location. It is known that many downstream barriers to gene delivery exist following entry into the cell, including escape from the acidic endosome, trafficking to the nucleus, and transcription and translation of the gene [29–31]. Although many studies of nucleic acid delivery use uptake as a measure of successful transfection, efficient uptake is likely necessary but not sufficient for high transgene expression. In particular, although polymers 455 and 447 were among the most successful polymers to cause transgene expression, most of the other polymers tested also cause uptake in as many cells as do 455 and 447 (Figure 2).
Figure 2.
PBAEs facilitate efficient uptake of fluorescently-labeled DNA by GB 319 astrocytes. Different polymer structures (indicated by the numbers in each set of images) are complexed at different weight ratios (w/w) with DNA and show high Cy3 signal 4 hours after transfection. In each group, left: Phase contrast merged with Cy3 signal (red); right: Cy3 signal only. Scale bar: 200 μm. High intracellular uptake is achieved with each of these different types of polymeric nanoparticles.
3.2. Gene Delivery to Glioblastoma Astrocytes
PBAEs similar to some of those studied here have been found to be effective in overcoming intracellular barriers to gene delivery [13, 29, 32] in addition to simply facilitating uptake. To identify the most effective of those polymers, they were used to transfect GB 319 astrocytes with EGFP plasmid DNA at polymer-to-DNA weight-to-weight (w/w) ratios from 30:1 to 120:1. Formulations of polymeric nanoparticles administered in the absence of serum are show variable transfection efficiency and viability in 319 GB astrocytes depending on polymer structure (Supplemental Figure S1). All were used at a dose of 1.5 μg DNA per condition unless otherwise stated. Because a simple combinatorial synthesis scheme was used to design the library of PBAEs used in this study, many distinct molecules could be tested. Although some trends were observed, it was also found that changes as small as one or two atoms in the base monomers could alter the polymers’ effects on cells. For instance, all polymers tested with the E7 end cap showed transfection efficiency above 15%; however, both 447 and 457 showed significantly higher transfection at the same w/w ratios than 657 (for 30 w/w, 38.4±3% and 45.1±5% for 447 and 457, respectively, compared to 18.4±1% for 657). The same is true of 454, which was more effective than 554, and of 455, which was more effective than both 545 and 655.
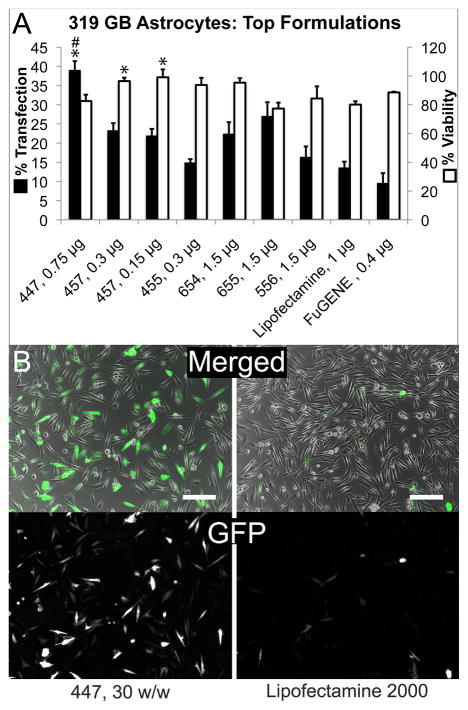
To further narrow the range of PBAEs most effective in GB gene delivery, top formulations were tested again, using a wider range of dosages (0.15 to 3.0 μg DNA per condition) and in the presence of 10% FBS. In agreement with studies by other groups on lipofection [33–35], the lipid-DNA complex formed by Lipofectamine 2000 was found here to be much less effective at gene transfer in the presence of serum, though its intrinsic toxicity was also lessened at lower doses (Figure 3B and C).
Figure 3.
PBAEs can transfect GB 319 astrocytes with comparable or superior efficiency and safety compared to leading commercial reagents. A: Top polymers were screened at a wider range of doses (indicated on the graph as μg DNA) in 10% serum and found to be more efficient at transfection than Lipofectamine 2000 and FuGENE HD while maintaining greater than 80% viability. *p<0.05 superior to Lipofectamine, #p<0.05 superior to FuGENE. In B, top: phase contrast merged with GFP (green) and DAPI (blue) signals; bottom: GFP only.
Many PBAEs, on the other hand, remained effective in the presence of serum. A subset, compared favorably to results from FuGENE HD and Lipofectamine 2000 transfection, yielding higher transfection efficiency at the optimal dose and still retaining 80% or greater viability (Figure 3B). For instance, one of the leading polymers found in this study was 447, used at a 30:1 w/w ratio to DNA, which transfected up to 60.6±5 % of cells in the absence of serum or, at a lower dose and in serum-containing medium, 40.0±2 % of cells while maintaining 82.5±4% viability. At doses low enough for Lipofectamine 2000 to maintain 80.1±2% viability, its transfection efficiency was decreased to 13.6±2%.
3.3. Duration of Transgene Expression
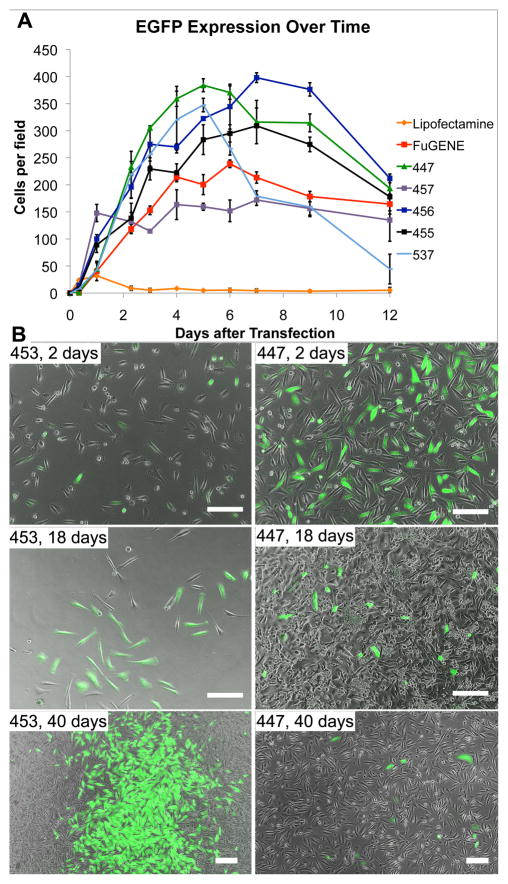
Polymeric methods of gene delivery are thought to be safer than virus-mediated delivery partly because of the reduced likelihood of inflammatory response and insertional mutagenesis. Because polymer-delivered plasmids integrate only rarely into the host cell’s genome, however, a potential concern is that expression of the transgene will be too transient. When GB 319 astrocytes were transfected with EGFP plasmids in the presence of 10% serum, expression Lipofectamine 2000 expression became visible first of all tested conditions (within 5 hr). However, it also declined the most quickly, decreasing to between 0–5 cells per microscope field (10x magnification) at the same time that FuGENE- and PBAE-mediated expression was still increasing. Of the reagents tested, PBAE 456 peaked latest in the number of cells per well (between 7–8 days), while the other PBAEs and FuGENE peaked slightly earlier, at 4–6 days (Figure 4). It should be noted that this number was not normalized to the number of total cells per field; however, the consistently increasing number of EGFP+ cells beyond the first few days suggests that GFP plasmids may be retained through the cell divisions during that time and are not completely silenced even after two weeks.
Figure 4.
EGFP expression peaks between 4–6 days for most PBAEs and FuGENE HD and at 1–2 days for Lipofectamine 2000 (graph, top). Few cells retain the EGFP gene for extended periods of time. All polymers tested led to persistent expression in some local areas of the plate (B), with FACS analysis showing 1.1% transfection for polymer 453 and 0.5% for 447 after 70 days. Scale bar: 200 μm.
Forty days later, all groups except Lipofectamine 2000 contained some GFP+ cells, including one of the most promising polymers from initial screens, PBAE 447 (Figure 4). Interestingly, in the experimental group transfected with PBAE 453, the number of GFP+ cells steadily increased up to the 40th day. Because of higher toxicity, 453 had not been included in optimization experiments; however, as a consequence of this toxicity, the increasing number of stably transfected GFP+ cells was visually clear, as shown in Figure 4. In each of the other PBAE formulations, a small group of cells (0.5–1.1%) appeared to be stably GFP+ after 70 days.
This raises the possibility that delivered transgenes could be continually expressed in a small number of cells that can act as long-term reservoirs for human brain cancer therapeutics. Instead of transfection lasting only a few days or weeks, as is typical of non-viral gene delivery, long-term protein expression could be achieved. Long-term expression may be important for diseases like brain cancer, where treatment will likely need to persist to completely eradicate all tumor cells or to continually slow tumor growth. At the same time, the low frequency (<1%) of chromosomal insertion greatly reduces the probability that a tumor suppressor gene or other essential gene will be disrupted and therefore cause de novo mutations.
3.4. Optimization and Characterization of Stable Nanoparticle Formulations
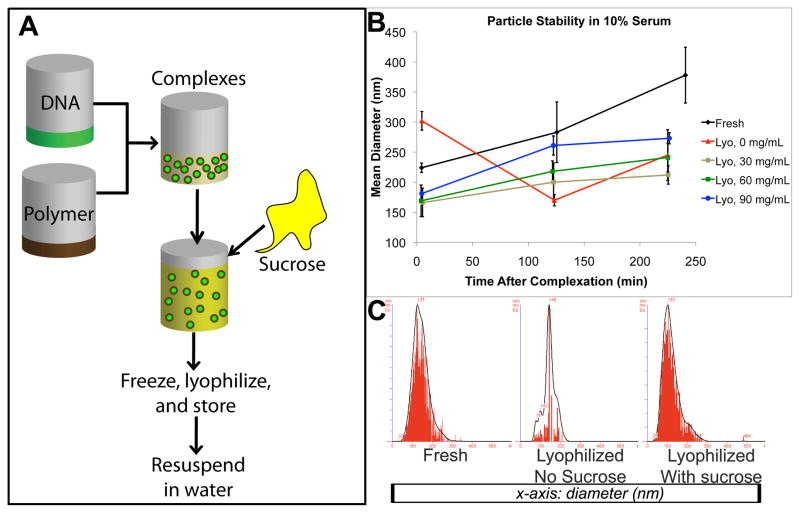
To optimize nanoparticle formulation for storage in dry form while maintaining stability, we used one of our top polymers as a test case, 447 at a 30:1 weight ratio to DNA. There was very little transfection and few particles remained when particles were lyophilized without modification; by adding 30–90 mg/mL sucrose as a lyoprotectant before freezing (final concentration 15–45 mg/mL sucrose), full functionality of the particles was retained after immediate reconstitution in water, measured by transfection efficiency, viability, and sizing (Supplemental Figure S2) in both the presence and the absence of serum. Moreover, freshly prepared particles usually begin to aggregate in suspension over time, particularly in media containing salts or serum, which can be measured by an increase in effective size and a wider size distribution (Figure 5). Lyoprotectants are known to help nanoparticles retain morphology and functionality after freezing and drying processes [17]. In this study, particles lyophilized with sucrose remained more stable in serum compared with freshly prepared particles over a period of four hours, a critical window of time for nanoparticle uptake during transfections.
Figure 5.
A: DNA nanoparticle lyophilization protocol. Particles in suspension are mixed with sucrose, frozen, lyophilized, and stored for quick and easy use. B: changes in mean particle diameter over 4 hr during incubation in 10% serum. C: Nanoparticle tracking analysis histograms show size distribution of freshly prepared particles and lyophilized particles immediately after drying with 0 or 60 mg/mL sucrose,
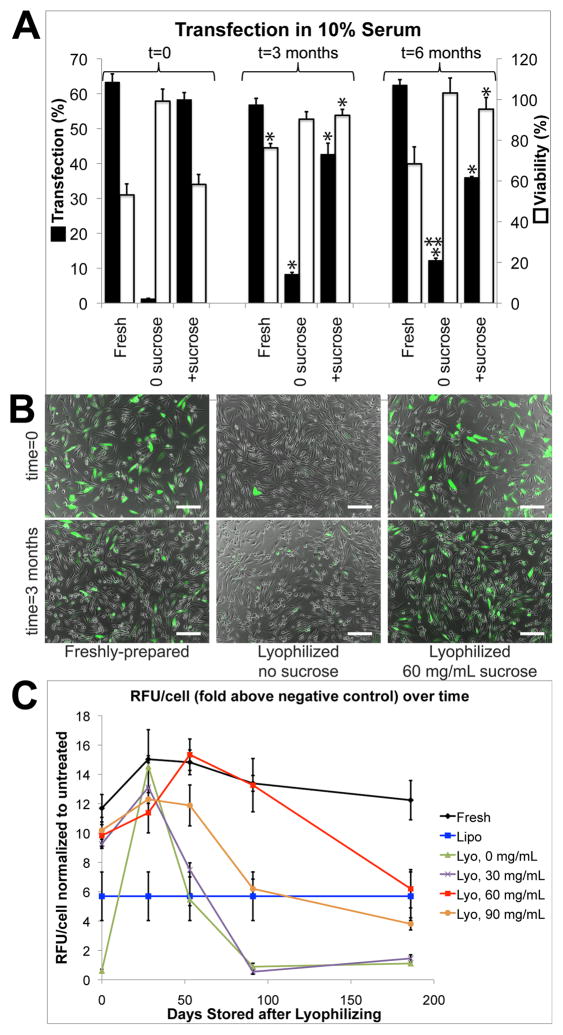
After 3 months, particles lyophilized with 60 mg/mL sucrose and stored at 4°C were not significantly different in transfection efficiency or toxicity compared to freshly prepared particles made with polymer stored at 4°C and fresh DNA stored at −20°C (p>0.05). Though the measured percent transfection efficiency of lyophilized particles was lower, the total fluorescence measured in each well after transfection did not show a significant decrease after 3 months compared to the starting value (Figure 6). Combined with the increase in measured viability over time, this may suggest that, while the proportion of GFP+ cells decreases up to 25% after 3 months, the absolute number of GFP+ cells or the total amount of GFP produced in each well remains approximately the same. Even after 6 months, approximately 50% of the original transfection efficiency and total fluorescence per cell is retained (Figure 6).
Figure 6.
A–B: Transfection efficiency of particles lyophilized with 60 mg/mL sucrose and stored at 4°C is comparable to that of freshly prepared particles at 0 and 3 months. Lyophilized particles have reduced efficacy after 6 months, though approximately 50% of the original efficacy is retained. *p<0.05 compared to t=0; **p<0.05 compared to t=3 months. Scale bar: 200 μm. C: Adding 60 mg/mL sucrose maintains function in lyophilized particles better than other sucrose concentrations after 3 and 6 months. Lipofectamine 2000 was prepared fresh and used according to the manufacturer’s instructions in each transfection. Values shown in the graph are averaged from all trials using Lipofectamine.
3.5. Gene Delivery to Adherent 551 GB BTSCs
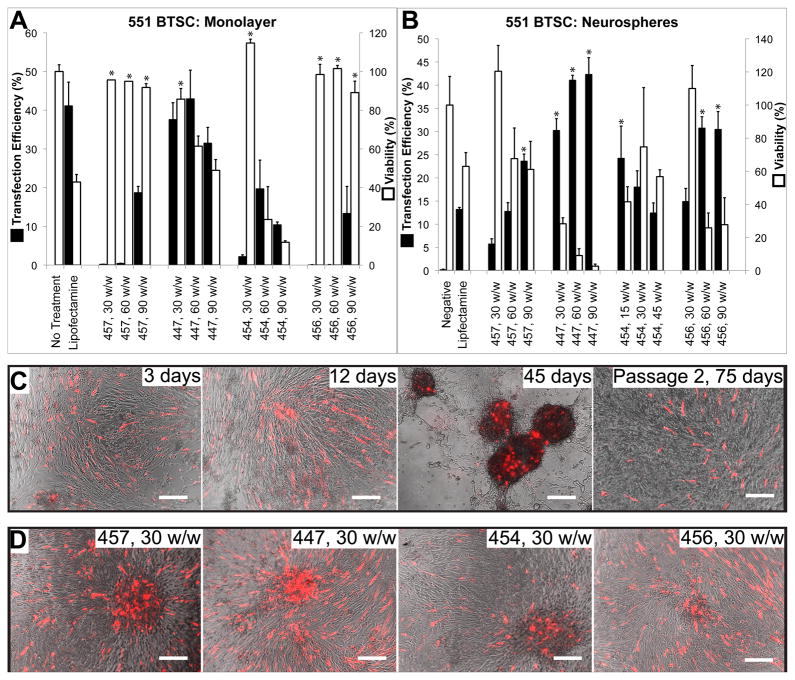
Using a smaller range of polymers shown to be effective in gene delivery to GB astrocytes, the same transfection screening experiments were carried out on stably transduced GFP+ BTSCs using plasmid DsRed-Max. Dissociated BTSCs plated in monolayer on laminin could also be transfected by PBAEs with high efficacy. As with the GB astrocytes, top PBAEs showed significantly less non-specific toxicity than Lipofectamine 2000 (ANOVA, post hoc Dunnett test, p<0.05) and caused up to 43.0±7% transfection efficiency while maintaining 61.4±5% viability, or 37.6±4% transfection with 85.7±6% viability (Figure 7).
Figure 7.
PBAEs facilitate efficient DNA delivery to BTSCs when plated in monolayer (A, C) or as 3–D neurospheres (B, D). Some polymers were less toxic than Lipofectamine 2000 while transfecting equally well or better (*p<0.05 superior to Lipofectamine). After 45 days, neurospheres that reform from overconfluent monolayer cultures remain brightly DsRed+, and a small number remain DsRed+ after 75 days and 1 passage (C). All conditions (C) are with polymer 447 and 30 w/w and all conditions (D) are shown 3 days after transfection.
When BTSCs transfected with PBAE 447 at a 30:1 w/w ratio were maintained in culture without passaging, the cells reformed adherent 3-D neurospheres over time, with DsRed-Max transgene expression still visible throughout the reformed neurosphere after 45 days (Figure 7B). Even after passaging the transfected BTSCs, mechanically dissociating all neurospheres, and re-plating on laminin, a subset of the cells remained stably DsRed+ at least 90 days after transfection.
Significantly, when BTSCs were plated on laminin as a mixture of single cells and neurospheres as a co-culture, PBAEs 454 and 456 were relatively effective in penetrating into the 3-D structure and transfecting cells throughout the neurosphere (up to 24.3±7% and 30.8±2% transfection, respectively), while DsRed was expressed in up to 40–45% of cells when 447 or 457 was used (Figure 7C).
3.6. Comparison of PBAE-mediated Gene Delivery to GB Cells and Healthy Cells
An eventual goal of this technology will be to deliver genes intracranially, where both GB cells and healthy brain cells could potentially be exposed to DNA-containing nanoparticles. Ideally, the polymer used for transfection should preferentially affect GB cells while having minimal or no effect on healthy cells. However, another possible therapeutic strategy is to deliver genes that are expressed only by GB cells using cancer-specific promoters or that have an effect only on cancer cells, such as through the induction of apoptosis. In the latter strategy, gene transfer of an exogenous paracrine factor to healthy cells would not be harmful and could even be advantageous, as these healthy cells could serve as a “factory” for secreted factors that would kill surrounding tumor cells.
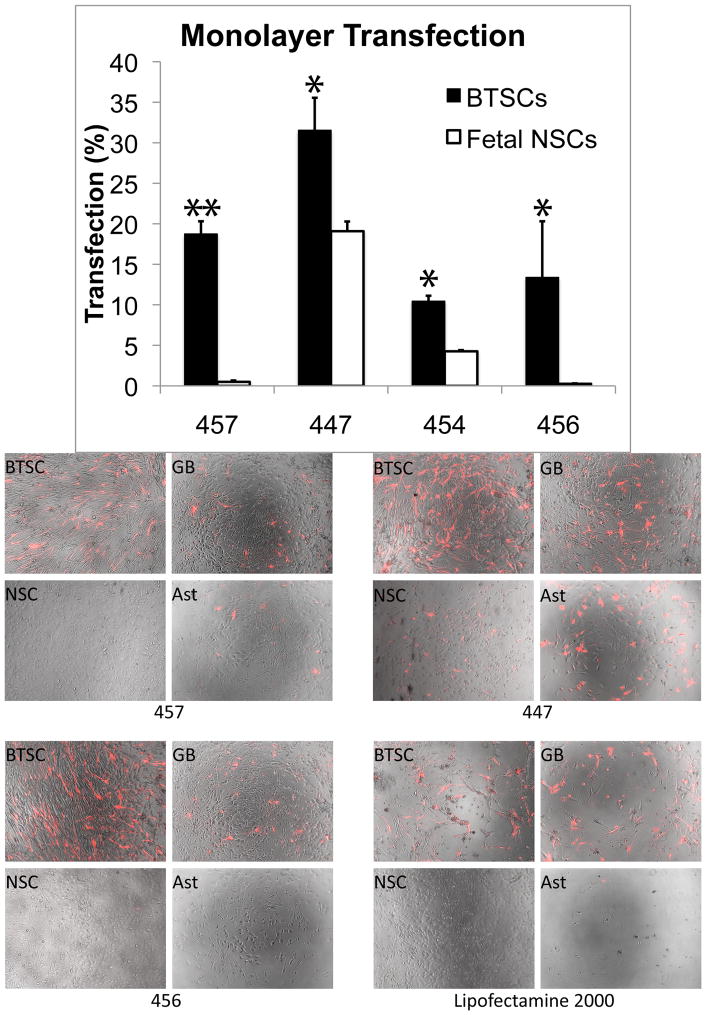
In this work, we identified PBAEs that could be effective for both potential strategies. Most of the top PBAE formulations for GB astrocytes or 551 BTSCs were significantly less effective (p<0.05) on F34 fetal cortical cells, especially when the healthy cells were maintained as undifferentiated NSCs rather than cultured as astrocytes (Figure 8). In particular, polymer 457 transfected nearly 20% of BTSCs but only 4.8% of F34 astrocytes, and it had no apparent effect on F34 NSCs. Polymer 456 had 13.4% efficacy on BTSCs with no quantifiable toxicity, but it had little to no effect on F34 cells (0.3% transfected with greater than 85% viability). On the other hand, 447’s efficacy in GB astrocytes and BTSCs, along with its moderate efficacy in healthy astrocytes and healthy NSCs (19.1±2% transfection), suggests that it may be particularly useful in delivery of genes whose products are secreted proteins that are specifically cytotoxic to cancer cells. Lipofectamine 2000 was almost completely ineffective on BTSCs at a standard dose (0.3 μg per well); by doubling the dose, transfection efficiency increased drastically to 41.2±6%, but viability plummeted to 42.9±4%. This effect was even more pronounced in fetal astrocytes, in which Lipofectamine 2000 treatment killed nearly all cells (Figure 8).
Figure 8.
In general, tumor stem cells (BTSC) and glioblastoma cells (GB) are transfected much more effectively by PBAEs than healthy fetal stem cells (NSC) and astrocytes (Ast). One exception is 447, which transfects all cell types tested, and Lipofectamine 2000, which, while having no visible effect on fetal NSCs, was highly toxic to fetal astrocytes. The difference between BTSC and fetal NSC transfection is statistically significant in all cases shown (*p<0.05, **p<0.0001).
Conclusions
End-modified versions of synthetic poly(β-amino esters) can be used for high levels of non-viral gene delivery to GB astrocytes and BTSCs. In this study, we identified polymers that are not only efficient in delivery to brain tumor cells but also in healthy cells (447), as well as polymers that affect only GB-derived cells and BTSCs with little or no apparent effect on human healthy cells and human neural stem cells (457 and 456). This shows potential for multiple strategies of treating GB through gene therapy, either using targeted delivery mediated by polymer structure or using delivered plasmids with differential expression in different cell types. We also identified formulations where PBAEs, once complexed with DNA, can be kept in a lyophilized form at 4°C and maintain function and size for at least 3 months, allowing for long-term storage and ease of use, both of which may be useful in a clinical setting. This work describes a promising biomaterials-mediated platform for the treatment of glioblastoma, a deadly disease in critical need of new therapeutic strategies. Future studies will examine the transfection efficiency and biological effects of PBAEs in an in vivo system to elucidate the potential role of PBAE-mediated gene delivery in the treatment of brain cancer.
Supplementary Material
Acknowledgments
The 551 BTSCs were transduced and provided by Dr. Tomás Garzón-Muvdi. This work was supported in part by the Institute for NanoBioTechnology at Johns Hopkins University and TEDCO MSCRF (2009-MSCRFE-0098-00). HG and AQ were funded by the National Institutes of Health (R01NS070024), the Howard Hughes Medical Institute, and the Robert Wood Johnson Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.DeAngelis LM. Brain tumors. N Engl J Med. 2001;344(2):114–23. doi: 10.1056/NEJM200101113440207. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen TD, DeAngelis LM. Brain metastases. Neurol Clin. 2007;25(4):1173–92. x–xi. doi: 10.1016/j.ncl.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 3.American Cancer Society. Cancer Facts & Figures 2010. Atlanta: American Cancer Society; 2010. Contract No.: March 16. [Google Scholar]
- 4.CBTRUS. CBTRUS Statistical Report: Primary Brain Tumors in the United States, 2004–2007. Central Brain Tumor Registry of the United States. 2011 Available from: www.cbtrus.org.
- 5.Klos KJ, O’Neill BP. Brain metastases. Neurologist. 2004;10(1):31–46. doi: 10.1097/01.nrl.0000106922.83090.71. [DOI] [PubMed] [Google Scholar]
- 6.McGirt MJ, Chaichana KL, Gathinji M, Attenello FJ, Than K, Olivi A, et al. Independent association of extent of resection with survival in patients with malignant brain astrocytoma. J Neurosurg. 2009;110(1):156–62. doi: 10.3171/2008.4.17536. [DOI] [PubMed] [Google Scholar]
- 7.McGirt MJ, Mukherjee D, Chaichana KL, Than KD, Weingart JD, Quinones-Hinojosa A. Association of surgically acquired motor and language deficits on overall survival after resection of glioblastoma multiforme. Neurosurgery. 2009;65(3):463–9. doi: 10.1227/01.NEU.0000349763.42238.E9. discussion 9–70. [DOI] [PubMed] [Google Scholar]
- 8.McGirt MJ, Than KD, Weingart JD, Chaichana KL, Attenello FJ, Olivi A, et al. Gliadel (BCNU) wafer plus concomitant temozolomide therapy after primary resection of glioblastoma multiforme. J Neurosurg. 2009;110(3):583–8. doi: 10.3171/2008.5.17557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA Cancer J Clin. 2002;52(1):23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 10.NCI. Cancer Advances in Focus: Brain and Other Central Nervous System Cancers. 2009. [Google Scholar]
- 11.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 12.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4(5):346–58. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 13.Putnam D. Polymers for gene delivery across length scales. Nat Mater. 2006;5(6):439–51. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 14.Bhise NS, Gray RS, Sunshine JC, Htet S, Ewald AJ, Green JJ. The relationship between terminal functionalization and molecular weight of a gene delivery polymer and transfection efficacy in mammary epithelial 2-D cultures and 3-D organotypic cultures. Biomaterials. 2010;31(31):8088–96. doi: 10.1016/j.biomaterials.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sunshine J, Green JJ, Mahon K, Yang F, Eltoukhy A, Nguyen DN, et al. Small molecule end group of linear polymer determine cell-type gene delivery efficacy. Adv Mater. 2009;21(48):4947–51. doi: 10.1002/adma.200901718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res. 2008;41(6):749–59. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdelwahed W, Degobert G, Stainmesse S, Fessi H. Freeze-drying of nanoparticles: formulation, process and storage considerations. Adv Drug Deliv Rev. 2006;58(15):1688–713. doi: 10.1016/j.addr.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 18.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 19.Fine HA. Glioma stem cells: not all created equal. Cancer Cell. 2009;15(4):247–9. doi: 10.1016/j.ccr.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7(2):150–61. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Zaidi HA, Kosztowski T, DiMeco F, Quinones-Hinojosa A. Origins and clinical implications of the brain tumor stem cell hypothesis. J Neurooncol. 2009;93(1):49–60. doi: 10.1007/s11060-009-9856-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmaggi A, Boiardi A, Gelati M, Russo A, Calatozzolo C, Ciusani E, et al. Glioblastoma-derived tumorospheres identify a population of tumor stem-like cells with angiogenic potential and enhanced multidrug resistance phenotype. Glia. 2006;54(8):850–60. doi: 10.1002/glia.20414. [DOI] [PubMed] [Google Scholar]
- 23.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–8. [PubMed] [Google Scholar]
- 24.Strack RL, Strongin DE, Bhattacharyya D, Tao W, Berman A, Broxmeyer HE, et al. A noncytotoxic DsRed variant for whole-cell labeling. Nat Methods. 2008;5(11):955–7. doi: 10.1038/nmeth.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295(5556):868–72. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 26.Guerrero-Cazares H, Chaichana KL, Quinones-Hinojosa A. Neurosphere culture and human organotypic model to evaluate brain tumor stem cells. Methods Mol Biol. 2009;568:73–83. doi: 10.1007/978-1-59745-280-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–80. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Green JJ, Shi J, Chiu E, Leshchiner ES, Langer R, Anderson DG. Biodegradable polymeric vectors for gene delivery to human endothelial cells. Bioconjug Chem. 2006;17(5):1162–9. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]
- 29.Akinc A, Langer R. Measuring the pH environment of DNA delivered using nonviral vectors: implications for lysosomal trafficking. Biotechnol Bioeng. 2002;78(5):503–8. doi: 10.1002/bit.20215. [DOI] [PubMed] [Google Scholar]
- 30.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92(16):7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene Ther. 2005;12(13):1023–32. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- 32.Green JJ, Zugates GT, Tedford NC, Huang Y, Griffith LG, Lauffenburger DA, et al. Combinatorial modification of degradable polymers enables transfection of human cells comparable to adenovirus. Adv Mater. 2007;19(19):2836–42. [Google Scholar]
- 33.Dodds E, Dunckley MG, Naujoks K, Michaelis U, Dickson G. Lipofection of cultured mouse muscle cells: a direct comparison of Lipofectamine and DOSPER. Gene Ther. 1998;5(4):542–51. doi: 10.1038/sj.gt.3300604. [DOI] [PubMed] [Google Scholar]
- 34.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A. 1987;84(21):7413–7. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofland HE, Shephard L, Sullivan SM. Formation of stable cationic lipid/DNA complexes for gene transfer. Proc Natl Acad Sci U S A. 1996;93(14):7305–9. doi: 10.1073/pnas.93.14.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.