Abstract
Changes in gene expression are thought to be important for morphological evolution, though little is known about the nature or magnitude of the differences. Here we examine Xenopus laevis and Xenopus tropicalis, two amphibians with very similar development, and ask how their transcriptomes compare. Despite separation for ~30–90 million years there is strong conservation in gene expression in the vast majority of the expressed orthologs. Significant changes occur in the level of gene expression but changes in the timing of expression (heterochrony) were much less common. Differences in level were concentrated in the earliest embryonic stages. Changes in timing were prominently found in pathways that respond to selective features of the environment. We propose that different evolutionary rates across developmental stages may be explained by the stabilization of cell fate determination in the later stages.
INTRODUCTION
In the 19th century, comparative anatomy – of embryos and adults – was the principal means for establishing the phylogeny of animal species (Gould 1977; Hennig 1979). With the rise of molecular methodologies over the last few decades, the study of animal evolution has increasingly relied upon comparison of molecular features (Graur and Li 2000; Carroll, Grenier et al. 2001). Following Darwin s expectation that species diverge by small steps, each built on another, it was initially expected by the architects of the neo-Darwinian synthesis that developmental processes at the molecular level would have diverged in proportion to the time of divergence from the last common ancestor (Mayr 1963). However, the results from genetic and molecular studies in Drosophila and subsequent molecular studies in several vertebrate and invertebrate species challenged this view when many key developmental signaling pathways and transcription factors showed deep evolutionary conservation (Carroll, Grenier et al. 2001). Although change does occur in the coding sequence of genes, a great deal of the innovation has arisen through regulatory reshuffling of common developmental pathways, altering the timing of transcriptional programs (Gerhart and Kirschner 1997; Davidson and Levin 2005; Carroll 2008).
Transcriptional regulation can affect the amount of a gene product, its timing, and its location of expression (Arthur 2000). Although we can easily document massive changes in intragenic DNA sequence, we cannot currently deduce by inspection the nature of its regulation. Hence, despite the new information from genome sequences, comparative studies of gene expression rely upon experiment. The introduction of high throughput methods now allows us to measure on a genomic scale when and how much a gene is expressed with great accuracy and sensitivity. In contrast, the location of expression, as measured by such methods as in situ hybridization, is still difficult to quantify and to scale to many genes. Finally, a more complete understanding would need to include posttranscriptional and posttranslational regulation. Yet given the power of the transcriptional methods, it is common and not unreasonable to use gene expression (in the form of steady-state RNA concentration) itself as a very approximate surrogate for generation of the phenotype.
While it has been long thought that transcriptional change is important to the divergence of metazoan species (Britten and Davidson 1971; King and Wilson 1975), recent work just has begun to query the global extent of this divergence, particularly in vertebrate lineages (Khaitovich, Enard et al. 2006; Blekhman, Oshlack et al. 2008; Nolte, Renaut et al. 2009; Renaut, Nolte et al. 2009; Xie, Chen et al. 2010). That is, although the function of certain transcription factors appears to have been highly conserved during evolution (e.g. Pax6, HOX), is this conservation an exception or the rule when one takes a global view of the developmental transcriptome? Does one expect to find massive changes in expression occurring in closely related lineages or strong overall conservation even in distantly related organisms? Are morphological and transcriptional change strongly coupled?
It has long been argued that a major source of evolutionary change is heterochrony, when whole modules of anatomy and physiology are temporally moved in ontogeny (Gould 1977). An oft-cited example of heterochrony is the axolotl, in which sexual maturation has been advanced to the larval state (Gould 1977). One could imagine an analogous process at the molecular level; this molecular heterochrony might not always be apparent at the anatomic or physiological level. In addition to changes in timing, two related species may differ during ontogeny in the relative size of their developing organs, a situation akin to allometry and which we will refer to here as heterometry. Again, one could imagine an analogous process at the molecular level, in which the absolute expression level of genes changes during evolution; this molecular heterometry may or may not be caused by changes to the size of the morphological area in which the gene is expressed.
Changes in gene expression between two related species could reflect evolutionary selection but could also reflect evolutionary drift, in which certain alleles become randomly more frequent and fixed in an interbreeding population (Khaitovich, Weiss et al. 2004; Yanai, Graur et al. 2004; Blekhman, Oshlack et al. 2008; Staubach, Teschke et al. 2010). Previous work highlighted both the roles of purifying selection and drift on the transcriptome. Rifkin et al. compared the transcriptomes of Drosophila species and strains and showed the signature of purifying selection at the start of metamorphosis (Rifkin, Kim et al. 2003). However, drift was also readily apparent and was proposed to be the main explanation for most divergence among transcriptomes, based upon tissue data in human and mouse (Khaitovich, Weiss et al. 2004; Yanai, Graur et al. 2004; Yanai, Korbel et al. 2006). Recently, a developmental time course comparing the transcriptomes of C. elegans and C. briggsae – a pair of organisms that diverged ~100 million years ago (Mya) but which maintain a common mode of development despite highly divergent genomic sequences – showed that a major fraction of the developmental transcriptome is evolving and that these changes correlate with changes in genomic location (Yanai and Hunter 2009). Other recent work has documented massive drift of TF binding sites during vertebrate evolution (Schmidt, Wilson et al. 2010).
Comparative temporal transcriptome studies afford new insights into evolution. Although lacking information about the location of gene expression, temporal transcriptome data can be comprehensive and quantitative and can provide new ways to evaluate changes in expression in general and heterochrony and heterometry in particular. In this study, we have examined the vertebrate transcriptomes of two Xenopus species, X. laevis and X. tropicalis. There is a wide range of estimates, from 30 to 90 Mya, for the time of the last common ancestor, depending on the methods used (Bisbee, Baker et al. 1977; Knochel, Korge et al. 1986; Evans, Kelley et al. 2004). Some phylogenies place X. tropicalis in the closely related genus Silurana. The coding DNA sequences of X. laevis and X. tropicalis are 90% identical within coding regions (Supplementary Methods), comparable to divergence between human and mouse sequences (85%). Xenopus laevis has a tetraploid genome thought to be derived from the hybridization of two parent species, in which roughly half of the duplicated genes have been maintained. The development of X. laevis and X. tropicalis appears to be largely conserved such that the same normal table of development is used for both organisms, although we know of no systematic attempts to characterize differences. Two well-appreciated differences are that X. tropicalis embryos will successfully develop over a temperature range that is higher than X. laevis (standard lab temperatures 28°C vs. 22°C, respectively) and that X. tropicalis embryos are about one-eighth the volume of X. laevis embryos.
In attempting to compare the embryonic transcriptomes of these two frogs, we encountered a number of obstacles: the lack of a genome sequence for Xenopus laevis, the different standard lab temperatures for development, the need to quantify mRNA levels in systems of different size, the need to quantify mRNA levels when total mRNA present in an embryo changes during development, the need to take into account variation between individuals within each species, the existence of alternative splice variants, and the lack of appropriate mathematical and statistical tools for these comparisons. As a by-product of this study, we have created an open-access browser (http://kirschner.med.harvard.edu/Xenopus_Transcriptomics.html) which reports the timing of expression of over 11,000 orthologs, allowing developmental biologists to compare the gene expression and to be aware of the transcriptional differences between the two species.
RESULTS
Comparative transcriptomics of Xenopus development
To compare the developmental transcriptomes of Xenopus laevis and Xenopus tropicalis we generated a large microarray dataset encompassing gene expression for 11,095 orthologs across 14 developmental stages spanning the blastula, gastrula, neurula, and tail-bud stages (Fig. 1a, and Supplementary Methods) in biological triplicates. For each species of frog, the data was collected three times, using clutches of embryos generated through in vitro fertilization from three distinct male-female combinations. The embryos were collected and aligned according to well-defined morphological stages based on their superficial anatomy. The embryos from each species were grown at their respective standard lab temperatures that have been found to be good for development (see Experimental Procedures). In X. laevis RNA was extracted from a single embryo, while in X. tropicalis RNA from three embryos was pooled to obtain comparable quantities (see Experimental Procedures). To allow a normalized comparison of expression, each RNA sample was spiked with a set of foreign RNAs at known relative concentrations. The expression data was normalized by linearly interpolating concentrations using these spike-in measurements for each sample. The final expression values are thus in log10 units of relative expression that are comparable across both developmental time points and species.
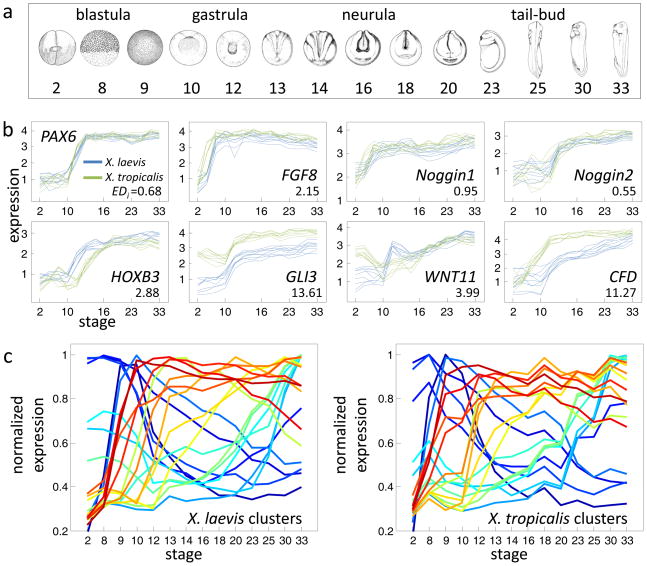
Figure 1. Comparative transcriptomics of Xenopus development.
a. Developmental stages assayed in this study, taken from Nieuwkoop and Faber (Nieuwkoop and Faber 1994). b. Microarray gene expression data for the eight indicated genes. For each gene, the nine profiles (3 clutches across 3 probes) are shown for both X. laevis (blue) and X. tropicalis (green). The y-axis indicates log10 relative concentrations of mRNA abundance (see Experimental Procedures). Also indicated for each gene is its EDi , a metric for divergence explained in the text. c. Summary of 20 gene expression clusters for X. laevis (left). Clusters were generated using QT clustering (Heyer, Kruglyak et al. 1999) with a maximum correlation distance of 0.85. Each summary profile is the mean of the member profiles, normalized by dividing by the maximum value. The average profile of the X. tropicalis orthologs in these same clusters are shown to the right. Orthologous clusters share the same color across plots.
Gene expression was assayed using a custom-designed microarray for each species. To enable comparison, each pair of orthologs was associated with probes at homologous locations across the sequences of both genomes. We used the X. tropicalis genome sequence (Hellsten, Harland et al. 2010) (which at the time was available in an unassembled form) and the extensive sequence data from the X. laevis ESTs and mRNAs. The 60-mer probes were selected based upon melting temperature, absence of low-complexity regions, nearness to the 3 end, and low probability of folding. Further, the probes were scored for specificity to the target genes. For 11,095 orthlogous sets, we detected bi-directional best hits, for which we designed three independent probes, identical or near-identical across the two species-specific microarrays. Therefore, for each species of frog, a given transcript is measured nine times –3 “technical” replicates for each of 3 “biological” replicates. In the presence of in-paralogs in either species (duplications following the divergence of the lineages), probes were selected in regions identical among duplicates to capture the sum of expression across duplicates.
We examined the overall quality of the data using the following measures. The reproducibility of the microarray data is high as assessed by a high correlation coefficient (R = 0.991) between technical replicates (Fig. S1a). The detected expression profiles of key developmental regulators (Table S1) are well conserved and are consistent with the previously reported expression profiles (Fig. 1b, top panels) (Fletcher, Watson et al. 2004; Eroshkin, Ermakova et al. 2006; Fletcher, Baker et al. 2006). Table S2 provides additional examples of conserved gene expression profiles. Furthermore, the dataset is generally consistent with a previously published genome-wide time-course of X. laevis expression (Baldessari, Shin et al. 2005) (Fig. S2a). The principal patterns of expression identified by clustering in either species are similar (Fig. 1c). In this analysis, the expression profiles of X. laevis genes were clustered; the average profile is shown for each of the 20 largest clusters (Fig. 1c). The X. tropicalis orthologs of each X. laevis cluster are also shown as an average profile. The similarity in these clusters provides an initial view of the overall similarity of the transcriptomes. We also estimated the quality of the data by comparing expression data across probes for the same gene (technical replicates) and clutches (biological replicates). A gene s expression profile is expected to be independent of the probe and clutch examined, except in instances of alternative splicing and clutch differences, respectively. Examining the distribution of correlation coefficients between the expression profiles of probes for the same gene but within the same clutches, we found that 97.5% have R>0.8 (Fig. 2). Likewise, comparing the expression profiles of the same probe across clutches produces a similar distribution of high correlations, relative to random comparisons (Fig. 2). According to inter-clutch differences (Fig. 2), the X. laevis data is noisier; attributed most likely to our pooling of three X. tropicalis embryos relative to single X. laevis embryos. Finally, despite isolating X. laevis and X. tropicalis staged embryos at different temperatures, we determined that this did not strongly affect the developmental transcriptome (Fig. S2b). From these controls, we concluded that our dataset provides a solid foundation for a developmental transcriptome comparison. The data is available through an online portal (www.kirschner.med.harvard.edu/Xenopustranscriptomics.html) where gene expression profiles can be examined and compared between species.
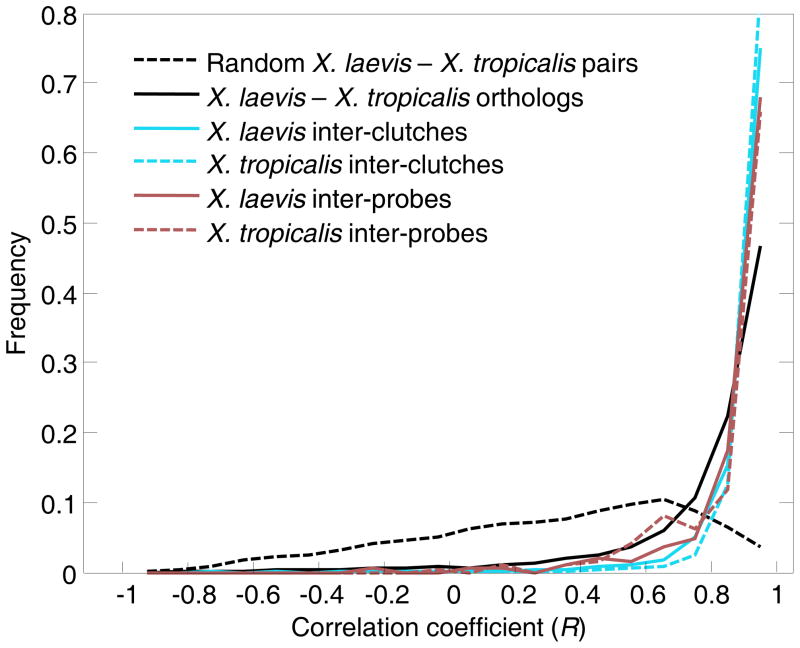
Figure 2. Dominant signal of gene expression conservation between the species.
Distributions of correlations coefficients when different classes of transcriptomes are compared. Dark red distributions indicate the median correlations between expression profiles of different probes (within the same clutch). Bright blue distributions indicate distributions of median correlations of expression profiles across clutches (within the same probes). These distributions are shifted towards higher correlations with respect to the distribution of median pairwise correlations between species (black) and randomly paired X. laevis and X. tropicalis genes (dashed line). Since this analysis tests the reproducibility of the data, we limited it to those genes showing dynamic expression in the developmental time course of either species (see Experimental Procedures).
Overall expression conservation between Xenopus species
The overall impression from a visual comparison of the two transcriptomes is that there has been remarkable conservation despite the >30 million years that passed since diverging from a common ancestor. Although one might argue that the morphologies of X. laevis and X. tropicalis are very similar and that this result would therefore be expected, there is evidence in Caenorhabditis that very similar species can vary considerably at the level of gene expression (Yanai and Hunter 2009). Table 1 indicates the number of genes with mRNA transcripts detected at each stage for each species. The large overlap (85%) of genes expressed throughout embryonic development between species suggests a strong level conservation in developmental gene regulation. Comparing the temporal profiles between orthologs, using Pearson s correlation coefficient, we also found a dominant signal of conservation (Fig. 2). The similarity in gene expression between orthologs was significantly higher than that of randomly paired genes. 89% of the orthologs have a high correlation coefficient (R>0.5), compared with 34% for randomly paired genes. The extent of correlation is higher than was observed in C. elegans vs. C. briggsae, with 66% orthologs having a R>0.5 (Yanai and Hunter 2009), and in human vs. rhesus macaque (diverging ~25 Mya) with 64% conservation on average between tissues (Blekhman, Oshlack et al. 2008). While we detected only 11% genes with R<0.5, assuming linear scaling, this divergence would amount to all genes diverging after 200 million years. Examining specifically a curated set of 248 known developmental regulators with dynamic expression in our data (Table S1), however, we found that 99% of them were highly correlated (R>0.5) suggesting that in particular for genes controlling development there is an overall strong conservation between the transcriptomes of these two species.
Table 1. Numbers of genes with detected expression during embryonic development in X. laevis and X. tropicalis.
The threshold for expression is 3 log10 units. See Experimental Procedures for definition of the set of dynamic genes.
| Stage | Expressed in X. laevis | Expressed in X. tropicalis | Expressed in both | Dynamic in X. laevis | Dynamic in X. tropicalis | Dynamic in both |
|---|---|---|---|---|---|---|
| 2 | 4,759 | 5,260 | 4,102 | 315 | 695 | 165 |
| 8 | 5,337 | 6,341 | 4,893 | 350 | 945 | 202 |
| 9 | 5,202 | 4,589 | 3,910 | 394 | 611 | 201 |
| 10 | 4,289 | 4,082 | 3,301 | 356 | 508 | 206 |
| 12 | 3,943 | 4,910 | 3,455 | 417 | 743 | 261 |
| 13 | 4,516 | 4,837 | 3,755 | 590 | 842 | 352 |
| 14 | 4,588 | 4,640 | 3,664 | 632 | 879 | 391 |
| 16 | 4,452 | 4,629 | 3,610 | 654 | 918 | 418 |
| 18 | 4,759 | 5,668 | 4,209 | 746 | 1,159 | 513 |
| 20 | 5,215 | 5,128 | 4,278 | 880 | 1,099 | 573 |
| 23 | 5,363 | 6,218 | 4,776 | 994 | 1,552 | 709 |
| 25 | 5,791 | 5,983 | 4,936 | 1,125 | 1,499 | 740 |
| 30 | 6,726 | 7,459 | 6,147 | 1,420 | 2,108 | 1,027 |
| 33 | 7,047 | 7,095 | 6,184 | 1,555 | 2,007 | 1,062 |
An important limitation of Pearson s correlation coefficient is that it registers only concordance in time of expression and is blind to changes in the absolute levels of expression. Thus it is possible for a pair of profiles to show a high correlation yet have large differences in expression level. Further, some of the variation could be due to variation within a species, which we can estimate by examining the level in the different clutches of the same species. A more comprehensive score of inter-species divergence should be sensitive to both time and level of expression and should be normalized to the degree of intra-species variation. For example, a gene with low intra-species differences and large cross-species differences should receive a high divergence score, reflecting the high confidence in the cross-species differences. We thus defined an expression divergence index, EDi, as the cross-species distance corrected for intra-species differences. EDi quantifies absolute profile differences between orthologs and is computed as the sum of differences between and within the sets of 9 available temporal profiles for each ortholog. This measure relies upon the additivity of the distances and is essentially a Euclidean distance between orthologs normalized to the internal species variation in expression. The EDi distributes log-normally (Fig. 3a, and Fig. S3a) with a mean and standard deviation of 3.3 and 3.4, respectively. We find that for 5% of the most divergent genes, the EDi is two standard deviations above the mean. For example, HOXB3 has an EDi =2.9, and GLI3 has a high divergence with an EDi of 13.6 (Fig. 1b). Table S1 gives the EDi of all the examined orthologs. We proceeded to use this divergence index to study the nature of the variation between the developmental transcriptomes.
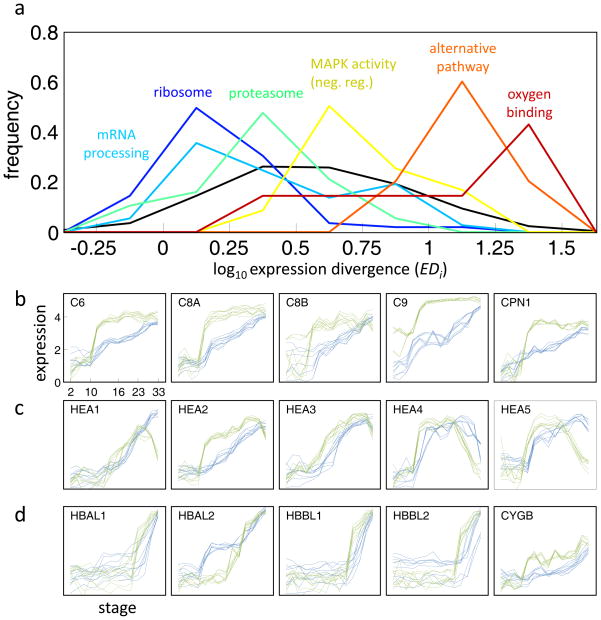
Figure 3. Conservation and divergence across pathways.
a. Expression divergence (EDi) distributions across functional gene sets and indicated by different colors. The y-axis indicates the normalized frequency. The black plot indicates the normalized distribution of divergences for all genes. A shift to the left/right indicates enrichment for conservation/divergence, respectively. b-d. Expression profiles of genes involved in the alternative pathway of the complement system (b), hatching enzymes (c), and oxygen-binding genes (d). See Fig. S3b for additional heterochrony in members of the membrane attack complex. Expression profiles are shown in log10 relative concentrations as in Figure 1.
Core cellular processes are broadly conserved, with heterochrony more prominently found in ecological adaptations
Developmental processes are comprised of gene products aggregated into pathways and arrays of pathways. To search for divergence in developmental pathways we examined entire pathways, complexes and functional modules. A concerted temporal shift in these modules could provide insight into heterochrony and general clues to the nature of developmental change. We thus first asked whether gene expression changes are biased for genes of particular functional categories. To be most inclusive, we used 2,225 previously collected gene sets (Subramanian, Tamayo et al. 2005) corresponding to curated pathways, functional groups, tissues and organs, and the results of specific gene expression experiments. For each set, we computed the levels of cross-species expression divergence in terms of EDi. Figure 3a shows the EDi distributions for a few gene sets, with respect to the distribution of EDi s of all genes. Gene sets with the least divergence comprise many core cellular processes; translation, protein degradation, and aspects of metabolism (Table S3). The great majority of ribosomal subunits, for example, are strongly induced during gastrulation in both species (Fig. S3c), perhaps reflecting the onset of cellular growth at the expense of yolk protein (Jorgensen, Steen et al. 2009). The signature of strong purifying selection for the expression pattern of core processes is also evident in many proteosome subunits and mitochondrial proteins (Table S3). On the other end of the spectrum, gene sets with divergent gene expression patterns are enriched in genes coding for transmembrane proteins such as GPCRs and kinases (Table S4). Genes involved in neurogenesis, hormone metabolism, Shh pathway, ion binding, myosin, and caspase regulation also show significant divergences.
Our EDi gene expression analysis (Fig. 3a and Table S4) provides insight into the changes that accompany specific environmental adaptations distinguishing the two frog species. Specifically, we considered the consequences of the more rapid growth of X. tropicalis at a higher temperature and its implications for predation, microbial pathogenesis and reduced oxygen solubility. We can expect that different environments, including temperature and general habitat, would be accompanied by changes in adaptive and innate immunity. Among the most rapidly evolving human genes, for example are genes involved in olfaction and pathogen protection (Voight, Kudaravalli et al. 2006; Barreiro and Quintana-Murci 2010). The membrane attack complex (MAC) is typically formed on the surface of intruding pathogenic bacteria as a result of the activation of the complement system, and is one of the deeply conserved elements of the immune system (Cole and Morgan 2003). We find a coordinated shift in expression of members of the alternative pathway of the complement system (Fig. 3a,b). Extensive expression of these molecules during gastrula/early neurula stage in X. laevis has been identified (McLin, Hu et al. 2008) suggesting an early developmental role in combating bacteria. The X. tropicalis members of this complex are induced earlier and more sharp (Fig. 3b) while keeping their respective order of induction intact with respect to X. laevis. Further, the MAC complex regulators protectin (CD59) and CPN1 also show a consistent and correlated pattern of divergent expression (Fig. S3b). We may speculate that this coordinated earlier induction of the MAC genes was selected in response to microbial conditions encountered in the environment of X. tropicalis, relative to X. laevis.
Despite the striking uniformity in developmental timing, X. laevis and X. tropicalis embryos are known to hatch at different development stages. X. laevis embryos kept in 21°C hatch approximately 48 hours after fertilization at stage 29/30 (Carroll and Hedrick 1974), while X. tropicalis embryos kept at 25°C hatch much earlier, at stage 26/27 approximately 25 hours after fertilization (Showell and Conlon 2009), where the stages are numbered according to the Xenopus laevis normal table (Nieuwkoop and Faber 1994). Morphologically these stages are clearly distinguished by the number of segregated somites, namely 16–17 somites at stage 25/26 compared to 24–25 somites at stage 28/29. Additionally, at stage 26 spontaneous movements begin. Consistent with this difference in the timing of hatching, the expression profiles of all six hatching enzyme genes exhibit a similar heterochronic displacement, where X. tropicalis hatching gene expression is induced earlier and begins to decrease sooner in comparison with the orthologs in X. laevis (Fig. 3c). Thus early hatching in X. tropicalis correlates with a concerted change in the expression of enzymes important for modulating its timing.
Aquatic vertebrates fine tune their hemoglobin levels and affinities to account for the temperature dependence of oxygen solubility. This is particularly well established for fish, where Antarctic fish can dispense with hemoglobin completely and temperate fish regulate different hemoglobin genes depending on water temperature (Ruud 1954; Borza, Stone et al. 2009). It was therefore not surprising that the oxygen-binding gene set showed widespread heterochronicity of expression (Fig. 3a). We have found five very clearly heterochronic hemoglobin genes (Fig. 3d). HBAL1, HBAL2, HBBL1, and HBBL2 are larval-specific alpha and beta hemoglobins, and their earlier expression in X. tropicalis suggests an adaptation to lower oxygen levels, requiring precocious delivery of oxygen rather than relying on diffusion from the ambient water. HBAL2 shows the opposite heterochrony in which X. laevis expression is precocious suggesting a unique adaptation here. To understand this completely we would have to know more about the oxygen saturation curves and physical features of the vascular bed. Another strong difference is in the expression of a globin gene, cytoglobin CYBG, which is involved in intra-cellular oxygen shuttling and which has been previously implicated in adaptation to hypoxia (Schmidt, Gerlach et al. 2004). In contrast to the hemoglobins, CYBG s is heterometric with an expression of 10-fold higher in X. tropicalis. This change may be one of the many adaptations to temperature.
Another especially clear example of changes in timing of expression involves a conserved group of developmental timing genes first discovered in C. elegans (Ambros 2003). This pathway includes two transcription factors, lin-28 and lin-41 and both show a coordinated shift in expression in our dataset. In X. laevis, both genes sharply increase in abundance by an order of magnitude after stage 10 compared to the level of maternal mRNA (Fig. S3d). In X. tropicalis these genes remain at levels close to maternal mRNA (Fig. 3d). The microRNA let-7 is a central member in this pathway in frog, fly, fish and worm (Kloosterman, Wienholds et al. 2004; Roush and Slack 2008). It is known that the let-7 ortholog is induced in the X. laevis gastrula/neurula (Watanabe, Takeda et al. 2005). Although miRNA expression is not queried in our dataset it seems likely that let-7 is shifted along with its regulator (lin-28) and target (lin-41). This finding is particularly interesting in light of previous reported conservation of let-7 temporal regulation is conserved across chordates (Pasquinelli, Reinhart et al. 2000).
Modeling expression of developmental genes reveals more heterometry than heterochrony
Although we discovered several clear examples of heterochronic switches, where whole pathways are moved forward and backward against the background of conserved developmental expression, such examples were relatively rare. This approach of examining the mean divergence of a gene set would be unlikely to identify situations where only a small subset of the genes in a particular pathway has shifted. We were interested in cataloguing all of the potential heterochronic (timing of transcriptional changes, Table S2) and heterometric (transcript abundance, Table S2) changes between the two developing frogs. To this end, we manually examined known developmental pathways involving ~400 genes, including HOX and other conserved transcription factors as well as common signaling pathways (e.g. TGF-beta, Hedgehog, Wnt), and found few cases of heterochrony (e.g. HOXB3, GLI3, Fig. 1b, and Table S2). We then turned to new statistical approaches to examine the remaining ~10,700 ortholog pairs for heterochrony and heterometry.
For the simplest patterns of gene expression, divergence may be distinguished as either heterochrony or heterometry. Of the 2,290 genes with large overall levels of divergence (EDi > 5), 67% have a correlation coefficient ≥0.6, suggesting that heterometry and not heterochrony is the dominant mode of divergence.
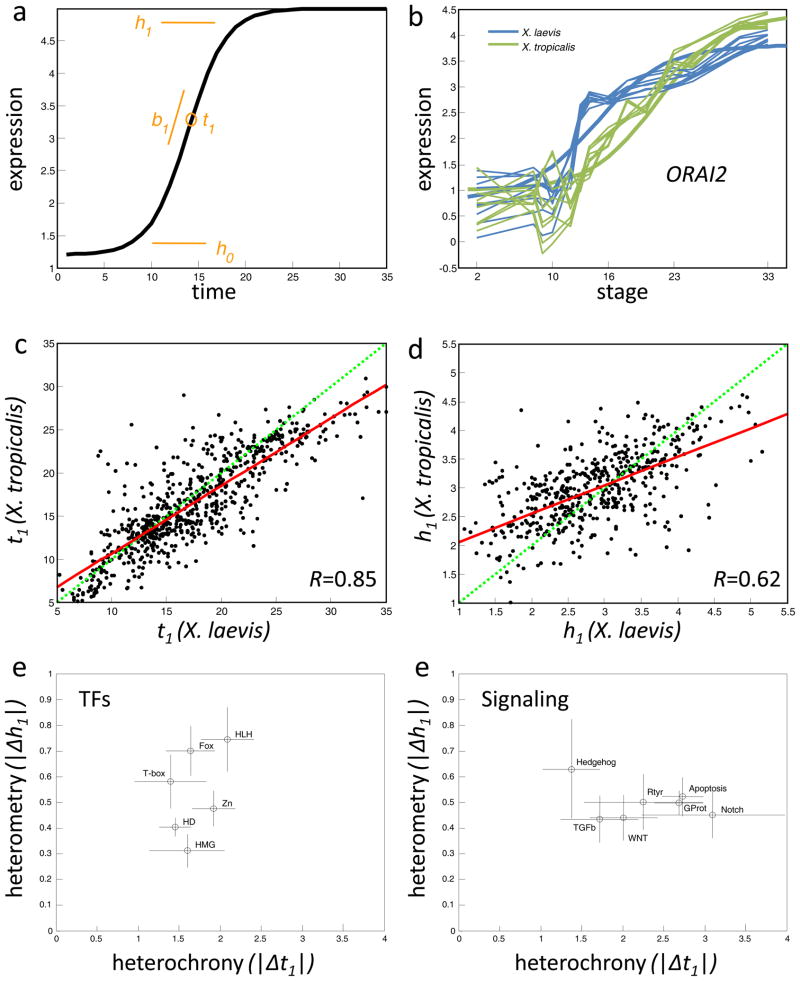
To gain further insight into the mode of divergences, we modeled expression profiles using sigmoid functions (see Experimental Procedures). A sigmoid curve is a well described function with four defining parameters corresponding to the slope (b1), time of half-rise (t1), level of initial expression (h0), and maximal or minimal expression (h1) (Fig. 4a). For 28% of the X. laevis and 21% of the X. tropicalis profiles we achieved an acceptable goodness of fit (≥0.8) with a sigmoidal function. Comparing the sigmoid parameters between orthologs allows us to compare specific features of the profiles, such as the levels of expression, the time of initiation of expression and the slope of the expression profiles. These enable an evolutionary comparison in terms of heterochrony or heterometry of individual genes. For example, the fitted sigmoids to the ORAI2 gene in both species reflect the observed heterochrony: a t1=21 in X. tropicalis compared to t1=15 in X. laevis, where 21 and 15 refer to stages (Fig. 4b). Plotting t1 pairings for 1,549 well-fitted orthologs we find a correlation of 0.85 (Fig. 4c), suggesting an overall conservation of developmental timing. Comparing the ranges of expression levels, h1, we found a weaker correlation (R=0.62), again suggesting a greater propensity of heterometries between the two species (Fig. 4d). This heterometry could not be generally attributed to differences in 3 UTR lengths influencing the preparation of the RNA samples (Fig. S4a). We conclude that most changes to gene expression between the two species involve heterometric changes rather than heterochronic changes. The trend also indicates that later X. laevis stages appear to be somewhat retarded relative to X. tropicalis. Notably, this advancement in X. tropicalis is not universal. The majority of values (65%) lie within a 5 hour time difference, and there are many genes advanced in X. laevis relative to X. tropicalis.
Figure 4. Heterochrony and heterometry in gene expression evolution.
a. A sigmoid is defined by b1, t1, h0, and h1 (see text). b. The ORAI2 gene expression profiles (log10 relative concentrations) in both X. laevis and X. tropicalis is shown fitted by sigmoids. c. Plots of t1 (time of induction) for pairs of genes with >0.8 goodness of fit in both species. The green line is unity and the red is fitted to the data. d. Plots of h1 (range of expression). Same format as c. e-f. Heterochrony/Heterometry phase-plane for families of transcription factors (e) and several signaling pathways (f). The circles and lines indicate the mean and standard deviation of each gene set s heterochronies and heterometries. The transcription factor families are helix-loop-helix (PF00010, 27 genes with sigmoids), Homeobox (IPR001356, 74 genes), Zinc finger (C2H2 type, IPR007087, 24 genes), T-box (IPR001699, 5 genes), Fox head (IPR001766, 16 genes), and HMG (IPR000910, 8 genes). The signaling pathways are Wnt receptor (GO:0016055, 18 genes with sigmoids), transforming growth factor beta receptor (GO:0007179, 13 genes), Hedgehog (GO:0007224, 6 genes), Transmembrane receptor protein tyrosine kinase (GO:0007169, 17 genes), Notch (GO:0007219, 9 genes), Apoptosis (GO:0006917, 40 genes), and G-protein coupled receptor protein (GO:0007186, 73 genes).
Do different groups of genes (representing pathways or molecular mechanisms) display biases in their t1 (heterochrony) and/or differences in h1 (heterometry)? We found that transcription factors as a group showed very little heterochrony while genes involved in signaling pathways show approximately three times more heterochrony (Fig. 4e); TFs showed a R=0.9 correlation in cross species t1 s, while R=0.79 for non-TFs. Of the transcription factors, those with the Homeodomain or HMG domain displayed the most constrained heterometry (h1 of 0.4 and 0.31, respectively), possibly reflecting the importance in timing in the patterning of the body plan relative to the morphological events used to stage the two species. On the other hand, genes with HLH and FOX domains – also critically important for development – are the most divergent in terms of heterometry (h1>0.7). With respect to signaling pathways, surprisingly some pathways – such as apoptosis, G-protein coupled receptor pathway, and receptor tyrosine kinases are considerably more heterochronic than the Hedgehog and TGF beta pathway genes (h1 of 3 relative to 1.5). Hedgehog signaling shows the greatest amount of heterometry among the signaling pathways (Fig. S4b). The observation that signaling pathways exhibit more heterochrony than TFs suggests that the same transcriptional programs may be triggered by different signals but at conserved stages. The reason TFs show a different heterochrony pattern than signaling pathways may be rooted in there simply being more transcription factors than types of signaling pathways; if pathways are collapsed by their common intracellular features, for example all MAP kinase pathways and GCPRs (Gerhart 1999).
The transcriptomes of X. laevis and X. tropicalis become increasingly similar as development proceeds
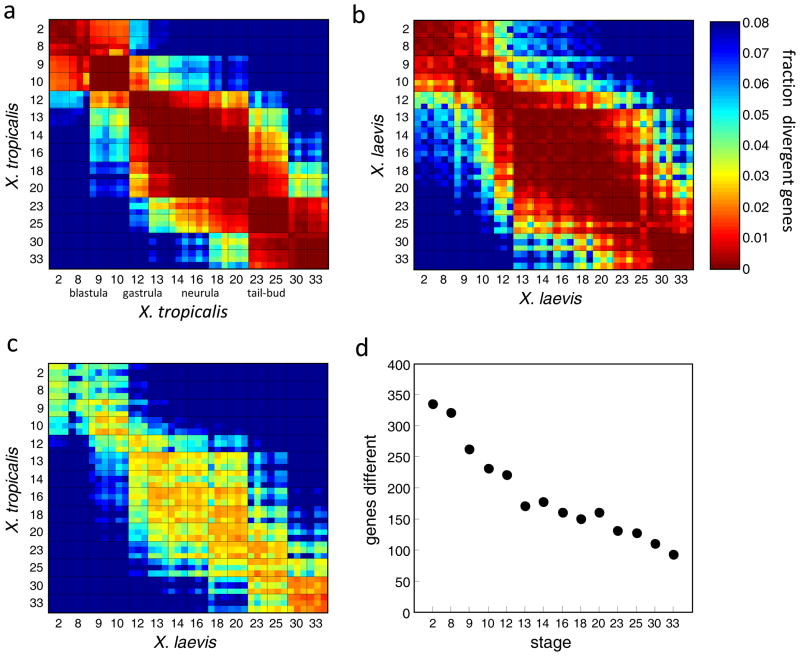
To examine features of conservation in the transcriptomic data at a global level we computed the number of genes with substantially different expression levels between pairs of transcriptomes (>1.5 log10 units). Figure 5a shows a matrix of distances between all X. tropicalis transcriptomes. By this means, we asked, for example, how many genes are differentially expressed between the blastula and neurula stages. Rather than a smooth continuum changing with developmental time, we observed striking clusters of correlated transcriptomic domains both within and between the two species (Fig. 5a–c). These clusters appear to define distinct transcriptional states: the maternal (st.2-8), blastula/early gastrula (st.9-10), late gastrula (st.12-13), neurula (st.14-20), and tailbud (st.23-33). The first transcriptomic domain is the maternal transcriptome shared by the 2-cell stage and the later blastula stages (st.8,9). In both species, there is a marked change after stage 8, almost certainly reflecting the mid-blastula transition during which the zygotic transcriptome becomes fully induced and many maternal RNAs are degraded (Fig. 5a–c). The major morphological changes undertaken by the developing embryo led early developmental biologists to roughly divide frog development into periods of blastocoel formation, gastrulation, neurulation, and organogenesis. These major morphological transitions appear to be mirrored by global changes in the transcriptome.
Figure 5. Global comparison of the X. laevis and X. tropicalis developmental transcriptomes.
a-c. The heat maps represent the fraction of genes significantly different between pairs of transcriptomes; the grid separates the replicates across the stages. The color of each square indicates the difference at the specified developmental stages. a and b represent X. tropicalis and X. laevis plotted against themselves. c. represent X. tropicalis plotted against X. laevis. The diagonal squares by definition have zero divergence. The fraction is computed as the number of genes with a difference of at least 1.5 log10 units out of the number of genes with a maximum expression of at least 2.5 log10 units in either transcriptomes. d. For 2,297 dynamically expressed genes, the plot indicates the number of genes with significantly different transcript abundance (>1.5 log10).
Are certain developmental stages more conserved than others? To address this question, we computed the fraction of differentially expressed genes between the transcriptomes from the same stage in the two frogs (e.g. the diagonal line in Figure 5c). Notably, the fraction of differentially expressed genes steadily decreased during development (Fig. 5c). About 4% of the paired orthologs in early transcriptomes are differentially expressed in the earliest stages, while only ~1.5% of the paired orthologs different in later stages (Fig. 5c). This trend was even more apparent when examining only a group of dynamically expressed genes (see Experimental Procedures): nearly four times more gene differences were detected in the first examined stage relative to the last (Fig. 5d). Average gene expression levels increase with developmental time (Table 1); nevertheless, we found the same relationship in maternally expressed genes in which expression levels decreased with time (Fig. S5a). Furthermore, enrichment for differences in early stages was also apparent among the different clutches in either species (Fig. S5b-c). From these analyses we conclude that different stages of development have unique levels of gene expression conservations between the two species.
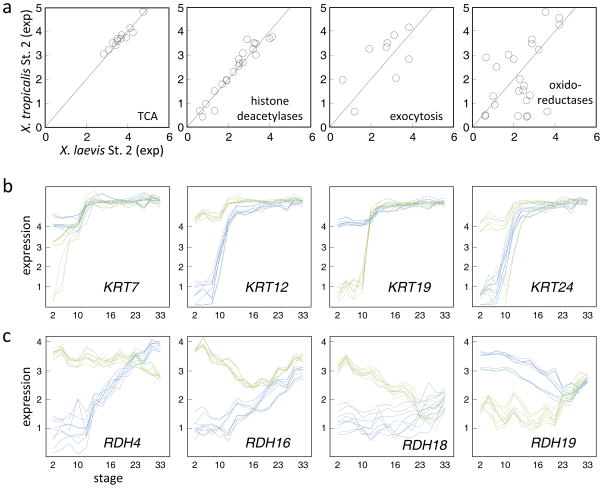
Very little transcription occurs in the embryo until the mid-blastula transition during stage 8 and consequently mRNA detected in stage 2 embryos mostly represents the maternal deposit. We observe a correlation of R=0.86 between the stage 2 mRNA levels across all examined genes between the species. Overall, 4,759 X. laevis and 5,260 X. tropicalis genes are detected at this stage above a threshold of 3 log10 relative concentration units (Table 1). Of these, 62 X. laevis and 52 X. tropicalis genes are not detected above a minimal threshold (1.5) in the other species. There may be some relationship between function and species differences. For examples, maternal genes involved in the TCA cycle and histone deacetylation were highly correlated between species (Fig. 6a) as compared to other pathways (Table S5). Conversely, genes involved in oxidoreductase and exocytosis were more divergent in their maternal contribution (Table S6). Keratin genes are an intriguing case, showing a sharp difference in the maternal transcripts of X. laevis versus X. tropicalis (Fig. 6b). While some keratin transcripts are deposited below detectable levels in one of the species but then subsequently increased by 4–5 orders of magnitude in abundance around gastrulation, in the other species the level remains nearly constant throughout the duration of experiment. Another striking example of expression difference in maternal deposit that is later compensated in development is the gene class of retinol dehydrogenases that catalyze the retinol/retinal exchange and play important role in cell differentiation (Fig. 6c). As further examples, Table S2 provides the expression profiles of 101 orthologs with convergent patterns.
Figure 6. Comparison of the maternal transcriptomes of X. laevis and X. tropicalis.
a. For each of four functional gene sets, the plot compares the expression levels (log10 relative concentrations) in stage 2 embryos between the two species. b. Comparative gene expression profiles of four keratin genes. These genes are less than 80% identical at the protein level allowing good resolution by the microarray data. KRT24 in X. tropicalis exhibits two patterns depending upon the probes examined: one with maternal expression and another with a profile heterochronic to the X. laevis ortholog. c. Comparative gene expression profiles of four retinol dehydrogenases.
DISCUSSION
Here we have compared two transcriptomes of amphibian species with very similar embryonic development, adult and larval morphologies and yet separated by over ~30–90 million years of independent evolution. Although similar, the embryos of the two frogs are clearly not identical, being marked by differences in overall size and sensitivities to environmental factors such as temperature. Using quantitative measurements of the timing and level of gene expression, we asked whether the extensive evolutionary genetic differences would dictate a large variation in the patterns of gene expression. Such an extensive comparison has not been made before in vertebrates, with the exception of a smaller study of early blastula in human, mouse and cow (Xie, Chen et al. 2010). Arguing for divergence is the evidence from nematodes of significant changes in expression patterns for morphologically similar species (Yanai and Hunter 2009). Furthermore in Drosophila species there is evidence for considerable conservation in the transcriptome in conserved developmental circuits and gratuitous and variable changes in expression in non-essential genes (Rifkin, Kim et al. 2003). Our overall conclusion from the X. laevis - X. tropicalis comparisons is that the timing of expression is very similar for most genes. Against this backdrop of conservation there is clear evidence for heterochrony in a few genes and more substantial variation in the level of expression of a much larger number of genes. Furthermore within each species, there is unexpected variation in the maternal storehouse of genes with differing levels of maternal transcripts.
We were interested in the nature of expression changes and their prevalence in different functional groups of genes. In particular, we were interested in changes in timing and level of gene expression, discussed in terms of heterochrony and heterometry. Heterochrony has long been suggested as an important source of phenotypic change in evolution (Huxley 1932; Gould 1977). We have found that heterochrony can be appreciated in genome-wide expression data by using sigmoid functions, which report the time, rate, and extent of transcription induction or repression. The movement en bloc of genes for innate immunity, cilia and the heterochronic genes let-28 and let-41 were very striking and enriched for differential expression between the two frogs, as reported by our divergence score EDi. However a much larger number of heterochronic or heterometric genes were discovered through the use of automated sigmoid fitting and the extraction of the parameters. These data suggest that gene expression heterochrony (where there has been a shift of more than two developmental stages) could be a bona fide path for evolutionary change. On the other hand, the apparent rareness of this process, even when using the more sensitive sigmoid function, suggests that developmental timing of expression of individual genes is generally under strong stabilizing selection.
Much more variable than heterochrony is the level of expression, or heterometry. Our use of a series of spike-in controls and identical or nearly identical probes between the two species allowed accurate quantitative comparisons between species. Though transcription factor classes had rather uniform and low heterochrony, the variation of heterometry was high. Overall, among the genes analyzed by sigmoid plots the level of expression was greater than 1 log10 unit for 9% of the genes and was more common in certain signaling pathways and transcription factor families (Fig. 4e,f). There are several reasons why heterometry might be more rapidly diverging than heterochrony. There are probably more ways to change the level of gene expression than to change the timing of gene expression. The former could be achieved on many levels without changing the complement of transcription factors binding to the promoter. The latter generally requires a different composition of transcription factors or a change in the timing of expression of the regulators. In addition, various posttranscriptional regulatory features could blunt the effects of the more rapidly evolving heterometry, including RNA splicing and RNA stability, translation efficiency, stability of the protein, protein modification, and various inhibitors of protein activity. Thus while the data presented here suggest strong stabilizing selection on timing of expression of most genes, the larger variation in levels could be moderated by other forms of protein regulation, allowing more drift in transcription, hence greater incidence of heterometry.
We found a strong inverse correlation between expression variation of orthologs and developmental time (Figs. 5,6). These data are consistent with the notion that the earliest stages of development are subject to rapid evolutionary change (Raff 1996; Gerhart and Kirschner 1997). Another feature of the variability in the egg was the clutch to clutch variability of certain transcripts within each of the frog species. Significantly, this clutch to clutch variation in the RNA levels strongly decreased after the mid-blastula transcription, suggesting that most of the maternal RNA variation is likely irrelevant to embryogenesis (Fig. S5b-c). We note that our observation of large differences between maternal transcriptomes is at odds with the results of a recent work comparing the preimplantation embryonic development in human, mouse, and cow in which it was observed that the mRNA pool deposited by the mother is more conserved than the zygotic activation gene set in the first few cell cycles (Xie, Chen et al. 2010). Yolk utilization starts after gastrulation and most of it is very late (Jorgensen, Steen et al. 2009). It is difficult to see why yolk differences would affect the distribution of transcripts between the egg and gastrula stages. It is also not clear that relative to volume that there is much difference in yolk content in X. laevis and X. tropicalis.
Differences in the level or timing of gene expression were concentrated in the earliest embryonic stages particularly the maternal RNA dowry – and these differences decline during embryogenesis. Our maternal and zygotic RNA comparisons could be consistent with an hour-glass view of the evolution of development (Raff 1996; Domazet-Loso and Tautz 2010; Kalinka, Varga et al. 2010), which posits that embryos within phyla are most similar at an intermediate stage of development. This phylotypic stage in vertebrates has been suggested to be the pharyngula stage, when embryos from fish, frogs, and mammals show axial segmentation around a notochord, pharyngeal (branchial) arches, and a post-anal tail. We propose that gene expression programs involved in the setting up of developmental domains are freer to vary; later stages may be more constrained due to the stabilization of cell fate determination pathways. The different evolutionary rates across developmental stages may be explained by buffering in the earlier stages. Our latest time point is stage 33, which finds the frog embryos at or just past the pharyngula stage and still very far from the fully-formed tadpole. It is therefore not possible to tell from our data if the tailbud stage is the low point of transcriptional variation between the two species. It should be noted that since the number of cells and the tissues specified increases with time, and the total number of expressed transcripts increases with time, an alternative explanation for the increase in expression conservation over time is an averaging of expression over multiple tissues.
This approach to comparative transcriptomics has some limitations. The temporal gene expression data does not reveal the spatial location of the expression within the embryo. However, changes in expression levels (heterometries) could suggest differences in the size of expression fields (allometries). Further we do not address gene expression changes among gene duplicates. 260 of the orthologs sets included duplicates in X. tropicalis and in X. laevis this problem is worse, although not quantifiable due to the lack of a completed genome, since many duplicates were retained following the genome duplication (Hellsten, Khokha et al. 2007). Since the profiles of duplicates are difficult to resolve with microarrays due to their similar sequence, we selected probes for conserved sequence regions in an attempt to collapse the expression patterns of duplicates (Supplementary Methods). Despite these efforts, expression conservation may disguise a subfunctionalization of expression between duplicates (Hellsten, Khokha et al. 2007). Next-generation sequencing may now be invoked to further resolve the evolution of gene expression following gene duplication.
Our study shows that there are maternal, zygotic and post-zygotic transcriptomes that have evolved somewhat independently against a background of evolutionary stability. In rapidly diverging species, these changes might have been obscured by the greater amount of selected variation. Furthermore, this analysis generated analytical and computational tools that may prove useful in other evodevomic studies. The browser we have provided will allow others to query the data for coexpressed genes, to correlate the timing of processes with gene expression, to explore further evolutionary divergence of transcriptomes among other clades of amphibians and perhaps further afield, and for workers with Xenopus laevis and X. tropicalis to know whether one can assume the identity of the transcriptional control processes in these two valuable laboratory models.
EXPERIMENTAL PROCEDURES
Microarray design
We designed two organism-specific microarrays which were then manufactured by Agilent as 44k arrays (4x44k). The guiding principle for the array design was for each cluster of X. tropicalis and X. laevis orthologs to be associated with three probes, each corresponding to homologous sequence regions. The probes on either organism-specific microarray are adjusted to the specific sequence of the respective genome (Fig. S1b,c). The details of designing the microarrays – which in general included clustering the available sequences and then identifying appropriate probes – are described in the Supplementary Methods.
Embryo collection
Standard Xenopus techniques were followed to induce ovulation , perform in vitro fertilization, and dejelly the eggs (Sive, Grainger et al. 2000). For the embryo collection of both X. tropicalis and X. laevis, the sperm of four individual males was used to individually fertilize clutches of eggs obtained from three females. These 12 clutches were then incubated at 22°C in the case of X. laevis and 28°C in the case of X. tropicalis, to provide each species with the optimal temperature for development (see Supplementary Materials). Embryos were staged according to Nieuwkoop and Faber (Nieuwkoop and Faber 1994) and 3 embryos were collected from each clutch at 14 developmental stages spanning the blastula (stages 2, 8, 9), gastrula (stages 10, 12, and 13), neurula (stages 14, 16, 18, and 20), and tailbud stages (stages 23, 25, 30 and 33). After collection, the three clutches with the highest rates of survival and normal development were selected for microarray analysis.
Sample preparation and microarrays
RNA was extracted from eggs using the RNaqueous kit (Ambion). In the case of X. laevis each sample comprised a single egg while in X. tropicalis three eggs were combined to generate RNA amounts comparable to X. laevis levels. 1μg of total RNA was amplified by one round of in vitro transcription (IVT) using the MessageAmpII kit (Ambion). We also added to this reaction RNA from a Spike-in kit (Agilent) that included 10 different species of RNA at various concentrations. 7μg of RNA was labeled with Cy-3 and 1.65μg of labeled aRNA was incubated on each microarray. Each embryonic stage was measured in biological triplicates (independent clutches). The hybridization and washing of the microarrays were performed following the Agilent protocol for single-channel arrays. Software provided by Agilent was used to extract the data. The log10 expression data was normalized by linearly interpolating to concentrations using the spike-in measurements for each sample. To summarize the replicate data for each gene, the mean for the nine values at each time points was computed. A gene was defined as having a dynamic expression profile if the range in levels throughout the stages ≥1.5 and the maximum level is ≥3. This set was used in all analyses requiring the set of dynamic genes.
Expression divergence index (EDi)
For each of the 9 X. tropicalis measurements (3 probes, 3 clutches) at each stage, we calculated the distance to each of the corresponding 9 X. laevis measurements. For each stage we also calculated the within-species distances among the 9 measurements. For a given gene, at a given stage, the EDi is the mean inter-species distance subtracted by the mean of the two mean intra-species distances. A gene s overall EDi is the sum of the EDi s of all stages.
Functional gene set analysis
Since there are no systematically collected gene sets for frogs, we used gene sets defined in other species. Thousands of gene sets representing known pathways, targets of particular transcription factors and micro-RNAs, structural models, cellular localization, etc. are available from the Broad Institute as part of the Gene Set Enrichment Analysis platform (Subramanian, Tamayo et al. 2005). Some gene sets are conserved across species (e.g. ribosomal protein encoding genes) while others are not (e.g. genes grouped by chromosome coordinates or by the tissue-specific expression). For each gene set we queried for a significant deviation from the expected divergence by comparing the EDi for that set with the EDi s for all genes using the Kolmogoron-Smirnov test.
Sigmoids
A sigmoid of the form , was fit to each gene s expression data. The data was in the form of 9 profiles (three probes, three clutches) across 14 stages. Stage was converted to time in hours past fertilization according to the X. laevis normal table. The data was pre-processed by aligning the profiles across the different probes in order to compensate for any variable probe specificity. The sigmoid was fit using Matlab s Curve Fitting Toolbox. Specifically, the Nonlinear Least Squares method was invoked with default optimization parameters twice for each gene: once to fit the best ascending sigmoid and once for the best descending sigmoid, with the corresponding limits to the search space set to the minimal and maximal expression value. The goodness of fit was measured by the coefficient of determination adjusted to the degrees of freedom and scaling from zero to one, with one corresponding to the perfect fit.
Supplementary Material
Acknowledgments
We thank Bob Freeman and Brian Frederick. IY acknowledges support from the Lorry I. Lokey Center for Life Sciences and Engineering. We thank Rob Grainger for providing adult male and female X. tropicalis. We thank Virginia Savova, Alexander Gimelbrant, and Martin Wuehr for a critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113(6):673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Arthur W. The concept of developmental reprogramming and the quest for an inclusive theory of evolutionary mechanisms. Evol Dev. 2000;2(1):49–57. doi: 10.1046/j.1525-142x.2000.00028.x. [DOI] [PubMed] [Google Scholar]
- Baldessari D, Shin Y, et al. Global gene expression profiling and cluster analysis in Xenopus laevis. Mech Dev. 2005;122(3):441–75. doi: 10.1016/j.mod.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010;11(1):17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- Bisbee CA, Baker MA, et al. Albumin phylogeny for clawed frogs (Xenopus) Science. 1977;195(4280):785–7. doi: 10.1126/science.65013. [DOI] [PubMed] [Google Scholar]
- Blekhman R, Oshlack A, et al. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 2008;4(11):e1000271. doi: 10.1371/journal.pgen.1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borza T, Stone C, et al. Atlantic cod (Gadus morhua) hemoglobin genes: multiplicity and polymorphism. BMC Genet. 2009;10:51. doi: 10.1186/1471-2156-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46(2):111–38. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- Carroll EJ, Jr, Hedrick JL. Hatching in the toad Xenopus laevis: morphological events and evidence for a hatching enzyme. Dev Biol. 1974;38(1):1–13. doi: 10.1016/0012-1606(74)90254-1. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134(1):25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, et al. From DNA to diversity: molecular genetics and the evolution of animal design. Malden, Mass: Blackwell Science; 2001. [Google Scholar]
- Cole DS, Morgan BP. Beyond lysis: how complement influences cell fate. Clin Sci (Lond) 2003;104(5):455–66. doi: 10.1042/CS20020362. [DOI] [PubMed] [Google Scholar]
- Davidson E, Levin M. Gene regulatory networks. Proc Natl Acad Sci U S A. 2005;102(14):4935. doi: 10.1073/pnas.0502024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature. 2010;468(7325):815–8. doi: 10.1038/nature09632. [DOI] [PubMed] [Google Scholar]
- Eroshkin FM, Ermakova GV, et al. Multiple noggins in vertebrate genome: cloning and expression of noggin2 and noggin4 in Xenopus laevis. Gene Expr Patterns. 2006;6(2):180–6. doi: 10.1016/j.modgep.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Kelley DB, et al. A mitochondrial DNA phylogeny of African clawed frogs: phylogeography and implications for polyploid evolution. Mol Phylogenet Evol. 2004;33(1):197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Fletcher RB, Baker JC, et al. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development. 2006;133(9):1703–14. doi: 10.1242/dev.02342. [DOI] [PubMed] [Google Scholar]
- Fletcher RB, Watson AL, et al. Expression of Xenopus tropicalis noggin1 and noggin2 in early development: two noggin genes in a tetrapod. Gene Expr Patterns. 2004;5(2):225–30. doi: 10.1016/j.modgep.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Gerhart J. 1998 Warkany lecture: signaling pathways in development. Teratology. 1999;60(4):226–39. doi: 10.1002/(SICI)1096-9926(199910)60:4<226::AID-TERA7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Kirschner M. Cells, embryos, and evolution : toward a cellular and developmental understanding of phenotypic variation and evolutionary adaptability. Malden, Mass: Blackwell Science; 1997. [Google Scholar]
- Gould SJ. Ontogeny and phylogeny. Cambridge, Mass: Belknap Press of Harvard University Press; 1977. [Google Scholar]
- Graur D, Li W-H. Fundamentals of molecular evolution. Sunderland, Mass: Sinauer; 2000. [Google Scholar]
- Hellsten U, Harland RM, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328(5978):633–6. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Khokha MK, et al. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biol. 2007;5:31. doi: 10.1186/1741-7007-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig W. Phylogenetic systematics. Urbana: University of Illinois Press; 1979. [Google Scholar]
- Heyer LJ, Kruglyak S, et al. Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 1999;9(11):1106–15. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley J. Problems of relative growth. London: Methuen; 1932. [Google Scholar]
- Jorgensen P, Steen JA, et al. The mechanism and pattern of yolk consumption provide insight into embryonic nutrition in Xenopus. Development. 2009;136(9):1539–48. doi: 10.1242/dev.032425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinka AT, Varga KM, et al. Gene expression divergence recapitulates the developmental hourglass model. Nature. 2010;468(7325):811–4. doi: 10.1038/nature09634. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, et al. Evolution of primate gene expression. Nat Rev Genet. 2006;7(9):693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Weiss G, et al. A neutral model of transcriptome evolution. PLoS Biol. 2004;2(5):E132. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188(4184):107–16. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, et al. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32(21):6284–91. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knochel W, Korge E, et al. Globin evolution in the genus Xenopus: comparative analysis of cDNAs coding for adult globin polypeptides of Xenopus borealis and Xenopus tropicalis. J Mol Evol. 1986;23(3):211–23. doi: 10.1007/BF02115578. [DOI] [PubMed] [Google Scholar]
- Mayr E. Animal species and evolution. Cambridge: Belknap Press of Harvard University Press; 1963. [Google Scholar]
- McLin VA, Hu CH, et al. Expression of complement components coincides with early patterning and organogenesis in Xenopus laevis. Int J Dev Biol. 2008;52(8):1123–33. doi: 10.1387/ijdb.072465v. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) : a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. New York: Garland Pub; 1994. [Google Scholar]
- Nolte AW, Renaut S, et al. Divergence in gene regulation at young life history stages of whitefish (Coregonus sp.) and the emergence of genomic isolation. BMC Evol Biol. 2009;9:59. doi: 10.1186/1471-2148-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Raff RA. The shape of life : genes, development, and the evolution of animal form. Chicago: University of Chicago Press; 1996. [Google Scholar]
- Renaut S, Nolte AW, et al. Gene expression divergence and hybrid misexpression between lake whitefish species pairs (Coregonus spp. Salmonidae) Mol Biol Evol. 2009;26(4):925–36. doi: 10.1093/molbev/msp017. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, et al. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet. 2003;33(2):138–44. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Ruud JT. Vertebrates without erythrocytes and blood pigment. Nature. 1954;173(4410):848–50. doi: 10.1038/173848a0. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328(5981):1036–40. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Gerlach F, et al. Cytoglobin is a respiratory protein in connective tissue and neurons, which is up-regulated by hypoxia. J Biol Chem. 2004;279(9):8063–9. doi: 10.1074/jbc.M310540200. [DOI] [PubMed] [Google Scholar]
- Showell C, Conlon FL. Natural mating and tadpole husbandry in the western clawed frog Xenopus tropicalis. Cold Spring Harb Protoc. 2009;2009(9):pdb prot5292. doi: 10.1101/pdb.prot5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, et al. Early development of Xenopus laevis : a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Staubach F, Teschke M, et al. A test of the neutral model of expression change in natural populations of house mouse subspecies. Evolution. 2010;64(2):549–60. doi: 10.1111/j.1558-5646.2009.00818.x. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, et al. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, et al. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579(2):318–24. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- Xie D, Chen CC, et al. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 2010;20(6):804–15. doi: 10.1101/gr.100594.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Graur D, et al. Incongruent expression profiles between human and mouse orthologous genes suggest widespread neutral evolution of transcription control. Omics. 2004;8(1):15–24. doi: 10.1089/153623104773547462. [DOI] [PubMed] [Google Scholar]
- Yanai I, Hunter CP. Comparison of diverse developmental transcriptomes reveals that coexpression of gene neighbors is not evolutionarily conserved. Genome Res. 2009;19(12):2214–20. doi: 10.1101/gr.093815.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Korbel JO, et al. Similar gene expression profiles do not imply similar tissue functions. Trends Genet. 2006;22(3):132–8. doi: 10.1016/j.tig.2006.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.