Abstract
Reovirus genome segment S1 encodes protein σ1, which is the receptor binding protein, modulates tissue tropism, and specifies the nature of the antiviral immune response. It makes up less than 2% of reovirus particles and is synthesized in very small amounts in infected cells. Any antiviral strategy aimed at reducing specifically the expression of this genome segment should, in principle, reduce the infectivity of the virus. To test this hypothesis, we have assembled two hammer-head motif-containing ribozymes (Rzs) targeted to cleave at the conserved B and C domains of the reovirus s1 RNA. Protein-independent but Mg2+-dependent sequence-specific cleavage of s1 RNA was achieved by both the Rzs in trans. Cells that transiently express these Rzs, when challenged with reovirus, were protected against the cytopathic effects caused by the virus. This protection correlated with the specific intracellular reduction of s1 transcripts that was due to their cleavage by the Rzs. Rz-treated cells that were challenged with reovirus showed almost complete disappearance of protein σ1 without significantly altering the levels of the other reovirus structural proteins. Thus, Rzs, besides acting as antiviral agents, could be exploited as biological tools to delineate specific functions of target genes.
Keywords: transcription, gene cleavage
Nucleic acids are no longer treated as passive molecules responsible for storing genetic information. The discovery of catalytic RNAs (1–3) capable of carrying out sequence-specific cleavage reactions, often with an efficiency comparable to protein enzymes, has attracted a great deal of attention as a means to selectively down-regulate the expression of target genes (4). Ribozymes (Rzs) have been constructed against a variety of viral and cellular genes; in particular against HIV-1 genes for which several monotarget and multitarget Rzs have been described (5–8). Recently, a functional hammer-head Rz targeted to cleave the HIV-1 coreceptor CCR5 was described (9). Because each target RNA will have its own secondary structure under physiological conditions, the efficacy of each potential Rz needs to be experimentally determined.
Cleavage by Rzs in trans has been reported for many viruses, but none against a double-stranded RNA-containing virus. We chose reovirus as a model to test the efficacy of a Rz approach as an antiviral strategy. The S1 genome segment encodes reovirus protein σ1, a minor outer capsid protein, as well as σ1s in a different reading frame (10–12). Reovirus protein σ1 is the cell attachment protein (13) and governs tissue tropism (14); it interacts with receptors on erythrocytes, thereby causing hemagglutination (15), and it elicits the formation of type-specific neutralizing antibodies (16). Differences in the capacity to induce apoptosis among the three serotypes of the reovirus have been found to be determined by the protein σ1 (17). Another important biological function that has been traced to the S1 genome segment is inhibition of host cell DNA replication (18). The function of protein σ1 is currently not known. Reovirus protein σ1 makes up less than 2% of the mass of reovirus particles and is synthesized in very small amounts in infected cells. An experimental proof that reovirus particles are noninfectious if they lack protein σ1 was provided elegantly by characterizing the 13 particle species that differ in their content of protein σ1 (19). Therefore, any antiviral strategy that selectively reduces the level of S1 gene expression should reduce the infectivity of the virus and may protect the cells from pathogenesis. With the aim of testing this hypothesis, we have assembled two hammer-head motif-containing Rzs targeted against two conserved regions in the S1 genome. We demonstrate sequence-specific cleavage of synthetic and authentic reovirus s1 RNA by both Rzs in trans. We also show that Rz-expressing cells are protected against the cytopathic effects caused by reovirus infection.
Materials and Methods
Growth of Cells and Viruses.
Reovirus serotype 3 (strain Dearing) was grown in HeLa cells and purified as described by Smith et al. (20). HeLa cells and mouse L cells were maintained in suspension culture at 37°C in Joklik's MEM (Grand Island Biological, Rockville, MD) supplemented with 5% FCS.
Selection of Target Sites.
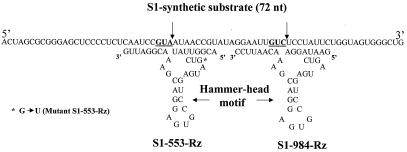
Two conserved sites, GUA and GUC, located at positions 553 and 984 were selected for designing the Rzs. Rz-553 was targeted to region B, and Rz-984 was directed against the conserved region C of genome segment S1(21). The cleavage of the target RNA is expected to take place after the A and C nucleotides, respectively (Fig. 1, shown by arrows). A 72-nt-long synthetic s1 substrate was chemically synthesized. When cleaved by Rzs, it generates RNA fragments of different sizes that could be resolved by gel electrophoresis. Synthesis of a 72-nt-long sequence was achieved by combining s1 RNA fragments 523–561 and 973-1005 (21) (Fig. 1).
Figure 1.
A 72-nt-long DNA was synthesized in a DNA synthesizer. Two regions (see text) of the s1 RNA (nucleotides 523–561 and 973-1005) (22) were fused. Transcripts of this DNA possess two Rz cleavage sites, GUA and GUC (underlined). The cleavage is expected to take place downstream of the A and C nucleotides of the target RNA (shown by arrows). The sequences of the two Rzs, 553 and 984, along with the hammer-head catalytic motif are also shown. Both the Rzs possessed the earlier described hammer-head catalytic motif. Eight bases on either side of the target site were synthesized to provide specificity for the s1 RNA. For Rz-553 the A nucleotide and C nucleotide (shown by arrows) for Rz-984 of the target RNA was left unpaired. The S1–553-mutant-Rz was constructed by changing a single nucleotide (G to U) in the catalytic motif (see text).
Cloning of Genome Segment S1 and Its Transcription.
We used two kinds of S1 substrates, a synthetic 72-nt-long fragment (Fig. 1) that possessed the two earlier described target sites and the full-length s1 RNA (1.4 kb) that was derived from the plasmid S1/S1/a as described (22, 23). The strategy for cloning the synthetic and truncated S1 genome fragment in a T-tailed vector (pGEM-T, Promega) was the same as described in ref. 9. Briefly, the 72-nt-long s1 fragment was synthesized chemically in a DNA synthesizer and amplified by using terminal primers in a PCR. The synthetic gene was cloned into the pGEM-T-Easy vector under the control of the SP6 promoter to yield plasmid pS1-syn. The sequence of the cloned synthetic S1 gene was confirmed by sequencing both strands. Plasmid pS1-Syn was linearized by NcoI digestion, using a restriction enzyme site that was present at the 3′ end of the cloned gene in the multiple cloning sites (MCS) before in vitro transcription. Eighty-one nucleotides in the 5′ end and 21 from the 3′ end of the synthetic gene derived from the MCS are expected to be transcribed, thereby generating a 174-base-long transcript. Plasmid S1/S1/a was linearized at the 3′ end of the S1 genome segment with SalI to obtain full-length (1.4 kb) S1 transcript.
Construction of S1 Rzs.
The trinucleotides GUA and GUC were selected as our target sites (Fig. 1). The strategy to construct a hammer-head Rz was same as described in ref. 9. Briefly, a sequence of eight bases complementary to the S1 genome segment was synthesized on either side of the target site (shown by arrows in Fig. 1) along with the central conserved catalytic domain. Two terminal primers were designed to amplify the 72-base-long synthetic S1 primer. PCR-amplified products were cloned into a T-tailed vector as described earlier. The following primers were synthesized for constructing S1–553-Rz: S1–553-Rz oligonucleotide, GgaattcTGTAATACGACTCACTATAACGGTTATctgatgagtccgtgaggacgaaACGGATTGggatccGG (the EcoRI and BamHI sites were engineered at the ends with one or two extra nucleotides; the hammer-head motif is written in lowercase, bold letters); forward terminal primer, 5′-GGAATTCTGTAATACGACTC; and reverse terminal primer, 5′-CCGGATCCCAATCCGTTTCG.
The PCR-amplified product (72 bp) was cloned into pGEM-T (Promega), such that the expression of this Rz was under the T7 promoter. This plasmid was named pS1–553-Rz. An essentially similar technique was used to construct S1–984-Rz. The three primers were: S1–984-Rz oligonucleotide, 5′-GAAGCTTGAATAGGActgatgagtccgtgaggacgaaACAATTCCGGATCCG (tt has a HindIII site at the 5′ end and a BamHI site at the 3′ end); forward primer, 5′-GAAGCTTGAATAGGAC; and reverse primer, 5′-CGGATCCGGAATTGTTTC.
The PCR-amplified product (52 bases) was cloned into pGEM-T-Easy vector (Promega), and this plasmid was named pS1–984-Rz. Both Rzs also were cloned directly into pTarge-T (Promega) under the control of the cytomegalovirus promoter. This vector contains a chimeric intron downstream of the promoter and a simian virus 40 polyadenylation signal. A point mutation (G to U: shown by an arrow, Fig. 1) in the catalytic motif of S1–553-Rz was constructed by introducing a single change in the 5′ terminal primer and the rest of the steps were same as followed for constructing S1–553-Rz. The mutant Rz also was placed under the control of the cytomegalovirus promoter of the expression vector pTarge-T (Promega).
In Vitro Cleavage of s1 RNA by Rzs.
Plasmids encoding Rzs or s1 RNA (substrate) were linearized at their 3′ ends and then subjected to in vitro transcription using either SP6 or T7 RNA polymerase as described (23). The cleavage reaction was carried out by mixing equimolar amounts (0.3 pmols) of labeled S1 substrate RNA and unlabeled Rz under standard conditions of cleavage (50 mM Tris⋅HCl, pH 7.5/10 mM MgCl2) in a volume of 10 μl as described (9). The cleaved products were analyzed by 7 M urea-6% PAGE using the MiniPROTEAN ll system from Bio-Rad.
Kinetic Parameters.
Kinetic parameters of the Rzs were determined by using varying concentration of labeled s1 RNA in the presence of excess amounts of Rz as reported earlier (24) and described recently (25). Kms and kcats were calculated from Lineweaver-Burk's plots according to standard procedures.
In Vivo Effects of Rz.
Cos-1 cells were grown to 60% confluency in 6-well plates and transfected with plasmid DNAs encoding Rzs or mutant Rz by using Lipofectin (GIBCO/BRL), following the manufacturer's directions. The pSV-β−galactosidase control (Promega) vector was used to ensure uniform transfection efficiencies of mammalian cells that varied less than 10%. Twelve hours later, the cells were washed once with medium and then infected with reovirus ST3 at a multiplicity of 1 plaque-forming unit/cell. The cells were harvested after 8 h of infection, and lysates were prepared for Western blot analysis using polyclonal antiserum against reovirus ST3 (kind gift from W. K. Joklik, Duke University Medical Center, Durham, NC) as described (26). Equal amounts of lysates were used from cells that received equivalent amounts of S1–553 mutant Rz-encoding DNA (control) or from cells that received plasmids encoding Rzs (1 or 4 μg) followed by reovirus infection as described earlier. RNA also was isolated from above cell lysates by using Trizol reagent (Promega) and purified according to the manufacturer's directions. Primers were designed to estimate the levels of intact s1 RNA by carrying out reverse transcriptase (RT)–PCR using the kit from Promega. Several dilutions were initially made to determine the linear range for the PCR-amplified products. Equal amounts of genomic DNA were used as determined by a 509-bp PCR-amplified product of the human glyceraldehyde phosphodehydrogenase (hGAPDH) gene (27). This DNA was amplified by using the primers: forward, 5′-ACCACCATGGAGAAGGCTGG, and reverse, 5′-CTCAGTGTAGCCCAGGATGC.
Cytopathic effects caused by the virus were monitored 8 h after virus infection and photographed by using an inverted microscope (Nikon Diaphot 300).
Results
In Vitro Cleavage of s1 RNA by Rzs in Trans.
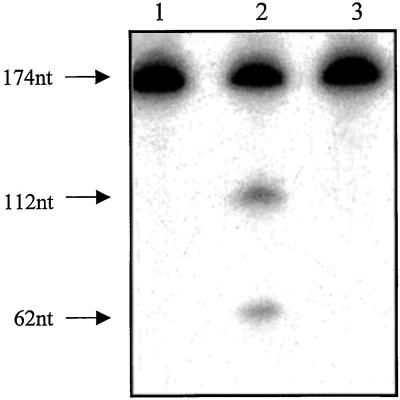
We first determined whether the correct size Rz transcript was synthesized. Rz-553-containing expression vector, when linearized with BamHI and subjected to in vitro transcription in the presence of labeled UTP (specific activity 3,000ci/mmol, NEN/DuPont), yielded a 118-nt-long Rz transcript (data not shown). The plasmid pS1-Syn, when linearized with NcoI and transcribed, directed the formation of 174-nt-long s1 RNA (substrate) (Fig. 2, lane 1). All of the cleavage reactions were carried out with UTP-labeled s1 RNA and unlabeled Rz in equimolar amounts (0.3 pmols each) in the presence of 10 mM MgCl2. Rz-553 cleaved the s1 RNA into specific products that were 112 and 62 nts long (Fig. 2, lane 2). These are the expected sizes of the cleaved products and demonstrate the sequence-specific cleavage activity of the Rz-S1–553. No cleavage was observed if MgCl2 was omitted from the reaction (data not shown). Mutant Rz-S1–553, under exactly similar experimental conditions, failed to cleave the target RNA (Fig. 2, lane 3). We conclude that the S1 mutant Rz with a single nucleotide change (G to U) in the catalytic motif (Fig. 1) was rendered completely inactive. This mutant Rz subsequently was used as the control for cellular experiments. Using similar experimental conditions, S1-Rz-984, also cleaved the s1 synthetic RNA into specific cleavage products (see Fig. 3B). Specific cleavage products (847- and 553-nt-long RNA fragments) also were obtained with S1-Rz-553 when full-length s1 RNA was used as the substrate (data not shown).
Figure 2.
Rz- or substrate-encoding plasmids were linearized at their 3′ ends as described in Materials and Methods and subjected to in vitro transcription using a kit from Promega. The synthesis of synthetic s1 RNA, which is 174 nts long, is shown in lane 1. When equimolar amounts (0.3 pmols) of unlabeled Rz and labeled substrate RNA are mixed in the presence of 10 mM MgCl2, specific cleavage products (112 and 62 nts long) could be seen (lane 2). Under exactly similar experimental conditions the mutant S1–553-Rz failed to cleave the target RNA (lane 3). Specific cleavage products also were observed with Rz-984 (see Fig. 3B).
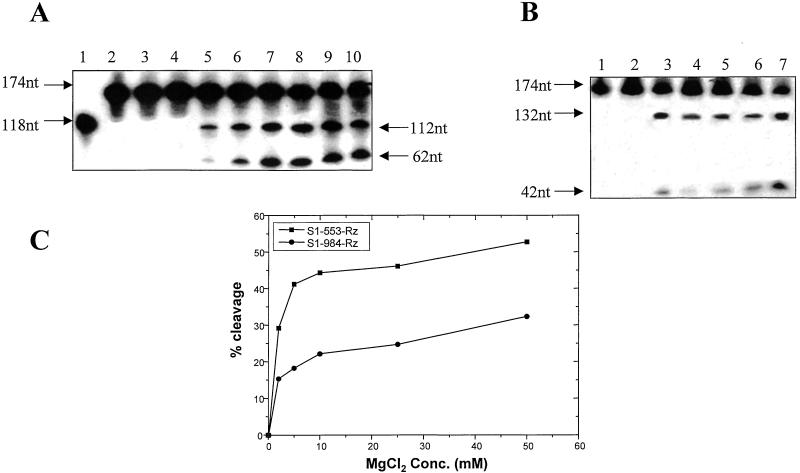
Figure 3.
The effect of varying the concentration of MgCl2 on cleavage efficiency. Synthetic s1 RNA (174 nts long) (A, lane 2 and B, lane 1) was used for the cleavage reaction in equimolar ratio (0.3 pmols) with the two Rzs separately. (A) The results obtained with Rz-553 are shown. Lane 1 depicts the synthesis of Rz-553 as described before. Lane 2 depicts the labeled s1 synthetic transcript that is 174 bases long. No cleavage was observed if the MgCl2 was omitted from the reaction (lane 3) or when it was present at a concentration of 0.1 mM (lane 4). Specific cleavage could be observed at 1 mM MgCl2 (lane 5). The cleavage efficiency increased significantly in the presence of increasing amounts of MgCl2 (lane 6, 3 mM; lane 7, 5 mM; lane 8, 10 mM; lane 9, 25 mM; and lane 10, 50 mM). (B) Lane 1 is same as lane 2 of A. No cleavage was observed in the absence of MgCl2 (lane 2) when equimolar amounts of Rz-984 were used. Lane 3 shows the extent of cleavage in the presence of 2 mM MgCl2. Specific cleavage products were also obtained under simulated physiological conditions (150 mM KCl, pH 7.5, 37°C, 2 mM MgCl2) (lane 4). The cleavage efficiency did not change in the presence of 5 mM (lane 5) or 10 mM MgCl2 (lane 6). Increased cleavage was, however, observed in the presence of 50 mM MgCl2. (C) A comparison of the cleavage efficiency of the two Rzs at varying concentration of MgCl2 is shown. It is evident from the graph that more than 40% cleavage of the target RNA is achieved by Rz-553 in presence of 5 mM MgCl2 as compared with only 20% with Rz-984 under identical experimental conditions. Also note that even in the presence of 50 mM MgCl2 the cleavage reaction did not proceed to completion.
Effect of MgCl2 on Cleavage Efficiency.
Synthetic s1 RNA (substrate −174 bases) was used for the cleavage reactions of both Rzs in trans. Rz-553-mediated cleavage is shown in Fig. 3A and that of Rz-984 in Fig. 3B. The efficiency of cleavage increased with increasing amounts of MgCl2 (Fig. 3A, lanes 5–10). As described earlier, 118-bases-long S1–553-Rz-specific RNA was synthesized from the plasmid pS1-Rz-553 (Fig. 3A, lane 1). Synthesis of the 174-nt-long synthetic s1 RNA that was derived from plasmid pS1-syn is shown in Fig. 3A, lane 2. All of the cleavage reactions were carried out by using labeled s1 RNA and equimolar amounts (0.3 pmols each) of cold Rz. No cleavage was observed in the absence of MgCl2 (Fig. 3A, lane 3) or in presence of 0.1 mM MgCl2 (Fig. 3A, lane 4). Specific cleavage products became visible at 1 mM MgCl2 (Fig. 3A, lane 5) and increased in the presence of 3 mM (Fig. 3A, lane 6) and 5 mM MgCl2 (Fig. 3A, lane 7). Further increases in the MgCl2 concentration (Fig. 3A, lane 8, 10 mM; lane 9, 25 mM; and lane 10, 50 mM) increased the cleavage efficiency only marginally. The most important observation was a significant cleavage activity at 1 and 3 mM concentrations of MgCl2 that is very close to physiological levels in humans (24). Rz-984 also was tested for its cleavage efficiency at varying concentrations of MgCl2, and the results are shown in Fig. 3B. Equimolar amounts (0.29 pmols) of labeled substrate RNA (174 bases, Fig. 3B, lane 1) and unlabeled 984-Rz RNA were used for the cleavage reaction. As expected, no cleavage products were observed in the absence of MgCl2 (Fig. 3B, lane 2). Specific cleavage products could be seen in presence of 2 mM MgCl2 concentration (Fig. 3B, lane 3). The cleavage efficiency remained more or less unchanged at simulated physiological conditions (Fig. 3B, lane 4), 5 mM (Fig. 3B, lane 5), or 10 mM (Fig. 3B, lane 6). A slight increase in the cleavage efficiency was observed at 50 mM MgCl2 concentration (Fig. 3B, lane 7). A comparative analysis of the cleavage efficiency of the two Rzs with respect to increasing concentration of MgCl2 is shown in the form of a graph (Fig. 3C). It is clear that Rz-553 is more efficient in its ability to cleave the s1 synthetic RNA under the experimental conditions used. We conclude that both Rzs cleave synthetic s1 RNA in a sequence-specific manner that is protein-independent but Mg2+-dependent and that Rz-553 is more efficient in cleaving the synthetic substrate RNA. Both Rzs retain the ability to cleave target RNA at physiologically relevant concentrations of MgCl2 (1–3 mM).
Kinetic Parameters.
Radiolabeled transcripts were quantitated by trichloroacetic acid precipitation protocol (Promega). The cleavage rate was determined by taking varying amounts of labeled synthetic s1 substrate under enzyme saturating conditions. The kinetic parameters were calculated by using the Lineweaver-Burk's plot for S1-Rz-553 and Rz-984. The Km and the kcat for the S1-Rz-553 and Rz-984 are shown in Table 1. The kinetic parameters suggest that the former Rz is more efficient.
Table 1.
Kinetic analysis of S1 Rzs
| Enzyme | Kcat, min−1 | Km, M | Kcat/Km, M−1⋅min−1 |
|---|---|---|---|
| S1-553-Rz | 30.3 × 10−4 | 1.8 × 10−7 | 1.68 × 104 |
| S1-984-Rz | 5.7 × 10−4 | 4.8 × 10−7 | 0.12 × 104 |
Kinetic measurements were carried out for the reovirus S1 Rzs by taking varying amounts of labeled s1-synthetic substrate RNA and carrying out cleavage reactions under Rz-saturating conditions as described before. The kinetic parameters were calculated from a Lineweaver–Burk's plot. Rate constants are averages of two separate experiments.
Inhibition of Full-Length s1 Gene Expression.
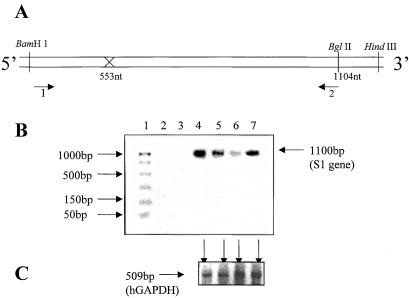
A comparison of the levels of full-length s1 RNA fragment from Rz-treated and control cells was carried out by using RT-PCR techniques. The linear range of amplification was determined by appropriate dilution of the target RNA and standardizing cycling conditions. The following primers were used. Primer 1, which spans the BamHI site of the s1 RNA, had the following sequence: 5′-GATCCTCGCCTACGTG. Primer 2, which spans the unique BglII site in the coding region of s1 RNA, had the following sequence: 5′-CAACGATAGATCTCCACC. The combination of primers 1 and 2 would generate a fragment of 1.1 kb by PCR (Fig. 4A). This was carried out in the following manner. Lysates from treated cells were divided into two equal proportions to estimate the levels of full-length s1 RNA and nonspecific control RNA (hGAPDH) by carrying out RT-PCR. The results are shown in Fig. 4 B and C. When equivalent amounts of lysate from only cells were analyzed, no s1 RNA-specific PCR product was observed (Fig. 4B, lane 2). Similar observations were made when the enzyme RT was omitted (Fig. 4B, lane 3). As expected, an intense band corresponding to s1 full-length RNA was amplified (Fig. 4B, lane 4) when cells were infected with reovirus (Fig. 4B, lane 4). When 1 μg of Rz-553 was used, a 4-fold reduction in the levels of s1 RNA was observed (Fig. 4B, lane 5), which was further reduced when 4 μg of the Rz-encoding DNA was used (Fig. 4B, lane 6). S1–553 mutant Rz-treated (4 μg) cells showed no significant reduction in the levels of s1 RNA (Fig. 4B, lane 7). This decrease was specific for s1 RNA as the corresponding levels of the control RNA (hGAPDH) remained unchanged (Fig. 4C, shown by arrows). We conclude that S1–553-Rz reduces the intracellular S1 gene expression specifically.
Figure 4.
To address the question of intracellular reduction of the reovirus s1 RNA by S1–553-Rz, primers were designed to amplify the full-length s1 RNA and the control hGAPDH RNA fragment (509 bases) from the same cell lysates by RT-PCR. Two terminal primers (1 and 2, A) were used to amplify the full-length s1 (1.4 kb) RNA. They had the following sequence: forward, 5′-GATCCTCGCCTACGTG and reverse, 5′-CAACGATAGATCTCCACC. Total cellular RNA was isolated from Cos-1 cells that were first transfected with 1 or 4 μg of plasmid encoding Rz-553 and then challenged with reovirus at 1 plaque-forming unit/cell. Serial dilutions of the target gene and number of cycles for PCR were optimized initially to determine the linear range. (B) Lane 1 depicts the size of standard DNAs as shown on the left. As expected the plain cells showed no evidence of the presence of s1 RNA (lane 2). The same was true if the enzyme RT was omitted from the reaction (lane 3). Reovirus infected cells, as expected, showed a prominent signal for s1 RNA (lane 4). The intensity of the signal diminished when either 1 μg (lane 5) or 4 μg (lane 6) of Rz-553 was used. Mutant-S1–553-Rz at a concentration of 4 μg failed to decrease the levels of s1 RNA (lane 7). This decrease was specific for the reovirus s1 RNA as the levels of the control RNA (hGAPDH) in all four corresponding lysates remained unchanged (C, shown by arrows).
Western Blot Analysis.
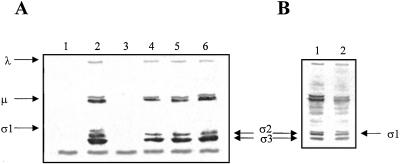
Equivalent amounts of lysates were used to estimate the level of reovirus protein σ1. The control consisted of Cos-1 cells infected with equivalent amounts of unrelated Rz (CCR5-Rz) (9) or mutant-Rz then followed by reovirus infection. Approximately 1/10 of the cell lysate was used to analyze the levels of reovirus protein σ1. Polyclonal antiserum against reovirus ST3 was used as described (26). Fig. 5A, lane 2 shows reovirus specific immunoreactive protein bands in the presence of unrelated Rz. Equivalent amounts of lysates of plain cells (Fig. 5A, lane 1) or only pS1–553-Rz DNA-transfected cells (Fig. 5A, lane 3) showed no reovirus-specific protein bands, but only one cross-reactive band of ≈20-kDa size that was common among all cell lysates. When Cos-1 cells were treated with 4 μg (Fig. 5A, lane 4) or 1 μg (Fig. 5A, lane 5) of Rz-553 encoding DNA, now under the control of the powerful cytomegalovirus promoter, no σ1 protein-specific band was detected. There was no difference in the levels of the other reovirus proteins when 1 μg of Rz-553 was used (Fig. 5A, compare lanes 2 and 5). When higher amounts of Rz-553 (4 μg) were used, a slight reduction in the levels of reovirus proteins (≈2-fold) was observed (Fig. 5A, lane 4). When 4 μg of Rz-984 was used in cells infected with reovirus ST3, all immunoreactive reovirus proteins could be identified except for protein σ1. We conclude that Rzs specifically interfere with the expression of the reovirus S1 genome segment and that the levels of other reovirus structural proteins remained essentially unchanged with Rz-984 but a slight reduction with Rz-553 was observed. Mutant S1–553-Rz encoding DNA at a 4 μg concentration showed no significant reduction in the intensity of the reovirus σ1 protein band (Fig. 5B, lane 2) when compared with cells infected with reovirus alone (Fig. 5B, lane 1). It therefore can be concluded that the specific reduction of reovirus protein σ1 is due to the catalytic nature of the Rz.
Figure 5.
Total extracts of transfected and infected cells (see text) were prepared, and one-tenth of the lysates collected from 6-well plates was subjected to Western blot analysis using polyclonal antiserum against reovirus ST3 raised in rabbits (kind gift from W. K. Joklik). No reovirus-specific immunoreactive proteins were detected in control cells (A, lane 1) except for a single cross-reactive band of similar intensity that was observed in all lanes. Lane 2 shows all reovirus structural proteins including protein σ1 in the lysates of cells treated with unrelated Rz. No reovirus-specific proteins were detected in the lysates of cells that were transfected with Rz-553 DNA alone (lane 3). Reovirus protein σ1 could not be detected when either 4 μg (lane 4) or 1 μg Rz-553 encoding DNA (lane 5) was used. Reovirus σ1 protein was also not detected when 4 μg of Rz-984-encoding DNA was transfected (lane 6). Mutant S1–553-Rz at a concentration of 4 μg showed no significant reduction in the levels of reovirus protein σ1 along with other structural proteins (B, lane 2) when compared with reovirus-infected cells not transfected with plasmids expressing Rzs (B, lane 1).
Inhibition of Cytopathic Effects Caused by Reovirus Infection by Rzs.
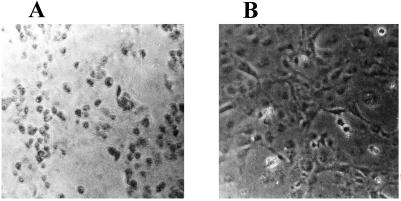
Cos-1 cells were treated with Rz-553 encoding DNA or control DNA (mutant S1-Rz-553) (4 μg each for 1 × 106 cells) for 8 h and then challenged with reovirus as described before. The infection was allowed to proceed for 8 h only. Mutant Rz-treated control cells showed extensive rounding of cells (Fig. 6A), whereas, Rz-553-treated cells showed remarkable protection (Fig. 6B). The protection was partial with Rz-984 under identical conditions. Also, the protection was not seen in Rz-treated cells when the reovirus infection was allowed to proceed for longer than 24 h (data not shown). Plasmid DNA concentrations above 4 μg for 1 × 106 cells could not be used, as they were toxic to cells.
Figure 6.
Cos-1 cells were grown to 60% confluency in 6-well plates. Rz-553 encoding DNA (4 μg) was transfected by using Lipofectin (GIBCO/BRL) (B). Control cells were transfected with equal amounts of mutant-S1–553-Rz (A) and then infected with reovirus as described earlier. The cells were observed under an inverted microscope (Nikon Diaphot 300) and photographed. (A) Extensive cytopathic effects caused by reovirus infection when cells were transfected with equivalent amounts of the mutant-Rz. The S1–553-Rz-treated cells showed remarkable protection from cytopathogenesis caused by reovirus infection (B). These are representative pictures from three separate experiments that gave similar results.
Discussion
We report on a functional Rz against a double-stranded RNA containing virus, namely reovirus (ST3, strain Dearing). With the aim of interfering specifically with reovirus gene expression, we chose the S1 genome segment with the hope that by selectively reducing the expression of protein σ1, it might be possible to reduce the infectivity of the virus and protect cells against the pathogenic effects caused by this protein during infection. We selected two target sites (GUX) from conserved regions B and C of the S1 genome segment (21) and constructed hammer-head Rzs through recombinant techniques. Rz-553 was ≈20% more efficient in cleaving the synthetic s1 substrate under in vitro conditions than Rz-984. This could be due to differences in the secondary structures at the two target sites of the s1 RNA. No cleavage was observed in the absence of MgCl2 and the cleavage efficiency of Rzs increased in presence of increasing amounts of MgCl2 as had been shown for other Rzs (9). The important feature of both Rzs was their ability to cleave the target RNA at 1 and 3 mM MgCl2 concentration at 37°C, which is close to the physiological level (24). The kinetic parameters of Rz-553 suggest that it is less efficient than what has been reported for other Rzs and DNA-enzymes (24, 25). One of the reasons could be the overall length of the Rz-containing RNA, which contained substantial proportions of vector sequences that were transcribed.
We next addressed the question of whether Rz could specifically reduce the amount of s1 RNA produced as a result of reovirus infection intracellularly and whether it could protect cells from reovirus-mediated cytopathic effects. Intracellular s1 RNA reduction was studied by quantitating the relative levels of full-length s1 RNA and the control housekeeping RNA (hGAPDH) from the same cell lysates. A dose-dependent decrease in the levels of full-length RNA but not of control RNA strongly suggests the specific effects of Rzs. Rz-984-treated cells showed less protection than Rz-553 under identical experimental conditions (data not shown). When equivalent amounts of lysates from Rz-treated cells were analyzed for the presence of reovirus proteins, all reovirus-specific protein bands could be detected except that of protein σ1. Mutant Rz-553 in equivalent amounts did not interfere with the formation of s1 RNA. Two important conclusions could be drawn. Reovirus protein σ1 is mainly responsible for causing the cytopathic effects. This is not surprising because the S1 genome segment was earlier shown to be a determinant for causing apoptosis (17) and inhibiting the cellular DNA replication (18). There was no effect on the levels of the rest of the reovirus proteins, which is not surprising because most of the structural reovirus proteins were earlier shown to assemble into reovirus core-like particles in the absence of protein σ1 using multiple recombinant vaccinia viruses that expressed reovirus structural proteins (28). A roughly 2-fold reduction in the amount of reovirus proteins in lysates of cells that were treated with 4 μg of Rz-553-encoding DNA suggested that further rounds of infection also were affected, presumably because of the generation of noninfectious reovirus particles that lacked σ1 protein. This reduction was not seen with 4 μg of Rz-984, which could be explained on the basis of its lower cleavage efficiency.
In summary, we have shown the efficacy of Rzs against one of the most important reovirus genes, namely S1. Sequence-specific cleavage was obtained with each of the Rzs in trans. We provide evidence for specific intracellular decreases in the levels of S1 RNA and reduction in the levels of protein σ1 in Rz-expressing cells. Rz-treated cells showed remarkable protection against the cytopathic effects caused by reovirus. Thus these Rzs not only can protect cells against reovirus challenge, but also, by selectively inactivating them, afford a unique opportunity for studying the role of individual genes in pathogenesis and reovirus morphogenesis.
Acknowledgments
We thank Dr. S. K. Basu, Director, National Institute of Immunology, New Delhi; Dr. M. Sharma, Secretary, Department of Biotechnology, Government of India; and Prof. V. Ramalingaswamy, National Research Professor, All India Institute of Medical Sciences, New Delhi, for their constant support and encouragement. The Rz project was funded by the Department of Biotechnology, Government of India, to A.C.B and to the National Institute of Immunology, New Delhi.
Abbreviations
- Rz
ribozyme
- RT
reverse transcriptase
- hGAPDH
human glyceraldehyde phosphodehydrogenase
References
- 1.Cech T R. Science. 1987;236:1532–1539. doi: 10.1126/science.2438771. [DOI] [PubMed] [Google Scholar]
- 2.Uhlenbeck O C. Nature (London) 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- 3.Haselholf J, Gerlach W L. Nature (London) 1988;334:585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- 4.Rossi J J. Trends Biotechnol. 1995;13:301–306. doi: 10.1016/S0167-7799(00)88969-6. [DOI] [PubMed] [Google Scholar]
- 5.Chen C-J, Banerjea A C, Harmison G G, Haglund K, Schubert M. Nucleic Acids Res. 1992;20:4581–4589. doi: 10.1093/nar/20.17.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paik S-Y, Banerjea A, Chen C-J, Ye Z, Harmison G G, Schubert M. Hum Gene Ther. 1997;8:1115–1124. doi: 10.1089/hum.1997.8.9-1115. [DOI] [PubMed] [Google Scholar]
- 7.Dropulic B, Lin N H, Martin M A, Jeang K-T. J Virol. 1992;66:1432–1441. doi: 10.1128/jvi.66.3.1432-1441.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarver N, Cantin E M, Chang P S, Zaia J A, Ladne P A, Stephens D A, Rossi J J. Science. 1990;247:1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- 9.Goila R, Banerjea A C. FEBS Lett. 1998;436:233–238. doi: 10.1016/s0014-5793(98)01137-5. [DOI] [PubMed] [Google Scholar]
- 10.Cashdollar L W, Chmelo R A, Wiener J R, Joklik W K. Proc Natl Acad Sci USA. 1985;82:24–28. doi: 10.1073/pnas.82.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basel-Duby R, Jayasuriya A, Chatterjee S, Sonenberg N, Maizel J V, Fields B N. Nature (London) 1985;315:421–423. doi: 10.1038/315421a0. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs B L, Atwater J A, Munemistsu J M, Samuel C E. Virology. 1985;147:9–18. doi: 10.1016/0042-6822(85)90222-3. [DOI] [PubMed] [Google Scholar]
- 13.Lee P W K, Hayes E C, Joklik W K. Virology. 1981;108:156–163. doi: 10.1016/0042-6822(81)90535-3. [DOI] [PubMed] [Google Scholar]
- 14.Weiner H L, Drayna D, Averill D R, Jr, Fields B N. Proc Natl Acad Sci USA. 1977;74:5744–5748. doi: 10.1073/pnas.74.12.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiner H L, Ramig R F, Mustoe T A, Fields B N. Virology. 1978;86:581–584. doi: 10.1016/0042-6822(78)90099-5. [DOI] [PubMed] [Google Scholar]
- 16.Finberg R, Weiner H L, Fields B N, Benacerraf B, Bankakoff S J. Proc Natl Acad Sci USA. 1979;76:442–446. doi: 10.1073/pnas.76.1.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyler K L, Squier M K, Rodgers S E, Schneider B E, Oberhaus S M, Grdina T A, Cohen J J, Dermody T S. J Virol. 1995;69:6972–6979. doi: 10.1128/jvi.69.11.6972-6979.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharpe H A, Fields B N. J Virol. 1981;38:389–392. doi: 10.1128/jvi.38.1.389-392.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larson S M, Antczak J B, Joklik W K. Virology. 1994;201:303–311. doi: 10.1006/viro.1994.1295. [DOI] [PubMed] [Google Scholar]
- 20.Smith R E, Zweerink H J, Joklik W K. Virology. 1969;39:791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- 21.Duncan R, Horne D, Cashdollar W L, Joklik W K, Lee P W K. Virology. 1990;174:399–409. doi: 10.1016/0042-6822(90)90093-7. [DOI] [PubMed] [Google Scholar]
- 22.Roner R M, Gaillard R K, Jr, Joklik W K. Virology. 1989;168:292–301. doi: 10.1016/0042-6822(89)90269-9. [DOI] [PubMed] [Google Scholar]
- 23.Banerjea A C, Joklik W K. Virology. 1990;179:460–462. doi: 10.1016/0042-6822(90)90315-i. [DOI] [PubMed] [Google Scholar]
- 24.Santoro S W, Joyce G F. Proc Natl Acad Sci USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dash B P, Harikrishnan T A, Goila R, Shahi S, Unwalla H, Husain S, Banerjea A C. FEBS Lett. 1998;431:395–399. doi: 10.1016/s0014-5793(98)00799-6. [DOI] [PubMed] [Google Scholar]
- 26.Banerjea A C, Brechling K A, Ray C A, Erikson H, Pickup D, Joklik W K. Virology. 1988;167:601–612. [PubMed] [Google Scholar]
- 27.Kwon H Y, Bultman S J, Loeffler C, Chen W J, Furdon P J, Powell J G, Usala A L, Wilkison W, Hansman I, Woychik R P. Proc Natl Acad Sci USA. 1994;91:9760–9764. doi: 10.1073/pnas.91.21.9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu P, Miller S E, Joklik W K. Virology. 1993;197:726–731. doi: 10.1006/viro.1993.1648. [DOI] [PubMed] [Google Scholar]