Abstract
The potential utility of the Collaborative Cross (CC) mouse resource was evaluated to better understand complex traits related to energy balance. A primary focus was to examine if genetic diversity in emerging CC lines (pre-CC) would translate into equivalent phenotypic diversity. Second, we mapped quantitative trait loci (QTL) for 15 metabolism- and exercise-related phenotypes in this population. We evaluated metabolic and voluntary exercise traits in 176 pre-CC lines, revealing phenotypic variation often exceeding that seen across the eight founder strains from which the pre-CC was derived. Many phenotypic correlations existing within the founder strains were no longer significant in the pre-CC population, potentially representing reduced linkage disequilibrium (LD) of regions harboring multiple genes with effects on energy balance or disruption of genetic structure of extant inbred strains with substantial shared ancestry. QTL mapping revealed five significant and eight suggestive QTL for body weight (Chr 4, 7.54 Mb; CI 3.32–10.34 Mb; Bwq14), body composition, wheel running (Chr 16, 33.2 Mb; CI 32.5–38.3 Mb), body weight change in response to exercise (1: Chr 6, 77.7Mb; CI 72.2–83.4 Mb and 2: Chr 6, 42.8 Mb; CI 39.4–48.1 Mb), and food intake during exercise (Chr 12, 85.1 Mb; CI 82.9–89.0 Mb). Some QTL overlapped with previously mapped QTL for similar traits, whereas other QTL appear to represent novel loci. These results suggest that the CC will be a powerful, high-precision tool for examining the genetic architecture of complex traits such as those involved in regulation of energy balance.
Keywords: genetics, wheel running, adiposity, body weight, quantitative trait loci
regulation of body weight and composition is dependent primarily on energy balance, the net sum of calories consumed and calories expended. Complex interactions between many genes and environmental factors modulate body weight, adiposity, and physical activity, contributing to the development of obesity, obesity-related disease, and individual differences in response to treatments and interventions to combat obesity (5, 20, 26, 34, 49) including exercise (26). Human genome-wide association studies have identified several loci associated with body weight and obesity, although the individual contributions of these loci to the genetic variance are relatively small (38, 41), and the functions of identified genes are often difficult to ascertain.
Studies in mice have identified hundreds of quantitative trait loci (QTL) for body weight, obesity, and physical activity traits (28, 29, 31, 33, 39, 46, 50). Most QTL studies have used intercross or backcross (BC) mouse populations to identify regions of the genome associated with metabolism- and exercise-related phenotypes. These studies have several limitations. First, QTL in these populations generally map to broad genomic intervals due to large regions of linkage disequilibrium (LD) (11, 12). Recently, the use of advanced intercross lines (AIL) and recombinant inbred lines (RIL) has increased QTL mapping precision, but identification of the underlying genetic variation remains elusive in almost all cases (14, 16, 25, 30). Second, the majority of QTL mapping crosses employ only the most commonly used inbred strains, which harbor a relatively small proportion of the genetic diversity of Mus musculus, greatly limiting the search space for genetic variation controlling complex traits (44). In addition, a large fraction of the genome are blind spots for genetic mapping because recent shared ancestry leads to identity by descent in crosses based on two parental strains (59).
The Collaborative Cross (CC) recombinant inbred panel was conceived as an improved resource for mammalian systems genetics and to overcome many of the limitations of traditional QTL mapping populations (10, 55). To investigate the utility and insights that can be learned from the CC, an experiment was performed involving incipient CC lines that had undergone at least five generations of inbreeding (pre-CC). CC lines originated from a cross of eight founder inbred strains, including three representing wild-derived strains from three of the Mus musculus subspecies and traditional inbred strains genetically predisposed to the development of complex disorders such as types 1 and 2 diabetes, obesity, and insulin insensitivity (36). Analysis of pre-CC genetic architecture revealed high diversity, balanced allele frequencies, and well-distributed recombination, all ideal qualities for a mapping panel (2).
In this study, we characterized phenotypic variation and mapped QTL in the pre-CC population for a suite of traits revolving around energy balance, including body weight and body composition, both before and after a 12-day period of voluntary exercise in running wheels. We found that the pre-CC lines adequately captured and often exceeded the phenotypic variation among the eight founder strains. In some instances, such as wheel running distances and body weight change in response to exercise, phenotypic variation in the pre-CC far exceeded the range of variability afforded by the progenitors. Importantly, many phenotypic correlations existing within the progenitor and also larger panels of extant inbred strains were disrupted in the pre-CC population with its randomly assorted variation. Despite the absence of full homozygosity in the pre-CC population and the small sample size, which limit statistical power in this proof-of-concept study, we identified five significant QTL and eight suggestive QTL for many energy balance traits, including body weight, body fat, levels of voluntary exercise, body weight change in response to exercise, and food intake during exercise. QTL mapping in the pre-CC population replicated existing QTL regions and identified novel QTL for the metabolism and exercise traits we evaluated.
EXPERIMENTAL PROCEDURES
Origin of the mice and development of the pre-CC lines and their husbandry prior to the current phenotyping screen are described elsewhere (2, 7). Briefly, five traditional (129S1/SvImJ, A/J, C57BL/6J, NOD/LtJ, NZO/H1Lt) and three wild-derived (CAST/EiJ, PWK/PhJ, and WSB/EiJ) inbred mouse strains were outcrossed for two generations, and the resulting G2:F1 population was inbred by sibling matings at the Oak Ridge National Laboratories (ORNL). The pre-CC lines used in the experiments described in this paper were a subset of 650 lines initiated at ORNL (7). Initial analysis of pre-CC lines demonstrated that all eight of the founder alleles contributed equivalently to the pre-CC population, suggesting that genetic contributions from each of the eight founder alleles were present at all loci (2). Recombination break points were distributed approximately evenly across the genome in our sample.
Mice were weaned at ORNL and transferred to a phenotyping facility at the University of North Carolina at Chapel Hill. One hundred eighty-four lines ranging from generation G2:F5 to G2:F12 were transferred to the phenotyping facility, and 176 were included in the final analyses. Animals were transferred according to availability. Exercise and metabolism data were collected as part of a high-throughput phenotyping screen (Table 1), and only these methods will be discussed herein.
Table 1.
Experimental time line
| Day | Phenotype | Procedure |
|---|---|---|
| Day 1 | Arrival | |
| Days 4–7 | Sedentary food intake (FI) | Mice housed in cages attached to running wheels with guillatine doors closed to prevent access to running wheel. |
| Day 7 | Initial BW and BFP | BW measured and BFP determined by MRI |
| Day 8 | RER | Indirect calorimetry (24 h) |
| Days 9–20 | Wheel running and food intake: D1, D56, D12 S1, S56, S12 FIRun | Mice returned to home cages. Guillotine doors opened to allow access to running wheels. FI measured every other day. |
| Day 21 | BWpost and BFpost | Guillotine doors closed. |
| BW measured and BFP determined by MRI | ||
| Day 22 | RERpost | Indirect calorimetry (24 h) |
| Day 23 | Euthanasia and dissection |
See text for definitions.
Adult male mice, 10–12 wk of age, from 176 pre-CC lines and the eight CC founder strains (129S1/SvImJ, n = 10; A/J, n = 10; CAST/EiJ, n = 11; C57BL/6J, n = 11; NOD/LtJ, n = 7; NZO/H1LtJ, n = 9; PWK/PhJ, n = 11; and WSB/EiJ, n = 7) were housed individually in standard laboratory cages with attached running wheels (1.1 m diameter; Lafayette Industries, Lafayette, IN) in a temperature- (23 ± 1°C) and humidity-controlled vivarium with a standard 12:12-h light-dark cycle (lights on at 0600). Access to the running wheels was blocked for the first 8 days after the mice arrived in the vivarium. Mice had ad libitum access to food and water. Ground standard laboratory chow that had been dyed blue for easier spill detection and collection (LabDiet 5053, blue; TestDiet, Richmond, IN) was presented in spill-resistant feeders (OYC Rodent Café; OYC International, Andover, MA). Any spillage was collected and weighed. Daily average food intake was calculated for 3 days prior to wheel access [sedentary food intake (FI)] by dividing the total amount of food consumed during this period by 3. During the 12 days of wheel running, food intake (FIRun) was measured every other day, and daily averages were calculated. Intakes were corrected for body weight (BW) by dividing average intakes by the average BW for the measurement period.
BW was measured at the beginning (BW) and end (BWpost) of the sedentary and wheel running phases of the experiment (Table 1). Body composition was evaluated prior to the start [body fat percentage (BFP)] of wheel running and again after the 12-day wheel trial (BFpost) using MRI (EchoMRI, Houston, TX) to determine fat and lean mass percentages. Change in BW and BFP in response to exercise was calculated by subtracting the preexercise value from the postexercise value (BWnet and BFnet, respectively). Negative values indicate a loss of BW or fat in response to wheel running. Indirect calorimetry was used to determine energy expenditure for each mouse before [RER (respiratory exchange ratio)] and after running wheel access (RERpost). The day before the mice were given access to the running wheels, mice were placed into calorimetric chambers (TSE calosys; TSE Systems, Germany) and O2 consumption, CO2 production, and respiratory exchange ratios were measured for 24 h.
Wheel running was recorded continuously over a 12-day period using an automated activity wheel monitoring program (AWM; Lafayette Industries, Lafayette, IN). For data analysis, distance and speed data were directly extracted for day 1(D1, S1), and running distances and speeds were averaged for days 5 and 6 (D56, S56) and days 11 and 12 (D12, S12) of wheel access.
Genotype data for QTL mapping were collected using intermediary models of the Mouse Diversity Array (60). This array contained 181,752 high-performing single-nucleotide polymorphisms (SNP) that spanned the entire mouse genome (2).
All procedures performed within this series of experiments were approved by the University of North Carolina - Chapel Hill Institutional Animal Care and Use Committee.
One-way ANOVA was used to assess strain effects among the founder strains on each of the phenotypes measured. Tukey's HSD (honestly significant difference) post hoc comparisons were used to determine significant differences among each of the strains. Pearson's correlations were performed to determine significant associations among the phenotypes for individual mice within the founder and pre-CC lines. Statistical analysis for QTL mapping is described by Aylor et al. (2). Briefly, QTL mapping was performed using a regression model (37) where each phenotype was regressed on expected haplotypes at each SNP interval. Resultant F-statistics were transformed to LOD scores. Genome-wide significance of LOD scores were determined by permutation.
RESULTS
Phenotypic variation among the pre-CC lines.
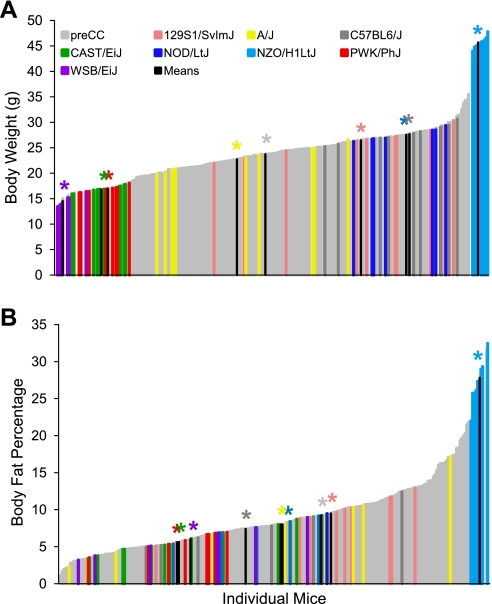
The genotypic diversity among the founder strains resulted in high phenotypic diversity. Metabolism- and exercise-related traits were measured over a 3-wk period. Phenotypic measurements included body weight and body fat percentage before and after 12 days of wheel running, respiratory exchange ratio, food intake while sedentary or during wheel running, and running phenotypes such as speed and distance measured on days 1, 5 and 6, and 11 and 12. There were highly significant differences among the eight strains in the 15 phenotypes tested [P < 0.001; Supplemental Table S1 (supplementary materials are found in the online version of this paper at the Journal website)]. Initial BW values ranged from 13.5 g in WSB/EiJ to 47.8 g in NZO/H1LtJ and averaged 26.5 ± 0.84 g across the eight strains. Similar widespread variability was observed in initial BFP, ranging from 2.4% in A/J to 32.6% in NZO/H1LtJ, and in wheel running distance and speed at all time points evaluated.
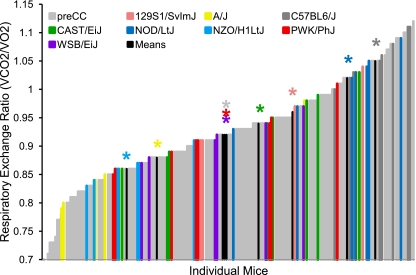
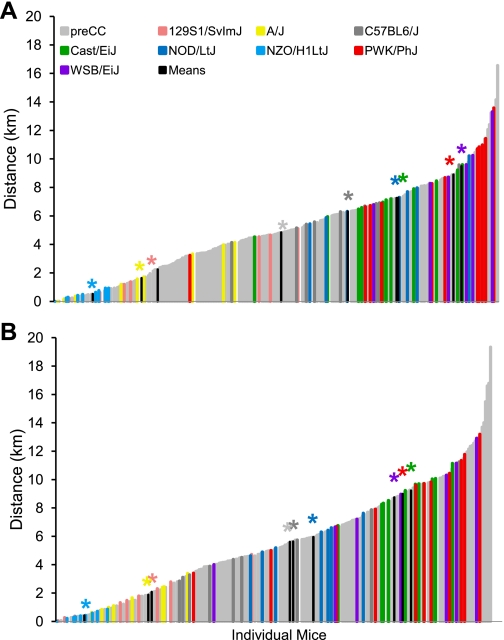
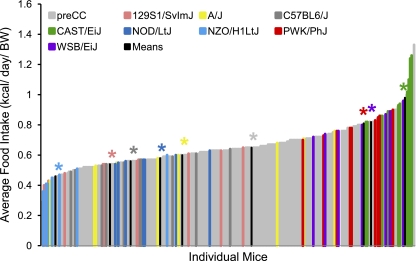
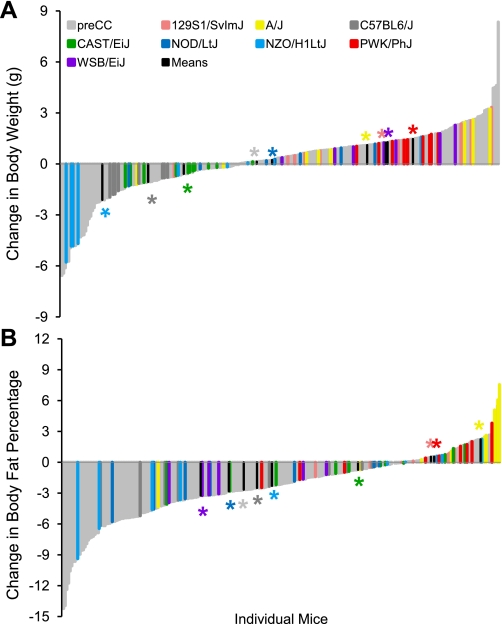
Phenotypic diversity in the pre-CC lines matched and often exceeded that seen in the founder strains. Ranges of phenotypic values for traits such as initial BW and initial BFP, although highly variable in the pre-CC population, did not exceed those observed in the founder strains (Table 2 and Fig. 1). However, variability in wheel running traits, food intake (while sedentary or while running), indirect calorimetry measures, and changes in BW and BF in response to wheel running often exceeded that seen in the founders (Table 2 and Supplemental Table S1). For instance, nocturnal RER, an indicator of energy utilization, measured prior to exercise ranged from 0.79 to 1.11 in the founder strains, whereas the pre-CC lines showed a broader range of variability with values from 0.69 to 1.12 (Fig. 2). Likewise, average wheel running distances on days 5 and 6 (D56) and days 11 and 12 (D12) of the running period far exceeded values in the founder strains, with maximum values of 13,600 and 13,200 m/day evident in the PWK/PhJ founder strain and 16,600 and 19,300 m/day in the pre-CC lines, respectively (Fig. 3). The pre-CC lines also demonstrated greater variability in average daily food intake during wheel access, with individual mice consuming 0.29–1.33 kcal·day−1·g BW−1, while individual intakes ranged from 0.36 kcal·day−1·g BW−1 in the NZO/H1LtJ to 1.26 g in CAST/EiJ (Fig. 4). Additionally, BW change after exercise (BWnet) ranged from a loss of 5.8 g BW in NZO/H1LtJ to a gain of 3.3 g BW in 129S1/SlvmJ and from a loss of 6.6 g BW to a gain of 8.4 g BW in the pre-CC mice (Fig. 5). Body fat loss (g) during exercise in the pre-CC far exceeded that of the founders, whereas body fat gain (g) was greater in the founders than in the pre-CC. Net changes in BFP ranged from a loss of 14.0% to a gain of 5.5% in the pre-CC and a loss of 9.4% in NZO/H1LtJ and a gain of 7.5% in A/J (Fig. 5).
Table 2.
Descriptive statistics for metabolic and exercise traits in pre-CC lines
| Trait | n | Mean (95% CI) | SD | Minimum | Maximum |
|---|---|---|---|---|---|
| Preexercise | |||||
| BW, g | 176 | 23.74 (23.15, 24.33) | 3.95 | 15.01 | 35.5 |
| BFP | 176 | 9.23 (8.56, 9.91) | 4.91 | 1.09 | 31.65 |
| FI, kcal/day/BW | 174 | 0.61 (0.58, 0.64) | 0.21 | 0.29 | 2.42 |
| Nocturnal RER | 130 | 0.92 (0.90, 0.93) | 0.09 | 0.69 | 1.12 |
| Exercise traits | |||||
| Running distances, m | |||||
| Day 1 | 176 | 4,096.73 (3,632.35, 4,561.11) | 3,121.51 | 0 | 14,940.84 |
| Days 5 and 6 | 176 | 4,850.18 (4,404.45, 5,295.91) | 2,996.16 | 0.97 | 16,557.57 |
| Days 11 and 12 | 175 | 5,610.96 (5,047.28, 6,174.63) | 3,778.03 | 10.91 | 19,333.33 |
| Running speeds, m/min | |||||
| Day 1 | 176 | 8.63 (8.01, 9.24) | 4.13 | 0 | 17.52 |
| Days 5 and 6 | 176 | 13.66 (12.92, 14.40) | 4.95 | 0.16 | 28.1 |
| Days 11 and 12 | 175 | 15.54 (14.64, 16.43) | 6 | 0.85 | 33.63 |
| FI, kcal/day/BW | 175 | 0.65 (0.63, 0.67) | 0.12 | 0.29 | 1.33 |
| Postexercise | |||||
| BW, g | 176 | 23.88 (23.32, 24.45) | 3.78 | 14.7 | 36.34 |
| BFP | 176 | 6.52 (5.92, 7.11) | 4.02 | 0.62 | 21.71 |
| Change in BW, g | 176 | 0.14 (−0.16, 0.45) | 2.08 | −6.57 | 8.35 |
| Change in BFP | 176 | −2.67 (−3.14, −2.19) | 3.17 | −13.95 | 5.5 |
CC, Collaborative Cross; RER, respiratory exchange ratio (Vco2/Vo2).
Fig. 1.
Body weights (BW) and body fat percentages (BFP) in pre-CC and founder lines prior to running wheel access. A: phenotypic variation among founder strains exceeded the variation among the pre-CC lines for BW. BW for all pre-CC lines fell within the low and high extreme values of WSB/EiJ and NZO/H1LtJ, respectively. B: variation in BFP in pre-CC lines was comparable to that of founder strains. Although several pre-CC lines held extreme low BFP values, extreme high BFP were determined solely by NZO/H1LtJ. Group means for each founder line are indicated in black with appropriate color-coded asterisk above to indicate strain being depicted.
Fig. 2.
Nocturnal respiratory exchange ration (RER) in founder and pre-CC lines prior to exercise. RER (V̇co2/V̇o2), a measure of energy utilization, was far more variable in pre-CC lines than in founders. RER values of 0.7 ad 1.0 represent using pure fat or carbohydrate, respectively, as energy. Mixed diets tend to produce RER values between 0.8 and 0.9. Extreme values beyond 1.0 may indicate hyperactivity, anxiety, or stress. Several pre-CC lines demonstrated extreme low or high RER vs. founder lines. Group means for each founder line are indicated in black with appropriate color-coded asterisk above to indicate strain being depicted.
Fig. 3.
Average running distances in pre-CC and founder lines on days 5 and 6 (D56) and days 11 and 12 (D12) of ad libitum running wheel access. Mice were given 12 days of ad libitum running wheel access. Running distances and speeds were recorded continuously over 24 h in 1-min increments. Mean distances for D56 (A) and D12 (B) were exceedingly more variable among pre-CC lines than founder lines. Group means for each founder line are indicated in black with appropriate color-coded asterisk above to indicate strain being depicted.
Fig. 4.
Average daily food intake in founder and pre-CC mice during wheel access. Food intake was measured every other day during 12 days of running wheel access. Intakes were averaged across the 12 days and corrected for BW. Intakes are expressed as kcal·day−1·BW−1. Although NZO/H1LtJ held the extremely low food intake values, food intake among pre-CC lines varied greatly, with a few of the pre-CC lines representing extremely high food intake values. Group means for each founder line are indicated in black with appropriate color-coded asterisk above to indicate strain being depicted.
Fig. 5.
Net change in BW and BFP in pre-CC and founder lines after ad libitum access to running wheels for 12 days. BW (A) and BF (B) change in response to exercise was again more variable in pre-CC lines than in founders. Group means for each founder line are indicated in black with appropriate color-coded asterisk above to indicate strain being depicted.
Disruption of phenotype correlations.
Recombination and assortment of the genomes of founder strains within the pre-CC lines led to disruption of phenotypic correlations commonly seen in the founder strains. For example, significant correlations were identified between most metabolism and exercise traits in the founder strains. Specifically, correlation analysis revealed significant correlations between BFP, both before and after exercise, and the traits BW, BWpost, BWnet, BF change after exercise (BFnet), food intake while sedentary (FI), food intake during the 12-day exercise period (FIRun), RER, and all wheel running distance and speed traits in the founder strains (Table 3). Likewise, BW was significantly correlated with all measured traits with the exception of RER and BFnet. In contrast, initial BW and BFP were uncorrelated with wheel running in the pre-CC mice, although they remained correlated with other metabolic phenotypes such as FI, RER, BWnet, and BFnet (Table 4). Nevertheless, running traits were correlated with each other and with food intake during running (Table 4). In the pre-CC lines, only running distance and speed on D1 of the running period were correlated with RER, which was measured on the night immediately prior to initiation of running (Table 4).
Table 3.
Correlations among phenotypes in progenitor lines
| BFP | FI | RER | D1 | D56 | D1112 | S1 | S56 | S1112 | FIRun | BWpost | BFpost | BWnet | BFnet | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Preexercise | ||||||||||||||
| BW | 0.864** | −0.395** | −0.095 | −0.591** | −0.620** | −0.668** | −0.448** | −0.447** | −0.447** | −0.719** | 0.984** | 0.798** | −0.496** | −0.200 |
| BFP | −0.246* | −0.409** | −0.483** | −0.557** | −0.549** | −0.416** | −0.444** | −0.450** | −0.552** | 0.840** | 0.917** | −0.474** | −0.265* | |
| FI | −0.231 | 0.354** | 0.333** | 0.456** | 0.201 | 0.172 | 0.215 | 0.792** | −0.450** | −0.246* | −0.103 | 0.011 | ||
| Nocturnal RER | 0.261* | 0.368** | 0.286* | 0.368** | 0.466** | 0.459** | −0.050 | −0.091 | −0.462** | 0.005 | −0.112 | |||
| Exercise traits | ||||||||||||||
| Running distances | ||||||||||||||
| D1 | 0.821** | 0.762** | 0.877** | 0.676** | 0.608** | 0.600** | −0.615** | −0.552** | 0.123 | −0.180 | ||||
| D56 | 0.905** | 0.783** | 0.844** | 0.793** | 0.591** | −0.640** | −0.638** | 0.129 | −0.169 | |||||
| D1112 | 0.675** | 0.717** | 0.775** | 0.718** | −0.694** | −0.629** | 0.101 | −0.163 | ||||||
| Running speeds | ||||||||||||||
| S1 | 0.796** | 0.711** | 0.408** | −0.464** | −0.528** | 0.126 | −0.273* | |||||||
| S56 | 0.893** | 0.345** | −0.480** | −0.562** | 0.011 | −0.269* | ||||||||
| S1112 | 0.407** | −0.482** | −0.594** | 0.005 | −0.326** | |||||||||
| FIRun | −0.739** | −0.502** | 0.109 | 0.082 | ||||||||||
| Postexercise | ||||||||||||||
| BWpost | 0.805** | −0.334** | −0.107 | |||||||||||
| BFpost | −0.279* | 0.137 | ||||||||||||
| BWnet | 0.557** |
See text for definitions.
P < 0.05,
P < 0.01.
Table 4.
Correlations among phenotypes in preCC lines
| BFP | FI | RER | D1 | D56 | D1112 | S1 | S56 | S1112 | FIRun | BWpost | BFpost | BWnet | BFnet | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Preexercise | ||||||||||||||
| BW | 0.487** | −0.243** | −0.180* | −0.129 | −0.074 | −0.125 | −0.027 | 0.046 | 0.003 | −0.524** | 0.856** | 0.311** | −0.341** | −0.354** |
| BFP | −0.271** | −0.276** | −0.120 | −0.055 | −0.062 | −0.119 | −0.036 | 0.024 | −0.378** | 0.314** | 0.762** | −0.355** | −0.572** | |
| FI | 0.069 | −0.011 | 0.007 | −0.002 | 0.051 | 0.001 | −0.018 | 0.445** | −0.149 | −0.181* | 0.191* | 0.190* | ||
| Nocturnal RER | 0.182* | 0.076 | 0.060 | 0.177* | 0.143 | 0.062 | 0.156 | −0.117 | −0.096 | 0.121 | 0.302** | |||
| Exercise traits | ||||||||||||||
| Running distances | ||||||||||||||
| D1 | 0.576** | 0.459** | 0.772** | 0.537** | 0.381** | 0.227** | −0.053 | −0.191* | 0.148* | −0.058 | ||||
| D56 | 0.842** | 0.559** | 0.842** | 0.748** | 0.256** | −0.114 | −0.266** | −0.068 | −0.252** | |||||
| D1112 | 0.442** | 0.738** | 0.852** | 0.285** | −0.178* | −0.327** | −0.086 | −0.318** | ||||||
| Running speeds | ||||||||||||||
| S1 | 0.602** | 0.450** | 0.139 | 0.076 | −0.165* | 0.189* | −0.026 | |||||||
| S56 | 0.839** | 0.115 | −0.014 | −0.202** | −0.114 | −0.200** | ||||||||
| S1112 | 0.109 | −0.055 | −0.207** | −0.105 | −0.299** | |||||||||
| FIRun | −0.477** | −0.306** | 0.125 | 0.195** | ||||||||||
| Postexercise | ||||||||||||||
| BWpost | 0.266** | 0.193* | −0.144 | |||||||||||
| BFpost | −0.107 | 0.096 | ||||||||||||
| BWnet | 0.410** |
P < 0.05,
P < 0.01.
QTL mapping metabolism and exercise traits.
QTL mapping of the phenotypes presented above revealed five significant QTL (P < 0.05) and eight suggestive QTL (P < 0.20) (Table 5). A single significant QTL was observed for D12 and FIRun. One significant and one suggestive QTL were identified for BW, and two significant and three suggestive QTL were identified for BWnet. Suggestive QTL were also identified for BF and D56 running distances.
Table 5.
QTL for metabolism and exercise traits
| Trait | LOD | Significance Threshold | Chromosome | Peak Position Mb (CI, 1.5 LOD drop) | Allele Effects |
|---|---|---|---|---|---|
| Preexercise | |||||
| BW | 7.90 | 0.05 | 4 | 7.54 (3.32–10.34) | C57BL6/J, NZO/H1LtJ |
| 6.16 | 0.20 | 15 | 86.72 (84.56–87.53) | Multiple classes | |
| BFP | 6.47 | 0.20 | 6 | 126.70 (122.37–129.55) | Multiple classes |
| Exercise traits | |||||
| Running distances | |||||
| D56 | 6.34 | 0.20 | 1 | 171.72 (167.05–174.40) | WSB/EiJ, A/J |
| 6.64 | 0.20 | 9 | 98.42 (94.73–102.43) | WSB/EiJ, A/J | |
| 6.44 | 0.20 | 16 | 33.25 (29.25–39.50) | WSB/EiJ, A/J | |
| D12 | 7.13 | 0.05 | 16 | 33.25 (32.54–38.34) | WSB/EiJ, A/J |
| FIRun | 7.39 | 0.05 | 12 | 85.13 (82.86–89.02) | C57BL6/J |
| Postexercise | |||||
| BWnet | 7.23 | 0.05 | 6 | 77.72 (72.74–83.45) | Multiple classes |
| 6.93 | 0.05 | 6 | 42.76 (39.42–48.13) | Multiple classes | |
| 6.17 | 0.20 | 3 | 47.63 (36.69–51.00) | Multiple classes | |
| 6.33 | 0.20 | 6 | 93.88 (69.55–98.01) | Multiple classes | |
| 6.43 | 0.20 | 10 | 111.13 (97.55–114.23) | Multiple classes |
QTL, quantitative trait loci.
The largest effect QTL we found was for initial BW and it mapped to proximal chromosome (Chr) 4 [peak at 7.54 Mb, named Bwq14 (2)]. This locus overlaps a previously reported QTL for BW in an F2 cross between C57BL6/J and KK/HIJ mice (52, 53), a polygenic type 2 diabetes model with moderate obesity that shares phenotypic similarity with NZO/HILtJ (22, 23). Similarly, a QTL for blood pressure associated with metabolic syndrome in an NZO/HILtJ × C3H/HeJ F2 population has been mapped in close proximity to this locus (40), as does a BW QTL in ILS × ISS (LXS) RI strains (Chr 4, 9.9 Mb). The BW QTL in the LXS was not identified upon initial analysis but was found to be significant using a multiple QTL model that incorporated additive and epistatic interactions (4). By analyzing SNP data and allele effects for each of the eight founder strains at this locus in the pre-CC population, positional candidate genes were identified that potentially underpin this body weight QTL (2). By further incorporating eQTL data, a high-priority candidate for future functional analysis, aspartate-β-hydroxylase (Asph), was identified from among the positional candidate genes (2).
Two significant and one suggestive QTL were identified for BWnet on Chr 6 (72.2–83.4 Mb, 39.4–48.1 Mb, and 69.5–98.0 Mb, respectively). Additional suggestive QTL for BWnet were identified on Chr 3 (47.63 Mb) and Chr 10 (111.13 Mb). A suggestive QTL for BF was also identified on Chr 6 (126.7Mb). These findings overlap with previously mapped BW- and BF-associated loci (1, 8, 9, 15, 16, 45, 48).
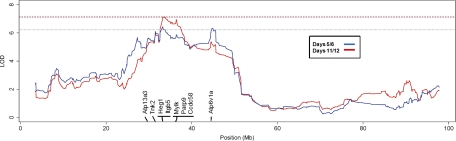
A significant QTL for D12 wheel running distance was identified on Chr 16 (33.2 Mb) that accounted for 17.0% of the observed phenotypic variance for this trait (Fig. 6). A similar peak was observed for D56 wheel running distance, although this peak failed to reach genome-wide significance thresholds. This likely pleiotropic effect on voluntary exercise at different ages is not surprising given the high degree of correlation between the two traits (r2 = 0.84). Chromosome 16 has not previously been associated with wheel running or locomotor behavior. Previous studies using F2 intercrosses identified loci associated with running distance on Chr 7 and 13 (27, 33, 39) and loci associated with running speed and duration on Chr 9 and 13 (33). In addition, recent haplotype association mapping revealed significant QTL for running distance on Chr 5, 6, 8, 11, 12, 13, 18, and 19 and for running speed on Chr 6 and 11 (32). Six of the strains used in the Lightfoot study were the same founder strains used to create the pre-CC population. However, significant differences in phenotypic expression exist across these two studies. For example, running distances for CAST/EiJ and WSB/EiJ were significantly shorter in the Lightfoot study than in the results reported here. This brings to light the potential effect of differences in experimental procedures and laboratory environments on the detection of QTL, especially for traits with behavioral influences. Moreover, running was presented as the total distance averaged over 21 days of wheel access in the Lightfoot study, whereas the current phenotypes were the average distances on D56 and D12 of wheel access.
Fig. 6.
Quantitative trait loci (QTL) maps for novel QTL identified on Chr 16 for average wheel running distances on days 5 and 6 (blue) and days 11 and 12 (red) in pre-CC mice. Dashed red and blue lines designate genome-wide significance threshold, P < 0.05. Select functionally relevant genes with colocalized expression (e)QTL are listed in their approximate locations within the region.
Suggestive QTL for spontaneous locomotor activity have been identified on Chr 3, 8, 12, 13, and 19 (56), while novelty- and stress-induced locomotor activity are associated with loci on Chr 1, 4, 5, 8, 9, 13, 14, and 19 (17). Mice examined in the aforementioned studies consisted of F2, BC, and AIL populations with only one strain (C57BL6/J) in common with those used to generate the pre-CC; thus, the suggestive QTL in the pre-CC may be attributed to allele-specific effects not present in the other genetic crosses. Additional suggestive QTL in the pre-CC study were identified for D56 on Chr 1 and 9. Several regions of Chr 1 are associated with stress- and anxiety-related locomotor activity in A/J × C57BL6/J (AXB/BXA) RI strains (17). Thus, stress and anxiety may have contributed to variability in wheel running. The locus on Chr 9 coincides with suggestive QTL for running distance in a C57L/J × C3H/HeJ F2 population and also overlaps with a speed QTL in that population (33).
Allele effect estimates and expression QTL (eQTL) analysis for the wheel running QTL on Chr 1, 9, and 16 were performed as described by Aylor et al. (2). Allele effect estimates suggest that WSB/EiJ alleles consistently contributed to increased wheel running distances at all QTL. Allele effect patterns were similar between the novel D56 and D12 peaks on Chr 16, with A/J and WSB/EiJ alleles contributing to increased wheel running distance at these two loci. Interestingly, when wheel running among the founder strains was assessed, WSB/EiJ ran the greatest distances, while A/J ran the second shortest distances of the eight founder strains. Moreover, wheel running distances in the pre-CC population revealed extreme phenotypes compared with the founder strains, providing evidence of transgressive segregation in this population (42, 43).
Expression QTL analysis in liver for the D12 QTL region on Chr 16 revealed 31 significant (P < 0.05 genome-wide) local eQTL (Supplemental Table S2). Several of the transcripts identified through eQTL analysis are involved with energy homeostasis (e.g.. Atp13a3, Parp9, Tnk2, Atp6v1a) (Fig. 6). Many of the identified genes are also implicated in smooth muscle function and mitochondrial and lung disease (e.g., Heg1, Mylk, Ccdc58, ltgb5). Comparisons of allele effect patterns specific to A/J and WSB/EiJ failed to refine the candidate region to allow for the identification of specific polymorphisms that might underpin the D12 QTL on Chr 16.
A significant QTL for food intake during exercise (FIRun) was identified on Chr 12 (85.1Mb), accounting for 17.6% of the observed phenotypic variance. Previously identified QTL overlapping with this locus has been demonstrated to interact with loci on Chr 2, exerting epistatic effects on BW and growth in a C57BL6/J backcross with Mus musculus castaneus trapped from the wild (24). Allele effect estimates indicate that the C57BL6/J allele was the predominant contributor to increased food intake during wheel running at this locus. Expression QTL analysis in liver revealed local eQTL (P < 0.05 genome-wide) for 18 genes within the FIRun QTL region (Supplemental Table S2). Three of these 18 genes (Acot2, Ptgr2, Dlst) are directly related to metabolic processing and energy homeostasis (21, 51, 58). Expression of an additional gene, Aldh6a1, is altered in response to fluctuations in energy states such as fasting (61). Although highly speculative, these findings suggest interesting targets for future investigations into the genetic architecture of energy homeostasis during exercise.
DISCUSSION
The CC was proposed as a model for systems biology and high-precision complex trait analysis. The CC is expected to capture unprecedented mouse genetic and phenotypic diversity similar to that seen in the human population (44). This assertion was validated in the present experiment. We demonstrated that the significantly broad genetic diversity captured in the pre-CC population manifested into similarly impressive phenotypic diversity in a host of metabolism- and exercise-related phenotypes. Moreover, mapping in the pre-CC study, examined as a proxy for eventual capabilities of the CC resource to dissect the genetic architecture of complex traits, uncovered novel QTL not previously observed in existing mouse resources while also replicating several existing loci.
Although significant phenotypic diversity in a broad array of metabolism and exercise traits exists among the genetically diverse founder strains of the CC, transgressive variation for many of these traits was observed in the pre-CC strains. For instance, there was a 19 km/day difference in running distance between the highest and lowest running pre-CC mice and only a 13 km/day difference among the highest and lowest running founders after 12 days of wheel running. Although this was not the case for all traits (e.g., no pre-CC strains exceeded NZO/H1LtJ for body weight), these findings complement an independent evaluation of pre-CC lines (7) to demonstrate that the CC will likely exhibit an extremely broad phenotypic diversity across many different sets of traits. Such diversity is rooted in the eight founder strains, but the existence of strong transgressive variation is likely based on new allele combinations and haplotypes within the pre-CC lines.
Residual heterozygosity within the pre-CC population may have contributed to transgressive variation observed in the pre-CC mice. Transgressive variation due to hybrid vigor has been demonstrated for decades across several agricultural, insect and mammalian species. Although hybrid vigor is usually diminished by the F2 generation, its effects on transgressive variation in the pre-CC mice cannot be completely ruled out (47).
Selective breeding experiments, which have been used for decades to gain an understanding of the genetic underpinnings of growth, fatness, and other traits related to energy balance, can expand the range of phenotypic variation to extremes far beyond the range of the founding population (6). For example, high-growth selection lines have been established with mean body weights of up to 77 g, far exceeding the body weights within the base populations from which these lines were selected, as well as body weight in 400 additional inbred mouse strains (6, 54). The range of variation in BW in the CC founder strains pales compared with variation produced with high selection lines, and no pre-CC mice exceeded the BW within the founder NZO/H1LtJ strain. Selective breeding for increased wheel running activity has also been successful in dramatically increasing exercise phenotypes. For example, 27 generations of selection led to a high runner line (HR) with running distances that were 2.7 times greater than randomly selected ICR controls (18). In contrast to the comparison with high body weight lines, running distances for several of the pre-CC mice significantly exceeds the mean distance run by HR lines (13). Thus, in this instance, the pre-CC population captured more variation and extremes in wheel running than either the founder strains or long-term selection lines. Given that the wild-derived founder strains, CAST/EiJ, PWK/PhJ, and WSB/EiJ, demonstrated greater levels of wheel running compared with the other founder strains, it is probable that allele combinations contributed by the three wild-derived strains may have driven expression of extreme running phenotypes in the pre-CC lines. Furthermore, it is tempting to speculate that phenotypic extremes in the pre-CC lines for any trait will be dictated primarily by the phenotypic characteristics of the wild-derived strains. For BW, where the wild-derived strains are small, the pre-CC mice do not exhibit extreme phenotypes on the high end of the phenotypic distribution. But for running distance, where the wild-derived strains have high phenotypes, the pre-CC lines exceed any other mice measured to date, including long-term selection lines.
Other methods for describing phenotypic diversity in laboratory mice include using the mouse phenome strains (19). Since these strains come from four Mus musculus subspecies, they represent a sampling of high genetic diversity (54). Phenotypic variation in BW and BF among the inbred strains listed in the Mouse Phenome Database is similar to that seen in the pre-CC founder strains. Phenotypic variation within surveys that included mice of similar age and strains known for extreme BF, such as NZO/H1LtJ, or those with low BF such as the wild-derived strains, was highly consistent with the variability demonstrated in the CC founders and in the pre-CC lines (3) (http://www.jax.org/cranio, MPD:115).
Correlations between metabolism and exercise traits traditionally seen in inbred strains or long-term selection lines were disrupted in the pre-CC lines. For example, nocturnal RER was positively correlated with running traits in the founder strains but not in the pre-CC lines. RER was correlated with running distance and speed only on the first day of running wheel access (D1) in the pre-CC mice. Wheel running on D1 may be an indicator of increased stress or anxiety (27, 57), which also increases respiratory exchange (35). Thus, the relationship between RER and wheel running may be evidence of stress and anxiety-induced locomotor activity in the pre-CC mice.
Transgressive variation and the breakdown in phenotypic correlations demonstrate the effects of new allele combinations in the pre-CC lines. Random genetic assortment disrupted long-range linkage disequilibrium in the founder strains as well as repeated SNP distribution patterns that resulted from the shared ancestry of the classical inbred strains. Congenic analysis has demonstrated that some major-effect QTL are actually due to the combined effect of multiple closely linked loci (e.g., Refs. 14, 25). Thus, the phase of closely linked genes may have been disrupted during the course of CC breeding. When a sufficient number of lines is available, the CC (in combination with the Diversity Outcross; http://jaxmice.jax.org/jaxnotes/514/514b.html) will have the power to resolve closely linked QTL and to support the identification of valid phenotypic correlations caused by pleiotropy rather than by nonrandom population structures.
Despite our limited power to map QTL in the pre-CC relative to expectations for completed CC lines (see Ref. 2), we replicated previously described QTL and identified novel QTL for metabolism and exercise traits. QTL mapping revealed five significant and eight suggestive QTL for metabolism- and exercise-related traits. QTL for BW, BW change after exercise, food intake during exercise, and wheel running distances primarily replicated previously identified locations for QTL for these traits, although for many such phenotypes a large number of QTL have been mapped, and thus it is hard to differentiate colocalization from coincidence. A novel QTL for wheel running distance was identified on Chr 16, with pleiotropic effects on both D56 and D12. This QTL is in a region that had previously been relatively devoid of QTL for any complex trait. In fact, there is little genetic variation on Chr 16 among the classical inbred mouse strains (59). This demonstrates the value of new variation contributed to this resource from the wild-derived strains.
The results of this experiment provide compelling evidence that the genetic diversity of the pre-CC population translates into equally broad phenotypic diversity. Phenotypic diversity in metabolism- and exercise-related traits among the pre-CC lines was equivalent to, or exceeded, that seen among the eight progenitor strains from which they were derived. QTL mapping in this population both replicated known loci and uncovered new loci important for metabolism and voluntary wheel running. These results suggest that the CC will be a powerful tool for examining the genetic architecture of complex traits.
Genetic analysis of complex traits is one of the methods by which novel pathways can be discovered through the use of a phenotype-driven, forward-genetics approach. Although a large number of mouse populations and mouse crosses have been used for the study of metabolism, obesity, and exercise, these resources have largely uncovered effects from a limited sample of mouse diversity (59). With the greatly expanded allelic diversity and allelic combinations present in the CC relative to conventional mouse resources, we now have the potential to uncover these pathways.
GRANTS
This project was partially supported by National Institutes of Health (NIH) Grants DK-076050 (D. Pomp), U01-CA-105417 and U01-CA-134240 (D. W. Threadgill), GM-076468 (G. A. Churchill), and MH-090338 and the Ellison Medical Foundation AG-IA-0202-05 (E. J. Chesler, F. P.-M. de Villena). W. F. Mathes was supported by NIH Grants T32-MH-07669403 and K12-HD-01441. D. L. Aylor was supported in part by Ruth L. Kirschstein National Research Service Award No. F32-GM-090667 from the National Institute of General Medical Sciences. Infrastructure support was provided by the Lineberger Comprehensive Cancer Center and the University Cancer Research Fund from the state of North Carolina. Phenotypes were collected using the Animal Metabolism Phenotyping core facility at UNC's Nutrition Obesity Research Center (funded by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK-056350). The production and maintenance of the Collaborative Cross was supported by the US Department of Energy, Office of Biological and Environmetnal Research, and performed at Oak Ridge National Laboratory (ORNL), managed by UT-Battelle, LLC, under contract DE-AC05-00OR22725. Initial support was provided to ORNL by The Ellison Medical Foundation.
DISCLOSURES
No conflicts of interest are reported by the authors.
Supplementary Material
REFERENCES
- 1. Anunciado RV, Ohno T, Mori M, Ishikawa A, Tanaka S, Horio F, Nishimura M, Namikawa T. Distribution of body weight, blood insulin and lipid levels in the SMXA recombinant inbred strains and the QTL analysis. Exper Animals/Jpn Assoc Laboratory Anim Sci 49: 217–224, 2000 [DOI] [PubMed] [Google Scholar]
- 2. Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, Gralinski LE, Bell TA, Didion JD, Hua K, Nehrenberg DL, Powell CL, Steigerwalt J, Xie Y, Kelada SN, Collins FS, Yang IV, Schwartz DA, Branstetter LA, Chesler EJ, Miller DR, Spence J, Liu EY, McMillan L, Sarkar A, Wang J, Wang W, Zhang Q, Broman KW, Korstanje R, Durrant C, Mott R, Iraqi FA, Pomp D, Threadgill D, Pardo-Manuel de Villena F, Churchill GA. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 2011. March 15 [Epub ahead of print] PMID: 21406540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bachmanov AA, Reed DR, Beauchamp GK, Tordoff MG. Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behav Genet 32: 435–443, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bennett B, Carosone-Link PJ, Lu L, Chesler EJ, Johnson TE. Genetics of body weight in the LXS recombinant inbred mouse strains. Mamm Genome 16: 764–774, 2005 [DOI] [PubMed] [Google Scholar]
- 5. Bouchard C. Gene-environment interactions in the etiology of obesity: defining the fundamentals. Obesity (Silver Spring, Md) 16, Suppl 3: S5–S10, 2008 [DOI] [PubMed] [Google Scholar]
- 6. Bunger L, Laidlaw A, Bulfield G, Eisen EJ, Medrano JF, Bradford GE, Pirchner F, Renne U, Schlote W, Hill WG. Inbred lines of mice derived from long-term growth selected lines: unique resources for mapping growth genes. Mamm Genome 12: 678–686, 2001 [DOI] [PubMed] [Google Scholar]
- 7. Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, Churchill GA, Johnson DK, Manly KF. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome 19: 382–389, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheverud JM, Ehrich TH, Hrbek T, Kenney JP, Pletscher LS, Semenkovich CF. Quantitative trait loci for obesity- and diabetes-related traits and their dietary responses to high-fat feeding in LGXSM recombinant inbred mouse strains. Diabetes 53: 3328–3336, 2004 [DOI] [PubMed] [Google Scholar]
- 9. Cheverud JM, Vaughn TT, Pletscher LS, Peripato AC, Adams ES, Erikson CF, King-Ellison KJ. Genetic architecture of adiposity in the cross of LG/J and SM/J inbred mice. Mamm Genome 12: 3–12, 2001 [DOI] [PubMed] [Google Scholar]
- 10. Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu HC, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, Kendziorski C, Kotb M, Kooy RF, Llamas B, Lammert F, Lassalle JM, Lowenstein PR, Lu L, Lusis A, Manly KF, Marcucio R, Matthews D, Medrano JF, Miller DR, Mittleman G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Morris DG, Mott R, Nadeau JH, Nagase H, Nowakowski RS, O'Hara BF, Osadchuk AV, Page GP, Paigen B, Paigen K, Palmer AA, Pan HJ, Peltonen-Palotie L, Peirce J, Pomp D, Pravenec M, Prows DR, Qi Z, Reeves RH, Roder J, Rosen GD, Schadt EE, Schalkwyk LC, Seltzer Z, Shimomura K, Shou S, Sillanpaa MJ, Siracusa LD, Snoeck HW, Spearow JL, Svenson K, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 36: 1133–1137, 2004 [DOI] [PubMed] [Google Scholar]
- 11. Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141: 1199–1207, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darvasi A, Weinreb A, Minke V, Weller JI, Soller M. Detecting marker-QTL linkage and estimating QTL gene effect and map location using a saturated genetic map. Genetics 134: 943–951, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eisenmann JC, Wickel EE, Kelly SA, Middleton KM, Garland T., Jr Day-to-day variability in voluntary wheel running among genetically differentiated lines of mice that vary in activity level. Eur J Appl Physiol 106: 613–619, 2009 [DOI] [PubMed] [Google Scholar]
- 14. Farber CR, Medrano JF. Fine mapping reveals sex bias in quantitative trait loci affecting growth, skeletal size and obesity-related traits on mouse chromosomes 2 and 11. Genetics 175: 349–360, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fawcett GL, Jarvis JP, Roseman CC, Wang B, Wolf JB, Cheverud JM. Fine-mapping of obesity-related quantitative trait loci in an F(9/10) advanced intercross line. Obesity (Silver Spring, Md) 18: 1383–1392, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fawcett GL, Roseman CC, Jarvis JP, Wang B, Wolf JB, Cheverud JM. Genetic architecture of adiposity and organ weight using combined generation QTL analysis. Obesity (Silver Spring, Md) 16: 1861–1868, 2008 [DOI] [PubMed] [Google Scholar]
- 17. Gill KJ, Boyle AE. Quantitative trait loci for novelty/stress-induced locomotor activation in recombinant inbred (RI) and recombinant congenic (RC) strains of mice. Behav Brain Res 161: 113–124, 2005 [DOI] [PubMed] [Google Scholar]
- 18. Girard I, McAleer MW, Rhodes JS, Garland T., Jr Selection for high voluntary wheel-running increases speed and intermittency in house mice (Mus domesticus). J Exper Biol 204: 4311–4320, 2001 [DOI] [PubMed] [Google Scholar]
- 19. Grubb SC, Maddatu TP, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res 37: D720–D730, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hetherington MM, Cecil JE. Gene-environment interactions in obesity. Forum Nutr 63: 195–203, 2010 [DOI] [PubMed] [Google Scholar]
- 21. Hunt MC, Nousiainen SE, Huttunen MK, Orii KE, Svensson LT, Alexson SE. Peroxisome proliferator-induced long chain acyl-CoA thioesterases comprise a highly conserved novel multi-gene family involved in lipid metabolism. J Biol Chem 274: 34317–34326, 1999 [DOI] [PubMed] [Google Scholar]
- 22. Igel M, Becker W, Herberg L, Joost HG. Hyperleptinemia, leptin resistance, and polymorphic leptin receptor in the New Zealand obese mouse. Endocrinology 138: 4234–4239, 1997 [DOI] [PubMed] [Google Scholar]
- 23. Igel M, Taylor BA, Phillips SJ, Becker W, Herberg L, Joost HG. Hyperleptinemia and leptin receptor variant Asp600Asn in the obese, hyperinsulinemic KK mouse strain. J Mol Endocrinol 21: 337–345, 1998 [DOI] [PubMed] [Google Scholar]
- 24. Ishikawa A, Hatada S, Nagamine Y, Namikawa T. Further mapping of quantitative trait loci for postnatal growth in an inter-sub-specific backcross of wild Mus musculus castaneus and C57BL/6J mice. Genet Res 85: 127–137, 2005 [DOI] [PubMed] [Google Scholar]
- 25. Jerez-Timaure NC, Eisen EJ, Pomp D. Fine mapping of a QTL region with large effects on growth and fatness on mouse chromosome 2. Physiol Genomics 21: 411–422, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Karavirta L, Hakkinen K, Kauhanen A, Arija-Blazquez A, Sillanpaa E, Rinkinen N, Hakkinen A. Individual responses to combined endurance and strength training in older adults. Med Sci Sports Exerc 43: 484–490, 2010 [DOI] [PubMed] [Google Scholar]
- 27. Kelly SA, Nehrenberg DL, Pierce JL, Hua K, Steffy BM, Wiltshire T, Pardo-Manuel de Villena F, Garland T, Jr, Pomp D. Genetic architecture of voluntary exercise in an advanced intercross line of mice. Physiol Genomics 42: 190–200, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kumar KG, DiCarlo LM, Volaufova J, Zuberi AR, Richards BK. Increased physical activity cosegregates with higher intake of carbohydrate and total calories in a subcongenic mouse strain. Mamm Genome 21: 52–63, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar KG, Poole AC, York B, Volaufova J, Zuberi A, Richards BK. Quantitative trait loci for carbohydrate and total energy intake on mouse chromosome 17: congenic strain confirmation and candidate gene analyses (Glo1, Glp1r). Am J Physiol Regul Integr Comp Physiol 292: R207–R216, 2007 [DOI] [PubMed] [Google Scholar]
- 30. Kumar KG, Smith Richards BK. Transcriptional profiling of chromosome 17 quantitative trait Loci for carbohydrate and total calorie intake in a mouse congenic strain reveals candidate genes and pathways. J Nutrigenet Nutrigenom 1: 155–171, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leamy LJ, Pomp D, Lightfoot JT. Genetic variation for body weight change in mice in response to physical exercise. BMC Genet 10: 58, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lightfoot JT, Leamy L, Pomp D, Turner MJ, Fodor AA, Knab A, Bowen RS, Ferguson D, Moore-Harrison T, Hamilton A. Strain screen and haplotype association mapping of wheel running in inbred mouse strains. J Appl Physiol 109: 623–634, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lightfoot JT, Turner MJ, Pomp D, Kleeberger SR, Leamy LJ. Quantitative trait loci for physical activity traits in mice. Physiol Genomics 32: 401–408, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loos RJ, Rankinen T. Gene-diet interactions on body weight changes. J Am Diet Assoc 105: S29–S34, 2005 [DOI] [PubMed] [Google Scholar]
- 35. McGregor IS, Lee AM, Westbrook RF. Stress-induced changes in respiratory quotient, energy expenditure and locomotor activity in rats: effects of midazolam. Psychopharmacology 116: 475–482, 1994 [DOI] [PubMed] [Google Scholar]
- 36. Morahan G, Balmer L, Monley D. Establishment of “The Gene Mine”: a resource for rapid identification of complex trait genes. Mamm Genome 19: 390–393, 2008 [DOI] [PubMed] [Google Scholar]
- 37. Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA 97: 12649–12654, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mutch DM, Clement K. Unraveling the genetics of human obesity. PLoS Genetics 2: e188, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nehrenberg DL, Wang S, Hannon RM, Garland T, Jr, Pomp D. QTL underlying voluntary exercise in mice: interactions with the “mini muscle” locus and sex. J Hered 101: 42–53, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nishihara E, Tsaih SW, Tsukahara C, Langley S, Sheehan S, DiPetrillo K, Kunita S, Yagami K, Churchill GA, Paigen B, Sugiyama F. Quantitative trait loci associated with blood pressure of metabolic syndrome in the progeny of NZO/HILtJxC3H/HeJ intercrosses. Mamm Genome 18: 573–583, 2007 [DOI] [PubMed] [Google Scholar]
- 41. Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring, Md) 14: 529–644, 2006 [DOI] [PubMed] [Google Scholar]
- 42. Rieseberg LH, Archer MA, Wayne RK. Transgressive segregation, adaptation and speciation. Heredity 83: 363–372, 1999 [DOI] [PubMed] [Google Scholar]
- 43. Rieseberg LH, Widmer A, Arntz AM, Burke JM. The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Phil Transact Royal Soc London 358: 1141–1147, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome 18: 473–481, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rocha JL, Eisen EJ, Van Vleck LD, Pomp D. A large-sample QTL study in mice: I. Growth. Mamm Genome 15: 83–99, 2004 [DOI] [PubMed] [Google Scholar]
- 46. Rocha JL, Eisen EJ, Van Vleck LD, Pomp D. A large-sample QTL study in mice: II. Body composition. Mamm Genome 15: 100–113, 2004 [DOI] [PubMed] [Google Scholar]
- 47. Rosas U, Barton NH, Copsey L, Barbier de Reuille P, Coen E. Cryptic variation between species and the basis of hybrid performance. PLoS Biology 8: e1000429, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shao H, Reed DR, Tordoff MG. Genetic loci affecting body weight and fatness in a C57BL/6J × PWK/PhJ mouse intercross. Mamm Genome 18: 839–851, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Silventoinen K, Kaprio J. Genetics of tracking of body mass index from birth to late middle age: evidence from twin and family studies. Obesity Facts 2: 196–202, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith Richards BK, Belton BN, Poole AC, Mancuso JJ, Churchill GA, Li R, Volaufova J, Zuberi A, York B. QTL analysis of self-selected macronutrient diet intake: fat, carbohydrate, and total kilocalories. Physiol Genomics 11: 205–217, 2002 [DOI] [PubMed] [Google Scholar]
- 51. Sokolovic M, Sokolovic A, Wehkamp D, Ver Loren van Themaat E, de Waart DR, Gilhuijs-Pederson LA, Nikolsky Y, van Kampen AH, Hakvoort TB, Lamers WH. The transcriptomic signature of fasting murine liver. BMC Genomics 9: 528, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Suto J. Coincidence of loci for glucosuria and obesity in type 2 diabetes-prone KK-Ay mice. Med Sci Monit 14: CR65–CR74, 2008 [PubMed] [Google Scholar]
- 53. Suto J, Matsuura S, Imamura K, Yamanaka H, Sekikawa K. Genetics of obesity in KK mouse and effects of A(y) allele on quantitative regulation. Mamm Genome 9: 506–510, 1998 [DOI] [PubMed] [Google Scholar]
- 54. Svenson KL, Von Smith R, Magnani PA, Suetin HR, Paigen B, Naggert JK, Li R, Churchill GA, Peters LL. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J Appl Physiol 102: 2369–2378, 2007 [DOI] [PubMed] [Google Scholar]
- 55. Threadgill DW, Hunter KW, Williams RW. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm Genome 13: 175–178, 2002 [DOI] [PubMed] [Google Scholar]
- 56. Toth LA, Williams RW. A quantitative genetic analysis of locomotor activity in CXB recombinant inbred mice. Behav Genet 29: 319–328, 1999 [DOI] [PubMed] [Google Scholar]
- 57. Uchiumi K, Aoki M, Kikusui T, Takeuchi Y, Mori Y. Wheel-running activity increases with social stress in male DBA mice. Physiol Behav 93: 1–7, 2008 [DOI] [PubMed] [Google Scholar]
- 58. Wu YH, Ko TP, Guo RT, Hu SM, Chuang LM, Wang AH. Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2. Structure 16: 1714–1723, 2008 [DOI] [PubMed] [Google Scholar]
- 59. Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat Genet 39: 1100–1107, 2007 [DOI] [PubMed] [Google Scholar]
- 60. Yang H, Ding Y, Hutchins LN, Szatkiewicz J, Bell TA, Paigen BJ, Graber JH, de Villena FP, Churchill GA. A customized and versatile high-density genotyping array for the mouse. Nat Methods 6: 663–666, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang YK, Saupe KW, Klaassen CD. Energy restriction does not compensate for the reduced expression of hepatic drug-processing genes in mice with aging. Drug Metab Dispos Biol Fate Chem 38: 1122–1131, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.