Abstract
Metals are key cofactors for many proteins, yet quantifying the metals bound to specific proteins is a persistent challenge in vivo. We have developed a rapid and sensitive method using electrospray ionization mass spectrometry to measure Cu,Zn superoxide dismutase (SOD1) directly from the spinal cord of SOD1-overexpressing transgenic rats. Metal dyshomeostasis has been implicated in motor neuron death in amyotrophic lateral sclerosis (ALS). Using the assay, SOD1 was directly measured from 100 μg of spinal cord, allowing for anatomical quantitation of apo, metal-deficient, and holo SOD1. SOD1 was bound on a C4 ZipTip® that served as a disposable column, removing interference by physiological salts and lipids. SOD1 was eluted with 30% acetonitrile plus 100 μM formic acid to provide sufficient hydrogen ions to ionize the protein without dislodging metals. SOD1 was quantified by including bovine SOD1 as an internal standard. SOD1 could be measured in subpicomole amounts and resolved to within two Daltons of the predicted parent mass. The methods can be adapted to quantify modifications to other proteins in vivo that can be resolved by mass spectrometry.
Keywords: Metals, Superoxide dismutase, Amyotrophic Lateral Sclerosis, Mass Spectrometry, Copper, Zinc
INTRODUCTION
Approximately one-third of all proteins are predicted to require a metal cofactor for activity [1] and as many as half of all the structures deposited in theProtein Data Bankcontain a metal ion [2]. Metal ion binding to proteins is critically important for many biological processes [3],but characterizing the metal ions boundto a protein in a biological context is difficult. Spectroscopic techniques, such as atomic absorption, electron paramagnetic resonance, and Mössbauer reveal metal coordination by proteins, but are insensitive and require purified protein in high concentrations[4]. Inductively-coupled plasma mass spectrometry and assays utilizing colorimetric chelators informon the total metal content in the sample, thus conveying only average metal ion stoichiometry[5; 6]. Electrospray ionization mass spectrometry (ESI-MS)3 is a well-established technique used in the study of non-covalent modifications and interactions of proteins such as metal binding[7; 8; 9; 10]. Once the ionization parameters are properly optimized, metal ionssuch as zinc and copper remain tightly associated with proteins during electrospray ionization and ESI-MS has been used to compare both relative and absolute metal ion affinities of many proteins[11; 12].
The activity of Cu,Zn superoxide dismutase (SOD1) is vitally dependent upon itscopper and zinc cofactors[13].The redox-active copper is the center of the protein's activity. SOD1 also contains a structural zinc atom closely coordinated to the copper that is essential for the proper folding of the protein[14]. Evaluating the metals bound to SOD1 is also important because mutations to the protein cause a toxic-gain-of-function that is responsible for 2-7% of all cases of amyotrophic lateral sclerosis (ALS) [15; 16]. ALS is a fatal neurodegenerative disorder characterized by the progressive death of motor neurons. However, the mechanism of the toxic function in vivo is still controversial. Most of the mutant SOD1 proteins bind one copper and one zinc ion per subunit and many retain almost the same level of enzymatic activity as the wild-type protein[17; 18; 19]. However, the mutant SOD1 proteinsseem to be more susceptible tothe loss of the zincion than the wild-type SOD1protein in vitro [20; 21; 22]. We have previously shown that purified zinc-deficient SOD1 delivered to cultured motor neurons is toxic [23]. Furthermore, endogenous expression of mutant SOD1 in motor neurons isolated from SOD1-transgenic animals kills by the same mechanism[24]. Consequently, the ability to assess the amounts of copper and zinc bound to SOD1 in the specific spinal cord regions affected by ALS may provide clues as to the nature of the toxic function in vivo.
Here we introduce a sensitiveESI-MS based method that allows the quantification of SOD1 directly from spinal cord tissue of transgenic ALS-model rats and concurrently reports on the zinc and copper ion content of the protein. We demonstrate the efficacy of the method for SOD1 from transgenic spinal cord tissue, although itcan be applied to other proteins in complex mixtures.
MATERIALS AND METHODS
Reagents and Equipment
Lyophilized bovine SOD1 (EC1.15.1.1, Sigma-Aldrich, St. Louis, MO, USA) was rehydrated with Milli-Q water (reverse-osmosis system), and the concentration was determined spectrophotometrically at 259 nm (ε= 5150 M-1cm-1for the Cu,Zn containing monomer)[13].
Two types of human SOD1 were used in the development of this assay. The first type was recombinant SOD1 expressed in Escherichia coli(recombinant SOD1) with coexpression of the copper chaperone for SOD1 (CCS).The protein was purified as described in Hayward et al.[25]. Ammonium sulfate was used in high concentrations for hydrophobic interaction chromatography, causing a sulfate adduct (+98 Da) to be often present in mass spectrometry samples. The concentration was determined spectrophotometrically at 265 nm (ε= 9200 M-1cm-1 for the Cu,Zn containing monomer)[13]. RecombinantSOD1was found to have the amino-terminal methionine cleaved and was not acetylated[26]. The second type was SOD1 from tissue samples of mutant G93A SOD1-overexpressing transgenic rats (referred to as tissue SOD1). These ratsdevelop fatal motor neuron disease and die around 120 days old[27]. In eukaryotic sources, SOD1 is acetylated on the amino-terminus, increasing the protein mass by 42.1 Da.
The solvent used for the mass spectrometric experiments was 30% acetonitrile, 70% water and 100 μM formic acid, delivered at a flow rate of 30 μL/min from a single isocratic pump (Shimadzu, Kyoto, Japan). The solvent was degassed for 10 minutes with helium and was changed each week. HPLC-grade solvents were used for all mass spectrometer experiments. Water was obtained from Burdick and Jackson (Muskegon, MI, USA) and 10% formic acid from MichromBioresources (Auburn, CA, USA). Universal grade acetonitrile was obtained from VWR (West Chester, PA, USA).
SOD1 was introduced into the mass spectrometer via one of two methods: by direct injection ofSOD1 in low ionic strength aqueous buffers, or by binding to a C4 reversed phase Ziptip® for tissue samples or higher ionic strength solutions as described below. For direct injection, solvent was pumped from the HPLC pump through PEEK® tubing with an interior diameter of 0.005 inches to a 2-way, 6-port Rheodyne injection port. The SOD1 was introduced into the sample loop of the injector via a 50 μL Hamilton syringe with a blunt tip.Solvent was then pumped through the loop and across a 1 μm inline stainless steel microfilter (Upchurch Scientific, Oak Harbor, WA, USA) to prevent clogging. The 0.005 inch inner diameter PEEK® tubing led from the microfilter into the mass spectrometer inlet.
Ziptip Method
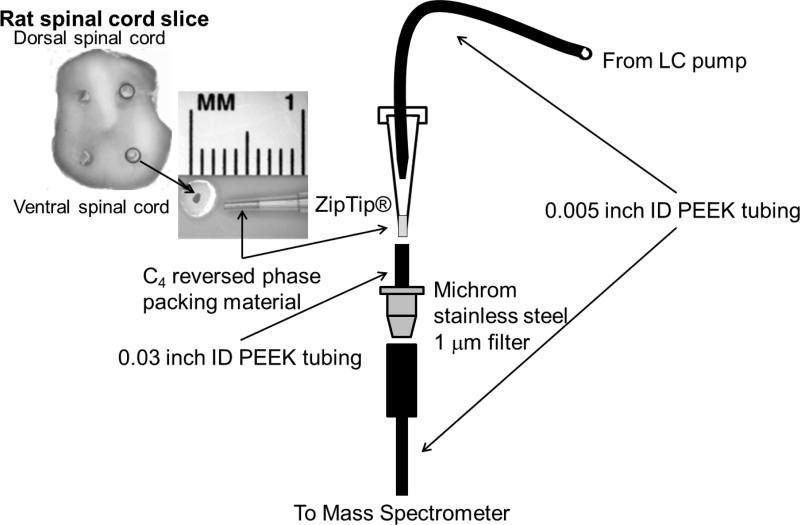
A C4Ziptip (Millipore, Billerica, MA, USA) was used as a disposable desaltingcolumn while binding SOD1. The Ziptip is a P10 pipette tip with 0.6 μL of reversed phase C4 packing material that is typically used to remove salts from peptide samples in preparation for mass spectrometry detection. For the assay, a Ziptip was prepared by rinsing three times (10 μL each) with acetonitrile, followed by four washes with water. The washes were accomplished by using a P10 pipette to repeatedly draw 10 μL of fresh solvent up through the Ziptip, and then to expelthe solvent. The SOD1 sample was then bound to the Ziptip by pipetting the protein solution three times, followed by rinsing six times with water to remove salts. The Ziptip was then inserted between the injection port and the microfilter(Figure 1).
Figure 1. Tissue preparation for Ziptip® assay with mass spectrometer interface.
A 500 micron biopsy punch is taken from a 1 mm spinal cord slice, thawed in buffer and adsorbed onto a Ziptip. Solvent from an HPLC pump flowed through 0.005 inch interior diameter PEEK tubing that was inserted snugly into the back of the Ziptip. The end of the Ziptip was inserted into a second short segment of PEEK tubing with an interior diameter of 0.03 inches that had been slightly expanded with a dissecting needleto fit the end of theZiptip snuggly. Astainless steel1 μm microfilterwas inserted between the Ziptip and the mass spectrometer inlet to trap large particulates released from the Ziptip.
Tissue Measurements
Fresh spinal cord tissues from transgenic G93A-SOD1+/- overexpressingrats housed in Institutional Animal Care and Use Committee-approved facilities were extracted via dissection of euthanized animals and frozen at -80 °C until use. The tissue was too brittle to section immediately at this temperature, and was placed on an insulating plastic plate on top of dry ice and allowed to warm to approximately -20 °C. Punches were taken from 1 mm thick transverse sections of tissueusing a 500 micron biopsy punch (Figure 1). An air-filled syringe was attached to the rearof the punch to expel the tissue (Zivic Instruments, Pittsburgh, PA, USA). The punches were weighed on a Cahn 25 Automatic Electrobalance with a sensitivity of 0.1 μg (Cerritos, CA, USA). SOD1 was released from the tissue punch by thawing the tissue in 5μL of 10 mM ammonium acetate,adjusted with acetic acid to pH 5.1,that included 200 nM bovine SOD1 (punch thawing buffer). The pH was chosen to reduce adventitious metal bindingandbecause interfering lipoproteins from central nervous system tissues precipitated, thereby preventing clogging of the tubing[28]. The thawed tissue was vortexed at maximum speed for 30 seconds then centrifuged for two minutes at 16,000 xg. The supernatant was adsorbed onto the Ziptip, eluted and analyzed in the mass spectrometer.
The amount of SOD1 released from tissue was verified by western blot analysis. The supernatants from three punches were combined to provide enough sample per lane for the western blot. The punches were thawed in 15 μL of 10 mM ammonium acetate at pH 5.1, vortexed and centrifuged as above. The tissue supernatant and pellet fractions were separated, the volume of each was adjusted to 30 μL withLaemmli buffer[29], and the samples were heated for five min at 95 °C. Samples were then run on a 10% SDS-PAGE at 150V for 60 min in a Bio-Rad Mini Protean II system. Gels were transferred to apolyvinylidene fluoride membrane(Bio-Rad product # 162-0177) for 60 min at 100V. SOD1 was detectedusing rabbit polyclonal antibodies raisedin the laboratory against human SOD1 (1:10,000).Affinity-purified antibodiesretained cross reactivity with mouse and rat SOD1. The secondary antibody was goat anti-rabbit IgGheavy plus light chain-horse radish peroxidase conjugate (Bio-Rad #170-6515).
Data Analysis
Proteins were detected using a LCT Premier Time-of-Flight mass spectrometer (Waters, Milford, MA, USA) with an electrosprayionization (ESI) interface in positive ion mode with capillary and sample cone voltages set to 3000 and 60 V, respectively. Nitrogen was used as a desolvation gas (400 L/hr), with a desolvation temperature of 120 °C. The scan range was set from 820 to 2450 m/z, which allowed for all SOD1 peaks to be recorded.
Data were collected using MassLynx software (Waters, v. 4.1) and mass spectrum deconvolution and integration was performed using the program MOP™ (Multiple Overlaying Pictures, SpectrumSquare Associates, Inc.) described below. Custom code written in Matlab™ (The MathWorks, Inc., Natick, MA, USA) was used to transfer the data from MassLynx to MOP™ and perform the peak integration.Deconvolution of electrospray m/z data to yield the zero charge parent mass spectrum was accomplished withthe Bayesian/maximum entropy algorithm MOP™. The program makes no assumptions about the types of proteins present in the spectrum. The only information provided to the program is1) that hydrogen ions are the source of charge, and2) the resolution of the mass spectrometer. The program assumes that noise is distributed according to a Poisson distribution and incorporates a mass-dependent point-spread function for a time-of-flight mass analyzer to account for instrumental blurring. The program uses an algorithm based on Bayes theorem to calculate the most probable parent mass distribution that would result in the observed multiply charged experimental spectrum with experimental noise.It incorporates a multinomial Bayesian prior and thus becomes the standard maximum entropy approach. The algorithm preserves the observed number of ion counts in the original spectrum and thus the output peak areas remain proportional to the concentration of the species present. SOD1 concentrations were determined by the use of bovine SOD1 as an internal standard.A comparison of the total integration of the human SOD1 peaks with the integration of the bovine SOD1 peak at a known concentration generated a ratio that allowed us to determine the human SOD1 concentration in the original solution. Bovine SOD1 was chosen because it behaves similarly to human SOD1 and binds the same metals, and therefore serves as an internal protein control in addition to a standard.
Molecular weights and theoretical isotope distributionswere calculated from the empirical formula of SOD1using the program iMass[30; 31]. Identical results were obtained using the isotopicdist function in the Bioinformatics Toolbox of Matlab™. To account for the two positive charges of each of the copper and zinc ions, two hydrogen atoms were subtracted for each metal ion bound so thatSOD1remained electrically neutral. In addition, two hydrogen atoms were subtracted for the internal disulfide bond in SOD1.
RESULTS
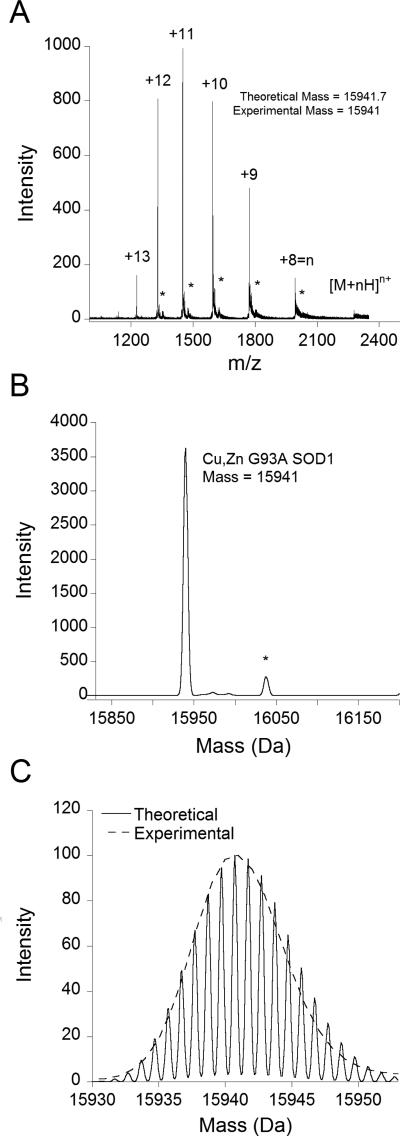
Copper and zinc remained associated with SOD1 during electrospray ionization in a aqueous solution containing 100 μM formic acid and 30% acetonitrile as shown by the mass spectrum of recombinant SOD1 ALS-mutant G93A (Figure 2a). Recombinant SOD1 protein was used to develop the assay before using tissue samples. The mass spectrometer displayed six prominent peaks corresponding to the 13th through 8th charge states, with the most intense being the 10th-12th charge states. Deconvolution using the Bayesian maximum-entropy method generated the parent mass spectrum (Figure 2b). The main peak was centered at 15941 Da, which agreed well with the average mass of 15941.7 calculated for the Cu,Zn G93A SOD1monomer (C680H1081N203O224S4CuZn).The width of the observed SOD1peak corresponds to the broadening predicted from the naturalisotope distribution using the empirical formula for Cu,Zn G93A SOD1 (Figure 2c).
Figure 2. Mass spectra of G93A SOD1.
A. The Ziptipm/z spectrum of 1 μM G93A SOD1 in 10 mM ammonium acetate, showing the charge state distribution of the protein, with the 10th-12th charge states being the most prominent. B. Mathematical deconvolution of the m/z spectrum yields the parent mass spectrum, showing a singular peak centered at a mass of 15941. *The spectrum also contained a small peak centered at 16039 (observable in A as well). This peak, which is 98 mass units larger than Cu,Zn G93A SOD1, likely corresponds to a sulfate (H2SO4) adductremaining from the purification process.C. Overlay of the experimental mass spectrum with the theoretical high resolution mass spectrum calculated from the empirical formula for G93A SOD1 with the program iMass. The high resolution theoretical spectrum is used to illustrate the effects of isotopic broadening, primarily due tothe natural isotope distribution.
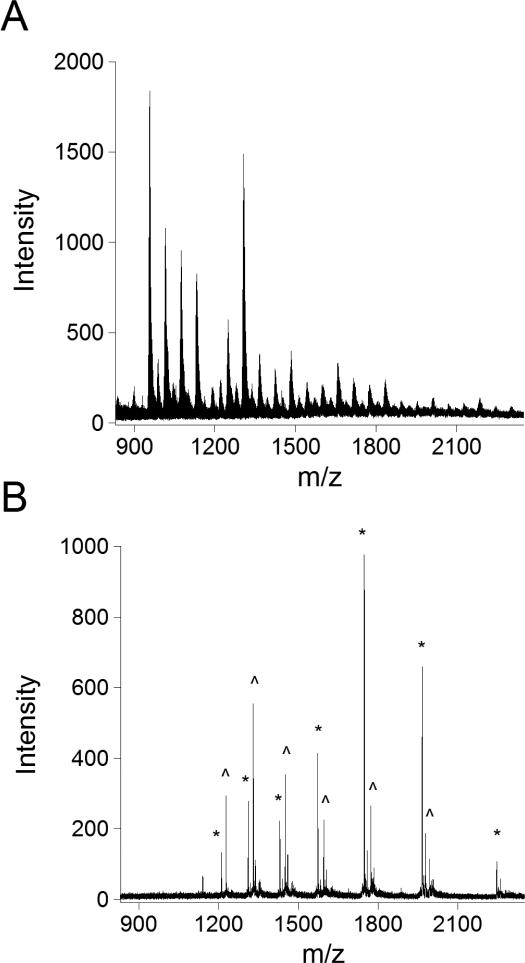
Physiological concentrations of sodium chloridecaused the mass spectrum to be unrecognizable as recombinant SOD1 and could not be deconvoluted (Figure 3a). AZiptipused as a rapid and disposable desalting column reversed the effects of the salt (Figure 3b). The Ziptip contained C4 reversed-phase packing material upon which the SOD1 could be adsorbed and rinsed with water to remove salts. An internal standard of bovine SOD1 was included in each sample to quantify the levels of SOD1 and control for any sample loss. Bovine SOD1 is shorter by two amino acids than human SOD1 and thus is well resolved from the three potential metal ion-containing forms of human SOD1. The charge state distribution for Cu,Zn G93A SOD1 in the presence of high salt could be deconvoluted after use of the Ziptip to give a mass of 15941 Da, identical to the spectrum of SOD1 with no salt present.
Figure 3. Elimination of salt interference using a Ziptip to bind G93A SOD1.
A. Mass spectrum of bovine and G93A SOD1directly injected in the presence of 100 mM sodium chloride. B. Mass spectrum of bovine (*) and G93A (^) SOD1 with 100 mM sodium chloride after washing with the Ziptip.
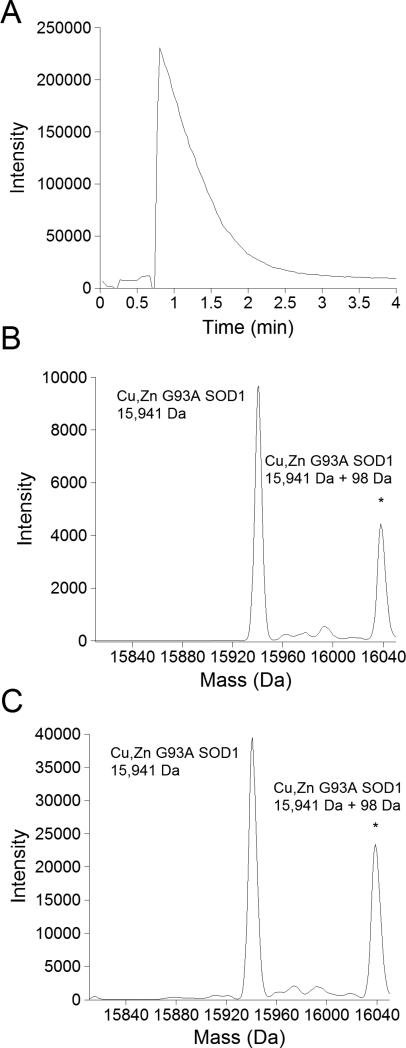
The chromatogram from a sample of bovine and recombinant G93A SOD1 showed that less thanthree minutes was neededfor SOD1 to isocratically elute from the Ziptip into the mass spectrometer (Figure 4a). The concentration of acetonitrile in the running solvent (30%) was optimized to be at the lowest level possible while still rapidly releasingSOD1 from the Ziptip. Dilute formic acid (100 μM) included in the running solventsubstantially improved ionization of the protein.Although acidic conditions are known to cause metal ion release from SOD1, the formic acid concentration was optimized to provide sufficient hydrogen ions for ionization while allowingcopper and zinc to remain stably bound to SOD1[32]. Metal ions were retained under the acid conditions over the period necessary to acquire the data, as shown by the deconvoluted mass spectrum of recombinant SOD1 incubated in water versus running solvent (30% acetonitrile/water with 100 μM formic acid) for 5 minutes at room temperature (Figure 4b,c).Without the rinse step that accompanies the use of a Ziptip, the sulfate adduct peak 98 mass units higher than the mass of the recombinant SOD1 monomer was more prominent.
Figure 4. Stability of metal binding of G93A SOD1 in elution solvent.
A. Chromatogram of G93A SOD1eluting from the Ziptip. B. Parent mass spectrum of G93A SOD1mixed immediately in elution solvent and directly injected into the mass spectrometer. C. Parent mass spectrum of G93A SOD1 incubated in elution solvent for 5 minutes to mimic the maximum exposure of SOD1 to solvent when bound to a Ziptip. *In both B and C, the sulfate-bound SOD1 peak was more prominent probably due to the absence of the washing step used with the Ziptip.
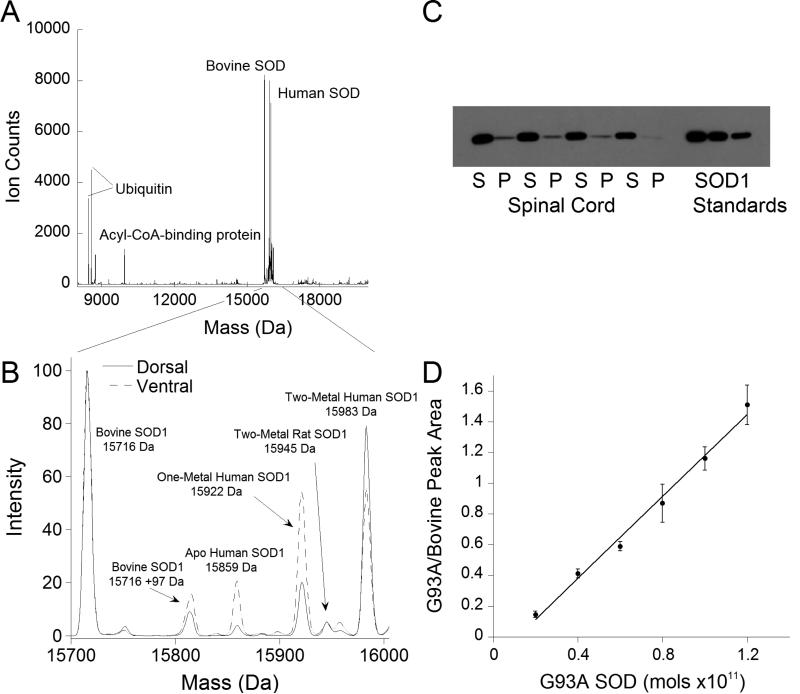
Tissue G93A SOD1could easily be picked out from the other abundant background proteins as it was significantly overexpressed in the spinal cord of G93A-SOD1 overexpressing transgenic rats (Figure 5a).Representative spectra from the dorsal and ventral (Figure 5b) grey matter of the spinal cord of an ALS-model rat showed three distinct metal-bound species: a Cu,Zn SOD1peak, a one-metal SOD1peak, and a small apoSOD1 peak. The one-metal peak was approximately 2-fold larger in the disease-affected ventral spinal cord as compared to that from the dorsal spinal cord.Thawing of frozen tissue punches was sufficient to release SOD1 and other soluble proteins from the punch into solution; this was verified via Western blots that showed less than 2% of SOD1 in the remaining pelletcompared to the supernatant fraction (Figure 5c). The SOD1 was quantifiedby use of the Ziptip and the inclusion of bovine SOD1 as an internal standard. To generate a standard curve, bovine SOD1 was kept at 800 nM, while the concentration of recombinant G93A SOD1 was varied over a range of 200 nM-1.2 μM (Figure 5d). The relationship was linear over the concentration range tested with a slope of approximately 1.3 (R2=0.995).
Figure 5. Metal content of G93A SOD1 in transgenic rats with symptoms of motor neuron degeneration.
A.Deconvoluted mass spectrum of a ventral tissue punch from the lumbar spinal cord of a G93A SOD1-overexpressing rat shown over the mass range 8000 to 24000 Da. Several other proteins, including ubiquitin, ubiquitin minus two glycine residues (-114 Da), and acyl-CoA binding protein were observable in addition to SOD1. B. Overlay of the mass spectra of dorsal and ventral tissue punches from the lumbar spinal cord of a G93A SOD1 overexpressing rat. Prominent peaks are identified in the spectrum, including the endogenous rat SOD1.C. Western blot of tissue punch supernatant (S) and pellet (P) fractions used to verify the release of SOD1 into the supernatant. The SOD1 standards were 4, 2, and 1 pmol, respectively. D. Standard curve using bovine SOD1 as an internal standard at a concentration of 800 nM and varying the G93A SOD1 over a concentration range of 200 nM-1.2 μM. The ratio of the peak integrations of the G93A SOD1 over the bovine SOD1 was plotted against the amount of G93A SOD1 in the sample to produce the standard curve.
DISCUSSION
Analysis of proteins in complex mixtures on a Ziptip via mass spectrometry provides a rapid, quantitative means to assay noncovalent modifications directly from tissues that is comparable and possibly more sensitive than western blotting. The remarkable sensitivity of the method allows for anatomical microdissection of the tissue. ALS affects motor neurons, which are localized in the ventral spinal cord (Figure 1). This assay allows SOD1 to be directly measured from the disease-affected regions, and is able to distinguish between one-, two-, and no-metal bound states; the one-metal bound species is significantly larger in the ventral compared to the dorsal spinal cord.This difference is lost if the entire spinal cord is used, as several previous methods have done[33; 34; 35].
Three main issues needed to be addressed in developing the assay. The first was to maintain metal binding to the protein while providing sufficient acidity to permit efficient electrospray ionization [36; 37]. We found that 0.1% formic acid, commonly used in ESI-MS coupled liquid chromatography, was too strong and could cause SOD1 to lose metals during the assay [32]. Hayward et al., used 0.1% formic acid to look at apo mutant SOD1 proteins [25].Leaving out formic acid resulted in poor ionization because the solvent could not provide enough hydrogen ions in solution to effectively ionize SOD1. The largest amounts of human Cu,Zn SOD1 eluting from the Ziptip were in the range of 1-10 pmol in a volume of a few microliters, resulting in a concentration of 1-10 μM. Because the charge states of SOD1 typically ranged from 8 to 13, the concentration of hydrogen ions needed to be 8-130 μM (Fig. 2a). We found that solvent containing 100 μM formic aciddelivered at a flow rate of 30 μL/minprovided enough protons for efficient ionization. The 100 μM formic acid concentration is approximately 200-fold more dilute than the standard 0.1% concentration, and was able to efficiently ionize SOD1 without altering metal binding after five minutes, twice the time needed to run the assay (Figure 4b,c).
The second issue to overcome was how to efficiently remove salts from the protein solution that would otherwise interfere with electrospray ionization. Salts suppress ionization of proteins, interfering with the deconvolution of raw m/z data (Figure 3a). Sodium ions in particular can associate strongly with an ionizing protein and sodium chloride is approximately 100 mM in the central nervous system, thereby presenting a significant potential interference to ionizing proteins directly from tissue. We used a C4Ziptipto desalt the samples. The Ziptip is a P10 pipette tip with a small amount of HPLC reversed-phase packing material. As tissue lysate is drawn over it, soluble proteins bind to the packing material. Once bound, the proteins could be rinsed multiple times with water, washing off salts and other small hydrophilic molecules while leaving the proteins bound to the packing material. The bound proteins could be eluted in the mass spectrometer with the minimal concentration of acetonitrile needed to elute SOD1 (30%) (Figure 4a).
The third issue was how to quantify the amount of human SOD1 released from the tissue. This was accomplished by including an internal standard, bovine SOD1, at a known concentration in the punch thawing buffer. Comparing the integration of the peak areas from MOP™ of the bovine and human SOD1 allowed us to calculate the amount of human SOD1 in the punch supernatant, which could be used to estimate the concentration of human SOD1 in the tissue.SOD1 was found to be 16pmol/mg tissue in the dorsal spinal cord and 26pmol/mg tissue in the ventral spinal cord. These amounts roughly translate to 16 and 26μM in the cells, respectively. A standard curve of recombinant human SOD1 compared to bovine SOD1 was used to determine if the ionization of the human and bovine proteins differed (Figure 5d). The slope of 1.3 on the standard curve suggests that human SOD1 may ionize more efficiently than bovine SOD1 due to the higher number of basic amino acid residues present in the human SOD1 sequence. This can also be seen in the differing charge state distributions of human and bovine SOD1, in which the human protein picks up two more positive charges on average than bovine SOD1 (Figure 3b).
One of the primary advantages of our method is the ability to monitor the metal-binding of SOD1 at the monomer level in complex mixtures, in contrast to other approaches that can determine the metal-binding of the population of SOD1 as a whole solution, thus giving only average stoichiometry [35]. This is an important distinction in being able to determine if metal-deficient SOD1 species are involved in ALS. An SOD1 dimer has 9 different metal-binding combinations distinguishable by mass. The intrasubunit disulfide bond further complicates matters, as it can be in an oxidized or reduced state, altering the mass of the protein by two Da. Indeed several groups have reported that as much as 20-30% of the SOD1 in the ALS model animals is reduced [38]. Combining these two phenomena, the SOD1 dimer has 27distinctcombinations of metal-binding and disulfide states, which would yield a complex and hard-to-interpret spectrum. An advantage of the described method is that the dimer dissociates in the course of the assayand only monomer is detected (Figure 2b). The SOD1 monomer has only eight distinct metal-binding/disulfide states, with the resulting spectrum being significantly easier to interpret, facilitating analysis of the metal-binding status of individual subunit populations.
A second advantage is that the method is highly adaptable, allowing monitoring of other proteins and post-translational modifications. We have used the method to measure recombinant proteins in solution such as CCS, hemoglobin, and di-iron mono-oxygenases. Other proteins were also observed when measuring SOD1 from tissue, such as ubiquitin and acyl-CoA-binding protein which were tentatively identified by their mass (Figure 5a). The endogenous rat SOD1 was also observable and demonstrates the enormous overexpression of human G93A SOD1 in transgenic rat tissue (Figure 5b). Taking advantage of other materials available for ZipTips (C18, etc.) could further extend the method to additional proteins. Modifying the solvent composition and the ionization parameters, such as the ionization voltage and capillary temperature, allows the method to be optimized for other proteins and post-translational modifications, possibly including small-molecule binding, nitration, metals, andphosphorylation. Furthermore, these modifications can be monitored in a dynamic fashion, as both modified and unmodified populations of a protein are observed at the same time.
The main limitation to our assay is the inability to distinguish between copper-containing, zinc-deficient SOD1 and zinc-containing, copper deficient SOD1. The isotopically broad mass spectrometer signature of a protein is wider than the mass difference between zinc and copper, and thus we cannot differentiate between the two (Figure 2c). However, we find a significant one-metal SOD1 peak that is substantially larger in the ventral gray matter, which provides additional evidence that non-natively metal-bound SOD1 species are involved in the progression of the disease (Figure 5b). Further work that is now ongoing will be needed to determine the composition of the one metal peak and elucidate whether either species has a role in the disease.
ACKNOLEDGEMENTS
We thank Keith Nylin for his early work in this area, and Dr. Mark Levy and Jared Williams for critical reading of the manuscript. This work was supported by the National Institute for Environmental and Health Sciences (NIEHS P30ES000210), the National Institutes of Neurological Disorders and Stroke(NINDS R01NS058628A) and the National Center for Complementary and Alternative Medicine (NCCAM P01AT002034), as well as the Amyotrophic Lateral Sclerosis Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used: ESI-MS, electrospray ionization mass spectrometry; SOD1, Cu,Zn superoxide dismutase; ALS, amyotrophic lateral sclerosis; CCS, copper chaperone for superoxide dismutase; MOP™, multiple overlaying pictures.
REFERENCES
- 1.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–18. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 2.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–30. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 3.Holm RH, Kennepohl P, Solomon EI. Structural and Functional Aspects of Metal Sites in Biology. Chem Rev. 1996;96:2239–2314. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]
- 4.Hann S, Obinger C, Stingeder G, Paumann M, Furtmuller PG, Koellensperger G. Studying metal integration in native and recombinant copper proteins by hyphenated ICP-DRC-MS and ESI-TOF-MS capabilities and limitations of the complementary techniques. Journal of Analytical Atomic Spectrometry. 2006;21:1224–1231. [Google Scholar]
- 5.Shi W, Chance MR. Metallomics and metalloproteomics. Cell Mol Life Sci. 2008;65:3040–8. doi: 10.1007/s00018-008-8189-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCall KA, Fierke CA. Colorimetric and fluorimetric assays to quantitate micromolar concentrations of transition metals. Anal Biochem. 2000;284:307–15. doi: 10.1006/abio.2000.4706. [DOI] [PubMed] [Google Scholar]
- 7.Ayed A, Krutchinsky AN, Ens W, Standing KG, Duckworth HW. Quantitative evaluation of protein-protein and ligand-protein equilibria of a large allosteric enzyme by electrospray ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1998;12:339–44. doi: 10.1002/(SICI)1097-0231(19980415)12:7<339::AID-RCM163>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 8.Ganem B, Henion JD. Going gently into flight: analyzing noncovalent interactions by mass spectrometry. Bioorg Med Chem. 2003;11:311–4. doi: 10.1016/s0968-0896(02)00537-0. [DOI] [PubMed] [Google Scholar]
- 9.Potier N, Rogniaux H, Chevreux G, Van Dorsselaer A. Ligand-metal ion binding to proteins: investigation by ESI mass spectrometry. Methods Enzymol. 2005;402:361–89. doi: 10.1016/S0076-6879(05)02011-2. [DOI] [PubMed] [Google Scholar]
- 10.Veenstra TD. Electrospray ionization mass spectrometry in the study of biomolecular non-covalent interactions. Biophys Chem. 1999;79:63–79. doi: 10.1016/s0301-4622(99)00037-x. [DOI] [PubMed] [Google Scholar]
- 11.Deng L, Sun N, Kitova EN, Klassen JS. Direct quantification of protein-metal ion affinities by electrospray ionization mass spectrometry. Anal Chem. 2010;82:2170–4. doi: 10.1021/ac902633d. [DOI] [PubMed] [Google Scholar]
- 12.Banci L, Bertini I, Ciofi-Baffoni S, Kozyreva T, Zovo K, Palumaa P. Affinity gradients drive copper to cellular destinations. Nature. 2010;465:645–8. doi: 10.1038/nature09018. [DOI] [PubMed] [Google Scholar]
- 13.McCord JM, Fridovich I. Superoxide dismutase: an enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 14.Kayatekin C, Zitzewitz JA, Matthews CR. Zinc binding modulates the entire folding free energy surface of human Cu,Zn superoxide dismutase. J Mol Biol. 2008;384:540–55. doi: 10.1016/j.jmb.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen PM. Genetics of sporadic ALS. Amyotroph Lateral Scler Other Motor Neuron Disord. 2001;2(Suppl 1):S37–41. doi: 10.1080/14660820152415726. [DOI] [PubMed] [Google Scholar]
- 16.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 17.Nishida CR, Gralla EB, Valentine JS. Characterization of three yeast copper-zinc superoxide dismutase mutants analogous to those coded for in familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci USA. 1994:9906–9910. doi: 10.1073/pnas.91.21.9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SM, Eum WS, Kang JH. Expression, purification, and characterization of a familial amyotrophic lateral sclerosis-associated D90A Cu,Zn-superoxide dismutase mutant. Mol Cells. 1998;8:478–82. [PubMed] [Google Scholar]
- 19.Winterbourn CC, Domigan NM, Broom JK. Decreased thermal stability of red blood cell glu100-->gly superoxide dismutase from a family with amyotrophic lateral sclerosis. FEBS Lett. 1995;368:449–51. doi: 10.1016/0014-5793(95)00708-h. [DOI] [PubMed] [Google Scholar]
- 20.Crow JP, Sampson JB, Zhuang Y, Thompson JA, Beckman JS. Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J Neurochem. 1997;69:1936–44. doi: 10.1046/j.1471-4159.1997.69051936.x. [DOI] [PubMed] [Google Scholar]
- 21.Goto JJ, Zhu H, Sanchez RJ, Nersissian A, Gralla EB, Valentine JS. Loss of in vitro metal ion binding specificity in mutant copper-zinc superoxide dismutases associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 2000;275:1007–1014. doi: 10.1074/jbc.275.2.1007. [DOI] [PubMed] [Google Scholar]
- 22.Museth AK, Brorsson AC, Lundqvist M, Tibell LA, Jonsson BH. The ALS-associated mutation G93A in human copper-zinc superoxide dismutase selectively destabilizes the remote metal binding region. Biochemistry. 2009;48:8817–29. doi: 10.1021/bi900703v. [DOI] [PubMed] [Google Scholar]
- 23.Estevez AG, Crow JP, Sampson JB, Reiter C, Zhuang Y, Richardson GJ, Tarpey MM, Barbeito L, Beckman JS. Induction of nitric oxide-dependent apoptosis in motor neurons by zinc-deficient superoxide dismutase. Science. 1999;286:2498–500. doi: 10.1126/science.286.5449.2498. [DOI] [PubMed] [Google Scholar]
- 24.Sahawneh MA, Ricart KC, Roberts BR, Bomben VC, Basso M, Ye Y, Sahawneh J, Franco MC, Beckman JS, Estevez AG. Cu,Zn-superoxide dismutase increases toxicity of mutant and zinc-deficient superoxide dismutase by enhancing protein stability. J Biol Chem. 2010;285:33885–97. doi: 10.1074/jbc.M110.118901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayward LJ, Rodriguez JA, Kim JW, Tiwari A, Goto JJ, Cabelli DE, Valentine JS, Brown RH., Jr. Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2002;277:15923–31. doi: 10.1074/jbc.M112087200. [DOI] [PubMed] [Google Scholar]
- 26.Leinweber B, Barofsky E, Barofsky DF, Ermilov V, Nylin K, Beckman JS. Aggregation of ALS mutant superoxide dismutase expressed in Escherichia coli. Free Radic Biol Med. 2004;36:911–8. doi: 10.1016/j.freeradbiomed.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 27.Howland DS, Liu J, She Y, Goad B, Maragakis NJ, Kim B, Erickson J, Kulik J, DeVito L, Psaltis G, DeGennaro LJ, Cleveland DW, Rothstein JD. Focal loss of the glutamate transporter EAAT2 in a transgenic rat model of SOD1 mutant-mediated amyotrophic lateral sclerosis (ALS). Proc Natl Acad Sci U S A. 2002;99:1604–9. doi: 10.1073/pnas.032539299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valentine JS, Pantoliano MW. Protein-metal ion interactions in cuprozinc protein (superoxide dismutase). In: Spiro TG, editor. Copper Proteins: Metal ions in biology. John Wiley and Sons; New York: 1981. pp. 291–358. [Google Scholar]
- 29.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Rockwood AL, Vanorden SL, Smith RD. Rapid Calculation of Isotope Distributions. Analytical Chemistry. 1995;67:2699–2704. [Google Scholar]
- 31.Rockwood AL, VanOrden SL, Smith RD. Ultrahigh resolution isotope distribution calculations. Rapid Communications in Mass Spectrometry. 1996;10:54–59. [Google Scholar]
- 32.Forman JH, Fridovich I. On the stability of bovine superoxide dismutase. J Biol. Chem. 1973;248:2645–2649. [PubMed] [Google Scholar]
- 33.Proescher JB, Son M, Elliott JL, Culotta VC. Biological effects of CCS in the absence of SOD1 enzyme activation: implications for disease in a mouse model for ALS. Hum Mol Genet. 2008;17:1728–37. doi: 10.1093/hmg/ddn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karch CM, Prudencio M, Winkler DD, Hart PJ, Borchelt DR. Role of mutant SOD1 disulfide oxidation and aggregation in the pathogenesis of familial ALS. Proc Natl Acad Sci U S A. 2009;106:7774–9. doi: 10.1073/pnas.0902505106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lelie HL, Liba A, Bourassa MW, Chattopadhyay M, Chan PK, Gralla EB, Miller LM, Borchelt DR, Valentine JS, Whitelegge JP. Copper and zinc metallation status of copper-zinc superoxide dismutase from ALS-transgenic mice. J. Biol. Chem. 2011;286:2795–2806. doi: 10.1074/jbc.M110.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huber CG, Premstaller A, Kleindienst G. Evaluation of volatile eluents and electrolytes for high-performance liquid chromatography-electrospray ionization mass spectrometry and capillary electrophoresis-electrospray ionization mass spectrometry of proteins. II. Capillary electrophoresis. J Chromatogr A. 1999;849:175–89. doi: 10.1016/s0021-9673(99)00533-6. [DOI] [PubMed] [Google Scholar]
- 37.Garcia MC. The effect of the mobile phase additives on sensitivity in the analysis of peptides and proteins by high-performance liquid chromatography-electrospray mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;825:111–23. doi: 10.1016/j.jchromb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 38.Jonsson PA, Graffmo KS, Andersen PM, Brannstrom T, Lindberg M, Oliveberg M, Marklund SL. Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain. 2006;129:451–64. doi: 10.1093/brain/awh704. [DOI] [PubMed] [Google Scholar]