Abstract
Combinatorial peptide chemistry and orthogonal high-throughput screening were used to select peptides that spontaneously translocate across synthetic lipid bilayer membranes without permeabilization. A conserved sequence motif was identified that contains several cationic residues in conserved positions in an otherwise hydrophobic sequence. This 9-residue motif rapidly translocates across synthetic multibilayer vesicles and into cells while carrying a large polar dye as a “cargo” moiety. The extraordinary ability of this family of peptides to spontaneously translocate across bilayers without an energy source of any kind is distinctly different from the behavior of the well-known, highly cationic cell-penetrating peptides, such as the HIV tat peptide, which do not translocate across synthetic bilayers, and enter cells mostly by active endocytosis. Peptides that translocate spontaneously across membranes have the potential to transform the field of drug design by enabling the delivery of otherwise membrane-impermeant polar drugs into cells and tissues. Here we describe the chemical tools needed to rapidly identify spontaneous membrane translocating peptides.
Introduction
In the development of new drugs, one of the most restrictive factors is the requirement that active compounds also be able to cross cellular membranes1. An active compound that is not lipophilic enough to spontaneously diffuse across membranes is unlikely to be a useful drug. In the drug development pipeline, many potential drugs are rejected for this reason. The discovery of the polycationic, cell penetrating peptides twenty years ago created excitement that this problem might be universally solved. Indeed, in recent years polar molecules, especially large ones such as peptides, proteins or RNA, have been transported into cells by the so-called cell penetrating peptides (CPP), exemplified by the highly cationic peptides penetratin and the HIV tat sequence2-5. However, there is a controversy in the literature about the mechanism of cell entry5,6 of this class of peptides. Some experimental approaches have suggested spontaneous membrane translocation into cells while others suggest endocytosis-dependent cell entry. In the latter case, there is the complicating question of how the peptide or cargo are released from the endosomal pathway, which can also be attributed, or not, to spontaneous membrane translocation. Despite the controversy, a consensus has emerged that the most commonly used cell penetrating peptides (i.e. the highly cationic ones, such as tat) are actively taken up into cells by one or more type of endocytosis2,4,6.
While the potential utility of cargo delivery via endocytosis has been demonstrated, especially for large cargo moieties2,4,7, the utility is limited by the capacity of cells for active endocytosis. Furthermore, highly cationic CPPs preferentially interact with particular cell types8, have limited plasma half-life9, do not cross multi-cellular barriers such as vasculature epithelia or the blood-brain barrier10 and often accumulate in cellular organelles such as endosomes5. Highly cationic cell penetrating peptides are therefore not ideal small molecule drug delivery vehicles. Instead, an ideal peptide delivery vehicle would be one that mimics small molecule drugs by crossing all cell membranes indiscriminately.
The goal of this work was to develop chemical tools and high-throughput screening methods to enable the discovery of peptides that spontaneously translocate across synthetic membranes and thus may be better suited for systemic small molecule drug delivery. Instead of characterizing the mechanism and efficiency of the membrane translocation of peptides derived from natural sequences, or designed by trial and error, here we take a very different approach. We use combinatorial chemistry and orthogonal high-throughput screening with synthetic lipid vesicles to identify peptides based, specifically, on their ability to spontaneously translocate across synthetic membranes. We show that the peptides discovered this way can efficiently carry polar cargo moieties across synthetic lipid bilayers and into living cells. Spontaneous membrane translocating peptides such as the ones we have discovered here could eliminate the membrane barrier in drug discovery and enable the systemic delivery of polar drugs to cells.
METHODS
Peptide Synthesis and Purification
Peptide library synthesis was done using a split and recombine method that we have described in detail elsewhere11,12. Library members were attached to Tentagel megabeads (Rapp Polymere) of 300 μm diameter using a photocleavable linker (Advanced Chemtech) through the side chain of a glutamate,which releases a glutamine upon photocleavage. This frees the C-terminus of the terminal phenylalanine for the aminomethylcoumarin (amc) moiety which we use to detect translocation via protease cleavage. For the library synthesis, completion of each coupling reaction was verified using the ninhydrin assay13, which is very sensitive to incomplete coupling. Ninhydrin assays were always performed on at least 30-50 beads at each step to be able to detect sequence-dependent coupling problems which might occur on only a fraction of beads. After completion of synthesis, cleavage of side chain protecting groups was done by reacting with trifluoroacetic acid:ethanedithiol:anisole:thioanisole (90:5:2.5:2.5) under nitrogen for four hours on ice followed by extensive washing in multiple solvents. To validate the synthesis and deprotection, we performed HPLC analysis, mass spectrometry and Edman degradation on the peptides extracted from individual beads. All techniques showed that the predominant sequence on all beads is full length peptide. Edman degradation was especially noteworthy in this regard because it was performed on dozens of individual beads, and showed full length peptides. Furthermore we found that there was no detectible free AMC dye associated with the beads. To prepare beads for screening, they were spread in a glass dish to a single sparse layer, dried completely, and then illuminated with a low power UV lamp (354 nm) for 5 hours. Control experiments showed no damage to the peptide by the UV light. We measured the peptide-AMC release from hundreds of individual beads and measured a consistent 0.5 – 1.0 nmol of extractable peptide from each bead.
Preparation of screening vesicles
For the high-throughput screen, we prepared large unilamellar vesicles of 0.1 μm diameter by extrusion14 essentially as described elsewhere15 except that we also included chymotrypsin at 10 mg/ml in the buffer in addition to 50 mM Tb3+. Separation of external enzyme and external Tb3+ from the vesicles was done by gel filtration chromatography. Verification of efficient entrapment/separation of chymotrypsin was done with non membrane permeable substrates, with and without detergent lysis of vesicles. The lipid composition used in all screening experiments was 10 mol% palmitoyloleoylphosphatidylglycerol (POPG) and 90% palmitoyloleoylphosphatidylcholine (POPC). Vesicles were prepared at 100 mM lipid to maximize total entrapment. For screening, vesicles were diluted to 1 mM lipid in buffer that also contained 50 μM DPA (for the leakage assay) and also contained 0.1 mg/ml α1-antitrypsin inhibitor.
High-Throughput Screening
Dried and photocleaved beads were placed in 96 well plates (one bead per well) followed by incubation overnight with 10 μl of DMSO to extract the free peptide from the bead. The DMSO was transferred to a new plate and 100 μl of buffer was added to each well to bring the peptide concentration to about 5 μM. This was followed by incubation at room temperature for at least 1 hour to allow precipitation of insoluble sequences. We estimate that the number of active peptides is reduced by more than half by this pre-incubation step. At this point the screening vesicles containing entrapped terbium and chymotrypsin and external DPA and trypsin inhibitor were added to bring the total lipid concentration to 1 mM. The rate of AMC cleavage was monitored in each well, starting immediately after vesicle addition, by measuring AMC fluorescence in all the wells every 2 minutes for 30 minutes. Most wells showed very little cleavage (<1%) in this time frame, but a few peptides had as much as 50% total cleavage of the AMC group in the well within 30 minutes. Cleavage, when observed, was always approximately linear with no lag or delay time. To normalize the results for library members, we express cleavage rate as a relative rate; defined as the initial rate of AMC fluorescence increase for the library peptide in the presence of intact vesicles, divided by the rate of change for the control peptide pool in the presence of lysed vesicles without inhibitor. The cleavage rate measured for vesicles lysed in the presence of inhibitor was negligible on these time scales. After 30 minutes of monitoring AMC cleavage, Tb3+/DPA fluorescence measurements were made to quantitate leakage from the vesicles. For these studies we used a Biotek Synergy fluorescence plate reader with a UV flash lamp. AMC cleavage measurements were done with excitation at 340 nm and emission at 440 nm, while Tb3+/DPA leakage measurements were done with excitation at 270 nm and emission at 475 nm. Once each day a plate would be analyzed containing control samples. Control samples for cleavage were Triton-X-100 lysed vesicles (with and without protease inhibitor) to which 1 nmol of a peptide pool from ~100 beads was added.
In a protease-dependent screen of this type, one must be sure that the inherent cleavage rates in the library are not sequence dependent or the results could be biased. We chose chymotrypsin for these experiments in part because it is known that residues on the N-terminal side of the cleavage site have little or no effect of cleavage rates16. In this library we have three fixed C-terminal residues after the combinatorially varied sites, and thus did not expect those varied residues, which are four or more positions away from the cleavage site, to affect the rate. Nonetheless, we verified the constancy of cleavage rates by measuring it directly for several hundred individual library members in the presence of lysed chymotrypsin vesicles. We observed no anomalous cleavage rates, high or low. They all were within a factor of 2 of the average. Given the small number of amino acids at each site, there are only 144 different sequences from residue 5 to the cleavage site at residue 12, thus in the few hundred library members tested we sampled every variation in peptide sequence from position 5 to the 12th residue chymotrypsin site. We conclude that inherent cleavage rates are constant within the library. Based on these data and on the fact that membrane-impermeant substrates are not cleaved by chymotrypsin entrapped in vesicles (see below) we conclude that only peptides that translocate are cleaved rapidly in the screen.
Multilamellar vesicle translocation experiments
Multilamellar vesicles (MLV) were prepared by aliquoting lipids from stock chloroform solutions into a glass tube and drying under nitrogen, followed by high vacuum for at least four hours. The dried lipid film was rehydrated with PBS by gentle mixing, followed by ten freeze-thaw cycles. In a translocation experiment 10 μl of lipid solution was added to a small eppendorf tube, followed by FD3 in PBS and peptide in DMSO to bring the total concentrations to 6 mM lipid, 10 μg/ml FD3 and 1 μM TAMRA-peptide. DMSO content was less than 5%, which we showed has no effect on vesicle integrity. Immediately after sample preparation, 3 μl of the mixture was spotted between a glass slide and cover slip and the slide was mounted on a Nikon laser scanning confocal microscope using a 60X oil immersion lens. After 10 minutes on the microscope (15 minutes total time) the slide was searched for multilamellar vesicles that were between 5 and 50 μm diameter and spherical in shape. Vesicles with these characteristics were imaged. The vesicles were imaged using a 488 nm laser and 520 nm bandpass filter for fluorescein and a 543 nm laser with a 580 nm bandpass filter for TAMRA. Under these conditions, bleed-through between channels was negligible. The focal plane was always adjusted to give the maximum vesicle diameter in the FD3 detection mode.
Cell translocation experiments
Chinese hamster ovary (CHO) cells were seeded on coverglass slides with 4 chambers. Typical seeding densities of CHO cells ranged from 1×104 to 1×105 cells per chamber with a 1.7 cm2 seeding area. Cells were grown in 500 μl complete media at 37°C with 5% CO2 for 24-48 hours prior to experimentation. To perform translocation experiments, slides with adherent cells were removed from the incubator, the cells were washed with PBS at 37°C and then with PBS at room temperature (22°C) and were allowed to incubate at room temperature for 30 minutes. At this point fluorescein dextran 3000 Da (FD3) was added as an aqueous phase marker to a concentration of 10 μg/ml. Dye labeled peptides were added from stock dimethylsulfoxide (DMSO) solution to a final concentration of 2 μM peptide. Final DMSO concentration in the cell culture was 1% or less which does not affect the cells. The cells were imaged using a 488 nm laser and 520 nm bandpass filter for FD3 and a 543 nm laser with a 580 nm bandpass filter for TAMRA. Under these conditions, bleed-through between channels is negligible. The focal plane was always adjusted to give the maximum cell diameter in the FD3 detection mode, thus avoiding focal planes too near the top or the bottom of the cell.
Statistical analysis of abundance
The binomial equation was used to compare the expected and observed number of occurrences of particular amino acids in specific position of the selected (positive) peptides. For each combinatorial site, or aggregate of sites, a p-value was calculated using the expected number of occurrences and the total number of possible occurrences of a single amino or a set of amino acids. Each p-value is the probability of observing at least as many (or at least as few) of an over/under represented amino acid by summing binomial probabilities for all values from the observed number to the highest possible abundance (for overrepresented amino acids) or to zero for underrepresented amino acids.
Results and Discussion
Orthogonal high-throughput screening for translocating peptides
We selected peptides from a combinatorial library based, specifically, on their ability to spontaneously translocate across synthetic lipid bilayer membranes. This was accomplished by using bilayer membrane vesicles as the basis for a high throughput screen that identifies only peptides that translocate across membranes spontaneously (Fig. 1A). We use an “orthogonal” high-throughput approach to select for multiple properties simultaneously and independently. In particular, we selected independently for aqueous peptide solubility, lack of membrane disruption, and rapid membrane translocation. Solubility pre-selection is accomplished by incubating library peptides in buffer for an hour before screening with lipid vesicles, such that insoluble library members are inactivated by precipitation15. Screening for membrane disruption is done with a vesicle-based screen described elsewhere12,15 that utilizes liposome-entrapped terbium(III) with an aromatic chelator, dipicolinic acid (DPA), in the external solution, so that a quantifiable luminescent metal-chelate complex forms only upon membrane disruption. Here we describe a third orthogonal selection criterion for translocation. To detect peptide translocation into terbium-containing vesicles, we also entrapped the protease chymotrypsin and added a protease inhibitor outside the vesicles to quench any external or released enzyme (Fig. 1B). A fluorescent chymotrypsin-sensitive aminomethylcoumarin (amc) moiety was attached to the carboxyl terminus of the peptides in the combinatorial library to report on peptide translocation (Fig. 1A). Using this orthogonal strategy, membrane disruption and peptide translocation can be measured independently in the same vesicles in a multiwell plate fluorescence format. Control experiments with membrane-impermeant (Fig. 1D) and membrane-permeant (Fig. 1E) chymotrypsin-sensitive probes were used to validate the design of the screen.
Figure 1.
Orthogonal High-throughput screen design. A: Library architecture: Peptide library members were attached to polystyrene resin microbeads through the side chain of a glutamate using a UV cleavable linker12, which releases an amidated glutamine upon irradiation. The C-terminal phenylalanine has an aminomethylcoumarin (AMC) moiety to report on chymotrypsin cleavage. There is a glycine residue linker to which the peptide library was added. Each 300 μm bead releases ~0.5 nmol of one peptide sequence. B: High throughput screen. Large unilamellar vesicles, 100 nm diameter, composed of 90% zwitterionic phosphatidylcholine and 10% anionic phosphatidylglycerol, were made by extrusion14. Tb3+ and chymotrypsin were entrapped and the external solution was exchanged with buffer using gel filtration chromatography. The aromatic chelator dipicolinic acid was added to the external solution to report on Tb3+ release, and α1-antitrypsin inhibitor was added to inhibit any released chymotrypsin. C: Simultaneous detection of Tb3+/DPA complex formation and the AMC cleavage is possible because of the lack of spectral overlap. The curves show the excitation and emission shift, respectively, of AMC upon cleavage. D: Intact chymotrypsin vesicles (1 mM lipid) without external inhibitor were incubated-with a membrane-impermeant substrate, succinyl-alanine-alanine-proline-phenylalanine-pnitroaniline. Little measurable cleavage occurred. When the same vesicles were disrupted with 0.1% v/v of the detergent Triton-X-100, AMC cleavage occurs with the same rate as a solution of 50 μg/ml chymotrypsin. E: Lipid vesicles (500 μM lipid, with entrapped chymotrypsin) were incubated with the membrane permeant chymotrypsin substrate glutaryl-phenylalanine-AMC. Two types of samples were used: Intact vesicles with chymotrypsin inhibitor, or lysed vesicles without inhibitor. Upon addition of detergent to the intact vesicles, or the addition of inhibitor to the lysed vesicles, cleavage stops completely. These experiments demonstrate the efficient entrapment of active chymotrypsin and the sufficiency of the external inhibitor to quench any released enzyme.
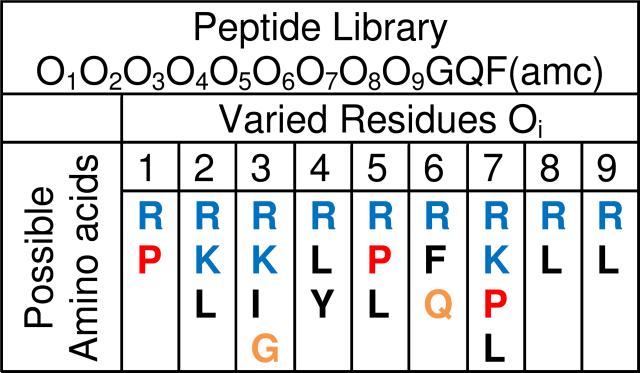
To test the sequence requirements for true spontaneous membrane translocation, a 12-residue peptide library was designed with nine variable residues to encompass the amino acid compositions of known cell penetrating peptides, ranging from fully cationic to fully hydrophobic (Fig. 2). An important practical consideration was to have a library with fewer than about 12,000 sequences so that it could be screened in a reasonable amount of time. Thus, each of the nine combinatorial sites contained only 2-4 possible amino acids. We designed the library so that each site had at least one basic and at least one hydrophobic residue, with the default residues being arginine and leucine. But we also designed the library to contain as many known cell penetrating peptides as possible from multiple classes, including polycationic, hydrophobic and proline-rich peptides. Therefore several positions contained lysine as an alternate basic residue, and some leucines were either augmented or replaced with other hydrophobes (P,I,Y,F) or other residues (e.g. K in position 3 and Q in position 6 derived from the HIV tat peptide) when that was needed to improve the match to a known CPP. The final library contained 10,368 members, and those members included the tat peptide sequence (RRKRRQRRR) the tat homolog Arg9 (RRRRRRRRR), and sequences that closely resembled the proline-rich cell penetrating peptides that are based on fragments of the antimicrobial peptide bactenecin 7. These sequences include Bac71-7 (RRIRPRP) and Bac715-24 (PRPLPFPRP) which have potent cell penetrating activity17. The library was also designed to have an N-terminal RRGY motif which we had identified as important for membrane activity in screening for antimicrobial peptides15. The three C-terminal amino acids were fixed, as we describe below. Glycine in position 10 constitutes a flexible linker. The side chain of a glutamate in positon 11 is used to link the peptides to the synthesis resin beads by a photolabile linker, which releases a glutamine upon cleavage. The C-terminal Phe has an aminomethylcoumarin (AMC) group for detection of chymotrypsin cleavage.
Figure 2.
Library design. The library screened in this work contained a rationally designed 9-residue combinatorial segment, followed by a fixed GQF(amc) (Fig, 1A). Each varied (Oi) position could contain one of the residues shown vertically below. Possible cationic residues are shown in blue, hydrophobic residues are shown in black, proline is shown in red. The glycine is a flexible linker. The glutamine is used to attach the library to synthesis beads and the phenylalanine(amc) is used as a probe for cleavage. The total library size is 10,368 sequences.
The library was synthesized by a split-and-recombine method as described previously12,15 in which members were linked to 300 μm diameter polystyrene beads. In this library each bead contained about 1 nmol of a single peptide sequence. High throughput screening was performed in 96-well plate format, with each well containing about 0.5-1.0 nmol of a single peptide sequence, extracted from a single photocleaved bead, along with 100 nmol total lipid in the form of 100 nm diameter single bilayer vesicles (Fig. 1B). At this stringency most library members are inactive, allowing the identification of extraordinarily active peptides. Upon addition of screening vesicles to a plate, the rate of AMC cleavage was monitored simultaneously in all 96 wells (Fig. 3A). When the cleavage rate was not zero, it was linear with time. The cleavage rates in wells containing library members were compared to control wells containing ~1 nmol peptide from a pool of library peptides added to lysed vesicles either with or without protease inhibitor. These wells serve as negative and positive cleavage controls, respectively. After 30 min of incubation, the amount of terbium leakage in each well was measured in the same plate (Fig. 3B) and was compared to control wells with either intact or detergent-lysed vesicles serving as zero and 100% leakage controls.
Figure 3.
A represented screening plate. Results for an example 96-well plate screened with the orthogonal high throughput screen. Each well contains about 5 μM of a single library peptide extracted from a single microbead, plus 1 mM lipid in the form of lipid vesicles prepared as described in Fig. 1B. A. The initial cleavage rate of AMC was measured in each of the 96 wells of a plate. Most wells have no measurable activity. This plate contains a very active translocating sequence in well G1. B. Tb3+/DPA leakage measured in the same plate as above after 30 minutes of incubation. A few wells have peptides with slight leakage activity. Well G1, containing the translocating peptide, had little leakage, thus it is a useful translocating peptide.
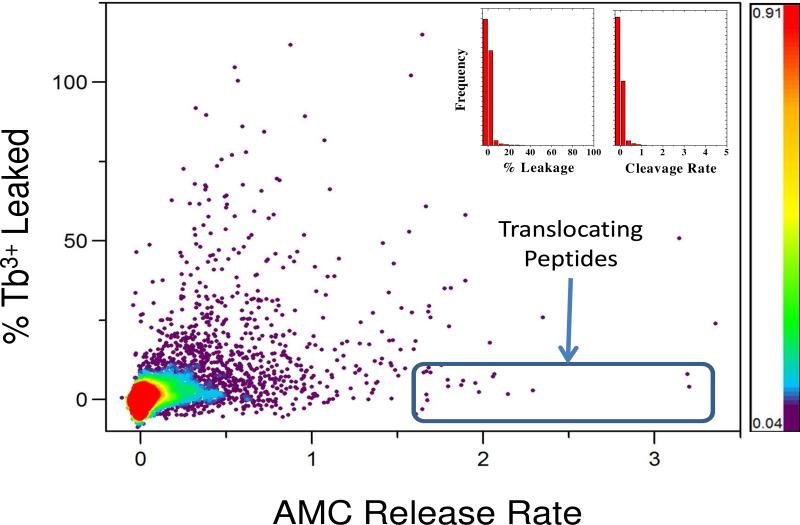
About 24,000 randomly selected sequences from the library were screened with this orthogonal high throughput approach. Because the library contains only 10,368 members, any particular sequence in the library could have been tested from once to several times, overall. Most library members (>99%) were inactive in both translocation and leakage assays (Fig. 4). A small number of peptides (<0.1%) had high leakage without protease cleavage, suggesting that they form large pores through which both terbium and the protease chymotrypsin can be released. An small number of peptides (<0.1%) showed high rates of AMC cleavage with little or no Tb3+/DPA leakage (Figs. 3 and 4). The median relative cleavage rate of library members was very close to zero and the average was ~0.03. Thus the majority of peptides are not cleaved by entrapped chymotrypsin at all. As shown in Fig. 4, the most active translocating peptides had cleavage rates at least 100 fold higher than the mean of the library. As we described above in methods, control experiments showed that the inherent cleavage rates were constant within the library, thus these rapidly cleaved peptides have to be translocating across the membranes into the vesicles in order to be cleaved by entrapped chymotrypsin. Eighteen such spontaneously translocating, non-permeabilizing library members were identified and sequenced (Fig. 5). Every translocating peptide identified in the screen was successfully sequenced. Several sequences were independently found multiple times within the 18 translocating sequences, demonstrating that the translocation-active peptides are a highly unique family of peptides.
Figure 4.
Screening results. 24,000 library members were screened using the orthogonal high throughput screen. Inset: Histogram of library screening results in the two axes of the orthogonal screen, leakage and chymotrypsin cleavage. More than 99% of all library members have no measurable activity in either assay. Main panel: Screening results plotted for all 24,000 randomly selected sequences screened. Each point represents the leakage and translocation data for a single peptide. Colors indicate point density, with over 23,000 points in the inactive (red to blue) zones, and only a few hundred points (<0.5%) in the black, high activity zones. Eighteen peptides (0.1%) with very high translocation rate and no leakage, highlighted by the rectangle, were sequenced.
Figure 5.
Analysis of translocating sequences. A. The amino acid sequences of eighteen selected translocating peptide sequences are shown. For visualization, the identified sequences are separated by dashes into a 4-residue and 5-residue segments. The fixed C-terminal -GQF sequence was present in all library members. Three of the sequences were observed multiple times independently. The number of observations are given by parentheses. B. The statistical preference for the observed abundance of amino acids compared to the expected abundance are plotted as p-values. The dotted lines indicate a cutoff of p=0.05 for statistical significance. Overall, cationic residues are significantly underrepresented and hydrophobic residues are overrepresented. The only exceptions are the arginines in the 5th and 9th positions which are overrepresented, indicating they are specifically selected for. C. Positive and negative peptides used in these studies. Three translocating peptides TP1, TP2 and TP3 were synthesized with a C-terminal cysteine for dye labeling and characterization. An all D-amino acid version of TP2 (TP2D) was also synthesized. DNEG is a “designed” negative of TP3 in which the three cationic arginine residues were replaced with aspartates. ONEG is an “observed” negative randomly selected in the screen from the many peptides with little activity. Arg9 is a known non-translocating, cell penetrating peptide. Arg9 was present in the library, but was not selected.
None of the selected membrane translocating sequences resembled any of the known cell penetrating peptides contained in the library. Instead, the selected translocating peptides contained a novel, conserved sequence motif (Fig. 5A). Ten of the 18 sequences contained a PLIL-XXXXX-GQF or PLIY-XXXXX-GQF sequence and 10 of the 18 contained an XXXX-LRLLR-GQF motif. Two nearly identical sequences, PLIL-LRLLR-GQF and PLIY-LRLLR-GQF, comprising 7 of the 18 translocating peptides, contained both motifs. Compared to the highly cationic cell penetrating peptides such as tat or Arg9, which were present in the library but were not selected, the spontaneously translocating peptides are much less polar, with only two or three cationic residues (range = 1 to 4, most common = 2), plus the N-terminal charge, in an otherwise hydrophobic sequence. Compared to the abundance in the library overall, the basic residues arginine and lysine were significantly underrepresented in the translocating sequences while hydrophobic residues leucine and isoleucine were significantly overrepresented (Fig. 5B). An important exception occurred at positions 6 and 9, part of the XXXX-LRLLR-GQF motif, where cationic arginine residues were found more often than expected by chance, and hydrophobes were found less often than expected by chance. The orthogonal high-throughput screen thus selected specifically for a novel, conserved membrane translocation motif that is mostly hydrophobic, with a few cationic residues at conserved positions.
Spontaneous peptide translocation across membranes
We synthesized peptide-AMC positives and measured the translocation/cleavage in the original assay compared to a random pool of library members. This experiment verified that the purified positives do, in fact, translocate under the original screen conditions, and that most library members do not. However, to fully test spontaneous membrane translocation we designed a translocation experiment with more stringent conditions than the original screen. Representative members of the translocating peptide family, including an all D-amino acid version of a translocating peptide, and a set of negative peptides (Fig. 5C) were synthesized. Each peptide had a C-terminal cysteine residue to which we attached a mock “cargo” moiety: 6-carboxytetramethylrhodamine (TAMRA), a zwitterionic, membrane-impermeant fluorescent dye of 430 Da molecular weight. TAMRA is larger and more polar than AMC, and thus allows us to test the peptides’ ability to deliver drug-like “cargo” across membranes. Furthermore, because TAMRA is a bright, visible-wavelength fluorophore we can use the same peptide-cargo conjugate to test translocation across both synthetic and cellular membranes by fluorescence microscopy. To conduct stringent tests of spontaneous membrane translocation we prepared multilamellar vesicles (MLV), which are 10-40 μm in diameter and have at least 10-15 concentric bilayers (Fig. 6A). Peptide translocation across MLVs was assessed using laser scanning confocal fluorescence microscopy. Soluble, polar probes such as free TAMRA dye and fluorescein dextran 3,000 Da (FD3) were excluded from the vesicles (Figs. 6B and 6C). When the selected translocating peptides and attached TAMRA cargo moiety were added to anionic multilamellar vesicles of the same lipid composition as used in the high-throughput screen (Fig. 1B) the peptides equilibrated across the multiple bilayers in within about 15-30 minutes (Figs. 6E and 6F) indicating very rapid, spontaneous translocation. In confocal images taken quickly (~10 min after addition of peptide), we could sometimes observe the last 10-20% of the transbilayer equilibration of the peptide-TAMRA conjugates suggesting a half-time of translocation on the order of 2-4 minutes (See supplemental information for more on the translocation rate). To test the generality of the peptide translocation we also measured translocation across multibilayer vesicles composed of 100% zwitterionic phosphatidylcholine (PC). The selected peptides rapidly translocated across multibilayer PC bilayers, indicating that translocation activity is a general property of these peptides and is not dependent on specific electrostatic interactions between the peptide arginines and anionic lipids
Figure 6.
Spontaneous translocation across multilamellar vesicles. A: Multilamellar vesicles were prepared by dispersing vacuum dried lipid in PBS followed by ten cycles of freezing and thawing. Vesicles were either made from 90% phosphatidylcholine (PC) + 10% phosphatidylglycerol (PG) to make anionic PC/PG vesicles or from 100% phosphatidylcholine (PC) to make zwitterionic vesicles. For initial characterization, vesicles were prepared with lipids that had been doped with a small amount of dye-labeled lipid for visualization by laser scanning confocal fluorescence microscopy. Examples of Multilamellar vesicles are shown in the composite image in panel. B: In this example experiment, PC/PG multilamellar vesicles were simultaneously incubated with free TAMRA dye (red) and with fluorescein dextran 3kD (FD3, green) for 30 minutes followed by confocal microscope imaging. The four images are of the same MLV (clockwise from upper left):Fluorescein channel, TAMRA channel, merged channel, phase contrast image. C: A translocation experiment with free TAMRA and FD3. After 30 minutes of incubation with a solution of MLVs, 4 μl of vesicles were placed between a slide and coverslip and imaged. D: In this image, four vesicles in the field were selected and the intensity was scanned across them. The low intensity inside the vesicles shows that both free TAMRA and FD3 are excluded. E: Translocating peptide TP1-TAMRA and FD3 were co-incubated with MLVs for 30 minutes and imaged using confocal microscopy. F: As shown by the intensity scans across the multilamellar vesicles in the image, the TAMRA-peptide (left) equilibrates completely across the multiple bilayers while FD3 (right) is excluded.
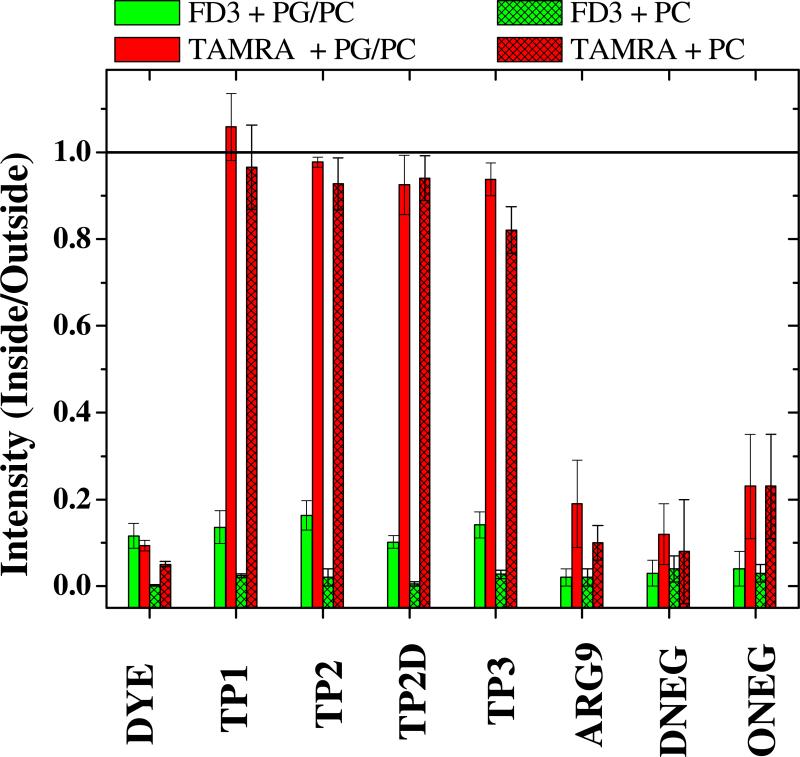
As we show quantitatively in Fig. 7 the translocating peptides and the covalently attached TAMRA moiety equilibrated across PC/PG bilayers and PC bilayers such that the ratio of interior to exterior fluorescence was about 1. The all D-amino acid version of TP2 (TP2D) behaved identically to TP2, indicating that there is no stereochemically specific interaction with lipids, which are chiral, in translocation. In each experiment, preformed vesicles were co-incubated with peptides and with fluorescein dextran, FD3. The dextran did not enter the vesicles measurably, giving only a background intensity ratio of about 0.1. Similarly, free TAMRA did not enter vesicles. For one negative control peptide we used the known cationic cell penetrating peptide, Arg9, which was contained in the library and was not found as a positive. We also used a randomly selected negative peptide from the screen (ONEG) and a negative peptide designed by replacing three arginine residues in the translocating peptide TP3 with aspartate (DNEG). The negative peptides did not translocate substantially across anionic or zwitterionic multibilayer vesicles even in experiments where we increased the incubation time to several hours. These translocation experiments showed that the peptides selected by the orthogonal screen have the remarkable ability to spontaneously translocate across multiple lipid bilayer membranes in just a few minutes while carrying a large polar cargo moiety.
Figure 7.
Compilation of MLV translocation data. The intensity ratio is the ratio of TAMRA fluorescent intensity inside and outside multilamellar vesicles after 30 minutes of simultaneous incubation with peptide or free TAMRA plus FD3. Experiments were done at 2-5 μM peptide, 10 μg/ml free FD3 and 6 mM lipid in PBS. Two lipid compositions were used, 10 mol % phosphatidylglycerol + 90% phosphatidylcholine and 100% phosphatidylcholine. The column labeled “Dye” refers to free TAMRA (red) and free FD3 (green) in the absence of peptide. The other red (TAMRA) bars are for TAMRA attached to peptide. The green (FD3) bars plotted along with each peptide are for FD3 that was present at the same time as the peptide. Measurements were made on vesicles that were spherical and were at least 5 μm in diameter. Images were always taken at the focal plane at which the vesicles have the largest area. Each bar is the result of measurements on at least 8 vesicles from at least two independent experiments. Error bars represent standard deviations.
The confocal fluorescence experiments showed no strong peptide binding to the multilamellar vesicle membranes, which is consistent with their low predicted membrane binding18,19. Because we observe little detectable membrane labeling, the steady state concentration of vesicle-bound peptide must be relatively low. To explain the observed overall translocation time, the rate of translocation across each bilayer must be high. We estimate that the first order translocation rate must be 0.1 sec-1 or faster. (See supplemental information for more on translocation rates).
It might seem surprising that peptides with multiple charged residues can spontaneously translocate across bilayer membranes. However, in our recent work on antimicrobial peptides3,15,20-23 we have discussed this issue in terms of an “interfacial activity model” where interfacial activity is defined as the ability of an amphipathic peptide to partition into the membrane interface and alter the vertical segregation of the lipid polar and nonpolar groups. We have proposed that there is a broad overlap between membrane permeabilizing and membrane translocating peptides. Arginine (and perhaps also lysine) has a special role in interfacial activity because it can interact with lipid phosphate groups when inserted into membranes. As we20,21 and others24,24-27 have recently discussed, the lipid phosphates may act as chaperones for the cationic amino acids and allow their deep penetration into the bilayer, or translocation across the bilayer. Biophysical studies of the molecular mechanism of translocation are currently underway in our laboratories.
Peptide translocation into living cells
In Figure 8 we show that the peptides discovered here translocate rapidly across the plasma membranes of living Chinese Hamster ovary (CHO) cells at room temperature in Dulbecco's phosphate buffered saline, which does not have a carbon source. These are conditions under which endocytosis and other active cellular processes are arrested (see below). Nonetheless, peptide translocation into cells is detectible within one minute of addition, and by ten minutes the translocating peptide and its TAMRA cargo moiety fill the cell cytoplasm with a bright, diffuse fluorescence, suggesting rapid translocation into cells. Multiple additional lines of evidence indicate that the translocating peptides described here are not entering cells by endocytosis at room temperature, and that endocytosis is inhibited under the conditions of Figure 8 (22°C in PBS). We know that the same peptides rapidly and spontaneously translocate across multiple synthetic lipid bilayers (Fig. 5) on the same timescale as they enter cells, thus they have an inherent capacity for translocation that has not been demonstrated for other “cell penetrating” peptide. Furthermore, when the translocating peptides are incubated with CHO cells at 22°C in PBS, no FD3-filled endosomes are seen in the cells (Fig.8). If any type of endocytosis were occurring we would have observed endosomes with entrapped FD3 which was always present in the external buffer. We also did not observe FD3 in endosomes in the absence of peptides. The classical, endocytosis-dependent5, cell penetrating peptide Arg9 binds to cells under the conditions of Figure 8 (22°C in PBS), but does not enter cells at all (see Figure 9, top left), confirming that endocytosis is inhibited. Also, at 22°C in PBS, Arg9-TAMRA does not trigger the entrapment of FD3 within endosomes, showing that no endocytosis can be triggered by Arg9 under these conditions. In contrast to these observations at 22°C in PBS, we show a very different behavior for Arg9-TAMRA and FD3 incubated with cells at 37°C in full growth media (Fig. 9, top right). Under those conditions, Arg9-TAMRA enters cells by endocytosis, as expected, and we observe large numbers of intracellular organelles which contain co-encapsulated Arg9-TAMRA and FD3. When the translocating peptide TP1-TAMRA and FD3 were similarly incubated with CHO cells at 37°C in full growth media (Fig. 9, bottom right), the behavior is very different from Arg9-TAMRA and 37°C, but the same as for TP1-TAMRA at 22°C in PBS (Fig. 8). Even at 37°C in full growth media, the translocating peptide fills the cytoplasm with a diffuse fluorescence. No punctuate fluorescence is observed, thus there is no entrapment of peptide in endosomes. Just as importantly, there is no entrapment of FD3 in endosomes when translocating peptides and free FD3 are co-incubated with cells at 37°C in full growth media. All the translocating peptides (Fig. 5) behave exactly like TP1-TAMRA shown in Figures 8 and 9. Furthermore, we have shown that translocation of our peptides into cells is not inhibited by metabolic poisons such as rotenone, as their behavior is identical to the behavior shown in Figure 8, even when ATP synthesis is blocked (not shown). Thus, it is clear that the translocating peptides discovered here can enter cells by non-endocytotic pathways, consistent with our findings that they spontaneously translocate across synthetic lipid vesicles.
Figure 8.
Example of cellular translocation. Cellular translocation was measured with living Chinese Hamster Ovary (CHO) cells that had been incubated for 30 minutes at room temperature (22°C) in phosphate buffered saline (i.e. without a carbon source) to arrest active cellular processes such as endocytosis. At 30 minutes 10 μg/ml of a small 3000 Da fluorescein labeled dextran (green) and 2 μM TAMRA-labeled translocating peptide TP1 (Red, Fig. 5) were added. Laser scanning confocal fluorescence microscopy was used to image CHO cells 1 minute after addition of peptide (top) and the same cells 10 minutes after the addition of peptide (bottom). Images were taken in situ without washing away free peptide outside the cells. From the upper left: a: Fluorescein dextran channel; b:TAMRA-peptide channel; c: Merged channel; d: Phase contrast channel.
Figure 9.
Control experiments for cellular translocation. Cellular translocation of TAMRA-labeled peptides was measured with living Chinese Hamster Ovary (CHO) cells prepared as described in Figure 8 and in the text. For room temperature experiments (left column, marked as 22°C) cells were incubated in PBS for 30 minutes, followed by addition of 10 μg/ml of a small 3000 Da fluorescein labeled dextran (FD3, green) and 2 μM TAMRA-labeled peptide (RED). Confocal microscopy was immediately begun on the samples without washing away free peptide or FD3. The left column (22°C) images were taken about 10 min after addition of peptide. For experiments at 37°C (right column images, marked as 37°C) cells were incubated in full growth media (including a carbon source and serum with growth factors) with 10 μg/ml FD3 and 2 μM TAMRA-labeled peptide for 20 minutes, followed by confocal microscope imaging without washing away free peptide or FD3. For all images we show four channels: from the upper left: a: Fluorescein dextran (FD3) channel; b:TAMRA-peptide channel; c: Merged channel; d: Phase contrast channel. In the top row of images we show results for the peptide Arg9-TAMRA a well-known cell penetrating peptide that enters cells by endocytosis and does not translocate spontaneously across membranes (Fig. 7). Top left is an experiment done at 22°C in PBS and the top right in an experiment is done at 37°C in full growth media. In the bottom left we show the results for DNEG-TAMRA(bottom) a negative peptide designed by replacing the three arginines of TP3 with aspartate. DNEG-TAMRA does not translocate across synthetic membranes (Fig. 7). In the bottom right we show results for a translocating peptide TP2-TAMRA incubated with CHO cells for 20 minutes, along with 10 μg/ml FD3, at 37°C in full growth media.
Conclusion
In summary, we have used combinatorial chemistry and orthogonal high-throughput screening to specifically identify a conserved family of related peptides within a 10,368-member library that can cross membranes spontaneously. We demonstrate the spontaneous translocation of the selected peptides and an attached polar cargo moiety across synthetic multibilayer lipid vesicles and across the membranes of living cells. The entry mechanism is distinctly different from the well known, highly cationic cell penetrating peptides, such as tat and Arg9 which do not translocate spontaneously into vesicles at all, and which probably enter cells by endocytosis5. Most importantly, the sequences identified here can readily carry a polar cargo moiety across synthetic and cellular membranes. The membrane barrier does not exist for these translocating peptides, and peptides with such properties could eliminate the membrane permeability barrier problem in drug design.
Supplementary Material
Acknowledgements
The authors thank Christopher M. Bishop for peptide synthesis, labeling and purification, Lirong Chen for assistance with cell culture and confocal microscopy and Thomas Freeman for writing data analysis and display software. Funded by NIH grant GM060000 (WCW), GM095930 (KH) and NSF grants DMR-1003411 (WCW) and DMR-1003441 (KH).
Footnotes
Supporting Information Available. A supplemental file is available which contains auxiliary data and images supporting the conclusion of this work. This information is available free of charge via the Internet at http://pubs.acs.org/.
Reference List
- 1.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv. Drug Deliv. Rev. 2001;46(1-3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 2.Heitz F, Morris MC, Divita G. Br. J. Pharmacol. 2009;157(2):195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rathinakumar R, Wimley WC. FASEB J. 2010;24:3232–3238. doi: 10.1096/fj.10-157040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt N, Mishra A, Lai GH, Wong GC. FEBS Lett. 2010;584(9):1806–1813. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 5.Langel U. Handbook of Cell Penetrating Peptides. Second Edition CRC Press; Boca Raton, FL: p. 2006. [Google Scholar]
- 6.Raagel H, Saalik P, Pooga M. Biochim. Biophys. Acta. 2010;1798(12):2240–2248. doi: 10.1016/j.bbamem.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Chugh A, Eudes F, Shim YS. IUBMB. Life. 2010;62(3):183–193. doi: 10.1002/iub.297. [DOI] [PubMed] [Google Scholar]
- 8.Olson ES, Jiang T, Aguilera TA, Nguyen QT, Ellies LG, Scadeng M, Tsien RY. Proc. Natl. Acad. Sci. U. S. A. 2010;107(9):4311–4316. doi: 10.1073/pnas.0910283107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Youngblood DS, Hatlevig SA, Hassinger JN, Iversen PL, Moulton HM. Bioconjug. Chem. 2007;18(1):50–60. doi: 10.1021/bc060138s. [DOI] [PubMed] [Google Scholar]
- 10.Simon MJ, Kang WH, Gao S, Banta S, Iii BM. Ann. Biomed. Eng. 2010 doi: 10.1007/s10439-010-0144-x. [DOI] [PubMed] [Google Scholar]
- 11.Lam KS, Lehman AL, Song A, Doan N, Enstrom AM, Maxwell J, Liu R. Methods Enzymol. 2003;369:298–322. doi: 10.1016/S0076-6879(03)69017-8. 298-322. [DOI] [PubMed] [Google Scholar]
- 12.Rausch JM, Marks JR, Wimley WC. Proc. Natl. Acad. Sci. U. S. A. 2005;102(30):10511–10515. doi: 10.1073/pnas.0502013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant GA. Synthetic Peptides:A User's Guide. WH Freeman and Company; New York: 1992. [Google Scholar]
- 14.Mayer LD, Hope MJ, Cullis PR. Biochim. Biophys. Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 15.Rathinakumar R, Wimley WC. J. Am. Chem. Soc. 2008;130(30):9849–9858. doi: 10.1021/ja8017863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keil P. Specificity of Proteolysis. Berlin-Heidelberg-New York; Springer-Verlag: 1992. [Google Scholar]
- 17.Sadler K, Eom KD, Yang JL, Dimitrova Y, Tam JP. Biochemistry. 2002;41(48):14150–14157. doi: 10.1021/bi026661l. [DOI] [PubMed] [Google Scholar]
- 18.White SH, Wimley WC. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 19.Wimley WC, White SH. Nature Struct. Biol. 1996;3(10):842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 20.Wimley WC, Hristova K. J. Membr. Biol. 2011 doi: 10.1007/s00232-010-9323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hristova K, Wimley WC. J. Membr. Biol. 2010 [Google Scholar]
- 22.Wimley WC. ACS Chem. Biol. 2010;5(10):905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rathinakumar R, Walkenhorst WF, Wimley WC. J. Am. Chem. Soc. 2009;131:7609–7617. doi: 10.1021/ja8093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schow EV, Freites JA, Cheng P, Bernsel A, von HG, White SH, Tobias DJ. J. Membr. Biol. 2010 doi: 10.1007/s00232-010-9330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krepkiy D, Mihailescu M, Freites JA, Schow EV, Worcester DL, Gawrisch K, Tobias DJ, White SH, Swartz KJ. Nature (London) 2009;462(7272):473–479. doi: 10.1038/nature08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freites JA, Tobias DJ, White SH. Biophys. J. 2006;91(11):L90–L92. doi: 10.1529/biophysj.106.096065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hessa T, White SH, von HG. Science. 2005;307(5714):1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.