Abstract
Red and white muscles are faced with very different energetic demands. However, it is unclear whether relative mitochondrial protein expression is different between muscle types. Mitochondria from red and white porcine skeletal muscle were isolated with a Percoll gradient. Differences in protein composition were determined using blue native (BN)-PAGE, two-dimensional differential in gel electrophoresis (2D DIGE), optical spectroscopy, and isobaric tag for relative and absolute quantitation (iTRAQ). Complex IV and V activities were compared using BN-PAGE in-gel activity assays, and maximal mitochondrial respiration rates were assessed using pyruvate (P) + malate (M), glutamate (G) + M, and palmitoyl-carnitine (PC) + M. Without the Percoll step, major cytosolic protein contamination was noted for white mitochondria. Upon removal of contamination, very few protein differences were observed between red and white mitochondria. BN-PAGE showed no differences in the subunit composition of Complexes I–V or the activities of Complexes IV and V. iTRAQ analysis detected 358 mitochondrial proteins, 69 statistically different. Physiological significance may be lower: at a 25% difference, 48 proteins were detected; at 50%, 14 proteins were detected; and 3 proteins were detected at a 100%. Thus any changes could be argued to be physiologically modest. One area of difference was fat metabolism where four β-oxidation enzymes were ∼25% higher in red mitochondria. This was correlated with a 40% higher rate of PC+M oxidation in red mitochondria compared with white mitochondria with no differences in P+M and G+M oxidation. These data suggest that metabolic demand differences between red and white muscle fibers are primarily matched by the number of mitochondria and not by significant alterations in the mitochondria themselves.
Keywords: proteomics, fast and slow-twitch muscle, iTRAQ, energetics, oxidative phosphorylation
skeletal muscle-specific oxygen consumption can increase over 100-fold from rest to maximal oxygen uptake (V̇o2max) (52). An additional threefold increase in power output can be achieved during maximal anaerobic exercise (43). Thus skeletal muscle is faced with a wide range of energetic demands dependent on both exercise intensity and duration. Slow and sustained activities such as maintaining posture or low-intensity exercise result in the recruitment of red, oxidative muscle fibers. Conversely, shorter bouts of high-intensity exercise are accomplished by the activation of white, glycolytic myocytes. Accordingly, protein expression in red and white fibers is tuned to meet their respective energetic demands. White muscle is characterized by a predominance of glycolytic enzymes and the fast isoforms of contractile proteins, whereas red muscle has greater abundance of contractile protein slow isoforms and higher oxidative enzyme content (16, 38, 51). The larger oxidative capacity in red muscle is due, at least in part, to a two- to threefold greater mitochondrial content compared with white muscle (19, 24). However, how these mitochondria in different muscle types are poised to perform different tasks by their relative mitochondrial protein expression is unclear.
Despite the large difference in mitochondrial content, red and white myocytes rest at the same tissue-specific oxygen consumption rate (32, 39) and, surprisingly, white muscle fibers maintain a higher resting energetic state (32–34). These results suggest qualitative differences in the regulation of oxidative phosphorylation between red and white muscle. Indeed, Jackman and Willis (24) found that mitochondria isolated from rabbit soleus had greater electron transport chain (ETC) activity and maximal (State 3) respiration with pyruvate + malate (P+M) or palmitoyl carnitine + malate (PC+M) as oxidative substrates than did mitochondria from gracilis muscle. However, Schwerzmann and colleagues (47) found no difference in State 3 respiration with P+M, glutamate + malate (G+M), or succinate in mitochondria isolated from cat soleus and gracilis. Furthermore, while Pande and Blanchaer (40) found no difference with P+M, they did find higher State 3 respiration with PC+M in mitochondria from red muscle compared with white. These conflicting results necessitate a deeper look into potential differences between mitochondria from red and white skeletal muscle.
The goal of this study was to evaluate the nuclear programming of mitochondrial protein expression in red and white skeletal muscle and to relate compositional differences with functional outcomes. We hypothesized that mitochondria from red muscle would have a greater capacity for fat oxidation and higher ETC activity. To test these predictions we examined the mitochondrial proteome using two-dimensional (2D) differential in gel electrophoresis (DIGE) and isobaric tag for relative and absolute quantitation (iTRAQ) labeling with mass spectrometry, assessed ETC complex content, composition, and activity by blue native (BN)-PAGE and optical spectroscopy, and related protein content to function by measuring State 3 respiration with different fuels as well as isocitrate dehydrogenase activity.
MATERIALS AND METHODS
Mitochondrial isolation.
All procedures were approved by the National Heart, Lung, and Blood Institute ACUC and performed in accordance with the guidelines described in the Animal Care and Welfare Act (7 USC 2142-13). Porcine vastus intermedius [red, 70% type I fibers (48)] and gracilis [white, 70% type IIb fibers (53)] muscles were excised upon death and immediately placed in ice-cold isolation buffer (in mM: 280 sucrose, 10 HEPES, 1 K2EDTA, 1 EGTA, pH 7.1). After fat and connective tissue were trimmed off, 60 g of muscle were weighed out, cut into small pieces on a glass Petri dish on ice, placed into 270 ml isolation buffer, and homogenized with a commercial food processor covered by ice for 2 min at 30% power. Protease (50 mg Subtilisin A) was added and the mixture continually stirred for 15 min when 50 ml of isolation buffer plus 1% wt/vol fatty acid-free BSA was added. The solution was centrifuged at 800 g for 10 min at 4°C to pellet down contractile protein and cellular debris. The supernatant was decanted through a double layer of cheesecloth, phenylmethylsulfonyl fluoride (3 mg/40 ml) was added, and the solution was centrifuged at 8,000 g for 10 min to pellet down the mitochondrial fraction. The supernatant was discarded, and 80 ml isolation buffer was added to the pellet. After gently being removed from the side of the centrifuge tube, the pellet was resuspended slowly with four to five passes of a tight-fitting pestle in a dounce homogenizer and centrifuged again at 8,000 g for 10 min. The supernatant was discarded and the pellet was resuspended in 3.4 ml of wash solution (250 mM sucrose, 10 mM HEPES, 0.1% wt/vol fatty acid-free BSA, pH 7.4). Four milliliters of the crude mitochondrial suspension were placed atop 32 ml of Percoll solution (30% vol/vol Percoll, 250 mM sucrose, 10 mM HEPES, 0.1% wt/vol fatty acid-free BSA, pH 7.4) and centrifuged at 68,000 g for 40 min. Typically, two layers formed during the Percoll gradient, a top layer containing cytosolic contaminants and a bottom layer containing the purified mitochondrial fraction. Occasionally, a small layer of blood would form below the mitochondrial layer. The mitochondrial layer was removed using a plastic transfer pipet, and wash solution was added until the volume was 40 ml. After being centrifuged for 10 min at 10,000 g, the supernatant was discarded and the pellet resuspended in 40 ml wash solution followed by a 10-min spin at 10,000 g. The final mitochondrial pellet was suspended in 500 μl solution B (in mM: 137 KCl, 10 HEPES, 2.5 MgCl2, 0.5 K2EDTA, pH 7.1) yielding a protein concentration of 20–35 mg/ml measured using a Bradford Assay (USB Quant Kit, Cleveland, OH).
Optical spectroscopy.
ETC cytochrome content was measured by adding 50 μl mitochondria to 950 μl of a 2% Triton, 100 mM phosphate buffer, pH 7.0, and recording absorbance from 300 to 800 nm (oxidized). Sodium hydrosulfite was then added and absorbance again recorded from 300 to 800 nm (reduced). Cytochrome contents were calculated using the optical absorbance differences between the oxidized and reduced spectra (6) using a custom-written IDL program (Research Systems, Boulder, CO) and millimolar extinction coefficients of 12, 14, and 21 for cytochromes a,a3, b, and c+c1, respectively. Cytochrome b and c spectra were separated using oxidized and reduced spectra from purified cytochrome c.
Native gel electrophoresis.
BN-PAGE was performed using the NativePAGE Novex Bis-Tris System (Invitrogen, Carlsbad, CA) with 4–16% 1 mm bis-Tris gels. Solubilized mitochondria (30 μg) were added to each lane and run at 4°C for 1 h at 150 V and 1.3 h at 250 V. For 2D BN-PAGE, gel lanes were cut and incubated in SDS equilibration solution [50 mM Tris·HCl (pH 8.8), 6 M urea, 30% glycerol, and 2% SDS] plus 0.1 g DTT for 10 min. After equilibration entire lanes or individual complex bands were applied to SDS-PAGE gels. Two whole lanes were placed atop an 8–16% Tris·HCl gel, sealed with 0.5% agarose plus bromophenol blue, and run in TGS buffer (25 mM Tris, 192 mM glycine, 0.1% wt/vol SDS, pH 8.3) at room temperature for 10 min at 50 V and 70 min at 180 V. Individual bands were placed in 16.5% Tris-Tricine gel lanes and sealed and run in TGS buffer as described for the whole gel lanes. After SDS-PAGE, gels were stained in Coomassie blue (CB) (0.75% wt/vol CB, 30% methanol, 3% phosphoric acid) overnight. The following morning, gels were destained in 30% methanol and 3% phosphoric acid and imaged.
In-gel activity assays were performed on BN-PAGE gels immediately after electrophoresis. Complex V activity was measured by incubating gel lanes in preincubation buffer (35 mM Tris·HCl, 270 mM glycine, 14 mM MgSO4, pH 7.8) for 30 min followed by 30 min in incubation buffer [preincubation buffer plus 0.2% wt/vol Pb(NO3)2 and 8 mM ATP, pH 7.8]. Gels were placed in water and then imaged against a CB background with infrared lights. Images shown below were converted to grayscale. Complex IV activity was measured by incubating gel lanes in 50 mM KPO4, pH 7.2, 0.05% wt/vol diaminobenzidine, and 0.0001% wt/vol bovine heart cytochrome c and imaging against a white light background. Activities per mole enzyme were quantified by comparing the optical density of the activity band to the respective protein band for both red and white mitochondria using ImageJ software (National Institutes of Health, Bethesda, MD).
Differential gel electrophoresis.
2D DIGE was performed on whole tissue homogenates and isolated mitochondria from red and white skeletal muscle. Protein (1 mg) was suspended in 250 μl lysis buffer [15 mM Tris·HCl, 7 M urea, 2 M thiourea, and 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) (wt/vol)] and kept on ice with frequent vortexing for 5 min. Samples were then spun at 16,000 g for 10 min at 4°C, and the supernatant was transferred to a fresh microcentrifuge tube. This was repeated two more times. Red (25 μg), white (25 μg), and red + white (12.5 μg each) samples were mixed with 0.3 nmol Cy5, Cy3, or Cy2 dyes, respectively (GE Healthcare, Piscataway, NJ) in 1 μl dimethylformamide and incubated in the dark for 30 min on ice. To quench protein binding to the CyDyes, 1 μl of 10 mM lysine was then added to each sample and incubated on ice for at least 15 min. Samples were combined and added to rehydration solution [7 M urea, 2 M thiourea, 4% CHAPS (wt/vol), 13 mM DTT, 1% pH 3–11NL Pharmalyte (vol/vol), and 2 μl of Destreak reagent] to a final volume of 210 μl and placed on ice for 5 min before being loaded onto 11 cm Immobiline DryStrip gels (pH 3–11NL, Sigma-Aldrich, St. Louis, MO). Isoelectric focusing (IEF) was achieved by active rehydration for 12 h at 30 V followed by stepwise application of 500 V (1 h), 1,000 V (1 h), a gradient to 6,000 V (2 h), and a final step at 6,000 V (1.2 h) for a total of ∼15,000 volt hours (Ettan IPG Phor2). After IEF, gel strips were incubated in SDS equilibration solution plus 0.05 g DTT for 10 min. The gel strip was then placed atop an 8–16% Tris·HCl gel and run as described above for 2D BN-PAGE. Upon completion of SDS-PAGE, gels were imaged on a Typhoon variable mode imager at a resolution of 100 μm.
iTRAQ labeling, mass spectrometry, and protein identification.
Samples were labeled using the iTRAQ Reagents 8plex Kit (Applied Biosystems, Foster City, CA). Mitochondria, 200 μg lysed proteins (see above), were added to six volumes of cold acetone (−20°C) and incubated for 1 h at −20°C. After being spun at 13,000 g for 10 min and the supernatant discarded, 20 μl dissolution buffer, 2 μl reducing reagent, and 1 μl denaturant were added. The samples were then incubated in a shaking water bath at 60°C for 1 h. Cysteine block (1 μl) was added, and the samples were incubated in a room temperature shaking water bath for 10 min followed by the addition of 20 μg trypsin. The trypsinized samples were incubated overnight at 37°C. Isopropanol (50 μl) was added to iTRAQ reagents 113–119 and 121, and each reagent was added to a different sample. Tetraethylammonium bicarbonate was added to bring the pH above 7.5, and the samples were incubated for 2 h at room temperature before combining all samples and drying with a Speedvac. After being reconstituted with 0.1% formic acid (FA), the digest was desalted on a Waters Oasis HLB column and eluted with 60% acetonitrile (ACN) + 0.1% FA. The eluted peptide mixture was then dried by Speedvac.
The sample was reconstituted with 100 μl strong cation exchange buffer A (10 mM KH2PO4, 20% ACN, pH 2.7) and separated on a PolyLC PolySULFOETHYL A column (200 × 2.1 m, 5 μm, 200 Å) with a linear 200 μl/min gradient of 0–70% buffer B (10 mM KH2PO4, 20% ACN, 500 mM KCl, pH 2.7) in 45 min on an Agilent 1200 LC device with Chemstation B.02.01 control software. Fractions were collected each minute and eventually pooled into 24 fractions. After being dried by Speedvac, the fractions were desalted and eluted on a Waters Oasis HLB column as described above. Samples were dried again and reconstituted with 0.1% FA. Liquid chromatography was performed on an Eksigent nanoLC-Ultra 1D plus system (Dublin, CA). Peptide digest was first loaded on a Zorbax 300SB-C18 trap (Agilent, Palo Alto, CA) at 6 μl/min for 5 min and then separated on a PicoFrit analytical column (100 mm long, ID 75 μm, tip ID 10 μm, packed with BetaBasic 5-μm 300 Å particles, New Objective, Woburn, MA) using a 40-min linear gradient of 5–35% ACN in 0.1% FA at a flow rate of 250 nl/min. Mass analysis was carried out on an LTQ Orbitrap Velos (Thermo Fisher Scientific, San Jose, CA) with data-dependent analysis mode, where MS1 scanned full MS mass range from m/z 300 to 2,000 at 30,000 mass resolution, and six HCD MS2 scans were sequentially carried out at resolution of 7500 with 45% collision energy, both in the Orbitrap.
MS/MS spectra from 24 fractions were searched against the Swiss Prot (Swiss Institute of Bioinformatics, updated August 10, 2010, 21241 entries) database, taxonomy Mammalia using a six-processor Mascot (Matrix Science, London, UK; version 2.3) cluster at NIH (http://biospec.nih.gov), with precursor mass tolerance at 20 ppm, fragment ion mass tolerance at 0.05 Da, trypsin enzyme with 2 miscleavages, methyl methanethiosulfonate of cysteine and iTRAQ 8plex of lysine and the NH2-terminus as fixed modifications, and deamidation of asparagine and glutamine, oxidation of methionine, and iTRAQ 8plex of tyrosine as variable modifications. The resulting .dat file was loaded into Scaffold Q+ (version Scaffold_3_00_04, Proteome Software, Portland, OR) to filter and quantify peptides and proteins. Peptide identifications were accepted at 80.0% or higher probability as specified by the Peptide Prophet algorithm (28) and an false discovery rate (FDR) of <1%. Protein identifications were accepted at 90.0% or higher probability and contained at least two identified peptides with FDR <1%. Protein probabilities were assigned by the Protein Prophet algorithm (37). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Peptides were quantified using the centroided reporter ion peak intensity, with minimum of 5% of the highest peak in the spectrum. Intrasample channels were normalized based on the median ratio for each channel across all proteins. The isobaric-tagged samples were normalized by comparing the median protein ratios for the reference channel. Quantitative protein values were derived from only uniquely assigned peptides. Protein quantitative ratios were calculated as the median of all peptide ratios.
Mitochondrial respiration.
Mitochondrial oxygen consumption (Jo) was measured using a Clark-type electrode in a water-jacketed chamber maintained at 37°C. Incubations were carried out in a 1.5-ml final volume of respiration medium containing (in mM) 100 KCl, 50 MOPS, 20 glucose, 10 K2PO4, 10 MgCl2, 1 EGTA, and 0.2% wt/vol BSA, pH 7.00. Mitochondria, 375 μg mitochondrial protein were added to the chamber with either pyruvate + malate (P+M, 1 mM each), glutamate (G) + M (10 mM + 1 mM), or palmitoyl-carnitine (PC) + M (10 μM + 1 mM) as oxidative substrates. Preliminary experiments concluded that 1.3 mM ADP was necessary to elicit maximal State 3 Jo. This amount of ADP results in the consumption of all the oxygen in the chamber while still at State 3. Thus State 4 (resting) respiration was determined before State 3 upon the phosphorylation of a 0.13 mM addition of ADP. The respiratory control ratio (RCR) was determined as the ratio of State 3/State 4 (13).
Isocitrate dehydrogenase activity.
NAD-linked isocitrate dehydrogenase (IDH) activity in red and white skeletal muscle mitochondria was measured by following the appearance of NADH at 340 nm in 0.05% Triton buffer containing (in mM): 50 Tris acetate, 8 isocitrate, 1.3 MnCl2, 0.7 ADP, and 0.33 NAD, pH 7.4 (11). Measurements were made on the same samples used for iTRAQ analysis.
Statistical analyses.
All data were analyzed using a paired Student's t-test. A P value of 0.05 was used to determine significant differences.
RESULTS
White muscle has lower mitochondrial content but similar mitochondrial composition.
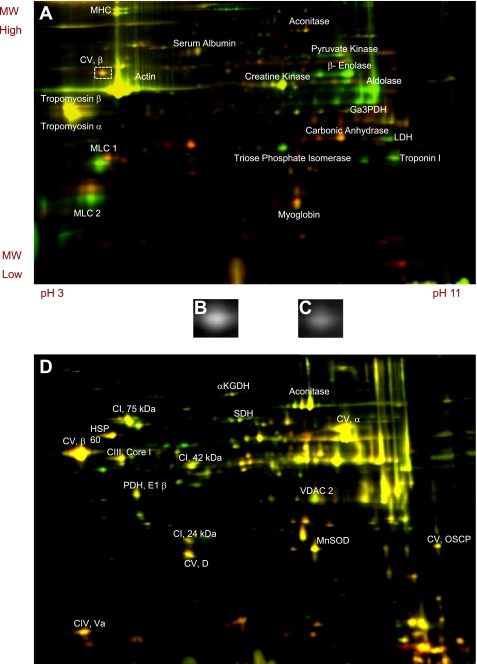
The large difference in protein content between red and white skeletal muscle is depicted in Fig. 1A as most protein spots are red or green, signifying higher protein content in red or white muscle, respectively. ATP synthase β-subunit, the most abundant mitochondrial protein in red and white muscle, is highlighted in Fig. 1, B and C, to demonstrate the twofold difference in mitochondrial content between muscle types. In contrast to the whole muscle DIGE, Fig. 1D demonstrates that mitochondria isolated from red and white skeletal muscle have very similar protein composition because nearly all the protein spots are yellow or the same between mitochondrial types. Proteins listed in Fig. 1, A and D, are those that have been identified many times by our laboratory (4, 5, 8, 20, 27, 41, 45) as well as by Kim et al. (30).
Fig. 1.
Two-dimensional (2D) differential in gel electrophoresis (DIGE) of whole muscle homogenates (A) and isolated mitochondria (D) from red and white porcine skeletal muscle. Red spots signify higher protein content in red muscle, green spots are higher in white muscle, and yellow spots mean equal protein content. Whole muscle ATP synthase, β subunit is highlighted (B is red, C is white) to represent the difference in mitochondrial content between fiber types.
Mitochondria from red and white skeletal muscle have similar oxidative phosphorylation components.
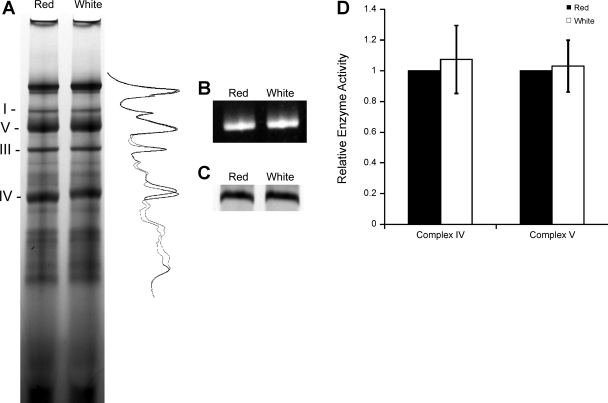
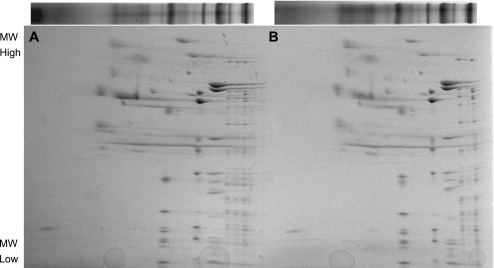
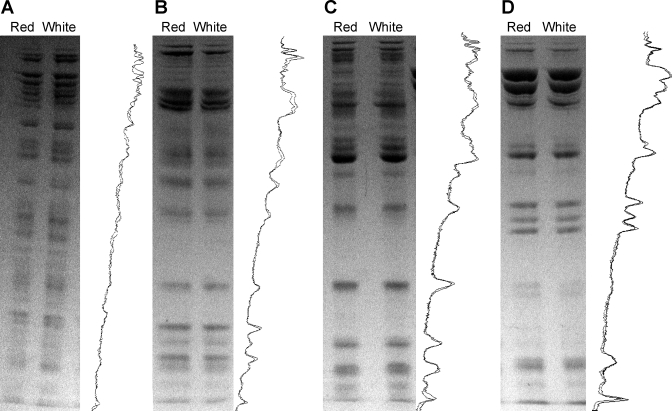
Native protein complexes were evaluated in red and white muscle mitochondria using BN-PAGE. The similarity between red and white mitochondrial protein complexes is shown in Fig. 2A. In addition to similar complex contents, the activities of ETC complexes IV and V were also no different between mitochondrial types (Fig. 2, B–D). Separation of whole BN-PAGE gel lanes by molecular weight (Fig. 3) suggests little difference in the composition of each native complex between red and white skeletal muscle mitochondria. Examining the composition of ETC Complexes I, III, IV, and V individually also yields no differences (Fig. 4).
Fig. 2.
Native protein complexes in red and white muscle mitochondria were resolved by blue native (BN)-PAGE (A). Whole gel lane density traces for both mitochondrial types are shown to the right of the gel. Activities of Complexes IV (C) and V (B) were assessed with BN-PAGE in-gel activity assays. Activities of Complex IV and Complex V per mole of enzyme relative to red mitochondria are shown (D).
Fig. 3.
Composition of native complex components by 2D BN-PAGE. Gel lanes as shown in Fig. 2 were separated in the second dimension by molecular weight (MW) with an SDS buffer. A: red skeletal muscle mitochondria. B: white skeletal muscle mitochondria.
Fig. 4.
Composition of oxidative phosphorylation components using 2D BN-PAGE. Individual bands corresponding to electron transport chain (ETC) Complexes I (A), III (B), IV (C), and V (D) were cut from gel lanes as shown in Fig. 2 and separated by molecular weight using an SDS buffer. Whole gel lane density traces for both mitochondrial types are shown to the right of each gel.
Contents of ETC cytochromes a,a3, b, and c+c1 per milligram of mitochondrial protein were quantified based on optical density. Table 1 shows that both red and white skeletal muscle mitochondria have about 1 nmol cytochrome a,a3/mg of mitochondria and that there is no difference in cytochrome b or c+c1 contents.
Table 1.
Cytochrome-to-protein ratios in isolated mitochondria
| Cytochrome a,a3, nmol/mg | Cytochrome b, nmol/mg | Cytochrome c+c1, nmol/mg | |
|---|---|---|---|
| Red muscle | 1.15 ± 0.07 | 0.80 ± 0.07 | 0.62 ± 0.05 |
| White muscle | 1.05 ± 0.05 | 0.73 ± 0.04 | 0.54 ± 0.03 |
Values are means ± SE. n = 19 and 20 for white and red muscle mitochondria, respectively.
iTRAQ quantification suggests modest protein differences between red and white skeletal muscle mitochondria.
There were 485 unique proteins identified and quantified with iTRAQ labeling and mass spectrometry. Of those proteins, 481 were identified in all six samples yielding three red-to-white protein ratios. Two ratios were obtained for the remaining 4 proteins. Of the 485 proteins, 358 (74%) were considered mitochondrial proteins. Though mass spectrometry was sensitive enough to detect 127 nonmitochondrial proteins, the abundance of most was low enough to not be seen in 2D DIGE gels (Fig. 1B). Indeed, 88% of the spectra originated from mitochondrial proteins. The largest sources of nonmitochondrial spectra were trypsin (2.4%) and myosin isoforms (1.6%). Nonmitochondrial contamination was no different between mitochondrial types.
There were significant differences in 69 proteins between red and white mitochondria. Of those 69, 48 proteins were at least 25% different and are shown in Table 2. The only apparent whole pathway difference between mitochondrial types, whether employing the 25% threshold or not, was in fat metabolism. Proteins associated with all four β-oxidation enzymes were at least 25% higher in red muscle mitochondria. A full list of identified proteins, ratios, and P values can be found in Supplemental Materials online at the AJP-Cell Physiol website.
Table 2.
iTRAQ ratios for significantly and ±25% different proteins in mitochondria from red and white muscle
| Identified Proteins | UniProtKB Entry Name | Taxonomy | Molecular Mass, kDa | White/Red Ratio | P |
|---|---|---|---|---|---|
| Fat Metabolism | |||||
| Acetyl-coenzyme A synthetase 2-like, mitochondrial | ACS2L_HUMAN | Homo sapiens | 75 | 0.60 ± 0.08 | 0.038 |
| Mitochondrial carnitine/acylcarnitine carrier protein | MCAT_HUMAN | Homo sapiens | 33 | 1.29 ± 0.10 | 0.035 |
| Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADV_BOVIN | Bos taurus | 71 | 0.74 ± 0.04 | 0.000 |
| Long-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADL_PIG | Sus scrofa | 48 | 0.67 ± 0.02 | 0.004 |
| Short-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADS_PIG | Sus scrofa | 45 | 0.79 ± 0.05 | 0.011 |
| Enoyl-CoA hydratase domain-containing protein 2, mitochondrial | ECHD2_BOVIN | Bos taurus | 32 | 0.71 ± 0.08 | 0.012 |
| Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | HCDH_PIG | Sus scrofa | 34 | 0.80 ± 0.04 | 0.031 |
| 3-ketoacyl-CoA thiolase, mitochondrial | THIM_BOVIN | Bos taurus | 42 | 0.79 ± 0.04 | 0.002 |
| Peroxisomal 3,2-trans-enoyl-CoA isomerase | PECI_MOUSE | Mus musculus | 43 | 0.73 ± 0.02 | 0.007 |
| Trifunctional enzyme subunit alpha, mitochondrial | ECHA_PIG | Sus scrofa | 83 | 0.76 ± 0.04 | 0.002 |
| Oxidative Phosphorylation | |||||
| NADH-ubiquinone oxidoreductase chain 2 | NU2M_PIG | Sus scrofa | 39 | 1.30 ± 0.01 | 0.002 |
| COX assembly mitochondrial protein homolog | COXAM_BOVIN | Bos taurus | 13 | 1.99 ± 0.17 | 0.028 |
| ATP synthase mitochondrial F1 complex assembly factor 2 | ATPF2_BOVIN | Bos taurus | 33 | 0.77 ± 0.02 | 0.007 |
| NADP-Linked Reactions | |||||
| NADPH-dependent carbonyl reductase/NADP-retinol dehydrogenase | DHRS4_PIG | Sus scrofa | 28 | 0.75 ± 0.04 | 0.021 |
| Isocitrate dehydrogenase [NADP], mitochondrial | IDHP_PIG | Sus scrofa | 48 | 0.79 ± 0.05 | 0.008 |
| NADP-dependent malic enzyme, mitochondrial | MAON_HUMAN | Homo sapiens | 67 | 0.80 ± 0.01 | 0.001 |
| TCA Cycle | |||||
| Pyruvate dehydrogenase protein X component | ODPX_BOVIN | Bos taurus | 54 | 0.80 ± 0.06 | 0.020 |
| Isocitrate dehydrogenase [NAD] subunit gamma, mitochondrial | IDH3G_PIG | Sus scrofa | 11 | 1.85 ± 0.24 | 0.016 |
| Amino Acid Metabolism | |||||
| Branched-chain alpha-ketoacid dehydrogenase kinase | BCKD_HUMAN | Homo sapiens | 46 | 0.80 ± 0.04 | 0.031 |
| Branched-chain alpha-ketoacid dehydrogenase E1 component beta chain | ODBB_BOVIN | Bos taurus | 43 | 1.77 ± 0.13 | 0.030 |
| Alanine aminotransferase 1 | ALAT1_BOVIN | Bos taurus | 55 | 0.72 ± 0.06 | 0.049 |
| 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial | HIBCH_BOVIN | Bos taurus | 43 | 1.54 ± 0.12 | 0.045 |
| Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial | MCCB_MOUSE | Mus musculus | 61 | 1.27 ± 0.03 | 0.012 |
| Kynurenine–oxoglutarate transaminase 1 | KAT1_HUMAN | Homo sapiens | 48 | 0.70 ± 0.06 | 0.041 |
| Ribosomal Proteins | |||||
| 28S ribosomal protein S6, mitochondrial | RT06_BOVIN | Bos taurus | 14 | 0.73 ± 0.05 | 0.001 |
| 28S ribosomal protein S16, mitochondrial | RT16_HUMAN | Homo sapiens | 15 | 0.74 ± 0.07 | 0.001 |
| 39S ribosomal protein L10, mitochondrial | RM10_BOVIN | Bos taurus | 29 | 2.03 ± 0.18 | 0.029 |
| 39S ribosomal protein L49, mitochondrial | RM49_BOVIN | Bos taurus | 19 | 1.30 ± 0.06 | 0.037 |
| Membrane/Transport | |||||
| ATP-binding cassette sub-family B member 8, mitochondrial | ABCB8_MOUSE | Mus musculus | 78 | 1.68 ± 0.09 | 0.018 |
| ATP-binding cassette sub-family B member 10, mitochondrial | ABCBA_HUMAN | Homo sapiens | 79 | 0.68 ± 0.04 | 0.018 |
| ATP-binding cassette sub-family C member 2 | MRP2_MOUSE | Mus musculus | 174 | 0.80 ± 0.03 | 0.019 |
| Calcium-binding mitochondrial carrier protein Aralar2 | CMC2_HUMAN | Homo sapiens | 74 | 0.61 ± 0.03 | 0.005 |
| Calcium-binding mitochondrial carrier protein SCaMC-1 | SCMC1_BOVIN | Bos taurus | 53 | 1.98 ± 0.10 | 0.010 |
| Metaxin-2 | MTX2_PIG | Sus scrofa | 30 | 1.33 ± 0.04 | 0.015 |
| MOSC domain-containing protein 2, mitochondrial | MOSC2_BOVIN | Bos taurus | 37 | 1.53 ± 0.19 | 0.037 |
| Mitochondrial inner membrane protein OXA1L | OXA1L_BOVIN | Bos taurus | 49 | 1.33 ± 0.02 | 0.003 |
| Other | |||||
| Glycerol-3-phosphate dehydrogenase, mitochondrial | GPDM_BOVIN | Bos taurus | 81 | 1.62 ± 0.15 | 0.009 |
| Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial | MMSA_RAT | Rattus norvegicus | 58 | 0.72 ± 0.07 | 0.010 |
| Phospholipid hydroperoxide glutathione peroxidase, mitochondrial | GPX4_HUMAN | Homo sapiens | 22 | 0.47 ± 0.01 | 0.001 |
| Glutaredoxin-related protein 5, mitochondrial | GLRX5_HUMAN | Homo sapiens | 17 | 1.36 ± 0.02 | 0.004 |
| Ubiquinone biosynthesis protein COQ4 homolog, mitochondrial | COQ4_HUMAN | Homo sapiens | 30 | 1.27 ± 0.03 | 0.012 |
| Histidine triad nucleotide-binding protein 2, mitochondrial | HINT2_BOVIN | Bos taurus | 17 | 0.79 ± 0.05 | 0.049 |
| ES1 protein homolog, mitochondrial | ES1_HUMAN | Homo sapiens | 28 | 0.80 ± 0.04 | 0.033 |
| Inorganic pyrophosphatase 2, mitochondrial | IPYR2_MOUSE | Mus musculus | 38 | 0.77 ± 0.05 | 0.045 |
| Complement component 1 Q subcomponent-binding protein, mitochondrial | C1QBP_BOVIN | Bos taurus | 31 | 1.40 ± 0.04 | 0.012 |
| Coiled-coil-helix-coiled-coil-helix domain-containing protein 10, mitochondrial | CHC10_HUMAN | Homo sapiens | 14 | 1.40 ± 0.09 | 0.049 |
| Polymerase I and transcript release factor | PTRF_HUMAN | Homo sapiens | 43 | 2.49 ± 0.14 | 0.008 |
| Brain protein 44 | BR44_HUMAN | Homo sapiens | 14 | 0.80 ± 0.04 | 0.031 |
Red and white muscle mitochondrial samples (n = 3 pairs) were isobaric tag for relative and absolute quantitation (iTRAQ) labeled and analyzed by mass spectrometry. Shown above are the 48 mitochondrial proteins that were significantly and at least 25% different between red and white skeletal muscle mitochondria. The full list of proteins and ratios can be found in Supplemental Materials online at the AJP-Cell Physiol website. White/red values are means ± SE.
Mitochondrial function correlates with protein abundance.
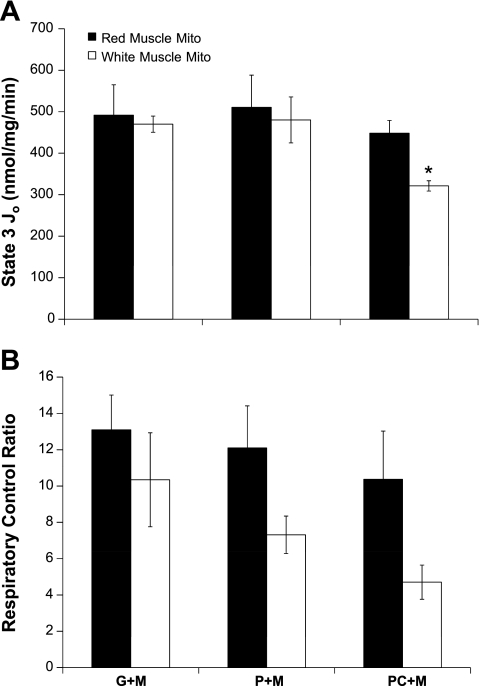
For red skeletal muscle mitochondria, there was no difference in maximal respiration or RCR between G+M (491.7 ± 73.2 and 13.1 ± 1.9), P+M (510.5 ± 77.7 and 12.1 ± 2.3), or PC+M (448.2 ± 30.9 and 10.4 ± 2.4). Conversely, white muscle mitochondria respiring on PC+M had lower State 3 Jo and RCR (321.2 ± 12.4 and 4.7 ± 0.9) than with G+M (470.0 ± 19.5 and 10.3 ± 2.6). Figure 5 also shows that white muscle mitochondria had a 40% lower State 3 Jo with PC+M compared with red muscle mitochondria. Differences in RCR between red and white mitochondria did not reach statistical significance.
Fig. 5.
Mitochondrial respiration in the presence of different oxidative substrates. Mitochondria (0.25–0.5 mg mito protein/ml) were incubated with 10 mM Pi and either glutamate + malate (G+M), pyruvate + malate (P+M), or palmitoyl-carnitine + malate (PC+M). A: maximal (State 3) respiration of red and white skeletal muscle mitochondria. B: respiratory control ratio (State 3/State 4) of red and white skeletal muscle mitochondria. *Significantly different from red muscle mitochondria.
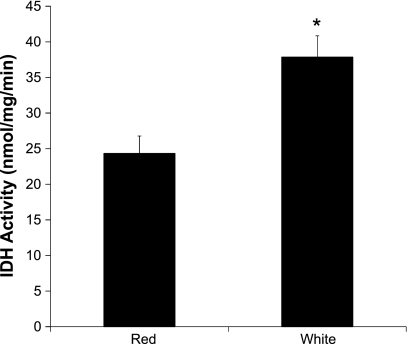
Previous reports showed higher NAD-linked IDH activity in white mitochondria compared with red (21, 24). iTRAQ analysis here showed a less than 12% difference in NAD-IDH subunits α and β, however, the γ subunit was 85 ± 24% higher in white mitochondria. We measured NAD-IDH activity on the same samples used for iTRAQ analysis and found that white muscle mitochondria had 57 ± 10% higher activity than red muscle mitochondria (Fig. 6).
Fig. 6.
NAD-linked isocitrate dehydrogenase (IDH) activity in red and white skeletal muscle mitochondria. NADH appearance was measured spectrophotometrically on the same samples used for isobaric tag for relative and absolute quantitation (iTRAQ) analysis. *Significantly different from red muscle mitochondria.
DISCUSSION
The current study demonstrates that protein content and function are similar between mitochondria from red and white porcine skeletal muscle. Red and white skeletal muscle mitochondria contain ∼1 nmol of cytochrome a,a3 per milligram of protein (Table 1), similar to that value found in porcine heart (22) and are capable of nearly identical maximal respiration rates (Fig. 5). The primary difference found between red and white muscle mitochondria was in fat oxidation. Protein expression of the entire β-oxidation pathway was upregulated in red mitochondria (Table 2), which resulted in a higher rate of maximal respiration with a fat fuel source compared with white mitochondria (Fig. 5).
A key feature of this study was the use of a Percoll gradient during the mitochondrial isolation process. Without the Percoll gradient step, major cytosolic protein contamination was noted in the 2D DIGE of the white but not red muscle mitochondria (results not shown). The disproportionate contamination between mitochondrial types before using the Percoll gradient may explain some of the discrepancies in the literature. Previous studies looking at differences in maximal respiration between mitochondria from red and white muscle have found State 3 respiration with P+M higher in red (24, 36) or no different (2, 7, 40, 47, 54); with G+M, higher in white (10) or no different (47); with succinate, higher in red (7) or no different (47); and with PC+M, higher in red (24, 36, 40). Of these studies, only Schwerzmann et al. (47) reported using a Percoll gradient, and, similar to our results (Fig. 5), they found no difference in State 3 respiration between red and white cat muscle mitochondria with P+M or G+M. These results suggest that great care must be taken when comparing skeletal muscle mitochondrial samples as results normalized to milligram of protein will always be skewed toward a lesser contaminated sample. Marker enzyme activities, such as citrate synthase, are often used in an attempt to avoid this problem; however, citrate synthase activity is reportedly different between red and white muscle mitochondria (24) making it a potentially unreliable marker for these types of studies. As such, the use of a purification step such as a Percoll gradient or a protein screen such as a 2D DIGE is recommended when comparing mitochondrial samples from muscles that may differ in fiber type composition.
Increased expression of glycolytic and fast isoforms of contractile proteins and decreased expression of oxidative enzymes in white compared with red skeletal muscle has been well-characterized previously (9, 16, 23, 29, 38, 51), a profile similar to which was found between the red and white porcine muscles used in this study (Fig. 1A). However, because mitochondria can comprise <3% of skeletal muscle volume density (19), it is difficult to discern relative mitochondrial protein expression differences from mitochondrial content differences in the whole muscle studies. Using our isolated mitochondrial preparation, we were able to compare and quantify the relative expression of 358 mitochondrial proteins from red and white skeletal muscle. Only 3 of 89 detected proteins associated with ETC Complexes I–V were significantly and 25% different between red and white mitochondria (Table 2). Further support for these results is shown by Figs. 1D and 2–4 in which no apparent differences in the abundance of oxidative phosphorylation complexes or their respective subunits are found. Moreover, the contents of cytochromes a, b, and c were no different between red and white muscle mitochondria (Table 1).
It is important to point out that this protein content data is only one of the elements in the complex control of mitochondria function. Though the protein contents might be very similar, the regulatory mechanisms including allosteric factors, substrates, and posttranslational modifications can completely change the flux patterns. Thus this analysis of protein content can only provide information on the potential flux of a given pathway. The actual physiological flux is likely also influenced by a host of other factors not fully understood in this complex network. One approach to look for posttranslational modifications is to assay extracted activity of the enzymes involved. To begin this process, we assayed the activity of Complex IV and V activity as well as maximal respiration of the isolated mitochondria. With the use of BN-PAGE in-gel activity assays, Complex IV and V activities per mole of enzyme were no different between mitochondria from red and white skeletal muscle (Fig. 2, B–D). Given that both Complex IV and V have been suggested to be sites of flux control (18, 55), perhaps it is not surprising then that State 3 respiration with both P+M and G+M were no different between red and white mitochondria (Fig. 5). It should be stressed that these assays were conducted on mitochondria extracted from anesthetized resting muscle. Thus any persistent posttranslational modifications of the complexes related to work load will be in the resting muscle state. As previously shown in the heart (46), extracted complex V activity increases with workload consistent with a posttranslational modification. Whether this occurs in skeletal muscle has yet to be established. These functional results combined with the proteomic data above and previous findings that the Km values for both ADP and Pi are no different between mitochondria isolated from red and white muscle (36, 44) suggest that many of the acute regulatory mechanisms within these mitochondria are also similarly poised when isolated from resting muscle. This similarity in protein content and activity level per mole enzyme, for those we queried, suggest that the configuration of mitochondria in these two muscle types is nearly identical to produce ATP as a function of mitochondrial volume. This might represent an “optimal” ratio of mitochondrial volume to ATP production to optimize the use of cell volume for energy support of fiber contraction. The use of a minimal volume of the “optimal” mitochondria would reserve the area within the cell to optimize the cross-sectional area of fibers for contractile power, independent of the muscle action.
Tissue-specific differences in function are generally met with upregulation of protein expression of entire pathways (26). The increased expression of all β-oxidation enzymes in red compared with white muscle mitochondria shown here falls in line with this theory. Only 2 of 22 detected tricarboxylic acid cycle and pyruvate dehydrogenase proteins were significantly and 25% different between mitochondrial types. These results combined with the similarities in oxidative phosphorylation machinery and State 3 respiration with P+M make the greater content and activity of NAD-linked IDH in white mitochondria a curious finding. Jackman and Willis (24) also found increased IDH activity in white muscle mitochondria, suggesting that regulation of NADH supply to the ETC may be different between red and white muscle mitochondria. Higher IDH activity would predict a greater NADH level and may explain why reactive oxygen species production is higher in white muscle mitochondria (3, 10, 42, 49). Flux through IDH also results in formation of α-ketoglutarate (AKG), which plays an important role in amino acid metabolism as the α-keto acid pair for glutamate in transaminase reactions occurring in both mitochondria and the cytosol. Hutson (22) showed that white skeletal muscle has greater relative activity of branched chain aminotransferase in the cytosol than in mitochondria, whereas red muscle has branched chain aminotransferase activity exclusively in the mitochondria. Hutson (22) also suggests that muscles with lower mitochondrial content may release more keto acids than those with higher mitochondrial content. Thus the higher IDH activity in white muscle mitochondria may be a mechanism for increased AKG production to be used by cytosolic transaminase reactions.
These studies were conducted using porcine vastus intermedius [70% type I (48)] and gracilis [70% type IIb (53)] muscles. We initially attempted to use mitochondria isolated from rabbit soleus and gracilis [98% and 99% type I and IIb, respectively (39)], but the yield after the mitochondrial purification process was too small to proceed with the full proteomic analysis. We were, however, able to measure the cytochrome a content in rabbit soleus and gracilis homogenates and isolated mitochondria. Just as with porcine muscle, cytochrome a content per milligram of mitochondria was similar in both muscle types in the rabbit. Cytochrome a content per gram wet weight, on the other hand, was 6.5-fold higher in rabbit soleus versus gracilis compared with a twofold difference between red and white porcine muscle. Thus it is likely that the differences detected here with regard to substrate oxidation would be three to four times as large in rabbit muscle mitochondria, though we would still predict similar content and activities of oxidative phosphorylation proteins between muscle types. In fact, in our initial studies on rabbit muscle, there were no differences in the stoichiometry between ETC complexes or in the activities of Complexes IV or V per mole of enzyme (as in Fig. 2) between soleus and gracilis mitochondria (results not shown). Furthermore, Leary et al. (35) reported no differences in oxidative phosphorylation enzyme activities or P+M State 3 respiration between red and white muscle mitochondria from rainbow trout as well as increased fat oxidation enzyme activity in red mitochondria when normalized to cytochrome a. The collective findings from the pig and rabbit here, and fish (35) and cat (47) previously, suggest that the similarity in oxidative phosphorylation components between red and white muscle mitochondria may be found across a wide range of species, though it is possible that smaller species, such as rats or mice, may hold larger differences.
This primary focus of this study was to assess differences in the nuclear programming of mitochondrial protein expression between red and white skeletal muscle. The similarities in protein composition found here suggest there is little difference in metabolic capacity between red and white mitochondria. However, differences in metabolic control may still exist. For example, it is fully possible, and perhaps likely, that there are differences in posttranslational modifications such as phosphorylation, acetylation, or S-nitrosylation between mitochondrial types. Indeed, Feng et al. (14) reported twice as many carbonylated proteins in white versus red muscle mitochondria. However, as the functional significance of any of these posttranslational modifications is still largely unclear, they were left outside the scope of this project. The metabolic poise of red and white skeletal muscle, respectively, may also contribute to differences in metabolic control, which has been shown to change as a function of rate, both within mitochondria (17, 31), as well as for the mitochondrial contribution to cellular ATP free energy (25). White muscle fibers are recruited for shorter durations and thus result in mitochondria spending more time near State 4 where the proton leak exerts greater control over respiration (17, 31). In fact, white muscle mitochondria are reported to have a higher rate of proton leak (35). Despite these probable differences in metabolic control in vivo, the similarity in metabolic capacity between red and white muscle mitochondria suggests that nuclear programming of mitochondrial protein expression may be configured to meet maximal energy demands per unit volume rather than the specific patterns of muscle activation.
Mitochondria do not exist only as isolated organelles in vivo, but also as reticular networks located both near the sarcolemma and within skeletal muscle fibers. The isolation procedure used here is designed to recover both intermyofibrillar (IMF) and subsarcolemmal (SS) mitochondria, though the percent contribution of each to the final yield is unknown. IMF mitochondria are reported to comprise a greater proportion of the total mitochondrial population in white versus red skeletal muscle (19). The lack of difference in protein composition between red and white mitochondria may indicate that either a similar proportion of IMF and SS mitochondria were isolated from both red and white mitochondria or that IMF and SS mitochondria are not largely different themselves, though the latter would contradict a number of previous reports (1, 12, 15, 50). In addition, while disruption of the mitochondrial reticulum during the isolation process may remove any potential effect fission and fusion events have on mitochondrial composition and/or function, the lack of difference in fission/fusion proteins OPA1, FIS1, PARL, and MTP18 (Supplemental Materials) suggests these processes may also be similar between red and white skeletal muscle mitochondria.
The remarkable similarity of the oxidative phosphorylation complexes on a milligram of protein basis between the red and white muscle mitochondria suggest that the volume packing of these energy conversion components are nearly identical despite the large difference in peak aerobic ATP demands of the muscles. Thus the differences in the ATP requirements of these different muscles are accomplished by varying the number of mitochondria with essentially identical ATP production capacities. This observation suggests that the volume packing of mitochondrial oxidative phosphorylation elements in these muscles is near optimal providing the maximum area for muscle contraction elements for power with minimal volume for energetic support.
In summary, protein content and function are similar between mitochondria from red and white skeletal muscle. Both red and white muscle mitochondria contain about 1 nmol cytochrome a per milligram of mitochondrial protein, though there is a slight upregulation in fat oxidation in red muscle mitochondria. This suggests that differences in metabolic need between fiber types are met by the number of mitochondria rather than modifying the composition of individual mitochondria. There may be an optimal mitochondrial configuration to support oxidative phosphorylation in muscle.
GRANTS
This study was supported by Intramural Funding of the Division of Intramural Research, National Heart, Lung, and Blood Institute.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Stephanie French, Darci Phillips, and David Chess for technical assistance and Yong Chen for work with the mass spectrometry samples.
REFERENCES
- 1. Adhihetty PJ, Ljubicic V, Menzies KJ, Hood DA. Differential susceptibility of subsarcolemmal and intermyofibrillar mitochondria to apoptotic stimuli. Am J Physiol Cell Physiol 289: C994–C1001, 2005 [DOI] [PubMed] [Google Scholar]
- 2. Alvarado Rigault MY, Blanchaer MC. Respiration and oxidative phosphorylation by mitochondria of red and white skeletal muscle. Can J Biochem 48: 27–32, 1970 [DOI] [PubMed] [Google Scholar]
- 3. Anderson EJ, Neufer PD. Type II skeletal myofibers possess unique properties that potentiate mitochondrial H2O2 generation. Am J Physiol Cell Physiol 290: C844–C851, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Aponte AM, Phillips D, Harris RA, Blinova K, French S, Johnson DT, Balaban RS. 32P labeling of protein phosphorylation and metabolite association in the mitochondria matrix. Methods Enzymol 457: 63–80, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aponte AM, Phillips D, Hopper RK, Johnson DT, Harris RA, Blinova K, Boja ES, French S, Balaban RS. Use of (32)P to study dynamics of the mitochondrial phosphoproteome. J Proteome Res 8: 2679–2695, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balaban RS, Mootha VK, Arai A. Spectroscopic determination of cytochrome c oxidase content in tissues containing myoglobin or hemoglobin. Anal Biochem 237: 274–278, 1996 [DOI] [PubMed] [Google Scholar]
- 7. Blanchaer MC. Respiration of mitochondria of red and white skeletal muscle. Am J Physiol 206: 1015–1020, 1964 [DOI] [PubMed] [Google Scholar]
- 8. Boja ES, Phillips D, French SA, Harris RA, Balaban RS. Quantitative mitochondrial phosphoproteomics using iTRAQ on an LTQ-Orbitrap with high energy collision dissociation. J Proteome Res 8: 4665–4675, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cai D, Li M, Lee K, Wong W, Chan K. Age-related changes of aqueous protein profiles in rat fast and slow twitch skeletal muscles. Electrophoresis 21: 465–472, 2000 [DOI] [PubMed] [Google Scholar]
- 10. Capel F, Buffiere C, Patureau Mirand P, Mosoni L. Differential variation of mitochondrial H2O2 release during aging in oxidative and glycolytic muscles in rats. Mech Ageing Dev 125: 367–373, 2004 [DOI] [PubMed] [Google Scholar]
- 11. Chen RF, Plaut GW. Activation and inhibition of Dpn-linked isocitrate dehydrogenase of heart by certain nucleotides. Biochemistry 2: 1023–1032, 1963 [DOI] [PubMed] [Google Scholar]
- 12. Cogswell AM, Stevens RJ, Hood DA. Properties of skeletal muscle mitochondria isolated from subsarcolemmal and intermyofibrillar regions. Am J Physiol Cell Physiol 264: C383–C389, 1993 [DOI] [PubMed] [Google Scholar]
- 13. Estabrook R. Mitochondrial respiratory control and the polarographic measurement of ADP/O ratios. Meth Enzymol 10: 41–47, 1967 [Google Scholar]
- 14. Feng J, Xie H, Meany DL, Thompson LV, Arriaga EA, Griffin TJ. Quantitative proteomic profiling of muscle type-dependent and age-dependent protein carbonylation in rat skeletal muscle mitochondria. J Gerontol A Biol Sci Med Sci 63: 1137–1152, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferreira R, Vitorino R, Alves RM, Appell HJ, Powers SK, Duarte JA, Amado F. Subsarcolemmal and intermyofibrillar mitochondria proteome differences disclose functional specializations in skeletal muscle. Proteomics 10: 3142–3154, 2010 [DOI] [PubMed] [Google Scholar]
- 16. Gelfi C, Vigano A, De Palma S, Ripamonti M, Begum S, Cerretelli P, Wait R. 2-D protein maps of rat gastrocnemius and soleus muscles: a tool for muscle plasticity assessment. Proteomics 6: 321–340, 2006 [DOI] [PubMed] [Google Scholar]
- 17. Hafner RP, Brown GC, Brand MD. Analysis of the control of respiration rate, phosphorylation rate, proton leak rate and protonmotive force in isolated mitochondria using the “top-down” approach of metabolic control theory. Eur J Biochem 188: 313–319, 1990 [DOI] [PubMed] [Google Scholar]
- 18. Harris D, Das A. Control of mitochondrial ATP synthesis in the heart. Biochem J 280: 561–573, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoppeler H, Hudlicka O, Uhlmann E. Relationship between mitochondria and oxygen consumption in isolated cat muscles. J Physiol 385: 661–675, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hopper RK, Carroll S, Aponte AM, Johnson DT, French S, Shen RF, Witzmann FA, Harris RA, Balaban RS. Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium. Biochemistry 45: 2524–2536, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Howlett RA, Willis WT. Fiber-type-related differences in the enzymes of a proposed substrate cycle. Biochim Biophys Acta 1363: 224–230, 1998 [DOI] [PubMed] [Google Scholar]
- 22. Hutson SM. Subcellular distribution of branched-chain aminotransferase activity in rat tissues. J Nutr 118: 1475–1481, 1988 [DOI] [PubMed] [Google Scholar]
- 23. Isfort RJ, Wang F, Greis KD, Sun Y, Keough TW, Bodine SC, Anderson NL. Proteomic analysis of rat soleus and tibialis anterior muscle following immobilization. J Chromatogr B Analyt Technol Biomed Life Sci 769: 323–332, 2002 [DOI] [PubMed] [Google Scholar]
- 24. Jackman MR, Willis WT. Characteristics of mitochondria isolated from type I and type IIb skeletal muscle. Am J Physiol Cell Physiol 270: C673–C678, 1996 [DOI] [PubMed] [Google Scholar]
- 25. Jeneson JA, Westerhoff HV, Kushmerick MJ. A metabolic control analysis of kinetic controls in ATP free energy metabolism in contracting skeletal muscle. Am J Physiol Cell Physiol 279: C813–C832, 2000 [DOI] [PubMed] [Google Scholar]
- 26. Johnson DT, Harris RA, Blair PV, Balaban RS. Functional consequences of mitochondrial proteome heterogeneity. Am J Physiol Cell Physiol 292: C698–C707, 2007 [DOI] [PubMed] [Google Scholar]
- 27. Johnson DT, Harris RA, French S, Aponte A, Balaban RS. Proteomic changes associated with diabetes in the BB-DP rat. Am J Physiol Endocrinol Metab 296: E422–E432, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392, 2002 [DOI] [PubMed] [Google Scholar]
- 29. Kim NK, Joh JH, Park HR, Kim OH, Park BY, Lee CS. Differential expression profiling of the proteomes and their mRNAs in porcine white and red skeletal muscles. Proteomics 4: 3422–3428, 2004 [DOI] [PubMed] [Google Scholar]
- 30. Kim NK, Park HR, Lee HC, Yoon D, Son ES, Kim YS, Kim SR, Kim OH, Lee CS. Comparative studies of skeletal muscle proteome and transcriptome profilings between pig breeds. Mamm Genome 21: 307–319, 2010 [DOI] [PubMed] [Google Scholar]
- 31. Korzeniewski B, Mazat JP. Theoretical studies on the control of oxidative phosphorylation in muscle mitochondria: application to mitochondrial deficiencies. Biochem J 319: 143–148, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kushmerick MJ, Meyer RA, Brown TR. Regulation of oxygen consumption in fast- and slow-twitch muscle. Am J Physiol Cell Physiol 263: C598–C606, 1992 [DOI] [PubMed] [Google Scholar]
- 33. Kushmerick MJ, Moerland TS, Wiseman RW. Mammalian skeletal muscle fibers distinguished by contents of phosphocreatine, ATP, and Pi. Proc Natl Acad Sci USA 89: 7521–7525, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kushmerick MJ, Moerland TS, Wiseman RW. Two classes of mammalian skeletal muscle fibers distinguished by metabolite content. Adv Exp Med Biol 332: 749–760, 1993 [DOI] [PubMed] [Google Scholar]
- 35. Leary SC, Lyons CN, Rosenberger AG, Ballantyne JS, Stillman J, Moyes CD. Fiber-type differences in muscle mitochondrial profiles. Am J Physiol Regul Integr Comp Physiol 285: R817–R826, 2003 [DOI] [PubMed] [Google Scholar]
- 36. Mogensen M, Sahlin K. Mitochondrial efficiency in rat skeletal muscle: influence of respiration rate, substrate and muscle type. Acta Physiol Scand 185: 229–236, 2005 [DOI] [PubMed] [Google Scholar]
- 37. Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658, 2003 [DOI] [PubMed] [Google Scholar]
- 38. Okumura N, Hashida-Okumura A, Kita K, Matsubae M, Matsubara T, Takao T, Nagai K. Proteomic analysis of slow- and fast-twitch skeletal muscles. Proteomics 5: 2896–2906, 2005 [DOI] [PubMed] [Google Scholar]
- 39. Pagliassotti MJ, Donovan CM. Influence of cell heterogeneity on skeletal muscle lactate kinetics. Am J Physiol Endocrinol Metab 258: E625–E634, 1990 [DOI] [PubMed] [Google Scholar]
- 40. Pande SV, Blanchaer MC. Carbohydrate and fat in energy metabolism of red and white muscle. Am J Physiol 220: 549–553, 1971 [DOI] [PubMed] [Google Scholar]
- 41. Phillips D, Aponte AM, French SA, Chess DJ, Balaban RS. Succinyl-CoA synthetase is a phosphate target for the activation of mitochondrial metabolism. Biochemistry 48: 7140–7149, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Picard M, Csukly K, Robillard ME, Godin R, Ascah A, Bourcier-Lucas C, Burelle Y. Resistance to Ca2+-induced opening of the permeability transition pore differs in mitochondria from glycolytic and oxidative muscles. Am J Physiol Regul Integr Comp Physiol 295: R659–R668, 2008 [DOI] [PubMed] [Google Scholar]
- 43. Sargeant AJ, Hoinville E, Young A. Maximum leg force and power output during short-term dynamic exercise. J Appl Physiol 51: 1175–1182, 1981 [DOI] [PubMed] [Google Scholar]
- 44. Scheibye-Knudsen M, Quistorff B. Regulation of mitochondrial respiration by inorganic phosphate: comparing permeabilized muscle fibers and isolated mitochondria prepared from type-1 and type-2 rat skeletal muscle. Eur J Appl Physiol 105: 279–287, 2009 [DOI] [PubMed] [Google Scholar]
- 45. Schieke SM, Phillips D, McCoy JP, Jr, Aponte AM, Shen RF, Balaban RS, Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem 281: 27643–27652, 2006 [DOI] [PubMed] [Google Scholar]
- 46. Scholz TD, Balaban RS. . Mitochondrial F1-ATPase activity of canine myocardium: effects of hypoxia and stimulation. Am J Physiol Heart Circ Physiol 266: H2396–H2403, 1994 [DOI] [PubMed] [Google Scholar]
- 47. Schwerzmann K, Hoppeler H, Kayar SR, Weibel ER. Oxidative capacity of muscle and mitochondria: correlation of physiological, biochemical, and morphometric characteristics. Proc Natl Acad Sci USA 86: 1583–1587, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sorensen MT, Oksbjerg N, Agergaard N, Petersen JS. Tissue deposition rates in relation to muscle fibre and fat cell characteristics in lean female pigs (Sus scrofa) following treatment with porcine growth hormone (pGH). Comp Biochem Physiol A Physiol 113: 91–96, 1996 [DOI] [PubMed] [Google Scholar]
- 49. Starkov AA, Fiskum G. Regulation of brain mitochondrial H2O2 production by membrane potential and NAD(P)H redox state. J Neurochem 86: 1101–1107, 2003 [DOI] [PubMed] [Google Scholar]
- 50. Takahashi M, Hood DA. Protein import into subsarcolemmal and intermyofibrillar skeletal muscle mitochondria. Differential import regulation in distinct subcellular regions. J Biol Chem 271: 27285–27291, 1996 [DOI] [PubMed] [Google Scholar]
- 51. Vitorino R, Ferreira R, Neuparth M, Guedes S, Williams J, Tomer KB, Domingues PM, Appell HJ, Duarte JA, Amado FM. Subcellular proteomics of mice gastrocnemius and soleus muscles. Anal Biochem 366: 156–169, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weibel ER, Hoppeler H. Exercise-induced maximal metabolic rate scales with muscle aerobic capacity. J Exp Biol 208: 1635–1644, 2005 [DOI] [PubMed] [Google Scholar]
- 53. Weiler U, Appell HJ, Kremser M, Hofacker S, Claus R. Consequences of selection on muscle composition. A comparative study on gracilis muscle in wild and domestic pigs. Anat Histol Embryol 24: 77–80, 1995 [DOI] [PubMed] [Google Scholar]
- 54. Willis WT, Jackman MR. Mitochondrial function during heavy exercise. Med Sci Sports Exerc 26: 1347–1353, 1994 [PubMed] [Google Scholar]
- 55. Wilson DF, Owen CS, Holian A. Control of mitochondrial respiration: a quantitative evaluation of the roles of cytochrome c and oxygen. Arch Biochem Biophys 182: 749–762, 1977 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.