Abstract
Prolactin (PRL) is known to play an essential role in mammary alveolar proliferation in the pregnant mouse, but its role in lactation has been more difficult to define. Genetic manipulations that alter expression of the PRL receptor and its downstream signaling molecules resulted in developmental defects that may directly or indirectly impact secretory activation and lactation. To examine the in vivo role of PRL specifically in lactation, bromocriptine (BrCr) was administered every 8 h to lactating mice on the second day postpartum, resulting in an ∼95% decrease in serum PRL levels. Although morphological changes in secretory alveoli were slight, by 8 h of BrCr, pup growth was inhibited significantly. Phosphorylated STAT5 fell to undetectable levels within 4 h. Decreased milk protein gene expression, β-casein, and α-lactalbumin, was observed after 8 h of treatment. To assess mammary-specific effects on lipid synthesis genes, we isolated mammary epithelial cells (MECs) depleted of mammary adipocytes. Expression of genes involved in glucose uptake, glycolysis, pentose phosphate shunt, de novo synthesis of fatty acids, and biosynthesis of triacylglycerides was decreased up to 19-fold in MECs by just 8 h of BrCr treatment. Glands from BrCr-treated mice showed a twofold reduction in intracellular cytoplasmic lipid droplets and a reduction in cytosolic β-casein. These data demonstrate that PRL signaling regulates MEC-specific lipogenic gene expression and that PRL signals coordinate the milk synthesis and mammary epithelial cell survival during lactation in the mouse.
Keywords: bromocriptine, de novo lipogenesis
mammals are defined by the presence of mammary glands that synthesize and secrete milk to sustain the growth and development of their progeny. It has long been known that development of the alveolar structures that synthesize and secrete milk depends on pituitary prolactin (PRL). During puberty, ductal elongation and branching of mammary epithelium into the mammary fat pad is stimulated by estrogen, in combination with epidermal growth factor and insulin-like growth factor I (14, 16). With the onset of pregnancy, PRL and progesterone stimulate development of side branches that grow and differentiate into the alveolar units responsible for milk secretion, eventually filling the mammary fat pad. As pregnancy progresses, alveoli synthesize both cytoplasmic lipid droplets (CLD) and milk proteins (23, 29), but secretory activation is held in check. The fall of progesterone at parturition triggers the copious synthesis of all milk components, including milk proteins, lactose, and triglycerides, which are synthesized and secreted under the control of PRL, glucocorticoid, and insulin. Cessation of suckling prompts the alveolar epithelium to undergo involution (44).
A pituitary hormone was first associated with regulation of milk secretion in the late 1920s when Stricker and Grueter observed enhanced mammary development and increased milk secretion in rabbits, dogs, sows, and cows following injection of pituitary extract (40). In 1932, the responsible hormone was purified and named prolactin (30). Extensive studies since that time have shown PRL to be associated with mammary gland development and lactation in many mammalian species. As summarized in a recent review, PRL can reestablish lactation in hypophysectomized rats, rabbits, guinea pigs, goats, and sheep (42). During established lactation, both cows and pigs appear to be refractory to exogenous PRL treatment and to the loss of pituitary PRL after administration of bromocriptine (BrCr). BrCr is a dopaminergic compound that blocks PRL secretion from the lactotrope cells of the hypothalamus (9). Abrogation of milk synthesis was achieved in lactating rats when BrCr was combined with an antiserum against growth hormone (41). Together, these studies using BrCr suggest an important role for PRL in milk synthesis/secretion in mammals, but definitive proof of its essential role in established lactation is lacking, most particularly for the mouse.
Studies of genetically modified mice lacking PRL (15), its receptor (PRLR) (26), or downstream signaling molecules such as STAT5 (21) provide solid evidence that PRL and signaling pathways activated by its receptor regulate mouse mammary gland development during pregnancy and potentially lactation. Unfortunately, the developmental defects of the mammary epithelium in these mice prevent unambiguous analysis of the specific role of PRL and its receptor in murine lactation. Further complicating results of these studies was the analysis of whole mammary tissue, which contains large quantities of mammary adipocytes (1). Adipocyte responsiveness to PRL significantly confounds results related to lipid synthesis and metabolism when whole tissues are examined (8, 34). To circumvent these problems and to clarify previous studies conducted in mouse models, we treated lactating FVB mice with a regimen of BrCr that completely blocked pituitary release of PRL within 4 h of treatment. Our goal was to identify PRL-dependent gene regulation in mammary epithelial cells without the confounding responses of mammary adipose cells. (36).
As would be predicted from other studies, the expression of milk protein genes decreased significantly after BrCr treatment. Our results also demonstrate rapid, coordinate decreases in the expression of genes encoding GLUT1, enzymes involved in glycolysis, the pentose shunt, de novo synthesis of fatty acids, and triglyceride biosynthesis specifically in adipose-depleted mammary epithelial cells (MECs) following 8 h of BrCr. Additionally, the number of cytoplasmic lipid droplets (CLD) was reduced twofold when serum PRL was withdrawn. This study provides in vivo evidence that PRL orchestrates expression of lipogenic genes specifically in MECs, and likely regulates the activity of lipogenic enzymes that leads to CLD formation.
MATERIALS AND METHODS
Animals.
Female FVB mice 6–8 wk of age were purchased from Taconic and maintained in the US Department of Agriculture-approved Center for Laboratory Animal Care of the University of Colorado Anschutz Medical Campus Center for Comparative Medicine under a 12:12-h light-dark cycle. Pregnancy day 1 was designated at appearance of a vaginal plug after overnight mating. Lactation day 1 (L1) was designated the day of parturition. Mice were fed breeder's chow (Teklad S-2335, no. 7004 mouse breeder chow; Harlan Teklad, Madison, WI) for 12 days and then a standard diet (Teklad 22/5, no. 8640 rodent diet; Harlan Teklad) until parturition and subsequent death. Two days after parturition (L2), litters were standardized to eight pups unless otherwise stated and weighed every 8 h for 24 h during BrCr treatment. All mice were anesthetized by intraperitoneal injection with Avertin (125–250 mg/kg) and euthanized by cervical dislocation. All procedures were approved by the University of Colorado Anschutz Medical Campus Institutional Animal Care and Use Committee.
BrCr and oxytocin treatment.
BrCr (2-bromo-α-ergocryptine methanesulfonate salt, Sigma B2134; Sigma Chemical, St. Louis, MO) was dissolved in absolute ethanol (EtOH) at 8 mg/ml and then diluted with sterile normal saline solution (0.9% NaCl) to a final concentration of 2 mg/ml (25% ETOH). Vehicle was 25% EtOH in sterile saline. Groups of four mice were injected intraperitoneally with 0.02 mg/g BrCr, and mice were euthanized at the following time points: 4, 8, 16, and 24 h. For the 16-h time point, mice were re-treated with BrCr after 8 h; for the 24-h point mice were re-treated after the 16-h point. In one study, a group of three mice were given a single dose of BrCr and euthanized at either 8 or 24 h postinjection.For weigh-suckle-weigh studies, mice were treated with BrCr or vehicle as above, and litters were removed for 2 h. After 2 h, all dams were injected intraperitoneally with 300 μl of 2 U/ml oxytocin, and pups were weighed and returned to the dams and allowed to suckle for 4 h, after which the pups were reweighed to determine the milk intake over 4 h of oxytocin add back.
Collection and measurement of serum PRL levels.
At the time of euthanasia, mice were anesthetized with Avertin, and blood was collected via terminal cardiac puncture. Blood was allowed to clot at room temperature for 15 min and was centrifuged at 14,000 rpm at room temperature, and the serum was collected and stored at −20°C. Quantitation of serum PRL levels was performed by Dr. A. F. Parlow at the National Hormone and Peptide Program (Torrance, CA).
Tissue collection and histological analysis.
Contralateral third and fourth mammary glands were excised. Four sections from each fourth mammary gland, excluding the lymph nodes, were divided: two portions proximal to the teat were for RNA analysis, and two portions distal to the teat were used for protein analysis. Tissue sections for RNA were weighed and placed in 1 ml of RNeasy (Qiagen, Valencia, CA), flash-frozen and stored at −80°C. Tissues for protein were placed in a microcentrifuge tube, weighed, flash-frozen, and stored at −80°C.
The third mammary glands were fixed overnight in 4% paraformaldehyde (diluted from 16% solution; Electron Microscopy Services, cat. no. 15710, Hatfield, PA) in Ringer's saline solution (138 mM NaCl, 8.1 mM Na2HPO4, 1.2 mM K2HPO4, 2.7 mM KH2PO4, 0.9 mM CaCl2, 0.5 mM MgCl2) with 25% sucrose. The right third mammary glands were embedded in paraffin, cut, and stained with hematoxylin and eosin. The left third mammary glands were cut vertically into halves and placed in cryomolds (TissueTek Cryomold Standard no. 4557; Sakura Finetek USA, Torrance, CA) filled with OTC embedding medium (Tissue-Tek O.C.T. Compound no. 4583; Sakura Finetek USA) and flash-frozen by immersion in isopentane bath in liquid nitrogen.
Adipocyte-depleted MECs.
MECs depleted of adipocytes were prepared as recently described (36). The digested cell suspension was transferred into a new 15-ml conical tube and brought up to 15 ml with cold PBS and spun at 300 rcf for 5 min. Supernatant was decanted and the MEC pellet resuspended in 10 ml of ice-cold PBS. Washing was repeated two additional times. Approximately 500 μl of cell pack was divided equally, and one-half of the cell pellet was lysed in 1 ml of TRIzol (Invitrogen) for RNA analysis.
RNA isolation.
Each sample was homogenized using a Brinkman polytron (www.brinkmann.com); lysates were cleared at 13,000 rcf for 15 min at 4°C. RNA was isolated using TRIzol according to the manufacturer's protocol. Washed RNA pellets were resuspended in 100 μl of nuclease-free water and contaminants removed using the Qiagen RNeasy Mini Plus protocol (www.qiagen.com, no. 74134). Total RNA was quantified using the Nanodrop 1000 spectrophotometer (www.nanodrop.com). Total RNA integrity was examined using the Agilent Bioanalyzer Nanoscale Microfluidics Chip Assay (www.chem.agilent.com, 2100 Bioanalyzer). All samples were of sufficient integrity for use in quantitative reverse transcriptase-real-time PCR (qRT-PCR) analysis.
qRT-PCR analysis.
A table listing the primers and probes for genes is in supplemental materials (online at Journal website, this issue). cDNA was prepared using the Verso cDNA synthesis kit according to the manufacturer's protocol (www.Thermofisher.com). cDNA was diluted 1:10, and 5 μl was used as input into the qRT-PCR reactions (50 ng total RNA/reaction). qRT-PCR master mix included 12.5 μl of Absolute Fast qRTPCR Mix-Lox Rox (www.ThermoFisher.com), 2.5 μl of 10× primer/probe mix (5.0 μM forward, 5.0 μM reverse, and 2.5 μM probe), 5.0 μl of nuclease free water, and 5.0 μl of 1:10 diluted cDNA per reaction. qRT-PCR data were collected with the 7500 fast thermocycler (www.appliedbiosystems.com). Transcript copy numbers were calculated using a standard curve method. To generate standard curves for every qRT-PCR target, the PCR product of four 50-μl test reactions per target were pooled and purified using a Qiagen PCR cleanup kit. The target-specific PCR product was isolated using a 2.5% agarose gel and then gel purified using a Qiagen extraction kit according to the manufacturer's protocol. The concentration for each purified PCR product was determined using a Nanodrop. Copy numbers for each target were calculated as follows:
(Supplemental Table S1)]. Standard Curves were generated using serially diluted target-specific PCR products to an initial concentration of 1.204 × 107 molecules/μl and stepping down fivefold to a final of 7.70 × 102 molecules/μl. All standard curves had near-optimal slopes of −3.3, y-intercepts near 40, R correlations near 1, and PCR efficiencies near 100%, and each target amplified within its standard curve.
Immunohistochemistry.
Paraffin-embedded sections were dewaxed in xylene twice for 5 min and rehydrated in a series of graded alcohols (100%, 90%, 70%, and 30% ethanol) for 1 min in each. Sections were rehydrated in phosphate-buffered saline (PBS) for 5 min, and antigen retrieval was performed for 20 min in antigen retrieval solution (Antigen Unmasking Solution no. H3300; Vector Laboratories, Burlingame, CA). Sections were permeabilized in 0.2% Triton X-100 in PBS, blocked with 5% normal goat serum (no. 005-000-121; Jackson Immunoresearch, West Grove, PA) and 100 μg/ml saponin (no. S4521; Sigma) in PBS. Sections were rinsed twice with PBS and incubated with a polyclonal anti-β-casein (37) and a guinea pig polyclonal anti-adipophilin (RDI-PROGP40; Research Diagnostics, Flanders, NJ) at 1:200 for 1 h. Sections were rinsed five times (5 min) with PBS and incubated with FITC-conjugated donkey anti-rabbit IgG and Alexa fluor 594-conjugated goat anti-guinea pig IgG antibodies (Molecular Probes, Eugene, OR) plus 0.5 μg/μl 4′,6-diamidino-2-phenylindole (DAPI) for 45 min. The sections were rinsed six times (5 min) with PBS, and then mounted (ProLong Antifade Kit, no. P-7481; Molecular Probes). Digital images were captured using a Nikon fluorescent confocal microscope and Slidebook software (Intelligent Imaging Innovations, Denver, CO).
TUNEL staining and analysis.
Paraffin-embedded sections were dewaxed, rehydrated, and permeabilized as above and stained according to the manufacturer's protocol of the In Situ Cell Death Detection Kit, TMR red (cat. no. 2 156 792; Roche, Indianapolis, IN). Coverslips were mounted as above. Ten images per sample were captured using a Nikon fluorescent confocal microscope under a ×60 oil immersion objective and Slidebook software using identical parameters. Each image was counted for total alveolar nuclei and TUNEL positive alveolar nuclei. The percentages of apoptotic cells were averaged and the standard error of the mean (SE) was calculated to determine significant differences between untreated and BrCr-treated animals.
Western blotting.
Protein was extracted from whole fourth mammary glands by homogenizing in cell lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 1% Trition X-100, 1% DOC, 0.1% SDS, 1 mM DTT, 5 mM sodium orthovanadate] containing protease inhibitor cocktail [10 μg/μl leupeptin, 10 μg/μl aprotinin, 10 μM pepstatin A, 1 mM PMSF (L2023, A1153, P4265, and P7626, respectively; Sigma)]. The homogenates were centrifuged at 13,900 rpm for 30 min at 4°C, and supernatants were collected, snap-frozen, and stored at −20°C until use. Total protein (100 μg) was resolved on 8, 10, and 12% SDS-PAGE and transferred to nitrocellulose membranes (no. 162-0114; Bio-Rad, Hercules, CA). Anti-phospho-STAT5A/B (no. 06-798; Upstate Biotechnology, Lake Placid, NY), anti-STAT5 (no. 9352), phospho-p44/42 MAPK (Thr202/Tyr204; monoclonal E10), and p44/42 MAPK antibodies (nos. 9106 and 9102) were obtained from Cell Signaling Technology (Beverly, MA). ECL images were scanned, and densitometry was quantitated using the Bio-Rad Gel Doc EQ system.
Morphological analysis of CLD number and epithelial width.
Sections of mammary gland from 8-h BrCr-treated mammary glands were prepared as for histological analysis and stained for ADPH, casein, and DAPI. Images were captured on an Olympus IX81 inverted motorized microscope using Olympus objectives, a 100-W mercury lamp, and a Hamamatsu ORCA IIER monochromatic charge-coupled device camera (1,344–1,024 full chip, 6.45 m pixel), with filter sets as follows: excitation at 360–370 nm and emission at 420–460 for DAPI, excitation at 450–480 nm and emission at 535 nm for FITC, and excitation at 535 nm and emission at 635 nm for CY3. The system is controlled by SlideBook software (version 4.067, Intelligent Imaging Innovations), which was also used to normalize and analyze immunofluorescence. Three sections from each of three mice per condition were used for quantitation. Because the autofluorescence of mammary tissue sections stained only with DAPI gave a clear representation of the extent of the mammary epithelium (Supplementary Fig. S2), these sections were used for quantitation of epithelial thickness and the number of intracellular lipid droplets. The measurement function of Slidebook was used to quantify the width of 10 epithelial segments per slide; lipid droplets were quantified by hand, counting the number of unstained round areas within the epithelium as well as the DAPI-stained nuclei.
Statistics.
All statistical analysis was done using the t-test function of Excel with a one-tailed distribution and equal variance of samples. For all analyses statistical significance was declared at P ≤ 0.05.
RESULTS
BrCr diminishes serum PRL levels to suppress pup growth.
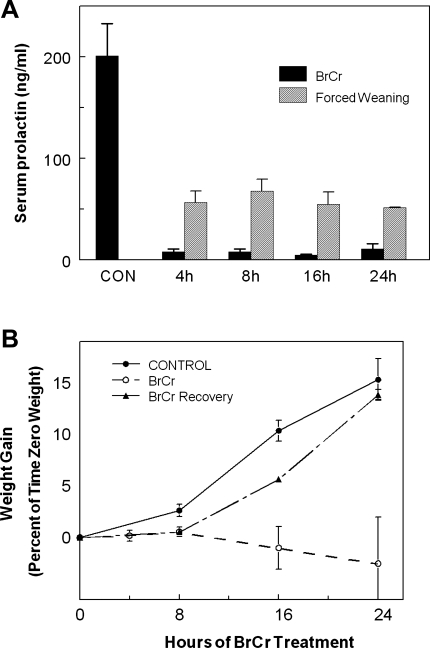
We conducted extensive dose response studies to determine a BrCr dose and injection schedule that consistently inhibited pituitary PRL secretion: injection of 0.02 mg/g body wt BrCr at 8-h intervals. This treatment regimen reduced serum PRL levels by more than 94% of control values at 4, 8, 16, and 24 h (Fig. 1A). Consistent with other studies, neither less frequent injections nor lower concentrations of BrCr was effective for reducing pup growth (19, 43). In contrast to BrCr treatment, forced weaning by litter withdrawal reduced serum PRL to levels 25–35% of untreated mice (Fig. 1A). BrCr injection at day 18 of pregnancy resulted in rapid death of all pups following parturition.
Fig. 1.
Effect of bromocriptine (BrCr) treatment on levels of serum prolactin (PRL) and growth of pups. A: serum was collected from controls or mice injected with BrCr every 8 h at indicated times (filled bars). Blood samples used for quantitation of serum PRL were drawn immediately prior to subsequent BrCr injection, demonstrating that this treatment regimen successfully inhibits pituitary secretion of PRL during the entire 8 h following treatment with BrCr. For comparison, serum was also collected from dams in which the pups had been withdrawn at time 0 (gray bars). Mean serum levels of PRL ± SE are shown; n = 4 mice per time point. B: litters were standardized to 8 pups and weighed at 4, 8, 16, and 24 h. Control mice were not injected with BrCr (●), and test mice were injected with BrCr every 8 h (○). One set of mice was injected with BrCr at time 0 and then allowed to recover without additional treatment (▴). Each point represents mean increase in weight as a %time 0 weight of that litter ± SE; n = 4 at 4 h, 15 at 8 h, 8 at 16 h, and 4 at 24 h; n = 3 for BrCr recovery study. Error bars indicate SE.
The growth of pups suckling BrCr-treated dams ceased within 4 h. By 8 h, pups suckling BrCr-injected dams had gained only ∼1.6% of their initial weight, whereas pups suckling the control dams gained ∼3.6% (Fig. 1B). Pups nursed by BrCr-treated dams continued to lose weight over 24 h despite observable suckling throughout the experiment, and milk accumulation was not observed in their stomachs. To determine whether the effects of BrCr treatment were reversible, additional dams were injected only once with BrCr and allowed to recover for the remaining 16 h (Fig. 1B). Consistent with other studies (19), 8 h after the initial BrCr injection, the weights of these pups increased in parallel with pups' weights from untreated control mice (Fig. 1B). Initial inhibition of pituitary PRL due to BrCr appears to be completely reversible because milk synthesis/secretion resumes if redosing does not occur.
Alveolar morphology in BrCr-treated mice.
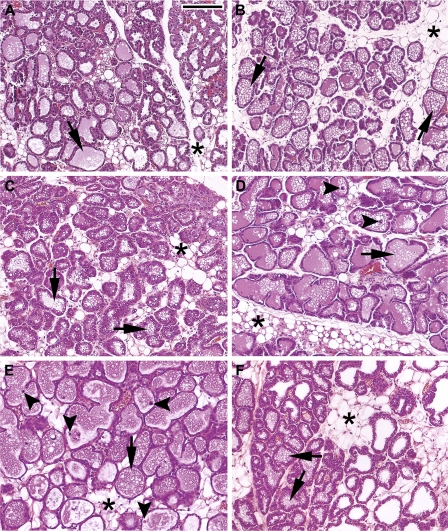
We examined the mammary gland histology at various times following BrCr treatment (Fig. 2). Nontreated mice displayed the normal characteristics of day 2 lactating mammary glands: organized epithelia, reduced area of adipose tissue (asterisks), thin layers of connective tissue surrounding the alveolus, very small or indiscernible lipid droplets within the epithelial cells, and fully expanded alveoli with lipid droplets and milk in the luminal space (arrows; Fig. 2A). Following 4, 8, 16, or 24 h of BrCr treatment (Fig. 2, B–E, respectively), the morphology of the mammary glands appeared similar to that of untreated controls (Fig. 2, A and F). Occasional apoptotic cells were observed in the luminal space by 16 and 24 h of treatment (Fig. 2, D and 2E, arrowheads). At 24 h, about 1% of the 4% total TUNEL-positive nuclei were luminal, and 3% remained intact within the epithelium, indicating only slightly increasing apoptosis following 24 h of BrCr treatment (Supplemental Fig. S1A). We also noted moderately increased expression of genes normally associated with involution, such as STAT3 and C/EBPδ, starting by 4 h of BrCr treatment (Supplemental Fig. S1B), possibly indicating the onset of early involution. This finding is consistent with the known role of PRL and its secondary signaling molecule STAT5 to promote mammary epithelial cell survival during lactation (44). However, the rapidity of these increases has not been previously noted.
Fig. 2.
BrCr-induced changes in mammary gland morphology. Representative images from hematoxylin and eosin-stained sections from mammary glands of control and BrCr-injected mice. A: control mice on day 2 of lactation. B: 4 h postinjection with BrCr. C: 8 h postinjection. D: 16 h postinjection. E: 24 h postinjection. F: untreated control mice on day 3 of lactation. *, adipose tissue; arrows, lipid droplets in lumina; arrowheads, luminal apoptotic cells. Bar = 200 μm.
Milk ejection.
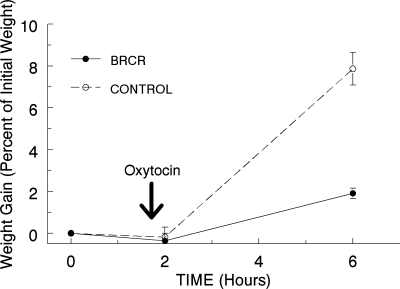
Due to the lack of pup weight gain following BrCr treatment, we reasoned that milk ejection might have failed. Oxytocin is needed for milk ejection, and previous work has shown that pituitary PRL is necessary for oxytocin secretion in the lactating rat (28). When we administered oxytocin directly to the exposed lactating gland of a BrCr-treated mouse, we observed milk filling the mammary ducts, indicating that myoepithelial function is not compromised in the absence of PRL (data not shown). To examine milk secretion in control and BrCr-treated dams, a weigh-suckle-weigh approach was coupled with oxytocin injection. The litters were weighed and then separated from both control and BrCr-treated dams for 2 h, after which the dams were injected with 300 μl of 2U/ml oxytocin. The pups were reweighed and returned to suckle oxytocin-treated dams for an additional 4 h. After 4 h of suckling (6 h post-BrCr injection), pups were weighed a final time. Pups nursed by the oxytocin-treated control dams showed an 8% increase in body weight, whereas pups nursed by the oxytocin-injected BrCr-treated dams increased 2% over the same period (Fig. 3). These data imply that the impaired lactation in BrCr-treated mice may be due to inhibited milk secretion and not to an inability of the gland to eject milk in response to oxytocin.
Fig. 3.
Oxytocin does not restore pup weight gain in BrCr-treated mice. Mice were treated with BrCr or vehicle (control) at time 0, and pups were removed for 2 h. All mice were then injected with 300 μl 2 U/ml oxytocin, and pups returned and were allowed to suckle for another 4 h. Pups were then weighed again to determine weight gain during the 4-h suckling period. Error bars indicate SE; n = 3 for both BrCr and control groups.
BrCr-mediated PRL loss eliminates Stat5 phosphorylation.
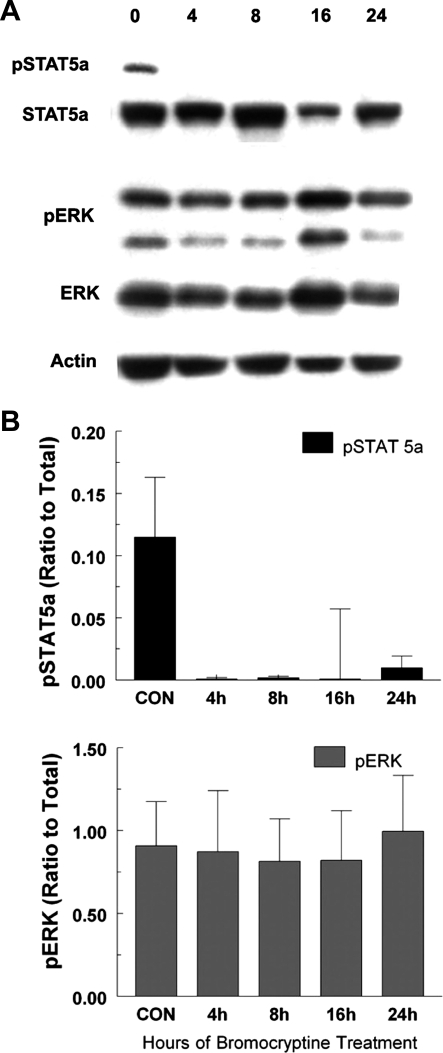
It is well known that PRL imparts many, if not all, of its lactogenic effects through the Jak2/STAT5 signaling pathway (12, 17). To begin to address the BrCr effects of decreased serum PRL at the molecular level, we performed immunoblotting for Jak2/STAT5 signaling using antibodies that recognize phosphorylated STAT5a, as well as one for total STAT5a. Figure 4A shows a representative blot for STAT5a and ERK activation. Within 4 h following BrCr treatment, phosphorylated STAT5a was reduced to undetectable levels, while total STAT5a remained constant. This finding is consistent with previous studies (2, 22). PRL also activates the mitogen-activated kinase ERK (7). Analysis of the same tissue lysates with antibodies recognizing either phosphorylated ERK or total ERK showed no change in ERK activation even after 24 h of BrCr treatment (Fig. 4A). These results are quantified in Fig. 4B (n = 4 mice/time point), showing that BrCr-mediated PRL loss does not affect downstream signal transduction pathways uniformly. STAT5a phosphorylation was PRL dependent, while stimuli other than PRL are likely to sustain ERK activation in the lactating mammary gland.
Fig. 4.
BrCr treatment inhibits phosphorylation of STAT5 but not ERK1/2. A: lysates were prepared from mammary glands isolated from mice treated with BrCr 0, 4, 8, 16, and 24 h after initial treatment with BrCr. Treatment with BrCr was repeated at 8 and 16 h to maintain low serum levels of PRL. Immunoblots were probed with anti-phospho-STAT5, anti-STAT5, anti-phospho-ERK, anti-ERK, and anti-actin to demonstrate equal phospho-STAT5 and phospho-ERK. Blots show representative data from individual mice. B: graphs show quantitative data from 4 mice per experiment.
PRL loss paused β-casein gene expression.
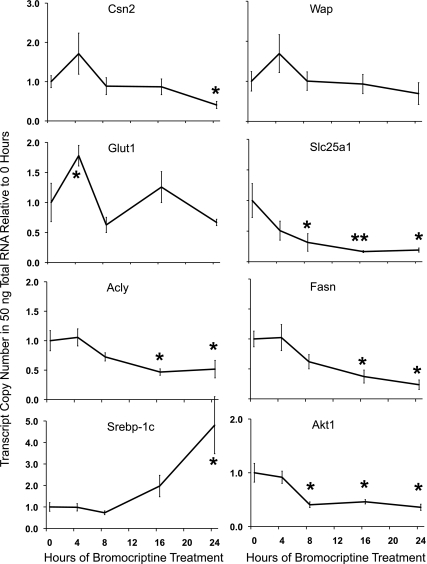
Milk protein genes are markedly upregulated at the transition from pregnancy to lactation and sustain this elevation throughout lactation (1). β-Casein (Csn2) and whey acidic protein (Wap) are well characterized as PRL/STAT5-regulated genes (31). As a measure of PRL/STAT5 signaling, we first examined Csn2 and Wap gene expression using standard curve qRT-PCR to measure transcript copy number relative to untreated controls (0 h). Our analysis revealed a transient increase in the amount of Csn2 and Wap gene expression at 4 h following BrCr treatment; however, Csn2 levels were suppressed after 8 h of BrCr, and the decrease reached statistical significance by 24 h (Fig. 5, top). The analysis of Csn2 and Wap gene expression is complicated at lactation day 2 because these genes are still in an inductive phase (32), but it is clear that BrCr treatment quells further induction of these milk protein genes by 24 h. Csn2 has been shown to have a long transcript half-life under conditions when PRL is present (i.e., lactation day 2) (10), but we do not know the in vivo decay rate (24). PRL is also known to specifically regulate the translation of Csn2 mRNA in mammary epithelial cells, which could also affect mRNA preservation (4). Additionally, we have some preliminary evidence that intracellular CSN2 protein is reduced following BrCr treatment (data not shown), suggesting that loss of PRL signaling alters CSN2 translation in vivo.
Fig. 5.
BrCr treatment decreases expression of milk protein and lipogenic genes in whole mammary gland. Standard curve method qRT-PCR was used to determine gene expression profiles for milk proteins CSN2 and WAP as well as for Glut1, mitochondrial citrate transporter (Slc25a1), de novo fatty acid synthesis enzymes Acly and Fasn, and regulatory factors Srebp1c and Akt1. Profiles are expressed as transcript copy number detected in 50 ng of total RNA relative to 0 time point and error bars are SE for 4 animals per time point. *Means differ significantly (P ≤ 0.05) from control.
PRL loss decreased lipogenic gene expression in whole mammary gland.
The expression of milk protein genes is known to be PRL dependent (13), but it remains uncertain whether gene expression for the lipogenic pathway is affected by loss of PRL in the mammary adipose or whether the response is specific to the mammary epithelium. To validate previous studies of PRL regulation on whole mammary gland, we used standard curve qRT-PCR to evaluate transcript copy numbers of genes thought to be associated with the lipogenic pathway (Fig. 5). Expression of the glucose transporter Glut1 increased at 4 h (Fig. 5), but the change was transient and not different at the following BrCr time points. Gene expression for the mitochondrial citrate transporter Slc25a1 rises dramatically at secretory activation (35); this protein supplies cytosolic citrate as a precursor substrate for fatty acids, making it critical for de novo synthesis of fatty acids (11, 33). Levels of Slc25a1 RNA fell to 20% of control by 8 h post-BrCr treatment and declined further by 24 h of BrCr (Fig. 5). ATP citrate lyase (Acly) did not fall initially but decreased by 16 h of BrCr treatment, reaching nadir at nearly 50% of control. ACLY generates the cytosolic acetyl-CoA required by fatty acid synthase (FASN) to initiate fatty acids. The principle de novo fatty acid synthesis gene Fasn also decreased by 16 h and diminished further by 24 h. Levels of Akt1 mRNA declined over 50% at 8 h and remained low over the time course. In contrast, the lipogenic transcription factor sterol-regulatory element-binding protein-1c (SREBP-1c) transcript decreased 25% at 8 h but then increased by 16 h of BrCr treatment, reaching statistical significance by 24 h. Consistent with previous studies of PRL signaling in lactation (22), these data indicate that PRL loss reduced gene expression of key lipogenic pathway components. However, complicating analysis of lipogenic pathway genes is the presence of mammary adipocytes, which express de novo fatty acid and tiacylglycerol (TAG) biosynthesis genes and proteins at high levels (34). The presence of mammary adipose cells might mask the BrCr effects that we wish to observe specifically in the mammary epithelium; therefore, to be certain these genes are PRL dependent in the MECs, we examined gene expression in preparations devoid of the contaminating adipocytes.
PRL controls lipogenic gene expression specifically in MECs.
Adipocyte-depleted MECs (36) were prepared from the mammary glands of control and BrCr-treated dams at the 8-h time point to address whether PRL regulates MEC specific de novo fatty acid synthesis pathway gene expression. We examined gene expression levels for a variety of functional categories, including milk proteins, glucose transport, the pentose phosphate shunt, de novo fatty acid synthesis, triglyceride synthesis, and β-oxidation of fatty acids as well as potential regulatory factors. Table 1 shows adipose-depleted MEC gene expression in transcript copy numbers, as determined by the standard curve method, relative to the mean value over all samples for that given gene. The fold changes included in Table 1 show the magnitude of decreases in the presence of BrC as [−1.0/(BrCr/Control)]. Expression of two major milk protein genes decreased, including Csn2 (1.65-fold decrease) and α-lactalbumin (Lalba; 2.01-fold decrease), but Wap did not change significantly in MECs following PRL loss. These results at the 8-h point verified the trend from the whole mammary gland gene expression study (Fig. 5).
Table 1.
Gene expression in adipose-depleted MECs following 8 h of BrCr treatment
| Adipose-Depleted MECs |
|||
|---|---|---|---|
| Functional Category | Control | BrCr | Fold change |
| Milk protein genes | |||
| Csn2 | 1.43 | 0.86 | −1.65c |
| Lalba | 1.36 | 0.67 | −2.01b |
| Wap | 1.10 | 0.88 | N/C |
| Glycolysis and pentose shunt | |||
| Glut1 | 1.95 | 0.54 | −3.59c |
| G6pd2 | 0.91 | 0.38 | −2.43b |
| AldoC | 1.69 | 0.37 | −2.93b |
| Gapdh | 1.24 | 0.55 | −2.25c |
| Me1 | 1.09 | 0.50 | −2.16b |
| De novo fatty acid synthesis | |||
| Slc25a1 | 1.69 | 0.37 | −4.59a |
| Acly | 1.65 | 0.30 | −5.45d |
| Acc1α | 1.44 | 0.22 | −6.43d |
| Fasn | 1.08 | 0.34 | −3.13b |
| Thrsp | 0.77 | 0.69 | N/C |
| TAG synthesis and β-oxidation | |||
| Dgat1 | 1.08 | 0.48 | −2.24b |
| Scd2 | 2.03 | 0.10 | −19.46c |
| Fads1 | 1.44 | 0.40 | −3.62c |
| Acaa2 | 0.50 | 0.52 | N/C |
| Acadl | 0.46 | 0.63 | N/C |
| Regulatory factors | |||
| Akt1 | 2.55 | 0.92 | −2.77c |
| Prlr-Long | 0.96 | 0.37 | −2.60d |
| Srebp1a | 0.77 | 0.38 | −2.05b |
| Srebp1c | 0.74 | 0.22 | −3.34b |
Gene expression in adipose-depleted mammary epithelial cells (MECs) following 8-h bromocriptine (BrCr) treatment. Genes are grouped into functional categories. Values are mean normalized transcript copy numbers for that gene using standard curve-based qRT-PCR; n = 5 mice per condition. Fold change is calculated as −1/(BrCr/Control). Statistical significance: aP ≤ 0.05, bP ≤ 0.01, cP ≤ 0.001, dP ≤ 0.0001. N/C, no statistical change, using Excel's t-test function.
We also examined expression of genes involved in lipogenesis (Table 1). Key genes in the glycolysis and pentose pathway shunt all decreased more than twofold when MECs were analyzed in the absence of mammary adipose. Particularly large decreases were observed for Glut1 (3.59-fold reduction) and aldolase C (AldoC, 2.93-fold reduction), a key glycolytic enzyme and known PRL target gene in the mammary epithelial cell (31). Large decreases for genes involved in de novo fatty acid synthesis were observed: Slc25a1 (4.59-fold reduction), Acly (5.45-fold reduction), and Acc1α (6.43-fold reduction) were observed; together, the proteins coded by these genes synthesize the precursor malonyl-CoA, the primary substrate for Fasn (3.13-fold reduction). Scd2, a Δ9 desaturase that modifies fatty acids made by FASN, decreased more than 19-fold, suggesting that expression of Scd2 is completely dependent on PRL. Although most of these changes mirror the results in whole tissue lysates (Supplemental Table S1), the fold changes are generally larger when the adipose tissue is excluded from the analysis. In some cases, notably Gapdh and Dgat1, the fold change in the MECs is masked by the adipose tissue, where expression is likely regulated differently. Note that these results make it clear that Gapdh is unsuitable as a reference gene for mammary epithelium expression studies in vivo. Altogether, PRL withdrawal led to a coordinate reduction in the lipogenic gene expression pathway, specifically in MECs, and it is interesting that this reduction is larger than that observed for milk protein genes, which have been extensively studied PRL target genes.
We also analyzed expression profiles for genes thought to be involved in regulating secretory activation and the “lipogenic switch” (1, 23, 35), including Akt1, Prlr (long form), and Srepf isoforms 1a and 1c. Expression of all these genes decreased more than twofold in MECs following 8 h of BrCr dosing. Interestingly, Thrsp (Spot 14), which is known to regulate de novo fatty acid synthesis in lipogenic tissues in the mouse (5, 45), showed no difference in either MECs or whole tissue. This result suggests that Thrsp is regulated by factors other than PRL at lactation day 2, even though Thrsp gene expression increases markedly at secretory activation. Cumulatively, BrCr-mediated PRL withdrawal led to rapid and coordinated changes in milk protein, glycolytic, lipid synthesis pathway, and regulatory factor gene expression. Although PRL clearly altered lipogenic gene expression following 8 h of BrCr exposure, we wished to know if the loss of PRL signaling curtailed posttranscriptional functions as well.
PRL loss decreases presence of casein and CDLs in mammary epithelium.
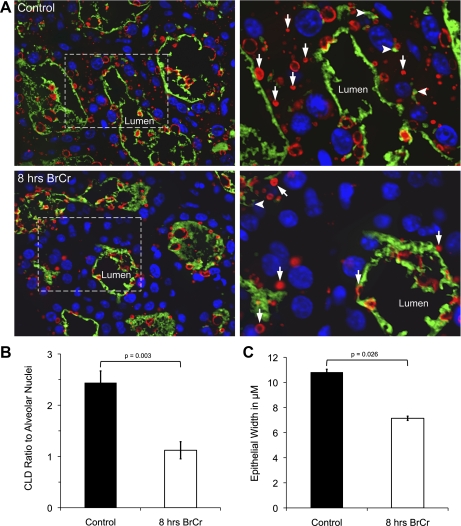
Both β-casein and adipophilin (ADPH) staining can be discerned in the epithelium of the lactating mammary gland; casein is associated with the protein-secretory pathway, while the ADPH protein coats CLDs in the mammary alveolar epithelium. ADPH abundance can be used as a surrogate measure of CLD abundance (38, 39). We noted that both ADPH and casein staining were reduced in the cytoplasmic regions of MECs in sections of the BrCr-treated mammary gland (Fig. 6). In Fig. 6, white arrows point to ADPH-stained CLD, whereas arrowheads point to regions of casein abundance in the cytoplasm. Although both are retained in the lumens of glands from BrCr-treated mice, the cytoplasm is notably devoid of both CLD and casein. We were able to quantify the difference in CLD number 8 h after FVB mice were injected with BrCr or vehicle. When the CLD were quantified, a twofold reduction was observed relative to the number of alveolar nuclei (Fig. 6B). The total number of nuclei per field as well as the number of acinar nuclei were quantified from each condition; both were increased slightly although the difference did not reach statistical significance. This finding indicates that acinar size was not increased after BrCr treatment, providing evidence that there is no alveolar distension even though oxytocin secretion was likely suppressed in the BrCr-treated animals. This finding is consistent with the decrease in milk synthesis, but not milk ejection, for which evidence is presented in Fig. 3. We also noted a significant 30% reduction in the width of the alveolar epithelium of BrCr-treated mice (Fig. 6C) despite similar milk accumulation in BrCr-treated acini relative to control acini. This observation suggests a potential loss of cellular volume due to a decrease in CLD and milk protein synthesis, although secretory activity may be preserved, since milk synthesized prior to loss of PRL is present in the lumens. Together, these data provide strong evidence that the lack of PRL signaling leads to a global cessation of milk synthesis by mammary alveolar epithelium.
Fig. 6.
Cytoplasmic secretory product are reduced following 8 h of BrCr treatment. A: histological sections of mammary glands from control (top) and 8-h BrCr-treated mice (bottom) were stained as described in materials and methods with anti-adipophilin to outline cytoplasmic lipid droplets (CLD; red), an anti-casein antibody, to show casein distribution (green), and DAPI (blue) to identify nuclei of stained cells. Light dotted box at left denotes magnified area at right. White arrows point to CLDs; arrowheads denote intracytoplasmic casein aggregates, likely in Golgi and secretory vesicles. B: CLD number was quantified and shown as the ratio to alveolar nuclei; a twofold decrease in CLD per cell was observed in mammary glands of BrCr-treated mice. C: epithelial width was decreased 35% in these sections. For B and C, data from 3 sections per mouse were averaged from Suppl. Fig. S1, in which it is possible to denote the extent of the cytoplasm from the autofluorescences of these secretory cells. Error bars show SE when data from 3 mice per condition were averaged.
DISCUSSION
In this study, we used BrCr to acutely inhibit pituitary PRL secretion in lactating mice to examine the effects of PRL loss specifically in the MEC. Although PRL loss was shown to inhibit oxytocin secretion and therefore milk ejection in rats (28), by applying exogenous oxytocin we were able to see that milk synthesis was the primary defect of BrCr treatment. This observation is supported by immunofluorescent analyses showing an acute decrease in the number of CLD and regions of casein staining in the mammary alveolar cells, as well as by a 30% reduction in the width of the epithelium. The second finding of interest is that PRL withdrawal leads to increases in gene expression characteristic of involution, which is supported by the appearance of TUNEL-positive apoptotic cells. In particular, loss of PRL signaling induced expression of mRNA for Stat3 and Cebpδ almost fourfold within 4 h and that for Tgf-β2 and Tgf-β3 more than fivefold within 8 h. These findings indicate that involution-associated gene expression increases take place rapidly in the absence of PRL; however, these changes were reversible over the 8-h window we examined. The increased involution-related gene expression was probably not due to alveolar distension, because acinar area (from nuclear number) was not significantly different after PRL withdrawal, which would not be the case if milk stasis were the prominent feature of BrCr treatment.
We have previously characterized changes in expression of lipogenic enzymes at secretory activation (1, 35) and have hypothesized what factors, such as PRL signaling, might regulate the lipogenic switch (32, 33). Previous efforts to define the role of PRL-disrupted signaling used genetic approaches in mice with deletions in the genes for PRL (15), the PRLR (25, 27), and STAT5 (6). Analysis of PRLR-null and heterozygous mice has clearly indicated a role for the PRLR in secretory activation, also called lactogenesis (18, 24, 25). Others used a regulator of pituitary PRL secretion, galanin (22); or a phosphomimetic mutant of PRL, PRL S179D (2, 3, 20, 22). All of these manipulations resulted in very useful characterization; but since PRL is essential for alveologenesis during pregnancy, it remained unclear whether effects on lactation resulted from changes in alveolar number and/or differentiation. For these reasons, we chose to use BrCr to inhibit serum PRL to investigate acute withdrawal of PRL signaling directly in lactating MECs.
The most striking finding of this study relates to changes both in the amount of mRNA-encoding proteins involved in lipid synthesis (Table 1) and in the number of CLDs (Fig. 6B). PRL signaling in the mammary gland is best defined for its role in gene expression events. The majority of studies investigating mammary gene regulation have focused on milk protein gene expression and disregarded that lipid biosynthetic enzyme genes are significantly upregulated at secretory activation (1). Analysis of PRLR heterozygous mice, galanin-null mice, and S179D PRL mice has clearly indicated a role for PRL in regulation of lipogenic gene transcription in whole mammary gland lysates (22). The data reported here extend previous results from Ormandy and colleagues by providing novel data on MEC-specific regulation for glucose transport, glycolysis, pentose shunt, de novo fatty acid synthesis, triglyceride synthesis, and regulatory genes under conditions where PRL availability is compromised for short duration only in the lactating animal (25, 27). Because these results are obtained from isolated MECs, lipogenic regulation can be reliably distinguished from data obtained in previous studies that used whole tissue lysates. Table 1 provides strong evidence that an entire suite of lipogenic genes is rapidly and coordinately decreased specifically in the lactating MEC due to suppression of serum PRL.
This study clearly shows that expression of lipid synthesis genes, specifically in MECs, decreased in BrCr-treated dams. Not all genes whose expression we analyzed after loss of PRL signaling decreased. For example, Thrsp (also known as Spot 14) expression remained unchanged. Thrsp has been shown to be necessary for the elevated rate of de novo fatty acid synthesis during lactation (45) but appears not to be regulated at the gene expression level by PRL at lactation day 2. For most genes, the decreases in expression were greater in MECs than in whole tissue alone (Supplemental Table S1), suggesting that both the mammary adipocytes and epithelium respond to PRL signaling for lipogenesis during lactation. Very striking is the difference in expression of Dgat1 and Gapdh in the whole gland vs. MECs. Expression was unchanged in whole mammary gland after 8 h of BrCr, whereas gene expression decreased approximately twofold in MECs. This observation shows that these two genes are differentially regulated in mammary adipocytes and in MECs.
At present, we cannot determine whether the decreased levels of transcripts for genes in the lipogenic pathway are due to loss of transcription initiation, increased mRNA turnover, or some combination of the two processes. Previous work, however, has shown that PRL signaling is required for transcriptional induction, mRNA stabilization, polysome loading, and translation of the prototypical mammary epithelial gene Csn2 (4, 10). We noted that loss of PRL signaling in mammary epithelium was accompanied by reduced cytosolic CSN2, implying that PRL is necessary for translation of CSN2 protein in vivo. This observation, coupled with the twofold decrease in CLD number following 8 h of BrCr treatment, suggests that PRL regulates multiple processes to synthesize milk. Such MEC processes necessary to sustain milk synthesis during lactation extend the in vivo role of PRL to include transcription of lipogenic genes, translation of milk protein CSN2, and formation of CLD.
In conclusion, this study demonstrates that, in mice, BrCr-mediated loss of serum PRL suppresses MEC-specific gene expression of milk protein genes, transporters for glucose and citrate, and both glycolytic and lipogenic enzyme transcripts. Morphological, immunohistochemical, and oxytocin add-back analyses suggest that milk synthesis is rapidly shut down following BrCr treatment but milk ejection may still function. During the same time frame, the loss of PRL induces molecular events that are associated with involution and MEC survival apparently in the absence of milk stasis. These findings provide a framework for future characterization of mammary epithelial-specific factors that modulate PRL signaling during lactation to regulate lipogenesis at the gene and enzyme levels.
GRANTS
S. M. Anderson and M. C. Neville are supported by NIH Grant PO1-HD-38129, which also contributed to the support of M. C. Rudolph and T. D. Russell. M. C. Rudolph is currently supported by Department of Defense Breast Cancer Research Predoctoral Fellowship BC 810596.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Mammary Gland Biology Program Project Grant at the University of Colorado Denver for on-going discussions regarding this research. We thank Lisa Litzenberger and Chiara Picorini for their contributions to the work herein.
REFERENCES
- 1. Anderson SM, Rudolph MC, McManaman JL, Neville MC. Key stages in mammary gland development. Secretory activation in the mammary gland: it's not just about milk protein synthesis! Breast Cancer Res 9: 204, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernichtein S, Kayser C, Dillner K, Moulin S, Kopchick JJ, Martial JA, Norstedt G, Isaksson O, Kelly PA, Goffin V. Development of pure prolactin receptor antagonists. J Biol Chem 278: 35988–35999, 2003 [DOI] [PubMed] [Google Scholar]
- 3. Bernichtein S, Kinet S, Jeay S, Llovera M, Madern D, Martial JA, Kelly PA, Goffin V. S179D-human PRL, a pseudophosphorylated human PRL analog, is an agonist and not an antagonist. Endocrinology 142: 3950–3963, 2001 [DOI] [PubMed] [Google Scholar]
- 4. Choi KM, Barash I, Rhoads RE. Insulin and prolactin synergistically stimulate beta-casein messenger ribonucleic acid translation by cytoplasmic polyadenylation. Mol Endocrinol 18: 1670–1686, 2004 [DOI] [PubMed] [Google Scholar]
- 5. Colbert CL, Kim CW, Moon YA, Henry L, Palnitkar M, McKean WB, Fitzgerald K, Deisenhofer J, Horton JD, Kwon HJ. Crystal structure of Spot 14, a modulator of fatty acid synthesis. Proc Natl Acad Sci USA 107: 18820–18825, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol 24: 8037–8047, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Das R, Vonderhaar BK. Activation of raf-1, MEK, and MAP kinase in prolactin responsive mammary cells. Breast Cancer Res Treat 40: 141–149, 1996 [DOI] [PubMed] [Google Scholar]
- 8. Flint DJ, Binart N, Kopchick J, Kelly P. Effects of growth hormone and prolactin on adipose tissue development and function. Pituitary 6: 97–102, 2003 [DOI] [PubMed] [Google Scholar]
- 9. Forsyth IA, Lee PD. Bromocriptine treatment of periparturient goats: long-term suppression of prolactin and lack of effect on lactation. J Dairy Res 60: 307–317, 1993 [DOI] [PubMed] [Google Scholar]
- 10. Guyette WA, Matusik RJ, Rosen JM. Prolactin-mediated transcriptional and post-transcriptional control of casein gene expression. Cell 17: 1013–1023, 1979 [DOI] [PubMed] [Google Scholar]
- 11. Halperin ML, Cheema-Dhadli S, Taylor WM, Fritz IB. Role of the citrate transporter in the control of fatty acid synthesis. Adv Enzyme Regul 13: 435–445, 1975 [DOI] [PubMed] [Google Scholar]
- 12. Hannun YA. Apoptosis and the dilemma of cancer chemotherapy. Blood 89: 1845–1853, 1997 [PubMed] [Google Scholar]
- 13. Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol 6: 715–725, 2005 [DOI] [PubMed] [Google Scholar]
- 14. Hinck L, Silberstein GB. Key stages in mammary gland development: the mammary end bud as a motile organ. Breast Cancer Res 7: 245–251, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horseman ND, Zhao W, Montecino-Rodriguez E, Tanaka M, Nakashima K, Engle SJ, Smith F, Markoff E, Dorshkind K. Defective mammopoiesis, but normal hematopoiesis, in mice with a targeted disruption of the prolactin gene. EMBO J 16: 6926–6935, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hovey RC, Trott JF, Vonderhaar BK. Establishing a framework for the functional mammary gland: from endocrinology to morphology. J Mammary Gland Biol Neoplasia 7: 17–38, 2002 [DOI] [PubMed] [Google Scholar]
- 17. Humphreys RC, Hennighausen L. Signal transducer and activator of transcription 5a influences mammary epithelial cell survival and tumorigenesis. Cell Growth Differ 10: 685–694, 1999 [PubMed] [Google Scholar]
- 18. Kelly PA, Bachelot A, Kedzia C, Hennighausen L, Ormandy CJ, Kopchick JJ, Binart N. The role of prolactin and growth hormone in mammary gland development. Mol Cell Endocrinol 197: 127–131, 2002 [DOI] [PubMed] [Google Scholar]
- 19. Knight CH, Calvert DT, Flint DJ. Inhibitory effects of bromocriptine on mammary development and function in lactating mice. J Endocrinol 110: 263–270, 1986 [DOI] [PubMed] [Google Scholar]
- 20. Kuo CB, Wu W, Xu X, Yang L, Chen C, Coss D, Birdsall B, Nasseri D, Walker AM. Pseudophosphorylated prolactin (S179D PRL) inhibits growth and promotes beta-casein gene expression in the rat mammary gland. Cell Tissue Res 309: 429–437, 2002 [DOI] [PubMed] [Google Scholar]
- 21. Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev 11: 179–186, 1997 [DOI] [PubMed] [Google Scholar]
- 22. Naylor MJ, Oakes SR, Gardiner-Garden M, Harris J, Blazek K, Ho TW, Li FC, Wynick D, Walker AM, Ormandy CJ. Transcriptional changes underlying the secretory activation phase of mammary gland development. Mol Endocrinol 19: 1868–1883, 2005 [DOI] [PubMed] [Google Scholar]
- 23. Oakes SR, Hilton HN, Ormandy CJ. The alveolar switch: coordinating the proliferative cues and cell fate decisions that drive the formation of lobuloalveoli from ductal epithelium. Breast Cancer Res 8: 207, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oakes SR, Rogers RL, Naylor MJ, Ormandy CJ. Prolactin regulation of mammary gland development. J Mammary Gland Biol Neoplasia 13: 13–28, 2008 [DOI] [PubMed] [Google Scholar]
- 25. Ormandy CJ, Binart N, Kelly PA. Mammary gland development in prolactin receptor knockout mice. J Mammary Gland Biol Neoplasia 2: 355–364, 1997 [DOI] [PubMed] [Google Scholar]
- 26. Ormandy CJ, Camus A, Barra J, Damotte D, Lucas B, Buteau H, Edery M, Brousse N, Babinet C, Binart N, Kelly PA. Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes Dev 11: 167–178, 1997 [DOI] [PubMed] [Google Scholar]
- 27. Ormandy CJ, Naylor M, Harris J, Robertson F, Horseman ND, Lindeman GJ, Visvader J, Kelly PA. Investigation of the transcriptional changes underlying functional defects in the mammary glands of prolactin receptor knockout mice. Recent Prog Horm Res 58: 297–323, 2003 [DOI] [PubMed] [Google Scholar]
- 28. Parker SL, Armstrong WE, Sladek CD, Grosvenor CE, Crowley WR. Prolactin stimulates the release of oxytocin in lactating rats: evidence for a physiological role via an action at the neural lobe. Neuroendocrinology 53: 503–510, 1991 [DOI] [PubMed] [Google Scholar]
- 29. Richert MM, Schwertfeger KL, Ryder JW, Anderson SM. An atlas of mouse mammary gland development. J Mammary Gland Biol Neoplasia 5: 227–241, 2000 [DOI] [PubMed] [Google Scholar]
- 30. Riddle O, Bates RW, Dykshorn SW. The preparation, identification and assay of prolactin—a hormone of the anterior pituitary. Am J Physiol 105: 191–216, 1933 [Google Scholar]
- 31. Rosen JM, Wyszomierski SL, Hadsell D. Regulation of milk protein gene expression. Annu Rev Nutr 19: 407–436, 1999 [DOI] [PubMed] [Google Scholar]
- 32. Rudolph MC, McManaman JL, Hunter L, Phang T, Neville MC. Functional development of the mammary gland: use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J Mammary Gland Biol Neoplasia 8: 287–307, 2003 [DOI] [PubMed] [Google Scholar]
- 33. Rudolph MC, McManaman JL, Phang T, Russell T, Kominsky DJ, Serkova NJ, Stein T, Anderson SM, Neville MC. Metabolic regulation in the lactating mammary gland: a lipid synthesizing machine. Physiol Genomics 28: 323–336, 2007 [DOI] [PubMed] [Google Scholar]
- 34. Rudolph MC, Monks J, Burns V, Phistry M, Marians R, Foote MR, Bauman DE, Anderson SM, Neville MC. Sterol regulatory element binding protein and dietary lipid regulation of fatty acid synthesis in the mammary epithelium. Am J Physiol Endocrinol Metab 299: E918–E927, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rudolph MC, Neville MC, Anderson SM. Lipid synthesis in lactation: diet and the fatty acid switch. J Mammary Gland Biol Neoplasia 12: 269–281, 2007 [DOI] [PubMed] [Google Scholar]
- 36. Rudolph MC, Wellberg EA, Anderson SM. Adipose-depleted mammary epithelial cells and organoids. J Mammary Gland Biol Neoplasia 14: 381–386, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Russell TD, Fischer A, Beeman NE, Freed EF, Neville MC, Schaack J. Transduction of the mammary epithelium with adenovirus vectors in vivo. J Virol 77: 5801–5809, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Russell TD, Palmer CA, Orlicky DJ, Bales ES, Chang BH, Chan L, McManaman JL. Mammary glands of adipophilin-null mice produce an amino-terminally truncated form of adipophilin that mediates milk lipid droplet formation and secretion. J Lipid Res 49: 206–216, 2008 [DOI] [PubMed] [Google Scholar]
- 39. Schwertfeger KL, McManaman JL, Palmer CA, Neville MC, Anderson SM. Expression of constitutively activated Akt in the mammary gland leads to excess lipid synthesis during pregnancy and lactation. J Lipid Res 44: 1100–1112, 2003 [DOI] [PubMed] [Google Scholar]
- 40. Stricker PG F. Action du lobe antérieur de l'hypophyse sur la montée laiteuse. Compt Rend Soc Biol 99: 1978–1980, 1928 [Google Scholar]
- 41. Travers MT, Barber MC, Tonner E, Quarrie L, Wilde CJ, Flint DJ. The role of prolactin and growth hormone in the regulation of casein gene expression and mammary cell survival: relationships to milk synthesis and secretion. Endocrinology 137: 1530–1539, 1996 [DOI] [PubMed] [Google Scholar]
- 42. Trott JF, Vonderhaar BK, Hovey RC. Historical perspectives of prolactin and growth hormone as mammogens, lactogens and galactagogues—agog for the future! J Mammary Gland Biol Neoplasia 13: 3–11, 2008 [DOI] [PubMed] [Google Scholar]
- 43. Vernon RG, Flint DJ. Control of fatty acid synthesis in lactation. Proc Nutr Soc 42: 315–331, 1983 [DOI] [PubMed] [Google Scholar]
- 44. Watson CJ. Involution: apoptosis and tissue remodelling that convert the mammary gland from milk factory to a quiescent organ. Breast Cancer Res 8: 203, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu Q, Anderson GW, Mucha GT, Parks EJ, Metkowski JK, Mariash CN. The Spot 14 protein is required for de novo lipid synthesis in the lactating mammary gland. Endocrinology 146: 3343–3350, 2005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.