Abstract
Iron homeostasis-related genes (e.g., Dmt1 and Dcytb) are upregulated by hypoxia-inducible factor 2α (HIF2α) during iron deficiency in the mammalian intestine. Menkes copper ATPase (Atp7a) gene expression is also strongly induced in the duodenum of iron-deficient rats. The current study was thus designed to test the hypothesis that Atp7a is regulated by HIF2α. Rat intestinal epithelial (IEC-6) cells were utilized to model the intestinal epithelium, and CoCl2 and 1% O2 were applied to mimic hypoxia in vitro. Both treatments significantly increased endogenous Atp7a mRNA levels; mRNA induction with CoCl2 treatment was blunted by a transcriptional inhibitor. The rat Atp7a promoter was thus cloned and studied. Various sized promoter constructs were inserted into a luciferase reporter vector and transfected into cells. A −224/+88 bp construct had full activity and was induced by CoCl2; this promoter fragment was thus utilized for subsequent analyses. Interestingly, this region contains three phylogenetically conserved, putative hypoxia response elements (HRE; 5′-NCGTGN-3′). It was further noted that HIF2α overexpression caused a significant upregulation of promoter activity while HIF1α overexpression had little effect. To determine whether Atp7a is a direct HIF target, three putative HREs were deleted individually or in combination; all were shown to be essential for transcriptional induction. Chromatin immunoprecipitation studies also demonstrated that HIF2α binds to the Atp7a promoter region. Lastly, Atp7a and HIF2α protein levels were shown to be increased by both treatments. In conclusion, the Atp7a gene is upregulated by direct interaction with HIF2α, demonstrating coordinate regulation with genes related to intestinal iron homeostasis.
Keywords: IEC-6 cells, iron deficiency, gene regulation, transcription
intestinal iron absorption is the result of the coordinated action of iron import and export proteins, located on the brush-border and basolateral membranes of enterocytes, mediated by divalent metal transporter 1 (Dmt1) (13, 15) and ferroportin 1 (Fpn1) (1, 11, 22, 29), respectively. Also required are a coupled reduction of dietary ferric iron by duodenal cytochrome B (Dcytb; or other proteins) (9, 21) to the import process and an oxidation event mediated by hephaestin (28) coupled to the export process. Absorption of iron is a regulated process, responding in the positive direction during states of iron deficiency. This enhancement of absorption is mediated partially via induction of genes related to enterocyte iron homeostasis; each of the genes mentioned above is modulated by physiological signals that increase expression during iron deprivation. Previous studies have also shown induction of copper transport-related genes in the gut of iron-deficient rats (5, 6), suggesting that alterations in copper homeostasis may be part of the compensatory mechanism to increase iron absorption. This is consistent with previous observations documenting increased copper in the intestinal epithelium and liver (25), and serum (12) of mammals during iron deficiency. The induction of iron and copper-related genes in the intestine during iron deprivation suggests a possible common regulatory mechanism.
During iron deficiency, when levels of the liver- derived, iron homeostasis-regulating hormone hepcidin (10) drop precipitously, other regulatory mechanisms likely come into play. In some cases, genes are induced via interaction of intracellular iron-sensing proteins (iron regulatory proteins) with stem loop structures [called iron response elements (IREs)] in the 5′- or 3′-untranslated regions of mRNA transcripts encoding iron homeostasis-related genes (Dmt1, Fpn1) (1, 18). But interestingly, not all genes induced during iron deprivation have IREs and some genes respond in opposite directions than predicted by the location of the IRE, suggesting that other mechanisms are involved. Indeed, recent studies have shown that Dmt1 and Dcytb (and possibly Fpn1) are upregulated during iron deficiency at the level of gene transcription via specific interaction with a hypoxia-responsive trans-acting factor, HIF2α (23, 26). These investigations and another recent study (16) suggested that, during iron deficiency, when many tissues become hypoxic, the intestinal epithelium responds by preferential stabilization of HIF2α and that the hypoxic response drives fundamental changes in gene expression intended to overcome the iron deficient phenotype.
The current study was thus undertaken to test the hypothesis that the Menkes copper ATPase (Atp7a) gene is upregulated by HIF2α in the intestine of iron-deficient rats. This supposition derives from the fact that Atp7a mRNA induction parallels that of Dmt1 and Dcytb, as noted in previous publications (6). Atp7a encodes an intestinal copper transporter, and its induction is particularly intriguing given the aforementioned potential link between iron and copper during conditions of low iron in the intestine. The induction of Atp7a was thus modeled in intestinal epithelial (IEC-6) cells in culture and hypoxia was applied (or mimicked). Data suggested transcriptional induction of Atp7a expression during conditions that mimic hypoxia, so the promoter was cloned and studied. Extensive mechanistic studies identified specific HIF binding sites in the Atp7a promoter, which were shown to drive induction during hypoxia. Moreover, a critical role for HIF2α in this induction is revealed, strengthening the supposition that there is coordinate regulation of iron and copper homeostatic genes in enterocytes during iron deficiency.
MATERIALS AND METHODS
Cell culture.
Rat intestinal epithelial (IEC-6) cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured as previously described (8), essentially according to the distributor's recommendations. In some experiments, cells were grown in a hypoxia chamber (BioSpherix, Lacona, NY) with 5% CO2 and 1% O2 balanced with 94% N2, according to the manufacturer's instructions, to set up low-oxygen conditions.
Plasmid construction.
The rat Atp7a promoter was amplified by PCR with a proof reading polymerase (Invitrogen, Carlsbad, CA), utilizing a rat bacterial artificial chromosome (BAC) genomic clone as a template (clone no. CH230-423 C20; Children's Hospital of Oakland Research Institute, Oakland, CA). The forward primer was ∼3,000 bp upstream of the transcriptional start site (which was previously identified) (8) and the reverse primer was in the first exon of the gene (5′ of the start codon). These primers were designed with overhanging KpnI (forward) and EcoRV (reverse) restriction enzyme cutting sites. Promoter fragments were cloned into pGL4.18 basic luciferase vector (Promega, Madison, WI). Further deletion constructs were created by PCR amplification using the 3-kb promoter fragment as a template, using the same reverse primer and different forward primers with overhanging KpnI restriction enzyme digestion sites. PCR products and vector were double digested with restriction enzymes KpnI and EcoRV (Fermentas, Glen Burnie, MD), and ligations were performed with LigaFast Rapid DNA Ligation System (Promega). Deletion of putative hypoxia response elements (HRE; 5′-ACGTG-3′) in the −224/+88 promoter construct was performed by amplifying the entire plasmid with AccuPrime Taq DNA Polymerase (Invitrogen) using primers flanking the putative HRE and going in opposite directions, thus deleting the HREs. For double and triple HRE deletions, plasmids having previously deleted HREs were used as templates in the same fashion. Linearized PCR products were subsequently circularized with the Quick Ligation Kit (New England BioLab, Ipswich, MA). All of the promoter constructs were sequenced by the DNA Sequencing Core in ICBR at the University of Florida. Sequences of each primer are shown in Supplemental Table S1 (Supplemental Material for this article is available online at the Journal website).
Transient transfection and luciferase assay.
IEC-6 cells were transfected in 24-well plates at ∼80% confluence with 1 μg of Atp7a promoter construct and 150 ng pRL cytomegalovirus (CMV) vector (containing the Renilla luciferase reporter gene) as an internal control using TurboFect in vitro Transfection Reagent (Fermentas, Glen Burnie, MD) according to the manufacturer's protocol. For HIF1α and HIF2α overexpression experiments, 1 μg Atp7a promoter construct, 1 μg HIF1α or HIF2α expression vector, and 150 ng pRL CMV vector were cotransfected into ∼80% confluent IEC-6 cells in 24-well plates. Empty pcDNA3.1 vector was used as a negative control (for the HIFs). Luciferase activity was measured using the Dual Luciferase Assay Kit (Promega) according to the manufacturer's instructions. Briefly, cells were lysed by incubating with 100 μl 1× Passive Cell Lysis Buffer for 20 min with gentle shaking. Cell lysates were collected and 20 μl was used to measure Firefly and Renilla luciferase activity in a tube luminometer (CAN/CAS STD C22.2; Berthold Technologies, Oak Ridge, TN).
Total RNA isolation and real-time quantitative RT-PCR.
Total cellular RNA was isolated by TRIzol reagent (Invitrogen) according to the manufacturer's protocol. RNA concentration was measured by spectrophotometry, and 1 μg RNA was reverse transcribed with the iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA) in a 20-μl reaction. After reverse transcription, the 20-μl reaction was diluted to 40 μl. One microliter was used for the quantitative RT-PCR (qRT-PCR) reaction with SYBR Green qRT-PCR master mix (Bio-Rad), as previously described (8). Sequences of each primer used for qRT-PCR are listed in Supplemental Table S1.
Cytosol and nuclear protein preparation and immunoblot analysis.
Cytosol and nuclear proteins were purified with the Nuclear Extract Kit (Active Motif, Carlsbad, CA) according to the manufacturer's protocol. Protein concentration was determined using the BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). Cytosol or nuclear proteins were resolved by 7% SDS-PAGE followed by electroblotting to a polyvinylidene difluoride membrane which was then blocked in 5% nonfat milk. Membranes were reacted with a commercially available antibody against HIF2α (NB100–132H; Novus Biologicals, Littleton, CO) and one previously developed and characterized against Atp7a (called 54–10) (25). Proteins on the membrane were visualized by enhanced chemiluminescence and autoradiography.
Chromatin immunoprecipitation.
Preconfluent IEC-6 cells with or without CoCl2 treatment grown in 10-cm cell culture dishes were cross-linked with 1× PBS containing 1% formaldehyde for 10 min and quenched with 2 M glycine at a final concentration of 0.2 M for 5 min at room temperature. Nuclei were isolated using the Active Motif kit and lysed with SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris·HCl pH 8.1, and protease inhibitor cocktail). Chromatin was sheered by a Bioruptor instrument (Diagenode, Sparta, NJ) following the manufacturer's instructions, using 45 cycles of 1-min sonication followed by 30 s at 4°C. The size of resulting fragments was determined by agarose gel electrophoresis. Nuclear samples were cleared by centrifugation at 16,000 g for 30 min at 4°C. The chromatin was subsequently immunoprecipitated with HIF2α antibody (NB100–122, Novus Biologicals) using a 1:100 dilution and incubation for 2 h at 4°C with mixing. Protein and chromatin were reverse cross-linked with 1 M NaCl for 6 h at 65°C following 1 h of incubation with RNase A (Fermentas) and overnight incubation with Proteinase K (Fermentas) at 37°C. Chromatin was extracted and purified with phenol/chloroform/isoamyl alcohol followed by precipitation with 100% isopropanol and glycogen. A 1-μl sample was used for qRT-PCR as described above, with primers specific for Atp7a (covering a region containing the HREs and another upstream region with no HREs), Dmt1, and ankyrin repeat domain protein 37 (Ankrd37). Phylogenetic footprinting and sequence alignments were utilized to identify regions in the rat Dmt1 and Ankrd37 genes that were homologous to the experimentally identified HRE-containing regions in the mouse Dmt1 (26) and human Ankrd37 (2) genes. Amplification was also performed with the same primer sets using input DNA (i.e., before immunoprecipitation with the antibody). All primers are listed in Supplemental Table S1.
RESULTS
Regulation of endogenous Atp7a gene expression by hypoxia.
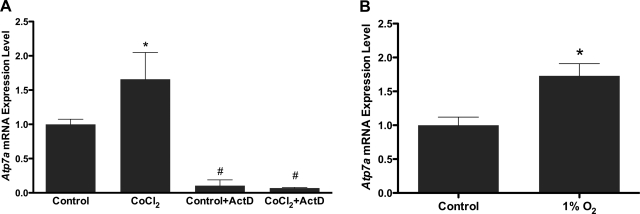
In an attempt to recapitulate the induction of Atp7a seen in the intestinal epithelium of iron-deficient rats, IEC-6 cells (which express Atp7a) (8) were treated with CoCl2 (to mimic hypoxia) or subjected to hypoxia (1% O2). qRT-PCR results demonstrated that endogenous Atp7a mRNA levels increased 1.8- and 1.6-fold following exposure to CoCl2 or 1% O2, respectively (Fig. 1). The induction of Atp7a by CoCl2 was abolished when cells were exposed to actinomycin D, a transcriptional inhibitor (1 μg/mg in H2O), 1 h before and during 16 h of CoCl2 exposure (Fig. 1A).
Fig. 1.
Real-time quantitative RT-PCR (qRT-PCR) analysis of Atp7a mRNA expression in rat intestinal epithelial (IEC-6) cells. Total RNA was extracted from IEC-6 cells that were treated with CoCl2 (A) or cultured in 1% O2 (B). Atp7a expression was normalized to 18S. Similar experiments were performed with actinomycin D (ActD; 1 μg/ml) pretreatment 1 h before and during CoCl2 treatment (A). Each bar represents the mean value ± SD. *P < 0.05, #P < 0.05, as compared with control, unpaired Student's t-test; n = 5.
Investigation of Atp7a transcriptional activity.
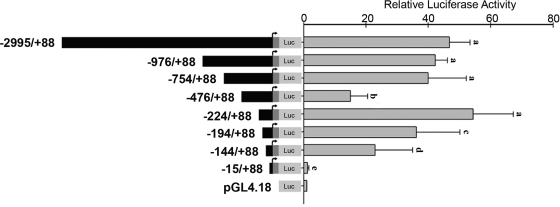
The previously mentioned data suggested that the mechanism of Atp7a induction by hypoxia was transcriptional; regulation of the Atp7a promoter was thus investigated. The following promoter constructs were generated as described in materials and methods: bps −2,995/+88, −976/+88, −754/+88, −476/+88, −224/+88, −194/+88, −144/+88, and −15/+88 (all relative to the transcriptional start site). Forward primers utilized to amplify these fragments were designed from DNA regions that did not have predicted cis-elements. Reporter gene assays demonstrated that the activity of the longest construct (−2,995/+88) had similar activity to the −224/+88 construct (Fig. 2). The −976/+88 and −754/+88 constructs showed similar activity levels. Interestingly, the −476/+88 construct had significantly diminished activity, suggesting that an inhibitory element exists in the region between bps −476 and −224; this element must be inactive or otherwise silenced by upstream elements because the −754/+88 construct retained full promoter activity. Promoter constructs with further 5′-deletions had decreased activity, with the −15/+88 construct having the same background activity as the empty plasmid. Because the −224/+88 construct had full activity, it was utilized for further studies.
Fig. 2.
Analysis of rat Atp7a promoter transcriptional activity. Firefly luciferase (Luc) reporter vectors with various length Atp7a promoter fragments were constructed as described in materials and methods. Each construct was transiently transfected into IEC-6 cells that were preseeded into 24-well plates at ∼80% confluency. pGL4.18 empty vector (1 μg) was used as a control. Twenty-four hours after transfection, firefly and Renilla luciferase activity was measured by a Dual Luciferase Assay System; firefly luciferase activity was normalized by Renilla luciferase activity. Each bar represents the mean value ± SD. Different letters next to bars indicate statistical significance (P < 0.05), between constructs, unpaired Student's t-test; n = 3.
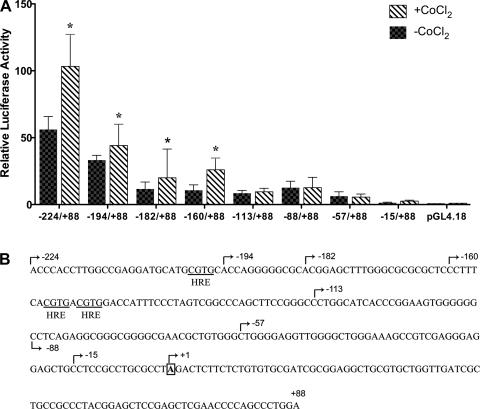
Additional experiments showed that the −2,995/+88 (data not shown) and the −224/+88 constructs were significantly induced by CoCl2 exposure (Fig. 3; sequence indicating 5′-ends of promoter constructs is shown in Fig. 3B). It was also noted that the −224/+88 construct contained three phylogenetically conserved, putative HREs (between bps −224 and −113). Additional 5′-deletions were thus generated to assess the potential role of the HREs in this induction. Promoter constructs containing 5′-ends from bp −224 to −160 all showed induction by CoCl2 exposure, while constructs ending from bp −113 to −57 were not responsive. These observations suggested that the HREs in the Atp7a gene may play a mechanistic role in induction by CoCl2.
Fig. 3.
Deletion analysis of the −224/+88 bp construct containing putative hypoxia response elements (HREs). A: further deletions of −224/+88 promoter construct were made as described in materials and methods. All of the constructs were transfected into preconfluent IEC-6 cells, followed by CoCl2 treatment for 16 h (200 μM). Each bar represents the mean value ± SD. *P < 0.05, +CoCl2 vs. −CoCl2 for each construct, unpaired Student's t-test; n = 3. B: sequence of the Atp7a promoter from −224 bp to +88 bp is shown. Putative HREs are indicated, and the 5′-endmost base of the different deletion constructs is shown.
HIFα overexpression and Atp7a transcriptional activity.
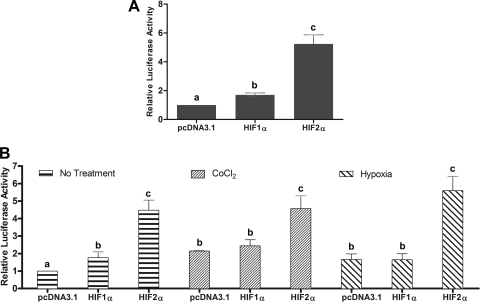
Since CoCl2 treatment and 1% oxygen exposure would presumably induce both HIFα isoforms, as has been described in many cell types (23, 24), and both isoforms bind to highly similar DNA sequences, it was important to determine experimentally which HIF was responsible for inducing the Atp7a gene. Additional experiments were thus performed in IEC-6 cells cotransfected with the −2,995/+88 and −224/+88 Atp7a promoter/luciferase constructs and HIF1α or HIF2α expression plasmids. The HIF1α plasmid contained the human cDNA while the HIF2α plasmid contained the mouse cDNA (26); cDNA expression was driven by the CMV promoter in the pcDNA3.1 plasmid (Invitrogen). HIF1α and HIF2α mRNA levels were significantly increased in cotransfected cells, as determined by qRT-PCR (data not shown). Results showed that HIF1α overexpression had only a minor effect on promoter activity of both constructs (Fig. 4; 1.7- and 1.8-fold, respectively). HIF2α overexpression however, resulted in a dramatic induction of promoter activity of both constructs, 5.2-fold for the −2,995/+88 construct (Fig. 4A) and 4.7-fold for the −224/+88 construct (Fig. 4B; No Treatment). Further studies were performed to determine the effects of HIFα overexpression with CoCl2 treatment or hypoxia (1% O2) on Atp7a promoter activity. This was important because even though there was a significant induction of HIF1α and HIF2α mRNA levels in transfected cells, it is likely that protein expression levels were not as significantly increased, as both HIFα subunits are unstable at normal oxygen tensions (21%). Studies described above were thus repeated using the −224/+88 promoter construct and CoCl2 or 1% O2. Basal promoter activity was increased 1.7- to 2.0-fold with these treatments (Fig. 4B; CoCl2 pcDNA3.1 and Hypoxia pcDNA3.1). HIF1α overexpression had no additional effect on Atp7a promoter activity, while HIF2α overexpression resulted in a significant increase (although it was not greater than activity with HIF2α overexpression under normoxic conditions).
Fig. 4.
Cotransfection of −2,995/+88 and −224/+88 bp constructs with hypoxia-inducible factor (HIF) expression vectors. A: the −2,995/+88 promoter construct was cotransfected with HIF1α or HIF2α expression plasmid, and luciferase activity was determined. B: the −224/+88 promoter construct was cotransfected with HIF1α or HIF2α expression plasmids. Twenty-four hours after transfection, cells were treated with CoCl2 (200 μM) or cultured in 1% O2. Each bar represents the mean value ± SD. In A and B, different letters above bars indicate statistical significance (P < 0.05), between transfection or treatment conditions, unpaired Student's t-test; n = 3.
HIF2α regulates Atp7a expression by direct binding to cis-elements in the promoter.
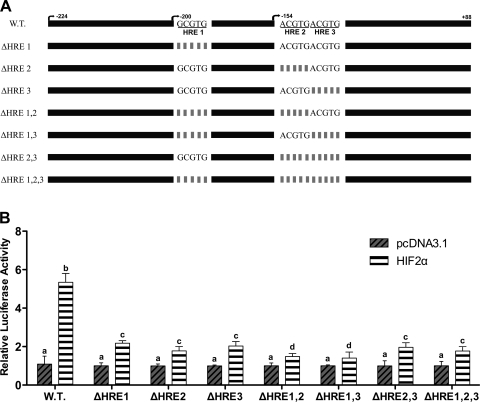
Since Atp7a promoter activity was induced by CoCl2 treatment, hypoxia, and HIF2α overexpression, the next logical step was to consider the role of the three putative HREs in the responsive promoter fragment. These sequences were thus deleted individually or in combination in the −224/+88 promoter construct, and transfection experiments were done in IEC-6 cells. The wild-type construct responded positively to HIF2α overexpression (∼5-fold increase over empty vector-transfected cells; Fig. 5). Deletions of the HREs, however, resulted in a significant decrease in the level of induction (∼2-fold), although there was no noticeable difference between individual deletions of an HRE, double deletions, or deletions of all three HREs. Next, to prove HIF2α regulation of the Atp7a promoter, chromatin immunoprecipitation (ChIP) assays were performed with cross-linked, soluble chromatin isolated from CoCl2-treated and nontreated IEC-6 cells. DNA fragments were found to be ∼500 bp in length after the sonication protocol. qRT-PCR was subsequently performed on DNA samples pulled down by a HIF2α-specific antibody (Fig. 6). As compared with input DNA (before pull down), there was a significant increase in the amount of DNA amplified from the CoCl2-treated samples representing the region of the Atp7a promoter containing the HREs [Fig. 6; Atp7a (+) HRE], while there was no amplification using primers targeting a region of the promoter upstream of the HRE region [Fig. 6; Atp7a (−) HRE]. Increased amplification was also noted from the CoCl2-treated samples for two positive controls, Dmt1 and Ankrd37 (Fig. 6). Previous studies demonstrated HIF2α regulation of Dmt1 in the mammalian intestine (19, 26), and Ankrd37 was also noted to be a HIF target (but regulated by HIF1α) (2).
Fig. 5.
Deletion analysis of the putative HREs. HREs in the −224/+88 promoter construct were individually deleted or deleted in combination. Wild-type (WT) or deleted Atp7a promoter constructs were cotransfected with HIF2α expression plasmid (or empty vector) into IEC-6 cells. Constructs are shown schematically in A, while luciferase assay results are shown in B. Each bar represents the mean value ± SD. Different letters above bars indicate statistical significance (P < 0.05), unpaired Student's t-test; n = 3.
Fig. 6.
Chromatin immunoprecipitation (ChIP) analysis of HIF2α binding to putative HREs in the rat Atp7a promoter. DNA fragments containing cross-linked nuclear proteins were pulled down by HIF2α antibody from IEC-6 cell nuclear extracts prepared from control (untreated) or CoCl2-treated cells. Primers were used to amplify the region of Atp7a containing the HREs [Atp7a (+) HRE] or an upstream region not containing HREs [Atp7a (−) HRE]. Divalent metal transporter 1 (Dmt1) and ankyrin repeat domain protein 37 (Ankrd37) were utilized as positive controls. Input indicates amplification from DNA before pull down with the antibody using Atp7a primers covering the putative HREs. Input samples were also run for all other primer sets, and identical results were obtained (data not shown). Data from two independent experiments are shown.
DISCUSSION
The hypothesis that common regulatory mechanisms are activated during iron deficiency stems from observations showing very similar patterns of gene expression in the intestine of iron-deficient rats (6). Several iron homeostasis-related genes (e.g., Dmt1, Dcytb, TfR1, etc.) were induced across several postnatal developmental stages, and in models of diet-induced and genetic iron deficiency (7). Genes related to copper homeostasis showed a parallel expression pattern. One particularly interesting gene, the Menkes copper ATPase encoding an intestinal copper transporter, was induced in a strikingly similar pattern to the gene encoding the predominant iron transporter, Dmt1 (6). Furthermore, recent published works have demonstrated that Dmt1 and other iron homeostasis-related genes are regulated positively by HIF2α in the intestine during iron deficiency (19, 26). This interesting observation leads to speculation that, during iron deficiency, hepcidin-independent mechanisms are invoked to increase iron absorption, and that identifying additional genes regulated by this mechanism is likely to reveal novel participants in the compensatory response of the intestinal epithelium to iron deprivation. Some of these participants may also play important, hitherto unrecognized roles in iron homeostasis as a part of normal physiology. The current study was thus undertaken to test the hypothesis that Atp7a is a novel HIF2α target in the intestine, which if true, would provide a mechanistic explanation for its strong induction during iron deficiency.
Atp7a is strongly expressed in IEC-6 cells (8), so this cell line was utilized as a model of the mammalian intestinal epithelium. Cells were exposed to 1% oxygen to recapitulate hypoxia which occurs in many tissues, including the intestine, during iron deficiency when hemoglobin levels are significantly reduced. Cells were also treated with CoCl2, which mimics hypoxia by binding to the oxygen-dependent degradation region of the HIFα subunits, which prevents oxygen from mediating their degradation (4). Initial observations demonstrated that expression of the endogenous Atp7a gene was increased in response to both treatments; this induction was abrogated by a transcriptional blocker, indicating regulation at the level of transcription. The rat Atp7a gene promoter was thus cloned and characterized. The transcriptional start site (TSS) had been previously mapped by 5′-RNA ligase-mediated rapid amplification of cDNA ends (RACE; 8); several 5′-endmost bases were identified, suggesting alternative start sites, all within a ∼30 bp region in exon 1 of the gene. Promoter constructs were generated representing almost 3,000 bp 5′ of the TSS, including 88 bp of the transcriptional unit covering all identified 5′-endmost bases (but not containing the putative start codon, which is in exon 2). Promoter constructs were transfected into IEC-6 cells, a well-studied model of the intestinal epithelium.
The longest promoter construct (−2,995/+88) and a shorter construct (−224/+88) both had similar reporter gene activity levels in transient transfection assays, so the latter construct was selected for further analyses. This construct was significantly induced by CoCl2 exposure; shorter deletion constructs containing 5′ upstream sequence to bp −160 also responded, but even shorter constructs were unresponsive. These data suggested the presence of the hypoxia responsive cis-elements (HREs) between bp −160 and −113. Interestingly, these putative HREs were conserved across three mammalian species (rat, mouse, and human), when 1,000 bp promoter sequences were used as input to run the FootPrinter web server (http://genome.cs.mcgill.ca/cgi-bin/FootPrinter3.0/FootPrinterInput2.pl). Further studies were designed to consider the role of these sequences in the induction of Atp7a gene expression by hypoxia.
Two HIFα subunits exist (HIF1α and HIF2α); either can heterodimerize with a binding partner (HIF1β) and translocate to the nucleus to regulate gene transcription. This only occurs under conditions where the α-subunit(s) are stabilized by hypoxia, or when cells are exposed to certain hypoxia mimics such as CoCl2 (24) or deferroxamine (26). To understand which HIFα subunit was important for the transcriptional induction of the Atp7a gene, experiments were performed to assess the effect of HIF1/2α overexpression on promoter activity. Data demonstrated that both the −2,995/+88 and −224/+88 bp constructs were significantly induced by overexpression of HIF2α, while HIF1α overexpression only had a marginal effect on the activity of both constructs. Interestingly, identical studies carried out after cells were exposed to 1% oxygen or CoCl2 demonstrated that there was no additional synergistic effect on promoter activity. This observation suggested that when HIF2α was overexpressed, even under conditions where it would normally be degraded (i.e., normoxia), protein levels must be sufficiently increased so as to overwhelm the degradative machinery. Both HIFα subunits are degraded via interaction with an accessory protein, von Hippel-Lindau (VHL), that targets them for destruction in the lysosome (4, 20). It is further speculated that hypoxia and CoCl2 exposure had no additional influence on Atp7a promoter activity due to the fact that the HIF2α-mediated induction of Atp7a promoter activity was already at a predetermined maximum. It would seem logical that Atp7a induction would have an upper limit so as to avoid inducing copper deficiency in cells, because one important role of the Atp7a protein is in copper efflux.
Additional studies considered the functional role of the HREs in the Atp7a promoter. The HIF2α overexpression system was thus utilized in cotransfection experiments using the wild-type −224/+88 bp promoter construct and additional constructs with specific deletions of the putative HREs. Results showed that deletion of individual HREs or combinatorial deletions in two or three of the HREs all had the effect of minimizing the induction of promoter activity to around 2-fold (as compared with a >5-fold induction of the wild-type construct). Surprisingly, none of the deletions, including deletions in all three sites in combination, resulted in complete loss of induction by HIF2α. This observation suggests that another trans-acting factor(s) is important in the response of the Atp7a gene to hypoxia. We previously speculated that Sp1 or a related G/C-rich binding trans-acting factor could play a role in the genetic response of the intestinal epithelium to iron deprivation (16, 17). Interestingly, the Atp7a promoter contains a phylogenetically conserved Sp1 binding site and it was noted that mutation of this site in the −224/+88 bp promoter construct led to a >80% decrease in promoter activity (data not shown). Further study is needed to investigate the role of this cis-element in controlling Atp7a gene transcription.
Although the HRE deletion studies described above strongly suggested that the Atp7a gene was a direct HIF target, further studies were necessary to independently confirm this observation. ChIP assays were thus performed utilizing a well-characterized HIF2α antibody and cross-linked DNA/protein isolated from control and CoCl2-treated cells. Results showed a substantial increase in Atp7a promoter sequences containing the HREs with CoCl2 exposure, while an upstream promoter region that did not contain the HREs was not detected in either condition. Dmt1 and Ankrd37, two known hypoxia-responsive genes, were utilized as positive controls. These data demonstrate that Atp7a is indeed a direct HIF2α target in the intestinal epithelium.
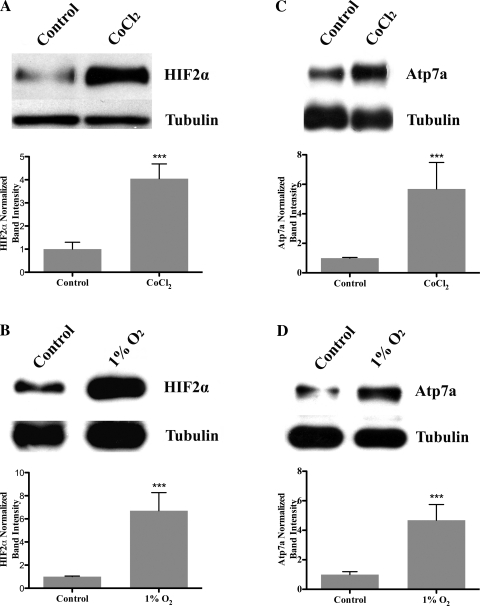
The last set of experiments was designed to address the issue of the potential physiological significance of the results reported in this manuscript. If the induction of Atp7a gene expression by HIF2α is of functional consequence, it would be predicted 1) that increased Atp7a mRNA expression would translate into increased protein levels, and 2) that the HIF2α protein would be stabilized in IEC-6 cells during hypoxia also resulting in increased protein levels. Well-established Atp7a (12, 19) and HIF2α antibodies were thus utilized to perform immunoblots of proteins isolated from control, CoCl2-treated, and hypoxia-exposed cells. In both cases (hypoxia and CoCl2), Atp7a and HIF2α protein levels increased four- to sixfold as compared with untreated cells grown under normoxic conditions (Fig. 7). These observations suggest that the induction of these genes during hypoxia is indeed of physiological relevance.
Fig. 7.
Immunoblot analysis of HIF2α (A and B) and Atp7a (C and D) protein expression in hypoxia and CoCl2-treated IEC-6 cells. Eighty percent preconfluent IEC-6 cells were treated with CoCl2 (200 μM), cultured in 1% O2, or cultured in normoxia (21% O2; control). Nuclear (for HIF2α) and cytosolic (including membrane for Atp7a) proteins were extracted and resolved by 7% SDS-PAGE, and blots were reacted with specific antibodies against these proteins. Images shown are from one representative experiment. Quantitative data from three independent experiments are shown below each representative blot. ***P < 0.05.
It is intriguing to note coordinate regulation of Dmt1, Dcytb, Fpn1, and Atp7a gene transcription by HIF2α in intestinal epithelial cells, representing iron and copper homeostasis-related genes. It has been previously suggested that alterations in copper levels during iron deficiency may be part of the compensatory physiological response to iron deprivation (25). Induction of Atp7a under these conditions, by a conserved regulatory mechanism of proven significance, provides impetus for further consideration of this potential role for copper. As copper increases in the intestinal epithelium during iron deficiency and metallothionein is induced (25), it is tempting to speculate that Atp7a induction could play a relevant role in cellular physiology of enterocytes. There is a documented increase in the production of reactive oxygen species (ROS) in mitochondria with CoCl2 exposure and hypoxia (3, 14), and copper may contribute to the production of ROS (27). It is thus possible that Atp7a plays an essential role to protect cells from the enhancement in membrane lipid peroxidation, DNA damage, and protein oxidation by ROS in the setting of iron deprivation when copper levels increase. This would be mediated by the copper efflux role for Atp7a in intestinal epithelial cells, which could partially mitigate ROS generation.
GRANTS
This work was supported by National Institutes of Health Grant 1R01 DK074867 (to J. F. Collins).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Yatrik Shah at the University of Michigan (Ann Arbor) for sharing the HIF1α and HIF2α expression plasmids.
REFERENCES
- 1. Abboud S, Haile DJ. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J Biol Chem 275: 19906–19912, 2000 [DOI] [PubMed] [Google Scholar]
- 2. Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ. An integrative genomics approach identifies hypoxia inducible factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res 37: 4587–4602, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci USA 95: 11715–11720, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cockman ME, Masson N, Mole DR, Jaakkola P, Chang GW, Clifford SC, Maher ER, Pugh CW, Ratcliffe PJ, Maxwell PH. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J Biol Chem 275: 25733–25741, 2000 [DOI] [PubMed] [Google Scholar]
- 5. Collins JF. Gene chip analyses reveal differential genetic responses to iron deficiency in rat duodenum and jejunum. Biol Res 39: 25–37, 2006 [PMC free article] [PubMed] [Google Scholar]
- 6. Collins JF, Franck CA, Kowdley KV, Ghishan FK. Identification of differentially expressed genes in response to dietary iron deprivation in rat duodenum. Am J Physiol Gastrointest Liver Physiol 288: G964–G971, 2005 [DOI] [PubMed] [Google Scholar]
- 7. Collins JF, Hu Z, Ranganathan PN, Feng D, Garrick LM, Garrick MD, Browne RW. Induction of arachidonate 12-lipoxygenase (Alox15) in intestine of iron-deficient rats correlates with the production of biologically active lipid mediators. Am J Physiol Gastrointest Liver Physiol 294: G948–G962, 2008 [DOI] [PubMed] [Google Scholar]
- 8. Collins JF, Hua P, Lu Y, Ranganathan PN. Alternative splicing of the Menkes copper ATPase (Atp7a) transcript in the rat intestinal epithelium. Am J Physiol Gastrointest Liver Physiol 297: G695–G707, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collins JF, Prohaska JR, Knutson MD. Metabolic crossroads of iron and copper. Nutr Rev 68: 133–147, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collins JF, Wessling-Resnick M, Knutson MD. Hepcidin regulation of iron transport. J Nutr 138: 2284–2288, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Donovan A, Brownlie A, Zhou Y, Shepard J, Pratt SJ, Moynihan J, Paw BH, Drejer A, Barut B, Zapata A, Law TC, Brugnara C, Lux SE, Pinkus GS, Pinkus JL, Kingsley PD, Palis J, Fleming MD, Andrews NC, Zon LI. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 403: 776–781, 2000 [DOI] [PubMed] [Google Scholar]
- 12. Ece A, Uyanik BS, Iscan A, Ertan P, Yigitoglu MR. Increased serum copper and decreased serum zinc levels in children with iron deficiency anemia. Biol Trace Elem Res 59: 31–39, 1997 [DOI] [PubMed] [Google Scholar]
- 13. Fleming MD, Romano MA, Su MA, Garrick LM, Garrick MD, Andrews NC. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc Natl Acad Sci USA 95: 1148–1153, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giordano FJ. Oxygen, oxidative stress, hypoxia, and heart failure. J Clin Invest 115: 500–508, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 388: 482–488, 1997 [DOI] [PubMed] [Google Scholar]
- 16. Hu Z, Gulec S, Collins JF. Cross-species comparison of genomewide gene expression profiles reveals induction of hypoxia-inducible factor-responsive genes in iron-deprived intestinal epithelial cells. Am J Physiol Cell Physiol 299: C930–C938, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu Z, Hu B, Collins JF. Prediction of synergistic transcription factors by function conservation. Genome Biol 8: R257, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee PL, Gelbart T, West C, Halloran C, Beutler E. The human Nramp2 gene: characterization of the gene structure, alternative splicing, promoter region and polymorphisms. Blood Cells Mol Dis 24: 199–215, 1998 [DOI] [PubMed] [Google Scholar]
- 19. Mastrogiannaki M, Matak P, Keith B, Simon MC, Vaulont S, Peyssonnaux C. HIF-2alpha, but not HIF-1alpha, promotes iron absorption in mice. J Clin Invest 119: 1159–1166, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399: 271–275, 1999 [DOI] [PubMed] [Google Scholar]
- 21. McKie AT, Barrow D, Latunde-Dada GO, Rolfs A, Sager G, Mudaly E, Mudaly M, Richardson C, Barlow D, Bomford A, Peters TJ, Raja KB, Shirali S, Hediger MA, Farzaneh F, Simpson RJ. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science 291: 1755–1759, 2001 [DOI] [PubMed] [Google Scholar]
- 22. McKie AT, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, Miret S, Bomford A, Peters TJ, Farzaneh F, Hediger MA, Hentze MW, Simpson RJ. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell 5: 299–309, 2000 [DOI] [PubMed] [Google Scholar]
- 23. Mukhopadhyay CK, Mazumder B, Fox PL. Role of hypoxia-inducible factor-1 in transcriptional activation of ceruloplasmin by iron deficiency. J Biol Chem 275: 21048–21054, 2000 [DOI] [PubMed] [Google Scholar]
- 24. Piret JP, Mottet D, Raes M, Michiels C. CoCl2, a chemical inducer of hypoxia-inducible factor-1, and hypoxia reduce apoptotic cell death in hepatoma cell line HepG2. Ann NY Acad Sci 973: 443–447, 2002 [DOI] [PubMed] [Google Scholar]
- 25. Ravia JJ, Stephen RM, Ghishan FK, Collins JF. Menkes copper ATPase (Atp7a) is a novel metal-responsive gene in rat duodenum, and immunoreactive protein is present on brush-border and basolateral membrane domains. J Biol Chem 280: 36221–36227, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shah YM, Matsubara T, Ito S, Yim SH, Gonzalez FJ. Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab 9: 152–164, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Videla LA, Fernández V, Tapia G, Varela P. Oxidative stress-mediated hepatotoxicity of iron and copper: role of Kupffer cells. Biometals 16: 103–111, 2003 [DOI] [PubMed] [Google Scholar]
- 28. Vulpe CD, Kuo YM, Murphy TL, Cowley L, Askwith C, Libina N, Gitschier J, Anderson GJ. Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat Genet 21: 195–199, 1999 [DOI] [PubMed] [Google Scholar]
- 29. Zoller H, Koch RO, Theurl I, Obrist P, Pietrangelo A, Montosi G, Haile DJ, Vogel W, Weiss G. Expression of the duodenal iron transporters divalent-metal transporter 1 and ferroportin 1 in iron deficiency and iron overload. Gastroenterology 120: 1412–1419, 2001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.