Abstract
Oxidative stress is a core abnormality responsible for disease progression in nonalcoholic steatohepatitis (NASH). However, the relevant pathways that contribute to oxidative damage in vivo remain poorly understood. Here we explore the gene-expression patterns related to oxidative stress, antioxidant defense, and reactive oxygen metabolism in an established dietary murine model of NASH. C57BL/6 mice were placed on either a methionine- and choline-deficient (MCD) or a control (CTL) diet for 6 weeks. Hepatic oxidative damage and the development of NASH were monitored by biochemical and histologic indices. Analysis of 84 oxidative stress–related genes was performed by real-time reverse transcription polymerase chain reaction (PCR) in the livers of the two groups of mice. Mice on the MCD diet showed increased ALT, histologic features of NASH, and oxidative liver damage with increases in 4-hydroxynonenal and 3-nitrotyrosine. Of the genes analyzed, the GPx family were most significantly upregulated, whereas SCD1 was most significantly downregulated. Other genes that were significantly upregulated included Fmo2 and peroxiredoxins, whereas genes downregulated included Catalase and Serpinb1b. Our data demonstrate that oxidative stress–related genes are differentially expressed in the livers of mice with diet-induced NASH. These findings have important implications for NASH pathogenesis and the development of novel therapeutic strategies for patients with this condition. Antioxid. Redox Signal. 15, 437–445.
Introduction
Nonalcoholic fatty liver disease (NAFLD) is currently the most common form of chronic liver disease affecting both adults and children and is strongly associated with obesity and insulin resistance (3, 32). One in three adults and one in 10 children or adolescents in the United States have hepatic steatosis, a stage within the spectrum of NAFLD that is characterized by triglyceride accumulation in liver cells and follows a benign nonprogressive clinical course (12). Nonalcoholic steatohepatitis (NASH) is defined as lipid accumulation with evidence of cellular damage, inflammation, and varying degrees of scarring or fibrosis (6). NASH is a serious condition, as approximately 25% of these patients progress to cirrhosis and its feared complications of portal hypertension, liver failure, and hepatocellular carcinoma (1, 2).
The precise mechanisms by which hepatic steatosis develops into the steatohepatitis phenotype remain incompletely understood. Oxidative stress due to generation of reactive oxygen species (ROS) or decreased antioxidant defenses or both are thought to play a pivotal role in this process and has been postulated to be a key component in the progression to NASH (9, 31). Experimental and human studies have demonstrated enhanced oxidative stress and lipid peroxidation products in the circulation, and in the liver of different animal models and patient cohorts with NASH (5, 16). The imbalance between generated reactive oxygen metabolites and antioxidant defense systems produces oxidative stress, with subsequent damage involving important cellular macromolecules (lipids, proteins, carbohydrates, and nucleic acids). Despite intense interest in the role of oxidation in the pathogenesis of NASH, the pathways that contribute to oxidative damage in vivo are poorly understood. In the present study, by using quantitative real-time polymerase chain reaction (PCR) microarrays, we catalogued the gene-expression levels of a total of 84 genes known to have pro- or antioxidative functions in liver tissue from an established murine dietary model of NASH.
Methods
Animal studies
These experimental protocols were approved by the Institutional Animal Care and Use Committee at Cleveland Clinic. Male C57BL/6 mice, 20 to 25 g body weight (Jackson Laboratory, Bar Harbor, ME), were placed on either a methionine and choline-deficient (MCD) diet (TD 90262; Teklad Mills, Madison, WI), which has been extensively shown to result in steatosis associated with significant inflammation and progressive fibrosis pathologically, similar to human severe steatohepatitis (23), or a control (CTL) diet for 6 weeks (TD 2918; Teklad Mills) (n=5–6 in each group). Total body weight was measured weekly. Animals in each group were killed after 6 weeks on the respective diets.
Real-time PCR microarrays
Total RNA was isolated from liver tissue of mice on either MCD or CTL diet by using RNeasy Mini kit (Qiagen, Valencia, CA). cDNA was transcribed by using an RT2 Reaction Ready First Strand Synthesis Kit (SuperArray Bioscience, Frederick, MD) and was analyzed, by using the mouse oxidative stress PCR array (Cat. no. PAMM-065, SuperArray Bioscience) focusing on gene families relevant to the induction and inhibition of ROS production and genes involved in ROS metabolism. The assay used a RT2 SYBR Green/Rox PCR master mix (Cat. no. PA-012, SuperArray Bioscience) on a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA). Data were normalized by using multiple housekeeping genes [β-actin, GAPDH (β-actin, GAPDH, glyceraldehyde-3-phosphate dehydrogenase), HPRT1, HSP90AB1], and analyzed by comparing 2ΔΔCt of the normalized sample. Five mice per group were used for this analysis.
Histopathology and serum assays
Blood samples and liver tissue were collected under deep anesthesia, as previously described in detail (14). Liver tissue was fixed in 4% paraformaldehyde and embedded in Tissue Path (Fisher Scientific, Pittsburgh, PA). Tissue sections (4 μm) were prepared, and hematoxylin-and-eosin, as well as Oil Red O–stained liver specimens were evaluated by light microscopy. Individual features, including degree of steatosis, inflammation, and ballooning, were assessed in MCD- and CTL-fed animals by an experienced pathologist in a blinded fashion. Steatosis, inflammation, and ballooning were scored based on NAFLD activity score (18). The presence of 4-hydroxynonenal, a representative lipid peroxide, and nitrotyrosine adducts, products of oxidative stress, were assessed by immunohistochemistry (4-HNE; Cayman Chemicals, Ann Arbor, MI) and immunofluorescence (3-nitrotyrosine; Cambridge, MA). Serum alanine aminotransferase (ALT) determinations were performed by using a commercially available kit (Sigma Diagnostics, St. Louis, MO).
Real-time PCR analysis
Total RNA was isolated from liver tissue by using RNeasy Lipid Tissue Mini kit (Qiagen, Valencia, CA). Reverse transcript (the cDNA) was synthesized from 1 μg total RNA by using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Real-time PCR quantification was performed. In brief, 25 μl reaction mix contained cDNA, Syber Green buffer, Gold Taq polymerase, dNTPs, and primers at a final concentration of 200 μM. The sequences of the primers used for quantitative PCR are as follows; Fmo2: 5′-AGTGGCCTAATCTCTCTGAAGT and 5′-CATCGGGAAGTCACTGAAACAG, Scd1: 5′-TTCTTGCGATACACTCTGGTGC and 5′-CGGGATTGAATGTTCTTGTCGT, Gpx3: 5′-TGGCTTGGTCATTCTGGGC and 5′-CCCACCTGGTCGAACATACTT, Gpx6: 5′-GCCCAGAAGTTGTGGGGTTC and 5′-TCCATACTCATAGACGGTGCC, 18S: 5′-ACGGAAGGGCACCACCAGGA and 5′-CACCACCACCCACGGAATCG. RT-PCR was performed in the Mx3000P cycler (Stratagene): 95°C for 10 min, 40 cycles of 15 sec at 95°C, 30 sec at 60°C, 30 sec at 72°C, followed by 1 min at 95°C, 30 sec at 55°C and 30 sec at 95°C. The fold change over control samples was calculated by using CT, ΔCT, and ΔΔCT values by using MxPro software (Stratagene). 18S rRNA was used as an endogenous control.
Sample preparation and Western blot analysis
Liver tissues were put in the RIPA buffer (Cell Signaling, Danvers, MA) and homogenized by using zirconium oxide beads (0.5 mm) in a Bullet Blender homogenizer (Next Advance Inc., Averill Park, NY). Tissues were incubated on ice for 15 min and then at 13,000 rpm for 15 min at 4°C. The cytosolic fractions (supernatants) were stored at −20°C until analysis. Then 30 μg of tissue lysate protein was separated by SDS-PAGE and transferred to nitrocellulose membrane (BioRad, Hercules, CA). The membrane was blocked with 5% BSA (Fisher Scientific, Pittsburgh, PA) in PBS buffer with 0.01% Tween20 for 1 h at room temperature. Blocking was followed by the incubation with either SCD1 antibody (Cell Signaling), Gpx3 antibody (Abcam, Cambridge, MA) or actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA) in 1% BSA. Membrane was then washed 3 times with PBS-T buffer, incubated with horseradish peroxidase anti-mouse secondary antibody (Cell Signaling) or anti-rabbit secondary antibody (KPL, Gaithersburg, MD) for 1 h at room temperature and washed 3 times with TBS-T buffer. The bands were visualized by using chemiluminescence system (SuperSignal West Pico Substrate; Thermo Scientific, Rockford, IL) and quantified with densitometry by using Image Quant RT ECL.
Data analysis
All data are expressed as the mean±SD unless otherwise indicated. Differences between groups were compared with an ANOVA analysis followed by a post hoc Bonferroni test to correct for multiple comparisons. Differences were considered to be statistically significant at p<0.05. The real-time PCR microarrays were analyzed with the use of appropriate cutoff criteria, a twofold induction or repression of expression, with a p value of <0.001, was considered to represent significantly up- or downregulated gene expression.
Results
MCD feeding results in NASH development and oxidative tissue damage
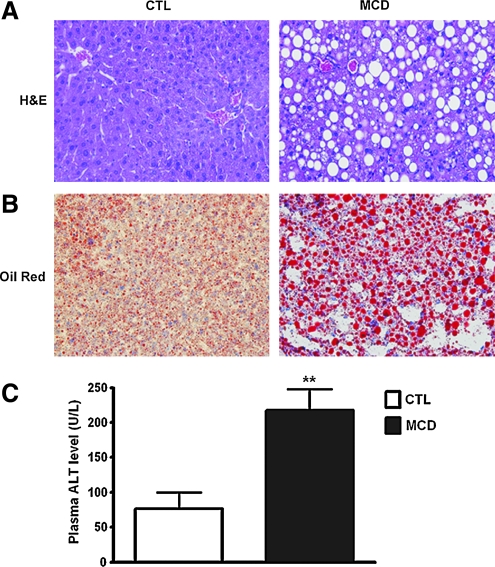
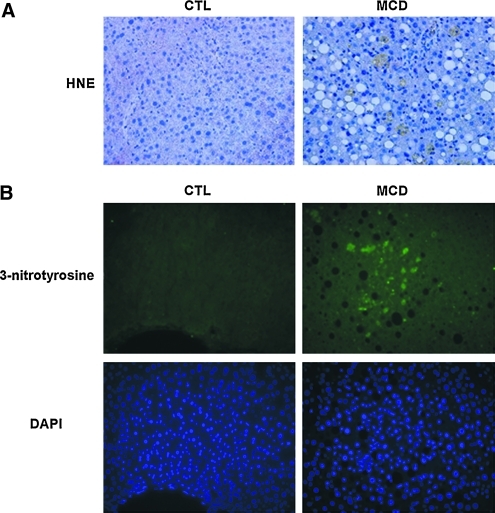
To investigate the role of oxidative stress in liver injury and development of steatohepatitis, we initially placed C57BL/6 mice on an MCD or CTL diet. After 6 weeks on the respective diets, we observed significant hepatic fat accumulation induced by MCD feeding (Fig. 1A and B). These changes were associated with significant increases in serum ALT levels (Fig. 1C), and increased histologic parameters of liver injury, including hepatic inflammation, and hepatocyte ballooning (Table 1). We further assessed the effects of the MCD diet on lipid and protein markers of oxidative stress. MCD feeding increased the hepatic presence of both 4-HNE and 3-nitrotyrosine modifications (Fig. 2A and B).
FIG. 1.
Liver steatosis and injury induced by MCD diet. (A) Representative photomicrograph of liver H&E staining (magnification, 40×) from mice on either control (CTL) or MCD diets. (B) Oil red O staining of livers from mice from the two groups. Plasma ALT levels used as a marker of liver injury. Data are presented as mean±SEM. **p<0.01 compared with control mice. (C) Plasma ALT levels in mice fed CTL and MCD diet. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Table 1.
Histopathologic Analysis of Liver Tissue from Mice Fed Chow and MCD Diet
| Chow | MCD | |
|---|---|---|
| Steatosis | 0.0 (±0.0) | 2.3 (±0.5) |
| Inflammation | 0.0 (±0.0) | 1.0 (±0.0) |
| Ballooning | 0.0 (±0.0) | 0.8 (±0.0) |
| NAS | 0.0 (±0.0) | 4.0 (±0.8) |
Hepatic histology was assessed by reviewing the H&E-stained sections from mice on either the MCD or the CTL diet (n=6 in each group). Individual histologic features were scored by using the NAS as detailed in the Methods section. Average (±SD). NAS, NAFLD activity score.
FIG. 2.
Oxidative stress caused by MCD feeding. (A) 4-HNE staining of liver tissue sections from mice fed CTL and MCD diets (magnification, 40×). (B) 3-Nitrotyrosine staining of liver sections from two groups of mice (magnification, 40×). The nuclear-binding dye DAPI was used to determine the total number of cells per 40×field. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Transcriptional profiles of genes involved in oxidative stress, antioxidant defense, and reactive oxygen metabolism are markedly affected in the livers of MCD-fed mice
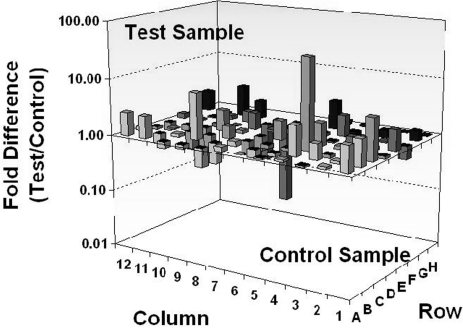
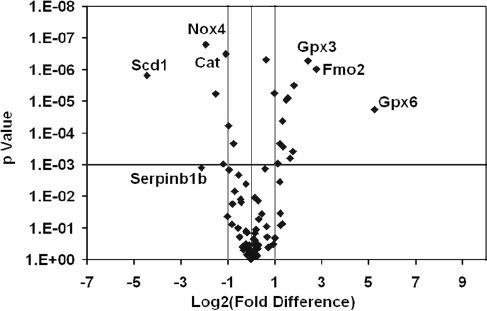
To explore the impact of the changes observed in the livers of MCD-treated animals on the transcriptional profile of genes involved in oxidative stress, antioxidant defense, and reactive oxygen metabolism, we used quantitative real-time PCR microarrays to catalogue gene-expression levels of a total of 84 genes in liver tissue from mice on either the MCD or the CTL diet (Fig. 3). Genes were considered to be expressed differentially in NASH livers induced by MCD feeding if greater than twofold difference was found in gene expression with a threshold of a p value of 0.001 (Fig. 4). As detailed in Table 2, of the 84 genes tested, 19 were uniquely differentially expressed (12 overexpressed and seven underexpressed). Genes that were significantly overexpressed included Glutathione peroxidises 3 and 6 (Gpx3 and Gpx6), EH-domain-containing 2 (Ehd2), peroxiredoxin 3 (Prdx3), cathepsin B (Ctsb), prostaglandin-endoperoxide synthase 2 (Ptgs2), α subunit of cytochrome b245 gene (Cyba), neutrophil cytosolic factor 2 (Ncf2), flavin-containing monooxygenase 2 (Fmo2), NAD(P)H dehydrogenase quinone 1 (Nqo1), thioredoxin-interacting protein (Txnip), cytoglobin (Cygb), and vimentin (Vim). Genes that were significantly underexpressed in NASH livers included catalase (Cat), peroxiredoxin 5 (Prdx5), serine peptidase inhibitor b1b (Serpinb1b), thyroid peroxidase (Tpo), NADPH oxidase 4 (Nox4), stearoyl-CoA desaturase-1 (SCD1), thioredoxin reductase 2 (Txnrd2), and Fanconi anemia complementation group C (Fancc).
FIG. 3.
Three-dimensional plot representing real-time RT-PCR microarray analysis. The 84 genes in the livers of mice associated with oxidative stress were analyzed. Data are presented as the fold difference from mice on the CTL diet, calculated from average ΔCT normalized to housekeeping genes [β-actin, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), HPRT1, HSP90AB1]. Columns pointing up (with z-axis values >1) indicate an upregulation of gene expression, and columns pointing down (with z-axis values <1) indicate a downregulation of gene expression in the test sample relative to the control sample.
FIG. 4.
Volcano plot representing real-time RT-PCR microarray analysis. The 84 genes in the livers of mice associated with oxidative stress were analyzed. This graph summarizes fold-change and t test criteria. The X axis represents fold difference from mice on the CTL diet. The Y axis represents the p value. The vertical line in the middle indicates a fold-change in gene expression of 1. The other two vertical lines indicate a threshold of two fold-changes in gene expression. The horizontal line indicates a threshold of p=0.001.
Table 2.
Microarray Analysis of 84 Genes Involved in Redox Signaling
| Gene name | Fold up- and downregulation | p-Value |
|---|---|---|
| Glutathione peroxidases | ||
| Glutathione peroxidase 1 (Gpx1) | −1.48 | 0.002 |
| Glutathione peroxidase 2 (Gpx2) | 1.18 | 0.773 |
| Glutathione peroxidase 3 (Gpx3) | 5.34 | 5.214 × 10−7 |
| Glutathione peroxidase 4 (Gpx4) | 1.54 | 4.959 × 10−7 |
| Glutathione peroxidase 5 (Gpx5) | −1.13 | 0.142 |
| Glutathione peroxidase 6 (Gpx6) | 38.06 | 1.829 × 10−5 |
| Glutathione peroxidase 7 (Gpx7) | −1.13 | 0.142 |
| Glutathione S-transferase kappa 1 (Gstk1) | −1.69 | 0.002 |
| Peroxiredoxins | ||
| EH-domain containing 2 (Ehd2) | 3.39 | 0.0004 |
| Peroxiredoxin 1 (Prdx1) | 1.22 | 0.014 |
| Peroxiredoxin 2 (Prdx2) | 1.10 | 0.151 |
| Peroxiredoxin 3 (Prdx3) | 1.98 | 5.61 × 10−6 |
| Peroxiredoxin 4 (Prdx4) | 1.04 | 0.662 |
| Peroxiredoxin 5 (Prdx5) | −1.17 | 0.004 |
| Peroxiredoxin 6 (Prdx6) | 1.09 | 0.322 |
| Other peroxidases | ||
| Aminoadipate-semialdehyde synthase (Aass) | 1.12 | 0.372 |
| Adenomatous polyposis coli (Apc) | 1.10 | 0.246 |
| Catalase (Cat) | −2.13 | 3.254 × 10−6 |
| Cathepsin B (Ctsb) | 2.49 | 4.288 × 10−5 |
| Dual oxidase 1 (Duox1) | 1.90 | 0.331 |
| Eosinophil peroxidase (Epx) | −1.14 | 0.702 |
| Kinesin family member 9 (Kif9) | −1.10 | 0.488 |
| Lactoperoxidase (Lpo) | −1.13 | 0.142 |
| Myeloperoxidase (Mpo) | 2.53 | 0.074 |
| Peroxiredoxin 6 related sequence 1 (Prdx6-rs1) | 1.21 | 0.364 |
| Prostaglandin-endoperoxide synthase 1 (Ptgs1) | 1.09 | 0.532 |
| Prostaglandin-endoperoxide synthase 2 (Ptgs2) | 2.37 | 0.035 |
| Recombination activating gene 2 (Rag2) | −1.13 | 0.142 |
| Serine (or cysteine) peptidase inhibitor, clade B, member 1b (Serpinb1b) | −4.38 | 0.001 |
| Solute carrier family 41, member 3 (Slc41a3) | 1.19 | 0.335 |
| Tropomodulin 1 (Tmod1) | −1.01 | 0.984 |
| Thyroid peroxidase (Tpo) | −2.03 | 0.044 |
| Other antioxidants | ||
| Glutathione reductase (Gsr) | −1.18 | 0.128 |
| Nucleoredoxin (Nxn) | 1.36 | 0.037 |
| Superoxide dismutase 1 (Sod1) | −1.06 | 0.444 |
| Superoxide dismutase 2 (Sod2) | −1.03 | 0.887 |
| Sulfiredoxin 1 homolog (Srxn1) | 1.09 | 0.601 |
| Thioredoxin reductase 1 (Txnrd1) | −1.93 | 0.001 |
| Thioredoxin reductase 2 (Txnrd2) | −2.88 | 5.914 × 10−6 |
| Thioredoxin reductase 3 (Txnrd3) | −1.19 | 0.343 |
| Superoxide dismutases | ||
| Superoxide dismutase 1 (Sod1) | −1.06 | 0.444 |
| Superoxide dismutase 2 (Sod2) | −1.03 | 0.887 |
| Superoxide dismutase 3 (Sod3) | 1.49 | 0.001 |
| Genes involved in superoxide metabolism | ||
| Copper chaperone for superoxide dismutase (Ccs) | −1.17 | 0.320 |
| Cytochrome b-245, alpha polypeptide (Cyba) | 2.54 | 2.701 × 10−4 |
| Neutrophil cytosolic factor 2 (Ncf2) | 2.16 | 0.001 |
| Nitric oxide synthase 2 (Nos2) | 1.15 | 0.693 |
| NADPH oxidase 1 (Nox1) | 2.34 | 0.082 |
| NADPH oxidase 4 (Nox4) | −3.84 | 1.595 × 10−7 |
| NADPH oxidase activator 1 (Noxa1) | −1.27 | 0.509 |
| NADPH oxidase organizer 1 (Noxo1) | −1.42 | 0.197 |
| RecQ protein-like 4 (Recql4) | −1.49 | 0.101 |
| Stearoyl-coenzyme A desaturase 1 (Scd1) | −21.94 | 1.537 × 10−6 |
| Genes involved in ROS metabolism | ||
| Flavin containing monooxygenase 2 (Fmo2) | 6.75 | 9.629 × 10−6 |
| Interleukin 19 (Il19) | 1.06 | 0.816 |
| Interleukin 22 (Il22) | −1.13 | 0.142 |
| Other oxidative stress responsive genes | ||
| Amyotrophic lateral sclerosis 2 (Als2) | −1.07 | 0.346 |
| Apolipoprotein E (Apoe) | −1.04 | 0.595 |
| Catalase (Cat) | −2.13 | 3.254 × 10−6 |
| Dual oxidase 1 (Duox1) | 1.90 | 0.331 |
| Eosinophil peroxidase (Epx) | −1.14 | 0.702 |
| Excision repair cross-complementing rodent repair deficiency, complementation group 2 (Ercc2) | −1.38 | 0.012 |
| Excision repair cross-complementing rodent repair deficiency, complementation group 6 (Ercc6) | 1.24 | 0.054 |
| GRB2-associated binding protein 1 (Gab1) | 1.13 | 0.113 |
| Isocitrate dehydrogenase 1 (Idh1) | −1.63 | 0.007 |
| Myeloperoxidase (Mpo) | 2.53 | 0.074 |
| Membrane protein, palmitoylated 4 (Mpp4) | 1.61 | 0.195 |
| NAD(P)H dehydrogenase, quinone 1 (Nqo1) | 2.31 | 2.207 × 10−4 |
| Nudix (nucleoside diphosphate linked moiety X)-type motif 15 (Nudt15) | −1.15 | 0.448 |
| Parkinson disease 7 (Park7) | −1.06 | 0.541 |
| Protein phosphatase 1, regulatory subunit 15B (Ppp1r15b) | −1.02 | 0.870 |
| Prion protein (Prnp) | −1.75 | 0.017 |
| Proteasome subunit, beta type, 5 (Psmb5) | 1.06 | 0.230 |
| Superoxide dismutase 1 (Sod1) | −1.06 | 0.444 |
| Thioredoxin interacting protein (Txnip) | 3.13 | 0.001 |
| Uncoupling protein 3 (Ucp3) | 2.01 | 0.208 |
| Xeroderma pigmentosum, complementation group A (Xpa) | −1.36 | 0.016 |
| Oxygen transporters | ||
| Aquarius homolog (Aqr) | 1.02 | 0.746 |
| Ataxia telangiectasia and Rad3 related (Atr) | 1.20 | 0.453 |
| Cytoglobin (Cygb) | 2.96 | 8.030 × 10−6 |
| Dynamin 2 (Dnm2) | 1.10 | 0.011 |
| Fanconi anemia, complementation group C (Fancc) | −2.29 | 0.001 |
| Hemoglobin, theta 1 (Hbq1) | 1.64 | 0.419 |
| Intraflagellar transport 172 homolog (Ift172) | −1.97 | 5.873 × 10−5 |
| Myoglobin (Mb) | 1.14 | 0.367 |
| Neuroglobin (Ngb) | −1.79 | 0.078 |
| Solute carrier family 38, member 1 (Slc38a1) | 1.56 | 0.091 |
| Vimentin (Vim) | 3.51 | 3.121 × 10−6 |
Values in bold are statistically significant.
Validation of differentially expressed genes detected on microarrays
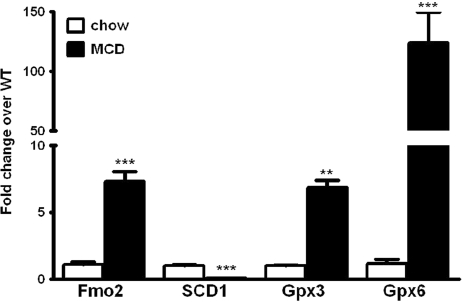
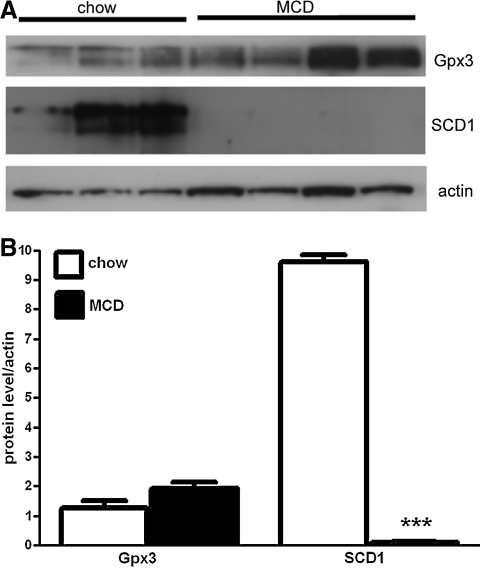
Changes of the most significantly differentially expressed genes observed in the microarrays were validated by using real-time RT-PCR for individual transcripts and Western blot analysis for the protein expression of the gene products. The data confirmed the significant upregulation of Gpx3 (6.8-fold increase), Gpx6 (19-fold increase), Fmo2 (7.3-fold increase), and the downregulation of SCD1 (10-fold decrease) in the livers of the MCD diet–fed animals (Fig. 5). Finally, Western blot analysis showed a markedly decreased expression of SCD1 and increased levels of Gpx3 in the mice fed MCD compared with CTL (Fig. 6A and B).
FIG. 5.
Real-time PCR data confirming microarray analysis for the most differentially expressed genes. RT-PCR measurements for individual transcripts were performed to validate the results observed on the microarrays. RT-PCR measurements are shown of the mRNA expression of the four mostly altered genes in the livers of mice on either the MCD or CTL diets. Data are presented as mean±SEM, n=5 mice per group. **p<0.005. ***p<0.001.
FIG. 6.
Measurement of protein levels of the most-altered genes analyzed by microarray. (A) Western blot analysis. (B) Densitometry of the protein bands measured with Image Quant RT ECL. Data are presented as mean±SEM, n=3–4 mice per group. ***p<0.001.
Discussion
Altered redox signaling due to overgeneration of reactive oxygen species (ROS) or decreased antioxidant defenses is thought to play a pivotal role in NASH development (10). Experimental and human studies have demonstrated enhanced oxidative stress and lipid peroxidation products in the circulation and in the liver of several different animal models and in patients with NASH (5, 16, 24, 26). The imbalance between generated reactive oxygen metabolites and antioxidant defense systems produces oxidative stress, with subsequent damage involving important cellular macromolecules (lipids, proteins, carbohydrates, and nucleic acids). Despite intense interest in the role of oxidation in the pathogenesis of NASH, the pathways that contribute to oxidative damage in vivo are poorly understood. The principal findings of this study relate to the identification of the transcriptional profile of genes associated with redox biochemistry in an established murine model of NASH. The results demonstrate that NASH induced by MCD feeding is characterized by altered expression in the liver of several key genes and gene families involved in oxidative stress, antioxidant defense, and reactive oxygen metabolism. Of the 84 genes examined by using the quantitative real-time PCR microarrays, 12 were significantly upregulated, and seven were significantly downregulated in the livers of MCD-fed animals.
Members of the glutathione peroxidase (GPx) family, GPx3 and GPx6, were the most significantly upregulated. GPxs are thought to be among the major components of human antioxidant defense. GPxs catalyze the reduction of hydrogen peroxide and organic hydroperoxides by using reduced glutathione. Increased expression of GPx3 and GPx6 suggests a compensatory mechanism that is triggered by enhanced ROS formation in the livers of mice fed an MCD diet. Thus, the genes involved in antioxidant defense systems are activated (11). The expression and role of both GPx3 and GPx6 in the liver, and particularly in the context of NASH, has not been previously explored and warrants further investigation. In contrast, MCD feeding led to a decrease in expression of two very important genes involved in the antioxidant defences: GPx1 and catalase. GPx1 is the most extensively characterized of the GPx family and is found in the cytosol and mitochondria (13, 15). It is thought to be the major enzyme inactivating hydrogen peroxide (27). One explanation for the variation found in GPxs expression levels that merits future investigation is that when the ability of GPx1 to deal with the burden of ROS is exceeded, the other members of the family, the expression of which is not usually associated with the liver, are upregulated as a secondary compensatory measure to deal with the increased production of peroxides and hydroperoxides. In keeping with this hypothesis, it is noteworthy that catalase was also downregulated. Catalase is regarded as one of the most efficient enzyme systems displaying zero-order kinetics (33). This enzyme regulates the breakdown of hydrogen peroxide and hydroperoxides. Decreased hepatic catalase expression suggests further abolished antioxidant defences in the liver. Our data are consistent with those of Bujanda et al. (7), who showed that, in the rat model of NAFLD, liver GPx1 and catalase expression also was decreased.
In addition to changes in the expression of catalase and Gpxs, we observed an upregulation of several peroxiredoxins, such as the EH domain containing 2 (Edh2), peroxiredoxin 3 (Prdx3), and reduced expression of peroxiredoxin 5 (Prdx5). Prdx3 and Prdx5 are mitochondrial peroxiredoxins that decompose hydrogen peroxide and become oxidized. They are reduced back to the native form by thioredoxin 2, which is reduced by thioredoxin reductase 2 (Txnrd2) (8). Upregulation of Prdx3 is another indicator of increase oxidative stress in the livers of mice fed the MCD diet. Downregulation of Prdx5 can suggest that the expression of these peroxiredoxins is regulated differentially. The other important observation we made is that Txnrd2 expression is significantly reduced in livers of MCD-fed mice. This can lead to impaired reduction of Prdx3 and Prdx5 because Txnrd2 is the main reductant of these two peroxireductases. These changes can lead to the accumulation of hydrogen peroxide, prolonged oxidative stress in the mitochondria causing mitochondrial damage, and liver injury.
Flavin-containing monooxygenase 2 (Fmo2) was the second most upregulated of the genes identified by microarray analysis. This monooxygenase is involved in the oxidative metabolism of various xenobiotics by adding oxygen to sulfur-, nitrogen-, phosphorus-, and selenium-containing compounds. In yeast, it catalyzes the oxidation of reduced glutathione to glutathione disulfide (25). It is possible that upregulation of Fmo2 may disturb the GSSG/GSH balance in murine liver. However, this hypothesis requires further investigation. The expression of glutathione reductase (Gsr), an enzyme that converts GSSG to GSH, was slightly downregulated, which suggests the depleted pull of reduced glutathione. An unbalanced GSSG/GSH ratio can in turn interrupt the function of GPxs that also use reduced glutathione to inactivate peroxides.
The most significantly downregulated of the genes identified by the microarray was stearoyl-CoA desaturase-1 (SCD1), with a 22-fold reduction in expression in the livers of MCD-fed mice. SCD1 is a key regulator of intracellular fatty acid composition. This enzyme facilitates the channeling of free fatty acids (FFAs) into triglyceride storage, phospholipids, and cholesterol ester synthesis (22). We and others previously demonstrated that MCD feeding results in a significant decrease in hepatic SCD1 protein expression and activity. Moreover, when we placed SCD1-knockout mice on the MCD diet, the mice accumulated less triglyceride in the liver compared with the wild-type animals on the same diet. However, the SCD1-knockout animals showed a pronounced decrease in the desaturation index of fatty acids that was accompanied by an increase in hepatocellular apoptosis, liver injury, and fibrosis (20). The current study extends these findings by demonstrating a marked reduction of SCD1 gene expression with real-time PCR in the livers of MCD-fed mice. Decreased hepatic SCD1 in this context would result in enhanced delivery of fatty acids to mitochondria and increased fatty acid oxidation, and in consequence, an overproduction of mitochondrial ROS. The molecular mechanism responsible for MCD-diet–induced downregulation of SCD1 as well as changes in SCD1 expression in human NAFLD, and its relation to liver damage and disease progression remain unknown and will require further investigation.
NADPH oxidase 4 (Nox4) expression was decreased in the livers of MCD-fed mice. Nox enzymes reduce molecular oxygen to superoxide that can be further converted to various secondary ROS (28). Reduction of Nox4 expression leads to reduced superoxide production by this enzyme. However, MCD feeding caused an increase of liver Cyba and Ncf2. Cyba encodes the subunit p22phox, which is required for function and stabilization of almost all Nox enzymes, including Nox4 (19, 30). Ncf2 encodes neutrophil cytosolic factor 2, a cytosolic subunit of the multiprotein NADPH oxidase complex found in neutrophils. Its role in the liver requires further investigation. Elevated expression of Cyba and Ncf2 and decreased expression of Nox4 indicate the different response of Nox genes to the MCD diet. It is possible that increased expression of Cyba could contribute to the upregulation of other Nox genes, which could therefore compensate for the reduced expression of Nox4. Further investigation is required to test this hypothesis.
The expression of one enzyme that may contribute to the inflammatory process is Ptgs2, also known as cyclooxygenase-2 (COX-2), was upregulated. The expression of COX-2 is induced by cell stressors, such as proinflammatory cytokines and oxidative stress (21). It has been shown that HNE, a lipid peroxidation product, can induce COX-2 (29). Prolonged increased COX-2 expression in the liver may lead to chronic inflammation and carcinogenesis. It has also been shown that, in livers of patients with chronic hepatitis and cirrhosis, COX-2 is upregulated, and inhibition of COX-2 ameliorates the severity of hepatitis in murine models of steatohepatitis. COX-2 can have both proinflammatory and antiinflammatory effects, depending on the duration of COX-2 activity and the site and nature of the inflammatory response. Proinflammatory products of COX-2 activity, leukotrienes and prostaglandin E2, are released early in the inflammatory response, causing hepatitis. Conversely, antiinflammatory products, lipoxins, and prostaglandins PGJ2 and PGD2, produced later during inflammation, can play a protective role (17). Thus, increased COX-2 expression in livers of MCD-fed mice is a marker of stress and, depending on the setting, can lead to steatohepatitis and liver damage or have a protective function.
SERPIN1B1 is a gene that encodes for a monocyte neutrophil elastase inhibitor (MNEI), a highly efficient protease inhibitor in humans, whereas in mice, it encodes EIA, which inhibits pancreatic and neutrophil elastases, cathepsin G, proteinase-3, and chymotrypsin (4). Downregulation of this protein seen in our model may be of direct relevance in terms of the pathogenesis of lipid-mediated hepatic damage, as without this potent protease inhibitor, the actions of monocyte and neutrophil proteases would not be regulated.
In summary, feeding mice an MCD diet for 6 weeks results in marked steatohepatitis and oxidative liver damage. The gene changes in this study show the imbalance between ROS production and antioxidant defence in livers of mice fed the MCD diet. We have shown that some antioxidant genes were upregulated (Gpx3, Gpx6, Prdx3), which suggests the presence of oxidative stress, and some were downregulated (Gpx1, Cat, Prdx5, Txnrd2), which leads to the impairment of antioxidant defense and thus to increased oxidative stress causing liver damage. We acknowledge the limitation of current experimental models of NASH. The MCD diet, although widely used as a model of fibrosing steatohepatitis, is not associated with obesity or other features of the metabolic syndrome characteristic of patients with NAFLD. We believe, however, that this model is particularly useful to investigate mechanisms of liver injury, fibrogenesis, and disease progression in NAFLD, as the liver histopathology closely resembles that of human NASH. Our findings provide novel evidence for the potential involvement of specific genes involved in redox homeostasis in NASH pathogenesis. Further studies are required to determine the mechanisms and pathophysiologic relevance of our findings in human NASH.
Abbreviations Used
- ALT
alanine aminotransferase
- Cat
catalase
- COX-2
cyclooxygenase-2
- Ctsb
cathepsin B
- Cyba
alpha subunit of cytochrome b-245 gene
- Cygb
cytoglobin
- Ehd2
EH-domain containing 2
- Fancc
Fanconi anemia complementation group C
- Fmo2
flavin-containing monooxygenase 2
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- Gpx
glutathione peroxidase
- HNE
4-hydroxynonenal
- HPRT1
hypoxanthine phosphoribosyltransferase 1
- HSP90AB1
heat-shock protein 90-kDa alpha
- MCD
methionine and choline deficient
- MNEI
monocyte neutrophil elastase inhibitor
- Ncf2
neutrophil cytosolic factor 2
- Nox4
NADPH oxidase 4
- Nqo
NAD(P)H dehydrogenase quinone 1
- PGD2
prostaglandin D2
- PGJ2
prostaglandin J2
- Prdx
peroxiredoxin
- Ptgs2
prostaglandin-endoperoxide synthase 2
- Scd1
stearoyl-CoA desaturase-1
- Serpin1b
serine peptidase inhibitor b1b
- Tpo
thyroid peroxidase
- Txnip
thioredoxin-interacting protein
- Txnrd2
thioredoxin reductase 2
- Vim
vimentin
Acknowledgment
This work was supported by NIH grants (DK076852) and (DK082451) to AEF.
Author Disclosure Statement
The authors verify that no competing financial interests exist.
References
- 1.Adams LA. Lymp JF. Sat SJ. Sanderson SO. Lindor KD. Feldstein A. Angulo P. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–121. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Adams LA. Sanderson S. Lindor KD. Angulo P. The histological course of nonalcoholic fatty liver disease: a longitudinal study of 103 patients with sequential liver biopsies. J Hepatol. 2005;42:132–138. doi: 10.1016/j.jhep.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 4.Benarafa C. Cooley J. Zeng W. Bird PI. Remold-O'Donnell E. Characterization of four murine homologs of the human ov-serpin monocyte neutrophil elastase inhibitor MNEI (SERPINB1) J Biol Chem. 2002;277:42028–42033. doi: 10.1074/jbc.M207080200. [DOI] [PubMed] [Google Scholar]
- 5.Bonnefont-Rousselot D. Ratziu V. Giral P. Charlotte F. Beucler I. Poynard T. Blood oxidative stress markers are unreliable markers of hepatic steatosis. Aliment Pharmacol Ther. 2006;23:91–98. doi: 10.1111/j.1365-2036.2006.02719.x. [DOI] [PubMed] [Google Scholar]
- 6.Brunt EM. Janney CG. Di Bisceglie AM. Neuschwander-Tetri BA. Bacon BR. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999;94:2467–2474. doi: 10.1111/j.1572-0241.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- 7.Bujanda L. Hijona E. Larzabal M. Beraza M. Aldazabal P. Garcia-Urkia N. Sarasqueta C. Cosme A. Irastorza B. Gonzalez A. Arenas JI., Jr Resveratrol inhibits nonalcoholic fatty liver disease in rats. BMC Gastroenterol. 2008;8:40. doi: 10.1186/1471-230X-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox AG. Winterbourn CC. Hampton MB. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem J. 2010;425:313–325. doi: 10.1042/BJ20091541. [DOI] [PubMed] [Google Scholar]
- 9.Day CP. Pathogenesis of steatohepatitis. Best Pract Res Clin Gastroenterol. 2002;16:663–678. doi: 10.1053/bega.2002.0333. [DOI] [PubMed] [Google Scholar]
- 10.Day CP. Saksena S. Non-alcoholic steatohepatitis: definitions and pathogenesis. J Gastroenterol Hepatol. 2002;17(suppl 3):S377–S384. doi: 10.1046/j.1440-1746.17.s3.31.x. [DOI] [PubMed] [Google Scholar]
- 11.De Oliveira CP. Stefano JT. De LV. de Sa SV. Simplicio FI. De Mello ES. Correa-Giannella ML. Alves VA. Laurindo FR. de Oliveira MG. Giannella-Neto D. Carrilho FJ. Hepatic gene expression profile associated with non-alcoholic steatohepatitis protection by S-nitroso-N-acetylcysteine in ob/ob mice. J Hepatol. 2006;45:725–733. doi: 10.1016/j.jhep.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 12.Ekstedt M. Franzen LE. Mathiesen UL. Thorelius L. Holmqvist M. Bodemar G. Kechagias S. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology. 2006;44:865–873. doi: 10.1002/hep.21327. [DOI] [PubMed] [Google Scholar]
- 13.Esworthy RS. Ho YS. Chu FF. The Gpx1 gene encodes mitochondrial glutathione peroxidase in the mouse liver. Arch Biochem Biophys. 1997;340:59–63. doi: 10.1006/abbi.1997.9901. [DOI] [PubMed] [Google Scholar]
- 14.Feldstein AE. Canbay A. Guicciardi ME. Higuchi H. Bronk SF. Gores GJ. Diet associated hepatic steatosis sensitizes to Fas mediated liver injury in mice. J Hepatol. 2003;39:978–983. doi: 10.1016/s0168-8278(03)00460-4. [DOI] [PubMed] [Google Scholar]
- 15.Flohe L. Gunzler WA. Schock HH. Glutathione peroxidase: a selenoenzyme. FEBS Lett. 1973;32:132–134. doi: 10.1016/0014-5793(73)80755-0. [DOI] [PubMed] [Google Scholar]
- 16.Gao D. Wei C. Chen L. Huang J. Yang S. Diehl AM. Oxidative DNA damage and DNA repair enzyme expression are inversely related in murine models of fatty liver disease. Am J Physiol Gastrointest Liver Physiol. 2004;287:G1070–G1077. doi: 10.1152/ajpgi.00228.2004. [DOI] [PubMed] [Google Scholar]
- 17.Holt AP. Adams DH. Complex roles of cyclo-oxygenase 2 in hepatitis. Gut. 2007;56:903–904. doi: 10.1136/gut.2006.115972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleiner DE. Brunt EM. Van NM. Behling C. Contos MJ. Cummings OW. Ferrell LD. Liu YC. Torbenson MS. Unalp-Arida A. Yeh M. McCullough AJ. Sanyal AJ. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 19.Lambeth JD. Kawahara T. Diebold B. Regulation of Nox and Duox enzymatic activity and expression. Free Radic Biol Med. 2007;43:319–331. doi: 10.1016/j.freeradbiomed.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li ZZ. Berk M. McIntyre TM. Feldstein AE. Hepatic lipid partitioning and liver damage in nonalcoholic fatty liver disease: role of stearoyl-CoA desaturase. J Biol Chem. 2009;284:5637–5644. doi: 10.1074/jbc.M807616200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin-Sanz P. Mayoral R. Casado M. Bosca L. COX-2 in liver, from regeneration to hepatocarcinogenesis: what we have learned from animal models? World J Gastroenterol. 2010;16:1430–1435. doi: 10.3748/wjg.v16.i12.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyazaki M. Kim YC. Gray-Keller MP. Attie AD. Ntambi JM. The biosynthesis of hepatic cholesterol esters and triglycerides is impaired in mice with a disruption of the gene for stearoyl-CoA desaturase 1. J Biol Chem. 2000;275:30132–30138. doi: 10.1074/jbc.M005488200. [DOI] [PubMed] [Google Scholar]
- 23.Nanji AA. Animal models of nonalcoholic fatty liver disease and steatohepatitis. Clin Liver Dis. 2004;8:559–574. doi: 10.1016/j.cld.2004.04.002. ix. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira CP. da Costa Gayotto LC. Tatai C. la Bina BI. Janiszewski M. Lima ES. Abdalla DS. Lopasso FP. Laurindo FR. Laudanna AA. Oxidative stress in the pathogenesis of nonalcoholic fatty liver disease, in rats fed with a choline-deficient diet. J Cell Mol Med. 2002;6:399–406. doi: 10.1111/j.1582-4934.2002.tb00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahman MF. Wang J. Patterson TA. Saini UT. Robinson BL. Newport GD. Murdock RC. Schlager JJ. Hussain SM. Ali SF. Expression of genes related to oxidative stress in the mouse brain after exposure to silver-25 nanoparticles. Toxicol Lett. 2009;187:15–21. doi: 10.1016/j.toxlet.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 26.Roskams T. Yang SQ. Koteish A. Durnez A. DeVos R. Huang X. Achten R. Verslype C. Diehl AM. Oxidative stress and oval cell accumulation in mice and humans with alcoholic and nonalcoholic fatty liver disease. Am J Pathol. 2003;163:1301–1311. doi: 10.1016/S0002-9440(10)63489-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spanier G. Xu H. Xia N. Tobias S. Deng S. Wojnowski L. Forstermann U. Li H. Resveratrol reduces endothelial oxidative stress by modulating the gene expression of superoxide dismutase 1 (SOD1), glutathione peroxidase 1 (GPx1) and NADPH oxidase subunit (Nox4) J Physiol Pharmacol. 2009;60(suppl 4):111–116. [PubMed] [Google Scholar]
- 28.Sumimoto H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 2008;275:3249–3277. doi: 10.1111/j.1742-4658.2008.06488.x. [DOI] [PubMed] [Google Scholar]
- 29.Uchida K. A lipid-derived endogenous inducer of COX-2: a bridge between inflammation and oxidative stress. Mol Cells. 2008;25:347–351. [PubMed] [Google Scholar]
- 30.Vibhuti A. Arif E. Mishra A. Deepak D. Singh B. Rahman I. Mohammad G. Pasha MA. CYP1A1, CYP1A2 and CYBA gene polymorphisms associated with oxidative stress in COPD. Clin Chim Acta. 2010;411:474–480. doi: 10.1016/j.cca.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Videla LA. Rodrigo R. Araya J. Poniachik J. Oxidative stress and depletion of hepatic long-chain polyunsaturated fatty acids may contribute to nonalcoholic fatty liver disease. Free Radic Biol Med. 2004;37:1499–1507. doi: 10.1016/j.freeradbiomed.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 32.Wieckowska A. Feldstein AE. Nonalcoholic fatty liver disease in the pediatric population: a review. Curr Opin Pediatr. 2005;17:636–641. doi: 10.1097/01.mop.0000172816.79637.c5. [DOI] [PubMed] [Google Scholar]
- 33.Zeisel SH. Blusztajn JK. Choline and human nutrition. Annu Rev Nutr. 1994;14:269–296. doi: 10.1146/annurev.nu.14.070194.001413. [DOI] [PubMed] [Google Scholar]