Abstract
It is unclear whether the muscle hypertrophy induced by loss of myostatin signaling in mature muscles is maintained only by increased protein synthesis or whether reduced proteolysis contributes. To address this issue, we depleted myostatin by activating Cre recombinase for 2 wk in mature mice in which Mstn exon 3 was flanked by loxP sequences. The rate of phenylalanine tracer incorporation into myofibrillar proteins was determined 2, 5, and 24 wk after Cre activation ended. At all of these time points, myostatin-deficient mice had increased gastrocnemius and quadriceps muscle mass (≥27%) and increased myofibrillar synthesis rate per gastrocnemius muscle (≥19%) but normal myofibrillar synthesis rates per myofibrillar mass or RNA mass. Mean fractional myofibrillar degradation rates (estimated from the difference between rate of synthesis and rate of change in myofibrillar mass) and muscle concentrations of free 3-methylhistidine (from actin and myosin degradation) were unaffected by myostatin knockout. Overnight food deprivation reduced myofibrillar synthesis and ribosomal protein S6 phosphorylation and increased concentrations of 3-methylhistidine, muscle RING finger-1 mRNA, and atrogin-1 mRNA. Myostatin depletion did not affect these responses to food deprivation. These data indicate that maintenance of the muscle hypertrophy caused by loss of myostatin is mediated by increased protein synthesis per muscle fiber rather than suppression of proteolysis.
Keywords: muscle hypertrophy, 3-methylhistidine, atrogin-1/muscle atrophy F-box, muscle RING finger-1, ribosomal protein S6
mice with constitutive absence of myostatin (Mstn−/−) develop muscles twofold larger than normal because of both fiber hyperplasia and fiber hypertrophy (28). Levels of phospho-Akt and phosphoribosomal protein S6 are increased in Mstn−/− muscles (8, 24, 31), which suggests increased activity of the Akt-mTOR-S6 kinase signaling pathway, a key regulator of muscle fiber size and protein metabolism (16). The rate of myofibrillar protein synthesis per whole muscle is increased in mature Mstn−/− mice, whereas the fractional myofibrillar synthesis rate is normal (41). Mstn−/− muscles have other abnormalities besides fiber hyperplasia and hypertrophy: slow-twitch fiber deficiency, reduced mitochondrial mass, reduced expression of genes encoding proteins involved in energy metabolism, and altered expression of more than 2,000 genes (2, 14, 38). In contrast, when myostatin is depleted or blocked after maturity, muscles enlarge because of fiber hypertrophy but are normal with respect to the number of fibers, fiber type distribution, and metabolic properties (6, 12, 14, 19, 26, 35, 36, 42, 45, 46). Mice with postdevelopmental inhibition or depletion of myostatin have normal muscle levels of phospho-Akt (18, 43, 44), and absence of Akt does not prevent the muscle hypertrophy induced by blocking myostatin activity (18). Nevertheless, S6 kinase activity and myofibrillar protein synthesis increased within a few days after blocking myostatin in mature mice (43). The effect of postdevelopmental myostatin deficiency on myofibrillar protein metabolism during the maintenance phase of muscle hypertrophy, described in the present report, has not been examined previously.
Protein synthesis rate in vivo can be determined readily by incorporation of an amino acid tracer. However, there is no accepted method to determine myofibrillar degradation rate in vivo. In the present study, the rate of proteolysis in vivo was estimated from the protein synthesis rates and changes in protein mass across different time points. This approach did not quantify proteolysis in an individual mouse at a single point in time, but it provided a measurement of the mean rate of proteolysis over long intervals in groups of mice. Other indices of myofibrillar proteolysis in the present study included muscle tissue concentrations of free 3-methylhistidine (3MH; a product of actin and myosin degradation) and expression of Trim63, a gene that encodes an E3 ubiquitin ligase [muscle RING finger-1 (MuRF1)] required for myofibrillar proteolysis (10, 11). Expression of F-box protein 32 [Fbxo32; the gene encoding the E3 ubiquitin ligase atrogin-1/muscle atrophy F-box (MAFbx)] was also determined because atrogin-1 can influence both muscle protein synthesis and proteolysis (3, 22, 40).
Food deprivation stimulates proteolysis and inhibits protein synthesis in muscle (4, 5, 9, 25, 30, 34, 47). Whether myostatin is required for the normal response to food deprivation is not entirely clear. A recent report demonstrated that Mstn−/− mice had normal increases in food deprivation-induced overexpression of MuRF1 and atrogin-1 mRNAs (1), but the effect of food deprivation on protein synthesis was not examined. Therefore, we determined whether postdevelopmental myostatin depletion affected the inhibition of myofibrillar protein synthesis induced by overnight food deprivation.
METHODS
Procedures were approved by the University of Rochester animal research committee. Mstn[f/f] mice with a C57BL/6J background (minimum of 7 crosses with this strain), in which exon 3 of the Mstn gene is flanked by loxP sequences (floxed) (41), and control mice with normal myostatin genes (Mstn[w/w], also with C57BL/6J background), were hemizygous for a CreER transgene driven by a cytomegalovirus/chicken β-actin promoter (42). Genotyping was done as described previously (42). Only male mice were studied. Mice had free access to food and water except for a 16-h period without food in the food deprivation experiment. The cages were in rooms with a 12:12-h light-dark cycle (lights on 0600–1800). Room temperature was maintained at 23°C.
Stimulation of Cre recombinase activity, which leads to excision of the floxed exon, was initiated in both Mstn[f/f] and Mstn[w/w] mice by feeding them tamoxifen [0.025% of chow, prepared by Harlan-Teklad as described previously (44)] when they were 4 mo old for a period of 2 wk. Myostatin mRNA levels are reduced to some extent in Mstn[f/f] mice prior to tamoxifen administration due to a low basal level of Cre recombinase activity (42). The exact time course of the further myostatin depletion induced by tamoxifen has not been defined. Thus, with this model there is no single point in time that can be declared as the starting point for myostatin deficiency. Because the present study was done to characterize chronic effects of myostatin depletion, this is not a critical issue. The acute effect of myostatin inhibition on myofibrillar protein synthesis was reported previously (43).
Two, five, or 24 weeks after the end of tamoxifen feeding, myofibrillar protein synthesis was measured between 9 and 10:30 AM, as described previously (41, 43). [2H5]phenylalanine was administered (ip) 30 min before mice were euthanized by CO2 inhalation, followed by cervical dislocation. Most of the mice had free access to food the night before and the morning of tracer injection, but some of the mice that were euthanized 2 wk after the end of tamoxifen administration were deprived of food for 16 h (± 1 h) before tracer injection. Gastrocnemius and quadriceps muscles were removed and weighed as quickly as possible and then immersed immediately in liquid nitrogen. These muscles were stored at −70°C until analysis. Tibialis anterior muscles from some of the mice euthanized at 5 wk were fixed in 10% neutral buffered formalin while still attached to the tibia.
A piece of gastrocnemius muscle (∼70–100 mg) was homogenized in 1 ml of water containing 1 nmol 3-[2H3]methylhistidine (Cambridge Isotopes). Myofibrils and other insoluble proteins were pelleted by centrifugation, and the supernatant containing free amino acids was used to determine the ratio of [2H5]phenylalanine to endogenous (unlabeled) phenylalanine and the ratio of 3-[2H3]methylhistidine to endogenous 3-methylhistidine. Ratios were determined by GC-mass spectrometric analysis of the t-butyldimethylsilyl derivatives of these amino acids. Myofibrillar proteins were extracted, washed, hydrolyzed, and analyzed for [2H5]phenylalanine enrichment, as described previously (41). The amount of myofibrillar protein recovered per milligram of tissue, determined with the Pierce BCA protein assay kit, was not affected by myostatin depletion (mean 6.9% of tissue mass in myostatin-deficient mice, 7.0% in controls, P > 0.5). We know that recovery of myofibrillar proteins is not 100% with this method; to compute absolute synthesis rate per muscle from the fractional rate, we used a value of 12 mg myofibrillar protein/100 mg tissue (41). The mean rate of myofibrillar protein degradation for the interval between two time points was computed as the mean myofibrillar synthesis rate for these time points minus the rate of increase of myofibrillar protein mass during the time interval. This value was divided by the myofibrillar mass during the interval (mean of initial and final value) to compute the mean fractional degradation rate. Because myofibrillar mass and synthesis rates could not be determined without euthanizing a mouse, it was not possible to compute degradation values for an individual mouse.
RNA was extracted from gastrocnemius muscle (∼75 mg) as follows. The frozen tissue was placed in 0.5 ml of ice-cold Trizol (Invitrogen) with ∼500 mg of zirconium oxide beads (0.5 mm in diameter; Next Advance) in a 1.5-ml microcentrifuge tube and immediately homogenized for 2 min at the maximum energy setting with a Bullet Blender (Next Advance). Another 0.5 ml Trizol was added to the tube, and then RNA was extracted per the instructions supplied by Invitrogen. Quality was confirmed by A260/A280 ratios >1.9, A260/A230 ratios >2, and strong ribosomal bands detected by ethidium bromide staining of RNA subjected to agarose gel electrophoresis. The RNA concentrations were determined fluorometrically (Quant-iT RNA assay kit and Qubit fluorometer from Invitrogen). RNA (2 μg) was reverse transcribed with a High-Capacity RNA-to-cDNA kit (Applied Biosystems). The cDNA was examined with the following Taqman assays (Applied Biosystems) as recommended by the manufacturer, using an amount of cDNA equivalent to 20 ng of input RNA in each well: Mstn Mm01254559_m1, Fbxo32 Mm00499523_m1, Trim63 Mm01188690_m1, Ube2b Mm00493998_m1. Ube2b cDNA was selected as a reference (to control for variations in the amount of cDNA synthesized per reverse transcription reaction) because a previous microarray study demonstrated no effect of myostatin knockout on Ube2b expression and low variation in Ube2b expression (45). In the present study, myostatin depletion had no effect on the number of PCR cycles required to reach a predefined fluorescence threshold (CT) in the Ube2b assay (mean 20.9 cycles in myostatin-deficient samples, 21.0 cycles in control samples). However, in the food deprivation experiment mean Ube2b CT was 0.3 cycles lower in food-deprived than in fed mice (P < 0.01), suggesting a slight increase in Ube2b expression. Thus, using Ube2b as the reference gene might have led to a small underestimation (<25%) of the effect of food deprivation on expression of MuRF1, atrogin-1, and myostatin mRNAs.
Quadriceps muscle concentrations of rpS6, total and phosphorylated, were determined by immunoblotting, as described previously (43). Formalin-fixed tibialis anterior muscles were removed from the tibia and agitated in 40% NaOH to release individual fibers (7). Fibers were washed in PBS, dried on microscope slides, and mounted in Vectashield Hard-Set Mounting Medium with 4,6-diamidino-2-phenylindole (which makes the myonuclei fluorescent). The fibers were examined at ×20 magnification with a fluorescence microscope. Diameters were measured at intervals of ∼400 μm, and all nuclei within the fibers were counted. The focal plane was moved through the whole depth of a fiber to ensure that all nuclei in the field of view were counted.
Data were analyzed by factorial analysis of variance (ANOVA) to determine the contributions of myostatin depletion, time point (or food deprivation), and the interaction between these factors to the total variance. R version 2.12.0 was used for ANOVA. If ANOVA indicated a significant (P < 0.05) main effect of myostatin depletion or an interaction between myostatin depletion and time, we assessed the significance of the effect of myostatin depletion at each time point using Bonferroni t-tests. The residual variance from ANOVA was used to compute the denominator for the t-tests, as described by Glantz (15). For analysis of the effect of myostatin deficiency on tibialis anterior muscle fiber characteristics, which were measured only at the 5-wk time point, we used standard t-tests. P < 0.05 was set as the threshold for statistical significance.
RESULTS
Effects of myostatin depletion in mice fed ad libitum.
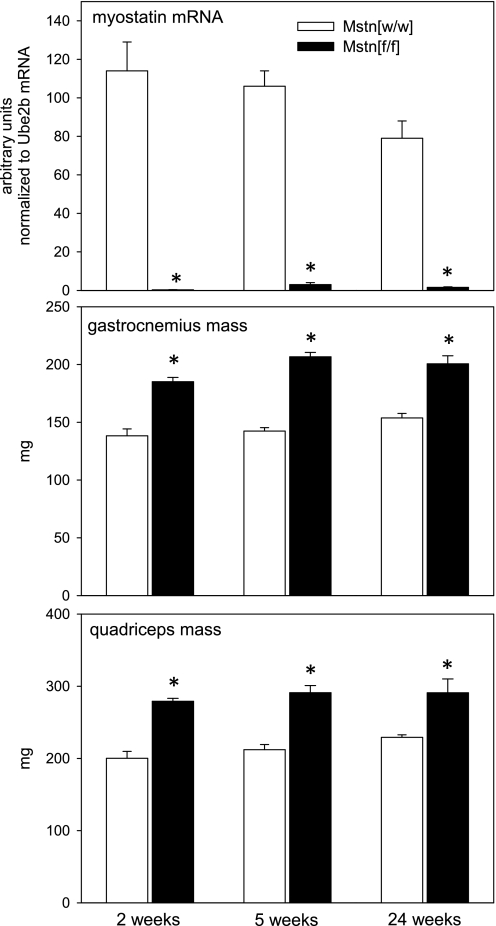
Table 1 shows the ANOVA results for all measurements. At all time points examined after tamoxifen administration, myostatin mRNA levels in Mstn[f/f] mice were <3% of the values in Mstn[w/w] mice (Fig. 1). Thus, in the remainder of this report we refer to Mstn[f/f] mice as myostatin deficient and to Mstn[w/w] mice as controls. As expected, myostatin depletion increased gastrocnemius and quadriceps muscle mass by ≥27% from 2 to 24 wk after tamoxifen treatment ended (Fig. 1). We did not determine muscle mass prior to tamoxifen administration in the present study; in a previous study of mice that did not receive tamoxifen, muscle mass was 15% greater in Mstn[f/f]/Cre+ mice than in Mstn[w/w]/Cre+ mice at 4 mo of age (42).
Table 1.
P values from ANOVA (mice fed ad libitum)
| Variable | Main Effect Myostatin Depletion | Main Effect Time Point | Myostatin Depletion × Time Point Interaction |
|---|---|---|---|
| Myostatin mRNA | <0.001 | 0.11 | 0.12 |
| Gastrocnemius mass | <0.001 | 0.004 | 0.11 |
| Quadriceps mass | <0.001 | 0.09 | 0.57 |
| Fractional synthesis | 0.16 | <0.001 | 0.11 |
| Synthesis per muscle | <0.001 | <0.001 | <0.001 |
| Synthesis/total RNA | 0.15 | <0.001 | 0.32 |
| Total RNA | 0.31 | <0.001 | 0.74 |
| Total rpS6 | 0.91 | 0.72* | 0.72 |
| Phospho-rpS6 | 0.01 | 0.35* | 0.36 |
| Phospho-rpS6/total rpS6 | 0.003 | 0.28* | 0.23 |
| 3MH | 0.69 | 0.36 | 0.23 |
| Atrogin-1 mRNA | 0.30 | 0.23 | 0.39 |
| MuRF1 mRNA | 0.007 | <0.001 | 0.14 |
rpS6, ribosomal protein S6; 3MH, 3-methylhistidine; MuRF1, muscle RING finger-1.
Main effect of time for these measurements has little meaning because of the normalization method (see legend to Fig. 5), but time was included in the ANOVA to test whether the effect of myostatin depletion varied over time (myostatin depletion × time point interaction).
Fig. 1.
Mean (+ SE) myostatin mRNA levels, gastrocnemius muscle mass, and quadriceps muscle mass after tamoxifen administration to Mstn[f/f] and Mstn[w/w] mice (n = 4–6 mice/condition). Time points refer to interval after the end of tamoxifen-induced Cre recombinase activation. *P < 0.001 for difference between Mstn[f/f] and Mstn[w/w] groups.
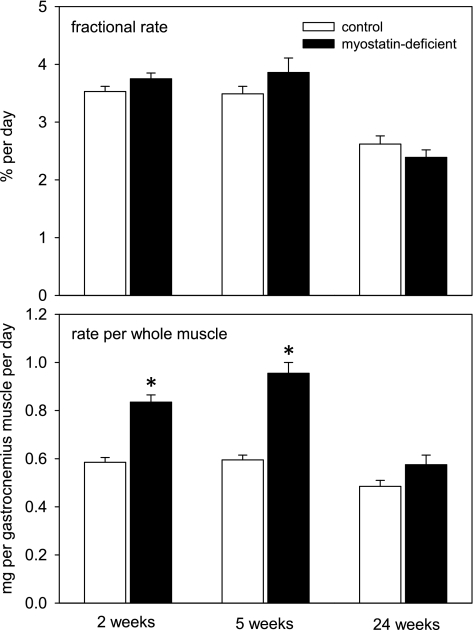
The fractional rate of myofibrillar protein synthesis, i.e., protein synthesized relative to the amount of protein present at the time of tracer injection, was not affected by myostatin depletion (Fig. 2). Total RNA concentrations (per mg muscle tissue and per mg myofibrillar protein mass) and rates of protein synthesis per milligram of total RNA also were not significantly affected by myostatin depletion (not shown, except for ANOVA results in Table 1). Myofibrillar synthesis rate per whole muscle was increased in myostatin-deficient mice. The magnitude of the effect varied across time points (significant myostatin depletion × time interaction), with larger effects observed at 2 and 5 wk after cessation of tamoxifen feeding than at 24 wk (Fig. 2).
Fig. 2.
Mean (+ SE) rate of synthesis of myofibrillar proteins in gastrocnemius muscle (n = 4–6 mice/condition). Time points refer to interval after the end of tamoxifen-induced Cre recombinase activation. Top: fractional rate of synthesis, i.e., the synthesis rate per mass of myofibrillar proteins already present. Bottom: rate per whole muscle. *P < 0.01 for difference between control and myostatin-deficient mice.
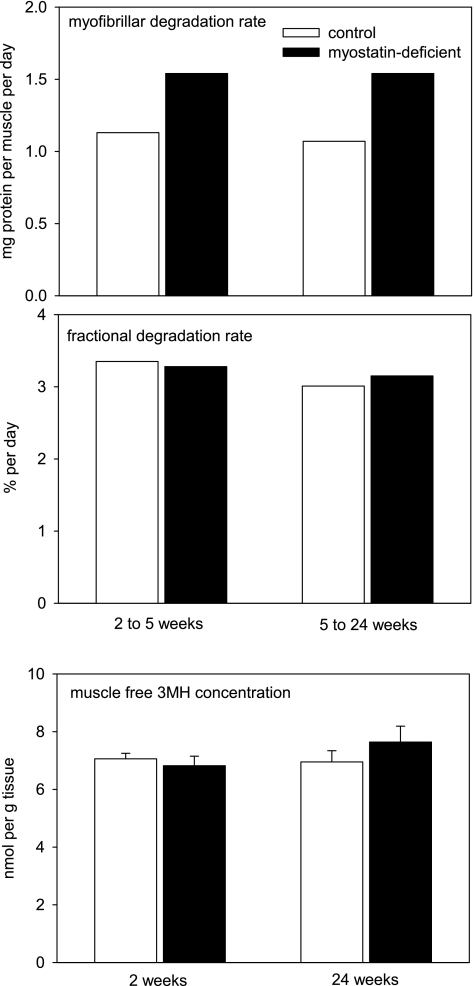
The mean rates of myofibrillar degradation in the time intervals between the measurements of protein synthesis rates are shown in Fig. 3. The higher absolute degradation rates in myostatin-deficient mice were proportional to their elevated muscle mass, so fractional myofibrillar degradation rates were similar in myostatin-deficient and control mice. This result is consistent with the observation that myostatin depletion did not affect free 3MH concentrations in muscle (Fig. 3).
Fig. 3.
Mean rates of myofibrillar protein degradation and free 3-methylhistidine (3MH) concentrations in control and myostatin-deficient gastrocnemius muscles. Time points refer to interval after the end of tamoxifen-induced Cre recombinase activation. Error bars at bottom represent 1 SE.
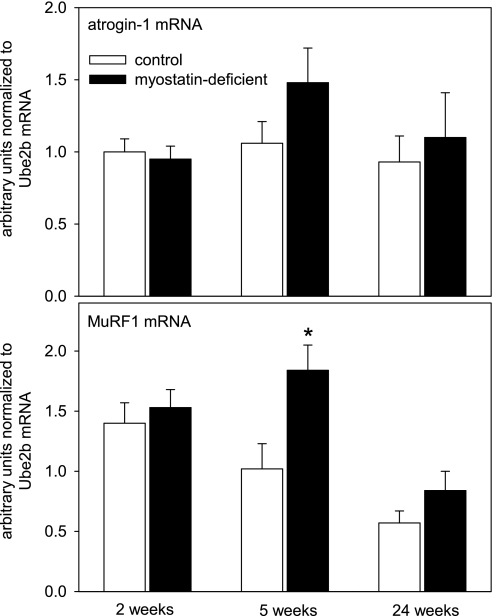
Myostatin depletion did not alter atrogin-1 expression (Fig. 4). MuRF1 expression tended to be higher in myostatin-deficient muscles (significant main effect of myostatin status according to ANOVA). Although there was not a significant interaction between myostatin status and time point, the 5-wk time point was the only one with a significant effect of myostatin depletion according to post hoc testing (Fig. 4).
Fig. 4.
Mean (+ SE) relative concentrations of atrogin-1 and muscle RING finger 1 (MuRF1) mRNAs in gastrocnemius muscle in mice fed ad libitum (n = 4–6/condition). Time points refer to interval after the end of tamoxifen-induced Cre recombinase activation. *P < 0.05 for comparison between control and myostatin-deficient muscles.
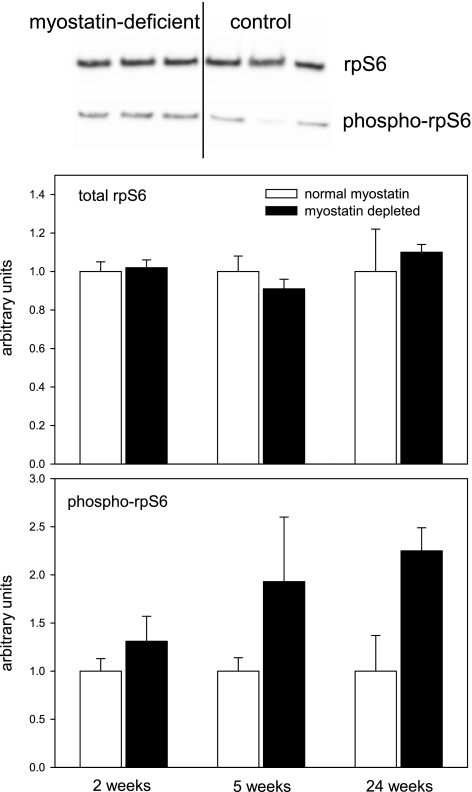
The muscle concentration of phospho-rpS6 and the phospho-rpS6/total rpS6 ratio were increased in myostatin-deficient muscles according to ANOVA, whereas the total rpS6 concentration was not (Table 1 and Fig. 5). There was not a statistically significant effect of myostatin depletion on the phospho-rpS6 concentration at P < 0.05 (adjusted for multiple comparisons) at any individual time point, but the ratio of phospho-rpS6 to total rpS6 was increased significantly at 24 wk (P < 0.05). The 24-wk time point was the one with the largest mean increase in phospho-rpS6 concentrations in myostatin-deficient mice, and this is the time point with the smallest mean effect of myostatin depletion on myofibrillar synthesis rate. This lack of correlation between phospho-rpS6 and protein synthesis also was evident in the data from individual mice (r = −0.21).
Fig. 5.
Representative immunoblots (separate blots for total and phospho-rpS6) and mean (+ SE) quadriceps muscle concentrations of total rpS6 and phospho-Ser235/236-rpS6 in mice fed ad libitum (n = 4–6 muscles/condition). Time points refer to interval after the end of tamoxifen-induced Cre recombinase activation. Equal amounts of total protein were loaded in each lane, and even transfer to the blotting paper was verified with Memcode (Pierce) staining (not shown). Values were normalized to the mean intensity of all control samples on a particular blot. Only samples from a single time point were analyzed simultaneously on the same blot.
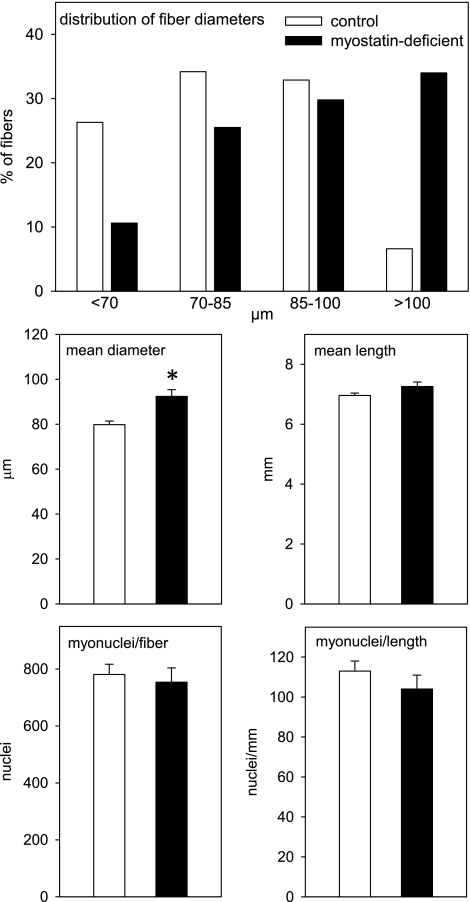
Previous research established that postdevelopmental myostatin depletion increased tibialis anterior mass to the same extent as quadriceps and gastrocnemius mass (35). A normal RNA concentration was maintained in the myostatin-deficient tibialis anterior muscles (mean 0.81 vs. 0.75 μg/mg in tibialis anterior muscles of control mice). To determine whether the maintenance of RNA concentrations in the enlarged muscles required an increase in the number of myonuclei per fiber, we isolated fibers from tibialis anterior muscles of several myostatin-deficient and control mice 5 wk after the end of tamoxifen feeding and counted all nuclei within each fiber. As expected, diameters of myostatin-deficient muscle fibers tended to be larger than control fibers (Fig. 6). Assuming that fiber cross-sectional area is proportional to the square of the diameter, the volume of the average myostatin-deficient fiber was ∼40% larger than the volume of the average control fiber. This fiber enlargement occurred without an increase in the number of myonuclei per fiber (Fig. 6). The number of myonuclei per myostatin-deficient fiber did not correlate with fiber diameter (r = −0.07).
Fig. 6.
Diameter, length, and myonuclear content of single fibers isolated from tibialis anterior muscles 5 wk after the end of tamoxifen-induced Cre recombinase activation. Data are from 76 fibers from 10 control mice and 47 fibers from 6 myostatin-deficient mice. Top: effect of myostatin depletion on fiber diameter distribution was significant (P < 0.001) according to a Chi-square test. Middle and bottom: bars represent means + SE. *P < 0.001 for control vs. myostatin deficient.
Effects of food deprivation.
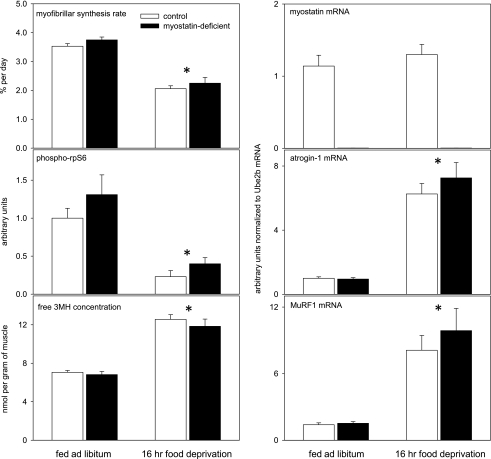
Table 2 shows the ANOVA results for the effects of myostatin depletion, overnight fasting, and their interaction 2 wk after the end of tamoxifen feeding. Food deprivation reduced RNA concentrations (by 15%; not shown), myofibrillar synthesis rate, and rpS6 phosphorylation, and it increased muscle concentrations of 3MH, atrogin-1 mRNA, and MuRF1 mRNA (Fig. 7). The key finding was the absence of a significant interaction between the effects of myostatin depletion and food deprivation, meaning that myostatin signaling does not contribute to these responses to short-term food deprivation.
Table 2.
P values from ANOVA (food deprivation experiment)
| Variable | Main Effect Myostatin Depletion | Main Effect Food Deprivation | Myostatin Depletion × Food Deprivation Interaction |
|---|---|---|---|
| Fractional synthesis | 0.21 | <0.001 | 0.93 |
| Total RNA | 0.60 | 0.02 | 0.09 |
| Protein synthesis/total RNA | 0.17 | <0.001 | 0.15 |
| Myostatin mRNA | <0.001 | 0.54 | 0.84 |
| Atrogin-1 mRNA | 0.62 | <0.001 | 0.54 |
| MuRF1 mRNA | 0.88 | <0.001 | 0.87 |
| 3MH | 0.46 | <0.001 | 0.70 |
| Total rpS6 | 0.58 | 0.23 | 0.83 |
| Phospho-rpS6 | 0.10 | <0.001 | 0.68 |
| Phospho-rpS6/total rpS6 | 0.10 | <0.001 | 0.72 |
Fig. 7.
Effects of overnight food deprivation in control and myostatin-deficient mice. Muscles were collected 2 wk after the end of tamoxifen-induced Cre recombinase activation. Phospho-Ser235/236-rpS6 concentrations were determined with quadriceps muscles; other measurements were made with gastrocnemius muscles. Means + SE are shown. *P < 0.001 for effect of food deprivation. The lack of myostatin expression did not affect any of the other measurements shown in the other parts of the figure.
DISCUSSION
Myostatin depletion did not affect the rate of myofibrillar synthesis relative to the myofibrillar mass at the time of measurement (“fractional” synthesis rate). However, it is the rate of synthesis per whole muscle rather than the fractional rate that determines the protein mass of the whole muscle at steady state (i.e., when synthesis and breakdown are in equilibrium). Synthesis rate per whole muscle was increased by myostatin depletion. A muscle can increase its production of myofibrillar proteins either by increasing the efficiency of translation of the mRNAs encoding myofibrillar proteins or by increasing the number of molecules of these mRNAs and the rRNAs needed to translate them. We have established in previous experiments that postdevelopmental myostatin knockout does not alter the muscle concentrations of rRNA or mRNAs encoding contractile proteins (42, 44, 45) and confirmed in the present study that the muscle concentration of total RNA is normal in myostatin-deficient muscles. A “normal” RNA concentration in the enlarged muscles is not really normal because the total amount of RNA per muscle is increased. This would be unremarkable except for the fact that it occurs without an increase in the number of myonuclei that generate the RNA based on our previous research (42, 44) and the myonuclei counting done in the present study. Thus, it appears that myostatin depletion either causes a global increase in transcription or inhibits RNA decay. In a previous study, we presented evidence for increased transcription (per myonucleus) of rRNA, actin, and myosin heavy-chain genes after myostatin depletion (44).
According to estimates based on myofibrillar synthesis rates and changes in myofibrillar protein mass over time, maintenance of hypertrophy in myostatin-deficient muscles did not require a reduction in the rate of myofibrillar protein degradation. A caveat regarding this approach is that synthesis rates were measured only at ∼9 AM, and we assumed that protein synthesis at this time of the day reflected the average synthesis rate for the whole day. The conclusion that myostatin depletion does not affect myofibrillar proteolysis is also supported by the observation that it did not affect free 3MH concentrations in muscle tissue. 3MH is formed by posttranslational modification of histidine residues in actin and myosin molecules, and once it is released by degradation of these proteins it cannot be reutilized for protein synthesis. Use of 3MH as a marker of myofibrillar proteolysis has been criticized, but the problems are related primarily to the use of urinary 3MH excretion. Actin and myosin are present in tissues other than skeletal muscle and turn over more rapidly in the nonmuscle tissues, 3MH can enter the body from ingestion of meat proteins, and mice do not dispose of all 3MH via urinary excretion (20, 29, 33). Local 3MH levels within a muscle should be a much better index than urinary 3MH excretion of changes in intramuscular actin and myosin degradation.
Several studies have examined the effect of myostatin excess or deficiency on expression of atrogin-1 and MuRF1. Both of these E3 ubiquitin ligases were thought to be critical determinants of muscle protein degradation, but there is more evidence that atrogin-1 interferes with protein synthesis via degradation of the translation factor eIF3F (3, 22). MuRF1 has a clear role in degradation of myofibrils (10, 11). High levels of myostatin stimulated atrogin-1 expression in both C2C12 myotubes and murine skeletal muscle in vivo and induced a more modest increase in MuRF1 expression in skeletal muscle (27). Activation of the myostatin signaling pathway increased atrogin-1 (but not MuRF1) promoter activity in murine skeletal muscle in vivo (36). In contrast, myostatin inhibited atrogin-1 and MuRF1 expression in myotubes grown from human myoblasts, which was attributed to a general inhibitory effect of myostatin on expression of genes upregulated during differentiation of myoblasts to form myotubes (39). Under normal conditions (no catabolic stressors), muscles of Mstn−/− mice have normal MuRF1 expression and either normal or increased atrogin-1 expression (1, 13, 31). Brief treatment with an anti-myostatin antibody reduced muscle atrogin-1 expression 1.5-fold (P < 0.01) (unpublished microarray data; available under accession number GSE13707 in Gene Expression Omnibus, www.ncbi.nlm.nih.gov/geo). A similar reduction in atrogin-1 mRNA levels occurred in muscles of old mice treated with an anti-myostatin antibody for 14 wk (32). However, we have not observed a significant reduction in muscle atrogin-1 or MuRF1 mRNA levels a few weeks or months after postdevelopmental myostatin knockout (Refs. 44 and 45 and the present study). MuRF1 mRNA levels actually tended to be higher in the myostatin-deficient muscles in the present study when mice were allowed free access to food. The present study demonstrated that postdevelopmental myostatin depletion, like constitutive myostatin knockout (1), does not influence the upregulation of atrogin-1 and MuRF1 mRNA levels induced by food deprivation.
The Akt-mTOR-S6 kinase pathway is one of the regulators of protein metabolism and cell size (16), so naturally there has been interest in whether myostatin affects the activity of this pathway. Mstn−/− mice have increased (∼2-fold) muscle concentrations of Akt1 mRNA, total Akt, phospho-Akt, and phospho-rpS6 (a marker of S6 kinase activity) (8, 24, 31). We reported that S6 kinase and rpS6 phosphorylation (but not total or phosphorylated Akt levels) increased after 4 days of myostatin inhibition in mature mice (43), but not 3 mo after postdevelopmental depletion of myostatin, to ∼10% of the normal level (44). More recent research has confirmed that postdevelopmental inhibition of myostatin activity does not affect the phospho-Akt concentration in muscle and that it causes muscle hypertrophy even in Akt1- and Akt2-null mice (18). However, Akt activity is not the only determinant of mTOR and S6 kinase activity, and in the present study rpS6 phosphorylation tended to be increased in myostatin-deficient muscles. The effect was statistically significant when data were pooled across all time points. There was not a correlation between rpS6 phosphorylation and the rate of myofibrillar synthesis. Together with the fact that rapamycin treatment eliminated S6 kinase and rpS6 phosphorylation but did not block the stimulation of myofibrillar protein synthesis induced by short-term myostatin inhibition (43), these data suggest that the increased muscle protein synthesis in myostatin-deficient muscle does not depend on increased S6 kinase activity.
The present study is consistent with previous reports that short-term food deprivation reduces the rate of muscle protein synthesis and that this decline is much larger than the modest decrease in total RNA concentrations (4, 5, 9, 30). The increases in expression of atrogin-1 and MuRF1 mRNAs in food-deprived mice were also expected based on previous research (1, 17, 21, 23). The relative increase in 3MH concentrations in muscle tissue after overnight food deprivation was similar to the effect of short-term food deprivation on release of 3MH and tyrosine from ex vivo muscle preparations (25, 34, 47). Allen et al. (1) observed an approximately threefold increase in myostatin mRNA and protein levels after 2 days of food deprivation in mice but only a nonsignificant trend for increased myostatin mRNA levels after 1 day of food deprivation. Food deprivation for 2 days increased myostatin protein levels in rat muscle even though myostatin mRNA levels were reduced (37). In the present study, overnight food deprivation did not affect myostatin mRNA levels. The fact that the food deprivation-induced changes in protein synthesis and measurements related to protein breakdown were not affected by myostatin knockout also supports the conclusion that myostatin does not contribute to these responses.
GRANTS
This research was supported by a grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR-054366).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
REFERENCES
- 1. Allen DL, Cleary AS, Lindsay SF, Loh A, Reed JM. Myostatin expression is increased by food deprivation in a muscle-specific manner and contributes to muscle atrophy during prolonged food deprivation in mice. J Appl Physiol 109: 692–701, 2010 [DOI] [PubMed] [Google Scholar]
- 2. Amthor H, Otto A, Vulin A, Rochat A, Dumonceaux J, Garcia L, Mouisel E, Hourde C, Macharia R, Friedrichs M, Relaix F, Zammit PS, Matsakas A, Patel K, Partridge T. Muscle hypertrophy driven by myostatin blockade does not require stem/precursor-cell activity. Proc Natl Acad Sci USA 106: 7479–7484, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Attaix D, Baracos VE. MAFbx/Atrogin-1 expression is a poor index of muscle proteolysis. Curr Opin Clin Nutr Metab Care 13: 223–224, 2010 [DOI] [PubMed] [Google Scholar]
- 4. Baillie AG, Garlick PJ. Responses of protein synthesis in different skeletal muscles to fasting and insulin in rats. Am J Physiol Endocrinol Metab 260: E891–E896, 1991 [DOI] [PubMed] [Google Scholar]
- 5. Bark TH, McNurlan MA, Lang CH, Garlick PJ. Increased protein synthesis after acute IGF-I or insulin infusion is localized to muscle in mice. Am J Physiol Endocrinol Metab 275: E118–E123, 1998 [DOI] [PubMed] [Google Scholar]
- 6. Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore LA, Ahima RS, Khurana TS. Functional improvement of dystrophic muscle by myostatin blockade. Nature 420: 418–421, 2002 [DOI] [PubMed] [Google Scholar]
- 7. Brack AS, Bildsoe H, Hughes SM. Evidence that satellite cell decrement contributes to preferential decline in nuclear number from large fibers during murine age-related muscle atrophy. J Cell Sci 118: 4813–4821, 2005 [DOI] [PubMed] [Google Scholar]
- 8. Chelh I, Meunier B, Picard B, Reecy MJ, Chevalier C, Hocquette JF, Cassar-Malek I. Molecular profiles of Quadriceps muscle in myostatin-null mice reveal PI3K and apoptotic pathways as myostatin targets. BMC Genomics 10: 196, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cherel Y, Attaix D, Rosolowska-Huszcz D, Belkhou R, Robin JP, Arnal M, Le Maho Y. Whole-body and tissue protein synthesis during brief and prolonged fasting in the rat. Clin Sci (Lond) 81: 611–619, 1991 [DOI] [PubMed] [Google Scholar]
- 10. Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, Glass DJ. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab 6: 376–385, 2007 [DOI] [PubMed] [Google Scholar]
- 11. Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, Latres E, Goldberg AL. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol 185: 1083–1095, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Foster K, Graham IR, Otto A, Foster H, Trollet C, Yaworsky PJ, Walsh FS, Bickham D, Curtin NA, Kawar SL, Patel K, Dickson G. Adeno-associated virus-8-mediated intravenous transfer of myostatin propeptide leads to systemic functional improvements of slow but not fast muscle. Rejuvenation Res 12: 85–93, 2008 [DOI] [PubMed] [Google Scholar]
- 13. Gilson H, Schakman O, Combaret L, Lause P, Grobet L, Attaix D, Ketelslegers JM, Thissen JP. Myostatin gene deletion prevents glucocorticoid-induced muscle atrophy. Endocrinology 148: 452–460, 2007 [DOI] [PubMed] [Google Scholar]
- 14. Girgenrath S, Song K, Whittemore LA. Loss of myostatin expression alters fiber-type distribution and expression of myosin heavy chain isoforms in slow- and fast-type skeletal muscle. Muscle Nerve 31: 34–40, 2005 [DOI] [PubMed] [Google Scholar]
- 15. Glantz SA. Primer of Biostatistics, Fourth Edition. New York: McGraw-Hill, 1997 [Google Scholar]
- 16. Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol 37: 1974–1984, 2005 [DOI] [PubMed] [Google Scholar]
- 17. Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA 98: 14440–14445, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goncalves MD, Pistilli EE, Balduzzi A, Birnbaum MJ, Lachey J, Khurana TS, Ahima RS. Akt deficiency attenuates muscle size and function but not the response to ActRIIB inhibition. PLoS One 5: e12707, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haidet AM, Rizo L, Handy C, Umapathi P, Eagle A, Shilling C, Boue D, Martin PT, Sahenk Z, Mendell JR, Kaspar BK. Long-term enhancement of skeletal muscle mass and strength by single gene administration of myostatin inhibitors. Proc Natl Acad Sci USA 105: 4318–4322, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harris CI, Rucklidge GJ, McDiarmid RM, Milne G. Sex- and phenotype-dependent metabolism of N tau-methylhistidine by mice. Biochem J 239: 229–232, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jagoe RT, Lecker SH, Gomes M, Goldberg AL. Patterns of gene expression in atrophying skeletal muscles: response to food deprivation. FASEB J 16: 1697–1712, 2002 [DOI] [PubMed] [Google Scholar]
- 22. Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet-Pichon S, Tintignac LA, Segura CT, Leibovitch SA. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J 27: 1266–1276, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J 18: 39–51, 2004 [DOI] [PubMed] [Google Scholar]
- 24. Lipina C, Kendall H, McPherron AC, Taylor PM, Hundal HS. Mechanisms involved in the enhancement of mammalian target of rapamycin signalling and hypertrophy in skeletal muscle of myostatin-deficient mice. FEBS Lett 584: 2403–2408, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lowell BB, Ruderman NB, Goodman MN. Regulation of myofibrillar protein degradation in rat skeletal muscle during brief and prolonged starvation. Metabolism 35: 1121–1127, 1986 [DOI] [PubMed] [Google Scholar]
- 26. Matsakas A, Foster K, Otto A, Macharia R, Elashry MI, Feist S, Graham I, Foster H, Yaworsky P, Walsh F, Dickson G, Patel K. Molecular, cellular and physiological investigation of myostatin propeptide-mediated muscle growth in adult mice. Neuromuscul Disord 19: 489–499, 2009 [DOI] [PubMed] [Google Scholar]
- 27. McFarlane C, Plummer E, Thomas M, Hennebry A, Ashby M, Ling N, Smith H, Sharma M, Kambadur R. Myostatin induces cachexia by activating the ubiquitin proteolytic system through an NF-kappaB-independent, FoxO1-dependent mechanism. J Cell Physiol 209: 501–514, 2006 [DOI] [PubMed] [Google Scholar]
- 28. McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-b superfamily member. Nature 387: 83–90, 1997 [DOI] [PubMed] [Google Scholar]
- 29. Millward DJ, Bates PC, Grimble GK, Brown JG, Nathan M, Rennie MJ. Quantitative importance of non-skeletal-muscle sources of N tau-methylhistidine in urine. Biochem J 190: 225–228, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Millward DJ, Garlick PJ, James WP, Nnanyelugo DO, Ryatt JS. Relationship between protein synthesis and RNA content in skeletal muscle. Nature 241: 204–205, 1973 [DOI] [PubMed] [Google Scholar]
- 31. Morissette MR, Cook SA, Buranasombati C, Rosenberg MA, Rosenzweig A. Myostatin inhibits IGF-I-induced myotube hypertrophy through Akt. Am J Physiol Cell Physiol 297: C1124–C1132, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murphy KT, Koopman R, Naim T, Leger B, Trieu J, Ibebunjo C, Lynch GS. Antibody-directed myostatin inhibition in 21-mo-old mice reveals novel roles for myostatin signaling in skeletal muscle structure and function. FASEB J 24: 4433–4442, 2010 [DOI] [PubMed] [Google Scholar]
- 33. Murray AJ, Nield MK, Jones LM, Galbraith N, Tomas FM. Metabolism of N tau-methylhistidine by mice. Biochem J 232: 409–413, 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nagasawa T, Kikuchi N, Ito Y, Yoshizawa F, Nishizawa N. Suppression of myofibrillar protein degradation after refeeding in young and adult mice. J Nutr Sci Vitaminol (Tokyo) 50: 227–230, 2004 [DOI] [PubMed] [Google Scholar]
- 35. Personius KE, Jayaram A, Krull D, Brown R, Xu T, Han B, Burgess K, Storey C, Shah B, Tawil R, Welle S. Grip force, EDL contractile properties, and voluntary wheel running after postdevelopmental myostatin depletion in mice. J Appl Physiol 109: 886–894, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sartori R, Milan G, Patron M, Mammucari C, Blaauw B, Abraham R, Sandri M. Smad2 and 3 transcription factors control muscle mass in adulthood. Am J Physiol Cell Physiol 296: C1248–C1257, 2009 [DOI] [PubMed] [Google Scholar]
- 37. Smith IJ, Aversa Z, Alamdari N, Petkova V, Hasselgren PO. Sepsis downregulates myostatin mRNA levels without altering myostatin protein levels in skeletal muscle. J Cell Biochem 111: 1059–1073, 2010 [DOI] [PubMed] [Google Scholar]
- 38. Steelman CA, Recknor JC, Nettleton D, Reecy JM. Transcriptional profiling of myostatin-knockout mice implicates Wnt signaling in postnatal skeletal muscle growth and hypertrophy. FASEB J 20: 580–582, 2006 [DOI] [PubMed] [Google Scholar]
- 39. Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol Cell Physiol 296: C1258–C1270, 2009 [DOI] [PubMed] [Google Scholar]
- 40. Wang H, Liu D, Cao P, Lecker S, Hu Z. Atrogin-1 affects muscle protein synthesis and degradation when energy metabolism is impaired by the antidiabetes drug berberine. Diabetes 59: 1879–1889, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Welle S, Bhatt K, Pinkert CA. Myofibrillar protein synthesis in myostatin-deficient mice. Am J Physiol Endocrinol Metab 290: E409–E415, 2006 [DOI] [PubMed] [Google Scholar]
- 42. Welle S, Bhatt K, Pinkert CA, Tawil R, Thornton CA. Muscle growth after postdevelopmental myostatin gene knockout. Am J Physiol Endocrinol Metab 292: E985–E991, 2007 [DOI] [PubMed] [Google Scholar]
- 43. Welle S, Burgess K, Mehta S. Stimulation of skeletal muscle myofibrillar protein synthesis, p70 S6 kinase phosphorylation, and ribosomal protein S6 phosphorylation by inhibition of myostatin in mature mice. Am J Physiol Endocrinol Metab 296: E567–E572, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Welle S, Burgess K, Thornton CA, Tawil R. Relation between extent of myostatin depletion and muscle growth in mature mice. Am J Physiol Endocrinol Metab 297: E935–E940, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Welle S, Cardillo A, Zanche M, Tawil R. Skeletal muscle gene expression after myostatin knockout in mature mice. Physiol Genomics 38: 342–350, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Whittemore LA, Song K, Li X, Aghajanian J, Davies M, Girgenrath S, Hill JJ, Jalenak M, Kelley P, Knight A, Maylor R, O'Hara D, Pearson A, Quazi A, Ryerson S, Tan XY, Tomkinson KN, Veldman GM, Widom A, Wright JF, Wudyka S, Zhao L, Wolfman NM. Inhibition of myostatin in adult mice increases skeletal muscle mass and strength. Biochem Biophys Res Commun 300: 965–971, 2003 [DOI] [PubMed] [Google Scholar]
- 47. Wing SS, Haas AL, Goldberg AL. Increase in ubiquitin-protein conjugates concomitant with the increase in proteolysis in rat skeletal muscle during starvation and atrophy denervation. Biochem J 307: 639–645, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]