Abstract
Previously, we demonstrated that the vacuolar-type H+-ATPase (V-ATPase) a2-subunit functions as an endosomal pH sensor that interacts with the ADP-ribosylation factor (Arf) guanine nucleotide exchange factor, ARNO. In the present study, we showed that ARNO directly interacts not only with the a2-subunit but with all a-isoforms (a1–a4) of the V-ATPase, indicating a widespread regulatory interaction between V-ATPase and Arf GTPases. We then extended our search for other ARNO effectors that may modulate V-ATPase-dependent vesicular trafficking events and actin cytoskeleton remodeling. Pull-down experiments using cytosol of mouse proximal tubule cells (MTCs) showed that ARNO interacts with aldolase, but not with other enzymes of the glycolytic pathway. Direct interaction of aldolase with the pleckstrin homology domain of ARNO was revealed by pull-down assays using recombinant proteins, and surface plasmon resonance revealed their high avidity interaction with a dissociation constant: KD = 2.84 × 10−10 M. MTC cell fractionation revealed that aldolase is also associated with membranes of early endosomes. Functionally, aldolase knockdown in HeLa cells produced striking morphological changes accompanied by long filamentous cell protrusions and acidic vesicle redistribution. However, the 50% knockdown we achieved did not modulate the acidification capacity of endosomal/lysosomal compartments. Finally, a combination of small interfering RNA knockdown and overexpression revealed that the expression of aldolase is inversely correlated with gelsolin levels in HeLa cells. In summary, we have shown that aldolase forms a complex with ARNO/Arf6 and the V-ATPase and that it may contribute to remodeling of the actin cytoskeleton and/or the trafficking and redistribution of V-ATPase-dependent acidic compartments via a combination of protein-protein interaction and gene expression mechanisms.
Keywords: ADP-ribosylation factor nucleotide site opener, vacuolar H+-ATPase, a-subunit isoforms, actin cytoskeleton, gelsolin, surface plasmon resonance, endosomal/lysosomal compartments
fructose bisphosphate aldolase (EC 4.1.2.13) is the fourth enzyme of glycolysis, which catalyses reversible cleavage of fructose 1,6-bisphosphate into glyceraldehyde 3-phosphate and dihydroxyacetone phosphate. Aldolase has been studied for over 70 years and is a very well-characterized protein. The success in studying this protein is in part due to its high abundance and relatively easy purification from natural sources. Several crystal structures of wild-type and mutant proteins are available now, which clarify the mechanism of its catalytic activity (2, 17, 18, 38). Although the role of aldolase in carbohydrate metabolism is well established, there is growing evidence for many alternative so-called “moonlighting” functions for this enzyme. In particular, aldolase interacts with various proteins unrelated to glycolytic enzymes, including F-actin, α-tubulin, dynein light chain 8, the actin nucleation promoting factor WASP, the endocytotic sorting protein nexin-9, and phospholipase D2 (10, 34, 55, 71, 73, 74). These multiple interactions indicate that aldolase is involved, probably as a scaffolder, in the coordination of membrane trafficking and cytoskeleton dynamics at the cell periphery. As a result, a novel emerging field for aldolase biology is its central role in a variety of vesicular trafficking events, including 1) endocytosis and parasite invasion, 2) cytoskeleton rearrangement and cell motility, 3) trafficking and recycling of membrane proteins, and 4) signal transduction.
Interaction of aldolase with vacuolar-type H+-ATPase (V-ATPase) was also reported previously (6, 35–37, 58). The V-ATPase is a multimeric proton-pumping protein complex that is found on plasma membranes and diverse endomembrane organelles. The biochemistry, cell biology, and pathophysiology of V-ATPase have been extensively reviewed by ourselves and others (9, 24, 42, 47, 72). The cytoplasmic V1 sector of the V-ATPase is composed of eight different subunits with defined stoichiometry (A3B3CDE3FG3H) and is responsible for ATP hydrolysis (20, 45, 75). The transmembrane V0 sector is composed of six different subunits (ac5c′′deAc45) and is responsible for proton translocation across the lipid bilayer. Consistent with the presence of V-ATPases in diverse subcellular compartments, a large spectrum of subunit isoforms and potential splice variants have been identified (9, 24, 42, 44, 47, 64, 72). The expression of these isoforms is tissue and cell specific. Intracellular targeting and assembly of V-ATPase in the functional holocomplex is regulated by a-subunit isoforms, four of which (a1, a2, a3, and a4) are found in mice and humans (49, 63, 65, 69). V-ATPase activity is controlled by different mechanisms, including the physical disassembly of V1 and V0 sectors of V-ATPase (31, 50). Disassembled V-ATPase is no longer able to hydrolyze ATP, and thus this nanomotor cannot pump protons across the membranes. Aldolase interacts with three different subunits of the V-ATPase: the transmembrane a-subunit of the V0 sector, as well as the soluble E- and B-subunits of the V1 sector (36, 37). In yeast and mammalian kidney cells, the interaction between aldolase and the V-ATPase is modified by glucose, suggesting that aldolase may act as a glucose sensor and mediate V-ATPase assembly/disassembly and, therefore, its function. However, further analysis demonstrated that aldolase enzymatic activity is not required for regulation of V-ATPase assembly/disassembly, suggesting an important role of physical association in the V-ATPase/aldolase complex (35). On the other hand, V-ATPase assembly/disassembly in yeast is also modulated by the Ras/cAMP/PKA pathway (6). In particular, upregulation of the Ras/cAMP/PKA pathway by expression of constitutively active PKA blocks glucose-dependent V-ATPase dissociation. Moreover, overexpression of Ras leads to decreased binding of aldolase to V-ATPase (6).
The Ras-superfamily small GTPases, which includes ADP-ribosylation factor small GTPase (Arf) proteins, function as “molecular switches” and regulate an extraordinary variety of cell functions (7). The transition between “on” and “off” states of this molecular device is mediated by a GDP/GTP cycle. Six Arfs have been identified in mammalian cells, and they are all critical for the regulation of vesicular trafficking both in exo- and endocytotic pathways (16, 21, 22). Whereas Arf1 is involved in regulating endoplasmic reticulum (ER)-Golgi trafficking (14, 60), Arf6 is targeted to the plasma membrane and/or endosomes and is involved in the regulation of endocytosis and membrane recycling (3, 46, 59). Activation of Arfs is achieved by GDP/GTP exchange in the presence of guanine nucleotide exchange factors (GEFs). ADP-ribosylation factor nucleotide site opener (ARNO) is one of four members of the cytohesin subfamily of Arf-GEFs, all of which share the following structural domains: 1) an NH2-terminal coiled-coil (CC), 2) a central Sec7 domain, 3) a pleckstrin homology (PH) domain, and 4) a COOH-terminal polybasic (PB) domain (11, 21, 30). Functionally, both Arf6 and ARNO have been implicated in regulation of the endocytotic pathway, organelle biogenesis, and actin cytoskeleton remodeling (11, 16, 21, 22, 56).
Recent work from our laboratory has uncovered a crucial link between V-ATPase and Arf-family small GTPases in regulation of the endosomal/lysosomal protein degradative pathway (29, 41). We demonstrated that V-ATPase has a novel function as endosomal pH sensor that scaffolds ARNO and Arf6. The transmembrane a2-subunit of the V-ATPase directly interacts with cytosolic ARNO, whereas the V-ATPase c-subunit specifically binds Arf6; these biochemical events are essential for vesicular trafficking between early and late endosomal compartments (41). However, the molecular mechanism and cell biological significance of the interactions between V-ATPase and small GTPases remain obscure. Recently, we mapped the interaction sites between the NH2-terminal tail of the V-ATPase a2-subunit (a2N) and ARNO and demonstrated the crucial role of the catalytic Sec7 domain in this interaction (43). Moreover, we also recently discovered a novel function of the V-ATPase as a regulator of the GEF activity of ARNO and, thus, as a modulator of Arf-small GTPase signaling (28).
Although this previous work uncovered a functional cross talk between V-ATPase, ARNO, and Arf small GTPases, other downstream effectors and related cell biological events have also been unraveled. In the present study, we hypothesize that since V-ATPase interacts with both ARNO (29) and aldolase (35), these two proteins could in turn interact with each other and coordinate endocytic vesicle trafficking and cytoskeleton rearrangements. To uncover the downstream effectors of ARNO and V-ATPase signaling, we have now applied a combination of protein-protein interaction techniques, and we indeed identify aldolase as a specific and high-affinity interaction partner of ARNO that could be involved in intracellular trafficking and cytoskeletal modulation.
MATERIALS AND METHODS
Reagents and antibodies.
If not otherwise specified, all reagents, including d(+)-glucose, bafilomycin A1, aldolase type IV from rabbit muscle (aldolase-A), bovine albumin-FITC, and iodixanol (OptiPrep density gradient medium) were obtained from Sigma (St. Louis, MO). Lipofectamine 2000 transfection reagent, Alexa Fluor 647 phalloidin, fluorescein-dextran (10,000 MW, anionic), and the acidic organelle probes DAMP and LysoTracker red DND-99 were obtained from Invitrogen (Carlsbad, CA). NuPAGE gels and buffers were also acquired from Invitrogen. EDTA-free Complete protease inhibitor tablets were obtained from Roche (Indianapolis, IN). Octylglucoside (n-octyl-β-d-glucopyranoside) was obtained from Anatrace (Maumee, OH). Western Lightning chemiluminescence reagent plus was obtained from PerkinElmer (Boston, MA). TALON metal affinity resin was acquired from Clontech (Mountain View, CA), and glutathione-Sepharose 4B beads were obtained from GE Healthcare (Piscataway, NJ). Metabolic labeling of recombinant proteins during in vitro translation was achieved by incorporation of either [35S]methionine purchased from GE Healthcare or BODIPY-lysine-tRNA purchased from Promega (Madison, WI). Thrombin cleavage capture kit was purchased from EMD-Biosciences/Novagen (Gibbstown, NJ). Bicinchoninic acid (BCA) protein assay kit was obtained from Thermo Fisher Scientific (Rockford, IL). Affinity-purified rabbit polyclonal anti-aldolase-B antibodies (HPA002198) and mouse monoclonal anti-gelsolin antibodies (GS-2C4) were purchased from Sigma. Affinity-purified goat polyclonal anti-actin (C-11) antibodies and affinity-purified goat polyclonal anti-aldolase-A/B (D-18) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The following affinity-purified antibodies against glycolytic enzymes were also purchased from Santa Cruz Biotechnology: goat polyclonal anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH; V-18), goat polyclonal anti-phosphofructokinase (anti-PFK; E-16), goat polyclonal anti-phosphoglycerate kinase (anti-PGK; E-20), and rabbit polyclonal anti-enolase (H-300). Rabbit polyclonal anti-FITC (A-889) and Alexa 488-conjugated anti-dinitrophenyl-KLH (anti-DNP) antibodies (A-11097) were obtained from Invitrogen. Mouse monoclonal anti-megalin (1H2) antibodies were previously described (26, 29). The following secondary antibodies were also used: goat anti-rabbit IgG-horseradish peroxidase (RPN4301) and sheep anti-mouse IgG-horseradish peroxidase (NA931V) were purchased from GE Healthcare, and donkey anti-goat IgG-horseradish peroxidase (sc-2020) was purchased from Santa Cruz Biotechnology.
DNA constructs, recombinant protein expression, and purification.
cDNAs encoding full-length wild-type mouse V-ATPase a1-isoform (832 aa, GI no. 7363245), a2-isoform (856 aa, GI no. 7363249), a3-isoform (834 aa, GI no. 7363247), and a4-isoform (833 aa, GI no. 13990958) were generously provided by Dr. Masamitsu Futai (Iwate Medical University, Iwate, Japan). cDNA constructs encoding the NH2-terminal cytosolic tail of mouse V-ATPase a-isoforms a1N (1–388 aa), a2N (1–393 aa), a3N (1–386 aa), and a4N (1–390 aa) were amplified using the Expand High Fidelity PCR system and subcloned into a pIVEX2.4d vector (Roche) as previously described (29). The resulting constructs contain a modified NH2-terminal 6XHis-tag (MSGSHHHHHHSSGIEGRGRLIKMT). All four constructs were in vitro translated and metabolically labeled with [35S]methionine using the RTS100 kit (Roche). For in vitro translation and pull-down experiments with aldolase, the human aldolase-B cDNA was obtained from ATCC (no. MGC-32618, I.M.A.G.E. clone ID 4593670, GenBank accession nos. BC029399, BG402660). This cDNA was cloned into pIVEX2.3 vector (Roche). This construct was in vitro translated using the RTS100 kit (Roche) with or without labeling by FluoroTect greenLys tRNA (Promega). The recombinant NH2-terminal 6His-tagged aldolase-B (6His-ALDO-B) for surface plasmon resonance (BIAcore) experiments was prepared as follows. DNA corresponding to amino acid residues 1–364 of human ALDO-B was amplified using normal human kidney cDNA (Invitrogen) as a template and PfuTurbo DNA polymerase (Stratagene). DNA was subcloned into NdeI/XhoI restriction sites of the pET28b vector (Novagen) in frame with a thrombin-cleavable NH2-terminal 6His tag. Recombinant human 6His-ALDO-B was expressed in Escherichia coli BL21(DE3) cells and purified by sequential chromatography on DEAE-Sepharose FF beads (GE Healthcare) and TALON beads (Clontech) according to the manufacturers' instructions. The construct for bacterial expression of the glutathione S-transferase (GST)-tagged triglycine variant of human wild-type ARNO [ARNO(wt)] was obtained from Dr. James Casanova (University of Virginia, Charlottesville, VA). Cloning and purification of recombinant GST-only and GST-ARNO(wt) proteins were performed as previously described (29). In addition, constructs corresponding to the four major domains of ARNO (CC, Sec7, PH, and PB) were also generated as previously described (43). The construct for bacterial expression of the 6His-tagged human ARNO(wt) was generously provided by Dr. Sylvain Bourgoin, and its expression and purification were performed as previously described (26). For aldolase-A overexpression in mammalian cells, a construct corresponding to amino acid residues 2–364 of human aldolase-A was amplified using normal human kidney cDNA (Invitrogen) as a template and PfuTurbo DNA polymerase (Stratagene). It was subcloned into XhoI/BamHI restriction sites of pEGFP-C1 and pDsRed-Monomer-C1 vectors (Clontech) in frame with NH2-terminal enhanced green fluorescent protein or monomeric red fluorescent protein, respectively.
Pull-down assay, in vitro translation, autoradiography, and Western blot analysis.
To study the specificity of ARNO interaction with enzymes of the glycolytic pathway, we performed pull-down assays as previously described (29). GST-ARNO(wt) was immobilized on glutathione-agarose beads and incubated with the 100,000 g cytosolic fraction prepared from mouse proximal tubule cells (MTC). Briefly, purified recombinant GST-ARNO(wt) fusion protein (20 μg) was immobilized on glutathione-agarose beads (65 μl) and incubated with MTC cytosol (1.5 mg of protein) for 2 h at 4°C in 650 μl of binding buffer. Unbound proteins were removed by washing the beads three times for 5 min each in 1 ml of ice-cold binding buffer. Proteins specifically bound during the pull-down assay were eluted using a thrombin cleavage capture kit (EMD-Biosciences/Novagen). Beads were incubated with 6 units of thrombin in 200 μl of cleavage-capture buffer overnight at room temperature. Interacting proteins were resolved by conventional NuPAGE and analyzed by Western blotting using anti-aldolase-A/B (D-18, 1:500), anti-GAPDH (V-18, 1:500), anti-PGK (E-20, 1:500), anti-PFK (E-16, 1:500), and anti-enolase antibodies (H-300, 1:500). To study direct interactions of ARNO with either V-ATPase a-isoforms or aldolase-B, we performed pull-down experiments with recombinant proteins. Four constructs of mouse V-ATPase (a1N, a2N, a3N, and a4N) were in vitro translated and metabolically labeled with l-[35S]methionine. These recombinant proteins were used in pull-down assays with GST-ARNO(wt) used as a bait immobilized on glutathione-Sepharose beads as follows. Recombinant a1N-[35S], a2N-[35S], a3N-[35S], and a4N-[35S] (25 pmol each) were incubated with 100 pmol of GST-ARNO(wt) overnight at 4°C in binding buffer (10 mM HEPES, 1 mM EDTA, 1 mM DTT, 100 mM NaCl, 10% glycerol, and 0.1% NP-40, pH 7.5). Next, 40 μl of glutathione beads were added, and the reactions were incubated at 4°C for 20 min and washed five times with ice-cold binding buffer. Bound proteins were eluted by NuPAGE sample buffer and resolved using NuPAGE gels (12 wells, 4–12% Bis-Tris). Gels were dried and analyzed by autoradiography. The pull-down experiments with aldolase-B were performed using the following purified ARNO-derived recombinant proteins: GST-ARNO(wt)-6His (1-400 aa, wild-type ARNO), GST-CC-6His (1-60 aa, CC domain of ARNO), GST-Sec7–6His (61-252 aa, Sec7 domain of ARNO), GST-PH-6His (253-378 aa, PH domain of ARNO), and GST-PB-6His (379-400 aa, PB domain of ARNO). For these experiments, human aldolase-B was in vitro translated as either unlabeled or labeled by BODIPY-lysine-tRNA using the RTS100 kit. Detection of the BODIPY-labeled aldolase-B was performed directly in-gel using a laser-based Typhoon 9410 fluorescent scanner (GE Healthcare). Unlabeled aldolase-B was detected by Western blot analysis with anti-aldolase antibodies (D-18, 1:500). In these in vitro translation assays, two aldolase bands were consistently observed, compared with one band detected in experiments with endogenous aldolase. We suggest that low molecular band represents an incompletely translated but interaction-competent version of recombinant aldolase. All pull-down experiments were repeated at least three times with the same results, and representative data are shown.
Real-time binding and kinetic analysis by surface plasmon resonance.
Surface plasmon resonance (SPR) binding assays were performed at 25°C on a BIAcore T100 instrument (GE Healthcare). All reagents, including buffers, sensor chips, and the amine coupling kit, were obtained from GE Healthcare. For kinetic analysis of the binding of ARNO(wt) with aldolase-B(wt), purified aldolase-B (20 μg/ml) in 10 mM HEPES (pH 7.4) was immobilized at 10,000 response units (RU) on a CM5 sensor chip using an amine coupling kit according to the manufacturer's instructions. The same kit was used to perform blank immobilization to create a reference surface on the same chip. For kinetic analysis, samples of purified 6His-ARNO(wt) at concentrations ranging from 0.25 to 5 μM were injected for 3 min over active and reference surfaces at a flow rate of 30 μl/min in NBS-EP, 1 mM DTT running buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.1% surfactant P20, and 1 mM DTT). Dissociation of the complexes was monitored for 10 min, and then the regeneration of the sensor surfaces was performed with 10 mM glycine-HCl (pH 1.5) with 1-min injection at a 30 μl/min flow rate. BIAcore T100 evaluation software was used to calculate the association and dissociation rate constants (ka and kd) with a bivalent analyte fitting model. The overall dissociation constant (KD) was determined as described in results.
Subcellular fractionation, separation, and identification of early endosomes from MTC cells.
To identify and purify early endosomes, MTC cells were grown in 150-mm cell culture dishes to confluency and then pulsed with 200 μg/ml albumin-FITC for 20 min. Cells were fractionated, and an endosomal fraction was separated using an iodixanol density gradient as previously described (13) and as previously used for early endosome fractionation (62). Briefly, after incubation with albumin-FITC, cells were washed twice with 20 ml of ice-cold Ringer solution (122.5 mM NaCl, 5.4 mM KCl, 1.2 mM CaCl2, 0.8 mM MgCl2, 1 mM NaHPO4, 10 mM HEPES, and 5 mM glucose, pH 6.0) and once with 10 ml of homogenization buffer (150 mM NaCl, 1.5 mM MgCl2, 5 mM EGTA, and 50 mM Tris, pH 7.5). Cells were then collected using a cell scraper into 2 ml of homogenization buffer containing Complete protease inhibitors. Cells were homogenized by being passed 7 times through a needle (25 gauge, 1.5 in.; Becton Dickinson) and broken by being passed 20 times through a ball-bearing cell cracker (8.020-mm internal diameter, 8.014-mm ball diameter; HGM, Heidelberg, Germany). Postnuclear supernatant (PNS) was collected by centrifugation at 1,500 g. PNS fraction (∼1.5 ml) was placed at the top of the 10–40% iodixanol gradient formed in buffer (78 mM KCl, 4 mM MgCl2, 8 mM CaCl2, 10 mM EGTA, and 50 mM HEPES, pH 7.5). The tubes were spun in a SW41Ti rotor for 20 h at 27,000 rpm (100,000 g) using a Beckman L8-80M ultracentrifuge. Fourteen fractions (0.8 ml each) were collected from the bottom (heavy to light), and iodixanol density was determined using a DTX 880 microplate reader (Beckman) with a 340-nm absorbance filter. Fractions were diluted with PBS and centrifuged using a TLA-55 rotor at 55,000 rpm (150,000 g) for 1 h to remove the soluble proteins. Pellets were collected and suspended into PBS (0.2 ml each), and protein concentration was determined using a BCA protein assay kit (Pierce). FITC fluorescence was determined using a DTX 880 microplate reader with 485-nm excitation and 525-nm emission filters (Beckman). Fourteen fractions of 20 μl each were subjected to 4–12% Bis-Tris NuPAGE followed by Western blot analysis. The following primary antibodies were used to detect proteins of interest: anti-megalin (1H2, mouse monoclonal at 1:1,000 dilution), anti-FITC (rabbit polyclonal at 1:2,000 dilution), and anti-aldolase-A/B (D-18, goat polyclonal at 1:500 dilution). The following secondary horseradish peroxidase-conjugated IgGs were also used: donkey anti-goat (1:5,000 dilution), goat anti-rabbit (1:10,000 dilution), and sheep anti-mouse (1:5,000 dilution). Purified proteins were used as positive controls in these experiments (100 ng of aldolase-A and 100 ng albumin-FITC).
Small interfering RNA knockdown and overexpression of aldolase, followed by Western blot analysis.
Stealth predesigned RNAi small interfering RNAs targeting two different regions of human aldolase-A (siRNA-1 and siRNA-2) and one nontargeting Stealth RNAi siRNA (negative control) were purchased from Invitrogen. The aldolase-A siRNA sequences are as follows: 5′-CCAACAGCCUUGCCUGUCAAGGAAA-3′and 5′-GGCGGUGUUGUGGGCAUCAAGGUAG-3′; the negative control siRNA sequence is 5′-GGAGUCACGCGAUCGUGACGCGCCA-3′. All three siRNAs were labeled with Alexa 555 dye at the 5′-end. siRNAs (100 nM) were delivered into HeLa cells using Lipofectamine 2000 transfection reagent. The transfection efficiency, monitored by fluorescent microscopy, was 100% at 48–72 h. The efficiency of aldolase protein knockdown was estimated at 72 h by semiquantitative Western blotting, followed by band densitometry with LabWorks Analysis software (UVP BioImaging Systems, Upland, CA). In addition, the efficiency of aldolase knockdown was evaluated using an enzymatic aldolase activity assay as described below. pEGFP-C1-AldoA and pDsRed-Monomer-C1-AldoA plasmids were delivered into HeLa cells using Lipofectamine 2000 transfection reagent according to the manufacturer's instructions.
Aldolase enzymatic activity assay in cell lysates.
HeLa cells were seeded in six-well cell culture plates. The next day, they were transfected with negative control siRNA, aldolase siRNA-1 or aldolase siRNA-2 as described above. Seventy-two hours after transfection, cells from each well were washed with PBS and lysed on ice in 100 μl of 20 mM HEPES-NaOH, pH 7.4, 150 mM NaCl, 1 mM EDTA, pH 8.0, 1% (vol/vol) Triton X-100, and Complete protease inhibitor tablets. Cell lysates were clarified by centrifugation at 15,000 g for 1 h at 4°C. Cleared cell lysates were kept on ice and used for aldolase activity assays on the same day. The total protein concentration of cleared cell lysates was determined spectroscopically using the BCA assay kit (Pierce). Aldolase activity with fructose 1,6-bisphosphate (Fru 1,6-P2) as a substrate was determined by measuring the decrease in absorbance per minute at 340 nm in an assay coupled to β-NADH oxidation by glycerol-3-phosphate dehydrogenase, essentially as described previously (52) with some modifications. Briefly, 20 μl (4–7 μg/μl total protein) of cleared cell lysates was added into 96-well plates, and the reaction was initiated by simultaneous addition to all wells of the reaction mixtures containing 50 mM triethanolamine-HCl, pH 7.4, 10 mM EDTA, 0.2 mM β-NADH, and 10 μl of glycerol-3-phosphate dehydrogenase/triose phosphate isomerase (1 mg/ml) with or without 2 mM Fru 1,6-P2 as an aldolase substrate to a final volume of 250 μl. The rate of the decrease in absorbance was recorded immediately and simultaneously for all wells for 10 min with 10-s intervals at 25°C using a SpectroMAX 190 spectrophotometer (Molecular Devices) or a Multimode Detector plate reader (model DTX 880; Beckman-Coulter, Fullerton, CA). The assay was performed twice in duplicate, with similar results. All data points obtained during reactions without 2 mM Fru 1,6-P2 substrate (representing a decrease in β-NADH absorbance due to enzymatic activities of proteins other than aldolase) were subtracted from all data points during reactions with substrate to obtain data specific for aldolase. All data points were normalized to total protein concentration, and linear portions of the graphs were plotted and analyzed with Microsoft Excel to obtain aldolase activity values in decrease of absorbance units per minute per milligram of total protein.
Immunocytochemistry and epifluorescence analysis.
HeLa (human cervix epithelium) cells were grown on 22 × 22-mm premium glass coverslips (Fisher Scientific, Pittsburgh, PA) in six-well plates. siRNA transfection is described above. Seventy-two hours after transfection, cells were incubated for 1 h in DMEM (without FBS) containing 50 μM DAMP. Cells were rinsed in PBS and fixed in PBS containing 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 20 min. After three PBS rinses, a permeabilization of cells was performed by treatment with 0.1% Triton X-100 in PBS for 10 min. After three PBS rinses, coverslips were blocked with 2% BSA in PBS for 1 h. For staining of DAMP-loaded acidic vesicles, the cells were incubated for 1 h with Alexa Fluor 488-conjugated anti-DNP antibodies, diluted 1:1,000 in 2% BSA/PBS. After three PBS rinses, coverslips were incubated for 20 min with the Alexa Fluor 647 phalloidin (20 U/ml), diluted 1:40 in 2% BSA/PBS to stain filamentous actin. After three final PBS rinses, the coverslips were mounted with Vectashield HardSet mounting medium (Vector Laboratories, Burlingame, CA) with 4,6-diamidino-2-phenylindole (1.5 μg/ml) to counterstain nuclei. Epifluorescence analysis was performed on a Nikon Eclipse 80i fluorescence microscope, and images were captured with an Orca C4742-95 digital camera (Hamamatsu Photonics, Bridgewater, NJ) using IPLab Spectrum (version 3.9.5) image processing software (Scanalytics, Fairfax, VA). All images were transferred into Photoshop CS4.0 and paginated using Illustrator CS4.0 (Adobe, Seattle, WA).
Quantitative fluorescent vesicle acidification assay.
HeLa cells were seeded in 48-well cell culture plates. The next day, they were transfected with 32 nM of negative control siRNA, aldolase siRNA-1 or aldolase siRNA-2, or with Lipofectamine 2000 transfection reagent only (“mock” transfection) in triplicate. For these experiments, siRNAs were not labeled with any fluorescent dye. Forty-eight hours after transfection, cells were rinsed briefly in DMEM (without FBS) and then incubated for 1 h in 1 μM LysoTracker diluted in DMEM (without FBS). For control experiments, nontransfected cells were preincubated with 1 μM bafilomycin (diluted in complete medium) for 1 h before being labeled with 1 μM LysoTracker. After incubation, cells from each well were washed twice in PBS and trypsinized, and cell suspensions (100 μl, ∼1 × 106 cells/ml) were transferred to a black 96-well plate (Corning, Corning, NY). Fluorescence was read immediately using a Multimode Detector plate reader (model DTX 880; Beckman-Coulter). The following settings were used: fluorescence intensity top method, 0.1-s integration time, 535-nm excitation filter, and 595-nm emission filter. Background autofluorescence values were obtained from unlabeled HeLa cell suspensions and were subtracted before further analysis. Data were normalized to LysoTracker labeling of mock-transfected cells, which were taken as 100%.
Estimation of endosomal, lysosomal pH.
HeLa cells were seeded and transfected as described in Quantitative fluorescent vesicle acidification assay. Forty-eight hours after transfection, cells were rinsed briefly in complete medium and then loaded with 2.5 mg/ml fluorescein-dextran (10,000 MW, anionic; Invitrogen) for 18 h at 37°C. For control experiments, nontransfected cells were loaded with fluorescein-dextran as described above and then treated with 1 μM bafilomycin (diluted in complete medium) for 1 h. Cells were then washed, trypsinized, and transferred to a black 96-well plate. pH calibration was performed with nontransfected HeLa cells loaded with fluorescein-dextran and harvested as described above and then resuspended in a series of MES calibration buffers (25 mM MES-KOH, 115 mM KCl, 5 mM NaCl, and 1.3 mM MgSO4) at pH 4.5, 5.0, 5.5, 6.0, 6.5, or 7.0 in the presence of 10 μM monensin and 10 μM nigericin, as described (48). The fluorescence of all samples was read simultaneously using a Multimode Detector plate reader. The following settings were used: fluorescence intensity top method, 0.1-s integration time, 370- and 485-nm excitation filters, and 535-nm emission filter. Background autofluorescence values were obtained from unlabeled HeLa cell suspensions and were subtracted before further analysis. The ratio of emission intensity at 535 nm and two different excitation wavelengths (485 or 370 nm) were then calculated for each sample, and a calibration curve was generated in Microsoft Excel. Finally, the pH values were calculated from a linear portion of this calibration curve.
RESULTS
Specific and direct interaction of wild-type ARNO with aldolase and a-subunit isoforms of V-ATPase.
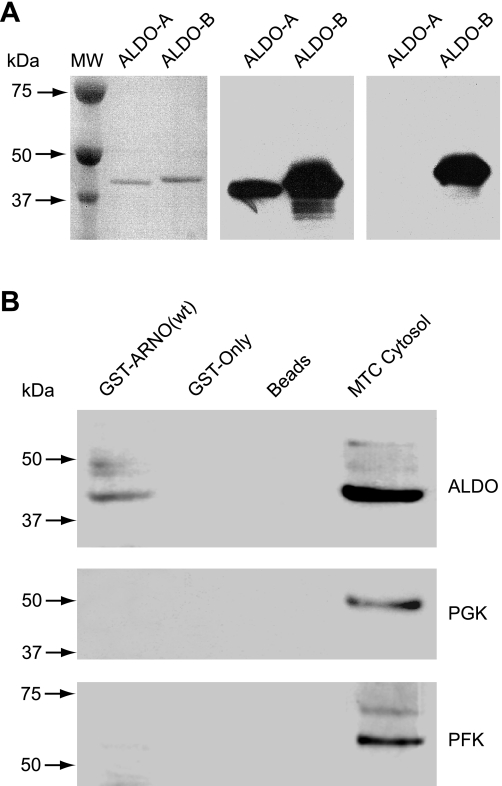
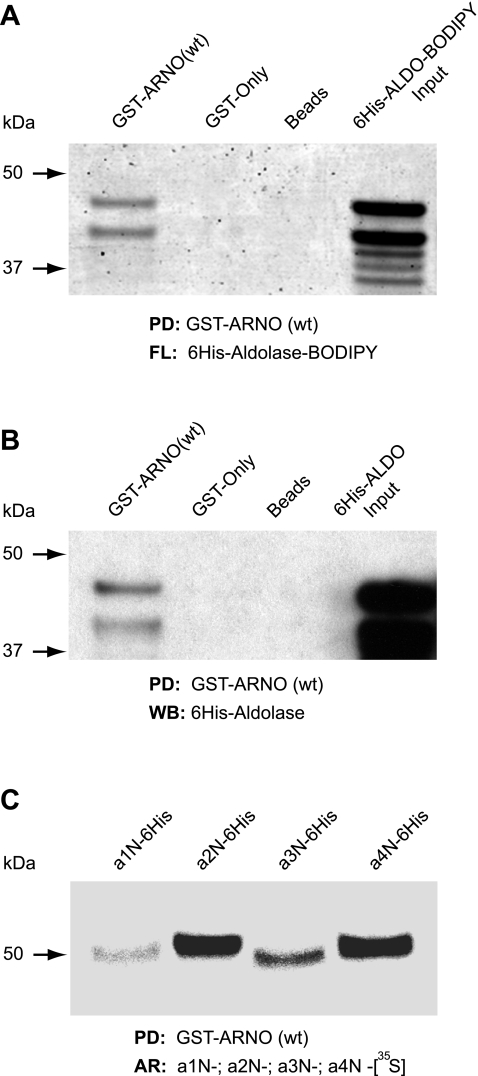
Since ARNO (8, 29, 41–43) and aldolase (35–37) were recently identified as important V-ATPase interactors, we hypothesized that ARNO could also directly and specifically interact with aldolase. To test this hypothesis, we performed the in vitro pull-down experiments as described in materials and methods. The specificity of anti-aldolase antibodies used in our studies was determined in experiments using highly purified aldolase-A and aldolase-B as shown in Fig. 1A. These experiments demonstrated that GST-ARNO(wt) specifically interacts with cytosolic endogenous aldolase-A/B expressed in MTCs but not with PGK or PFK (Fig. 1B). Interaction of GST-ARNO(wt) with endogenous GAPDH or enolase was also undetectable (not shown). To determine whether ARNO directly interacts with aldolase, we performed pull-down experiments with recombinant proteins. For these experiments, human 6His-tagged aldolase-B was in vitro translated as either unlabeled protein (6His-ALDO) or a fluorescent protein labeled with BODIPY-lysine-tRNA (6His-ALDO-BODIPY). After incubation of 6His-ALDO-B with GST-ARNO(wt) and glutathione beads, eluted proteins were examined by both fluorescence (Fig. 2A) and Western blotting (Fig. 2B), revealing a direct interaction between these proteins. In addition, the pull-down experiments with in vitro translated and metabolically labeled recombinant a1-, a2-, a3-, and a4-subunit isoforms demonstrated that ARNO directly interacts not only with the V-ATPase a2-subunit, as we previously reported (8, 29, 41), but also with the three other a-subunit isoforms of the V-ATPase (Fig. 2C).
Fig. 1.
Cytosolic aldolase specifically interacts with ADP-ribosylation factor nucleotide site opener (ARNO) in pull-down experiments. A: evaluation of specificity of anti-aldolase antibodies used in this study. Left image shows 1 μg of aldolase-A (ALDO-A) from rabbit muscle and 1 μg of 6His-tagged recombinant human ALDO-B resolved by NuPAGE, transferred to Immobilon membrane, and stained with Ponceau-S. Middle image shows Western blotting performed with the anti-ALDO-A/B (D-18) antibody, which recognizes both ALDO-A and ALDO-B. Right image shows Western blotting performed with anti-ALDO-B (HPA002198) antibody, which selectively recognizes ALDO-B. B: specific interaction of full-length wild-type (wt) ARNO with aldolase but not with other enzymes of the glycolytic pathway. Pull-down with glutathione S-transferase (GST)-ARNO(wt) from cytosolic fractions of cultured mouse proximal tubule cells (MTC) was followed by Western blot analysis using anti-ALDO-A/B (D-18) antibody. There was a specific interaction of ARNO with aldolase but not with 2 other enzymes of the glycolytic pathway, phosphofructokinase (PFK) or phosphoglycerate kinase (PGK).
Fig. 2.
Direct interaction of full-length ARNO with aldolase and all 4 a-subunit isoforms of vacuolar-type H+-ATPase (V-ATPase). A and B: direct interaction of ARNO with aldolase was further confirmed in pull-down (PD) experiments with recombinant proteins. First (A), ALDO-B was in vitro translated and metabolically labeled with fluorescent BODIPY (6His-ALDO-BODIPY; fourth lane). In this pull-down experiment, the ALDO-B was detected by laser scanning fluorescence analysis (FL) in the first lane (GST-ARNO) but not in the second and third lanes (GST-only and beads as controls). Second (B), ALDO-B was in vitro translated without metabolic labeling (6His-ALDO). In this pull-down experiment, the ALDO-B was detected only in the first lane (GST-ARNO), but not in the control lanes (GST-only and beads as controls), by Western blot analysis (WB) using an anti-ALDO-A/B (D-18) antibody. C: direct interaction of ARNO with all 4 a-subunit isoforms demonstrated in pull-down experiments using recombinant proteins. Cytosolic tails of a1-, a2-, a3-, and a4-subunit isoforms (a1N, a2N, a3N, and a4N, respectively) were in vitro translated and metabolically labeled with [35S]methionine and used in pull-down assays with GST-ARNO(wt). Detection of a1N, a2N, a3N, and a4N was performed by autoradiography (AR). Representative data of 3 independent experiments are shown.
ARNO interacts with aldolase via its PH domain.
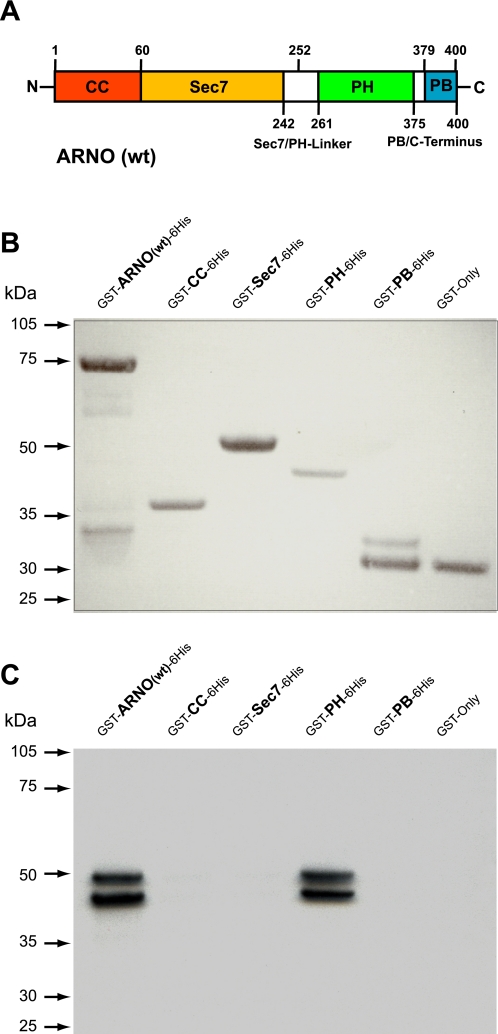
To identify which domain of ARNO mediates the interaction with aldolase, we subcloned all four ARNO domains (Fig. 3A) as doubled-tagged GST-6His fusion proteins and purified them as described in materials and methods. Recombinant glutathione transferase (GST only) was used as a negative control in these experiments. The integrity and loading of purified recombinant proteins was analyzed by Ponceau staining (Fig. 3B), and pull-down experiments were performed using in vitro translated, unlabeled aldolase-B and detected by Western blot analysis (Fig. 3C). These experiments demonstrated a very strong interaction of aldolase with the PH domain and lack of interaction with all other domains of ARNO (Fig. 3C).
Fig. 3.
Interaction of ARNO with aldolase is mediated via its pleckstrin homology (PH) domain. A: schematic representation of ARNO's structural domains. Boundaries of the domains used for cloning of human ARNO-derived recombinant proteins are indicated as amino acid numbers: GST-ARNO(wt)-6His (1–400 aa, wild type), GST-CC-6His (1–60 aa, coiled-coil domain), GST-Sec7–6His (61–252 aa, Sec7 domain), GST-PH-6His (253–378 aa, PH domain) and GST-PB-6His (379–400 aa, polybasic domain). N, NH2 terminus; C, COOH terminus. B: Ponceau-S staining of purified GST-6His double-tagged ARNO(wt) and its domains was used as a protein quality and loading control. C: GST pull-down assay using in vitro translated ALDO-B as a prey. Recombinant ARNO(wt) and its domains were immobilized on glutathione beads and incubated with in vitro translated ALDO-B, and eluted complexes were analyzed by Western blot analysis using an anti-ALDO-A/B (D-18) antibody.
Estimation of the binding affinities using SPR analysis.
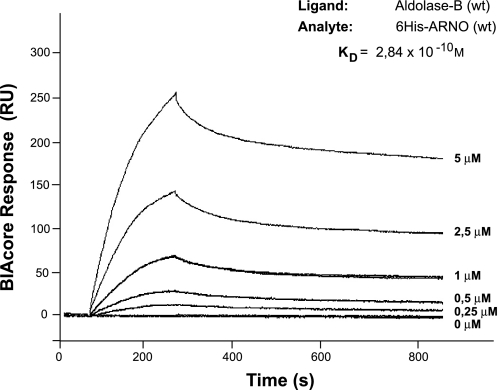
To determine the dissociation constants (KD) of ARNO(wt) and aldolase-B(wt) binding, we performed a SPR kinetic analysis using a BIAcore T100 system. For these experiments, highly purified aldolase-B was prepared as described in materials and methods. When various concentrations of 6His-ARNO(wt) were applied to aldolase-B(wt) immobilized on a sensor chip, a specific real-time binding between the two proteins was observed and quantified (Fig. 4). Binding curves were globally fitted to various binding models, provided by the BIAcore T100 evaluation software. The best-fitting models derivated from simple 1:1 kinetics. The bivalent analyte binding model was chosen for evaluation, since dimerization was previously demonstrated for ARNO (39, 57). The molecular weight of the ARNO dimer was considered in all calculations. Apparent rate constants obtained from the bivalent analyte model were ka1 = 1.9 × 102 M−1s−1, kd1 = 2.1 × 10−2 s−1, and ka2 = 7.1 × 10−6 RU−1s−1, kd2 = 2.0 × 10−4 s−1. The unit of ka2 in RU−1s−1 was converted to M−1s−1 using the following formula: ka2 (M−1s−1) = ka2 (RU−1s−1) × 100 × molecular weight of analyte, yielding ka2 = 7.2 × 101 M−1s−1. The overall dissociation constant for ARNO binding to aldolase-B was calculated as KD = (kd1/ka1) × (kd2/ka2), yielding 2.84 × 10−10 M (33).
Fig. 4.
Real-time and high-affinity interaction between ARNO and aldolase revealed by surface plasmon resonance (SPR). Kinetic analysis and estimation of a dissociation constant (KD) of full-length ARNO(wt) and aldolase(wt) interaction. ALDO-B was immobilized as ligand on the sensor chip, and 6His-ARNO(wt) at increasing concentrations was used as the analyte. Sensorgrams showing the binding of recombinant ALDO-B and ARNO are expressed in resonance units (RU). Evaluation was performed using the bivalent analyte binding model, since ARNO dimerization was previously demonstrated. The molecular weight of an ARNO dimer was considered in all calculations. The interaction isotherms were obtained, and the value for overall KD = 2.84 × 10−10M was determined using BIAcore T100 evaluation software.
Aldolase is associated with membranes of endosomal vesicles fractionated from MTC cells.
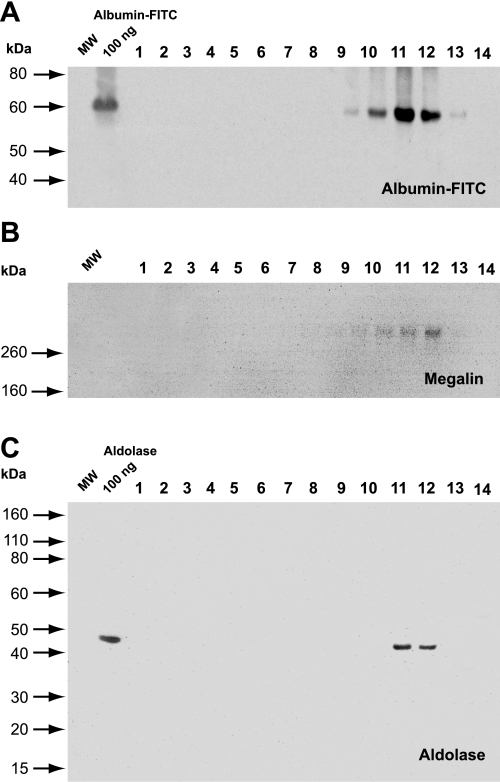
Previously, we demonstrated the direct interaction of ARNO with the V-ATPase in early endosomes of proximal tubule cells (29). Thus we evaluated whether aldolase, generally accepted as a cytosolic protein, could also be associated with endosomal membranes. To test this hypothesis, MTCs were pulsed with albumin-FITC for 20 min, to label early endosomes, followed by cell fractionation and Western blot analysis of albumin-FITC, megalin, and aldolase distribution in different fractions. The formation of the iodixanol (OptiPrep) density gradient is shown in Supplemental Fig. S1A. (Supplemental Material for this article is available online at the American Journal of Physiology-Cell Physiology website.) Protein concentration in the fractions (Supplemental Fig. 1SB) and the FITC fluorescence (Supplemental Fig. 1SC) were also determined (see Supplemental Material). In these cells, albumin is internalized by megalin/cubilin receptors and trafficked via the endosomal/lysosomal protein degradation pathway (1, 5, 29). Accordingly, both the ligand albumin-FITC (Fig. 5A) and the receptor megalin (Fig. 5B) were associated with light endosomal fractions 9–12, being more abundant in fractions 11 and 12. Importantly, aldolase was also found in the same fractions 11 and 12 and, therefore, is associated with membranes of endosomal vesicles fractionated from MTCs (Fig. 5C).
Fig. 5.
Cell fractionation and localization of endogenous aldolase in subcellular membrane fractions. For each gradient, the MTCs were grown in 150-mm plates, pulsed with albumin-FITC for 20 min to label early endosomes, and then fractionated as described in materials and methods. A: Western blot analysis and detection of endocytosed albumin-FITC in light fractions 9–12 of the gradient using an anti-FITC antibody. B: Western blot analysis and detection of the endocytotic receptor megalin in the same fractions 9–12 of the gradient. C: endogenous aldolase is associated with endosomal fractions 11 and 12 containing albumin-FITC and megalin receptor markers. Loading of 100 ng of albumin-FITC (A) and 100 ng of recombinant ALDO-A (C) were used as a positive controls.
Aldolase knockdown induces morphological changes and the formation of cell protrusions.
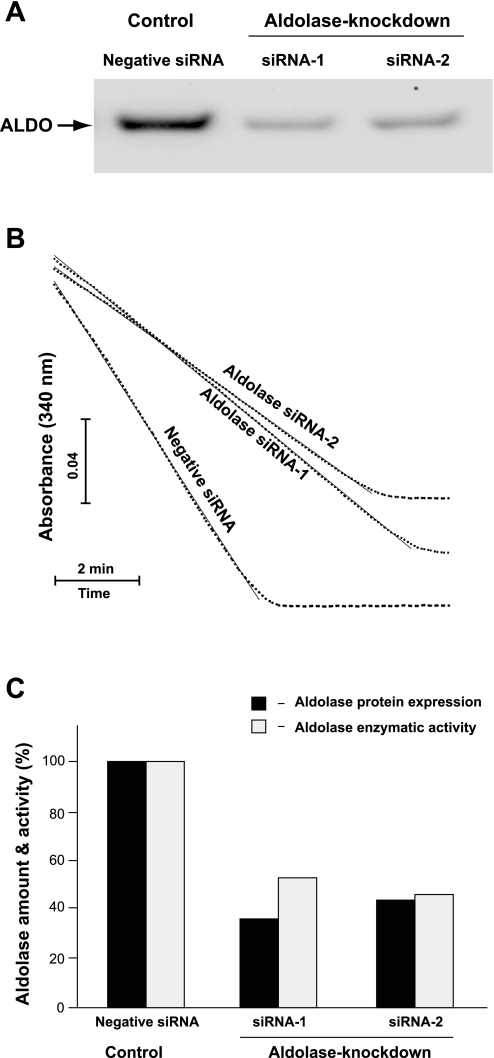
It was previously demonstrated that aldolase directly interacts with F-actin and V-ATPase and regulates their functions (35–37, 73, 74). Similarly, ARNO and Arf6 have been implicated in the regulation of the actin cytoskeleton (11, 16, 21, 56), and their acidification-dependent interaction with V-ATPase in endosomal membranes was also demonstrated in our previous studies (29). Together, these findings suggest that perhaps aldolase acts as a scaffolding protein that coordinates a dynamic interplay between actin cytoskeleton function and V-ATPase-dependent vesicular trafficking events. Thus we next investigated the phenotypic changes associated with aldolase knockdown at the level of 1) actin cytoskeleton dynamics, 2) acidic vesicle distribution, and 3) V-ATPase-driven vesicular acidification. Since in HeLa cells only aldolase-A is expressed, two sets of siRNAs fluorescently labeled by Alexa 555 targeting two different regions of human aldolase-A (aldolase siRNA-1 and aldolase siRNA-2) were designed and used in our studies as described in materials and methods. A nontargeting siRNA was used as negative siRNA control. The transfection efficiency with these siRNAs was monitored by fluorescence microscopy and was detected in 100% of cells inspected (data not shown). The efficiency of aldolase knockdown was estimated after 72 h of transfection at the protein level, using semiquantitative Western blot analysis (Fig. 6A), and by quantitative enzymatic aldolase activity assay (Fig. 6B). The slopes of linear portions of the graphs were used to compare initial velocities (Vi) of enzymatic activity of aldolase with 2 mM Fru 1,6-P2 as a substrate and were determined as follows: 1) −0.0056 for negative control siRNA-transfected cells, 2) −0.0029 for aldolase siRNA-1-transfected cells, and 3) −0.0026 for aldolase siRNA-2-transfected cells. Because these slopes are directly proportional to the initial velocities of the aldolase reaction, they were compared with each other to obtain relative aldolase activities in cell lysates, which were estimated as 1) 100% for negative control siRNA-transfected cells, 2) 53% for aldolase siRNA-1-transfected cells, and 3) 46% for aldolase siRNA-2-transfected cells. These experiments demonstrated that the results of two methods correlated well with each other, showing an ∼50% aldolase knockdown in HeLa cells (Fig. 6C).
Fig. 6.
Efficiency of aldolase knockdown by specific small interfering RNA (siRNA). A: Western blot analysis of aldolase expression levels was performed after 72-h transfection of HeLa cells with negative control siRNA (left lane), ALDO-A-selective siRNA-1 (middle lane), or ALDO-A-selective siRNA-2 (right lane). In each lane, 40 μg of total cell lysate proteins were loaded. Results of a representative experiment are shown. B: estimation of aldolase enzymatic activity in cell lysates after 72-h transfection of HeLa cells with negative control siRNA, ALDO-A-selective siRNA-1, or ALDO-A-selective siRNA-2. Data are presented as the time course of enzymatic cleavage of aldolase substrate fructose 1,6-bisphosphate (Fru 1,6-P2), normalized to total protein concentration as described in materials and methods. Results of a representative experiment are shown. C: histograms show relative amounts of aldolase protein expression and enzymatic activity in siRNA-transfected cells. Normalized densitometric quantification of Western blot signals from A is shown (solid bars). Relative aldolase enzymatic activities from B were calculated as described in materials and methods and are shown (shaded bars).
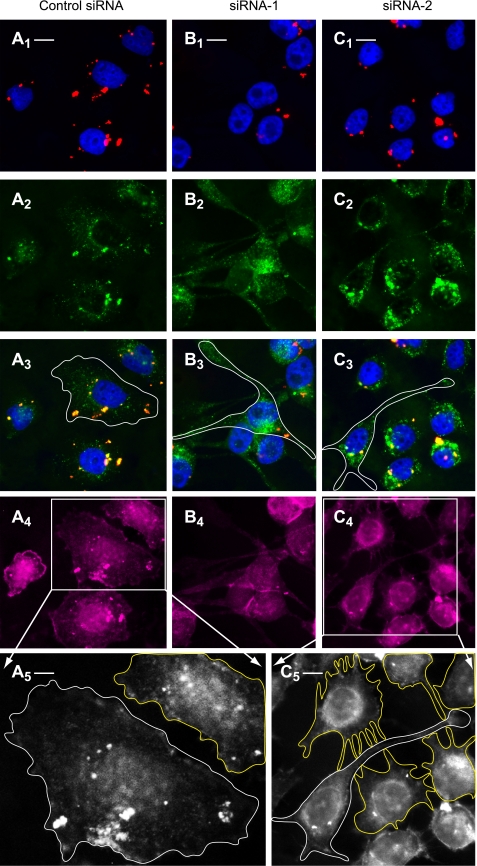
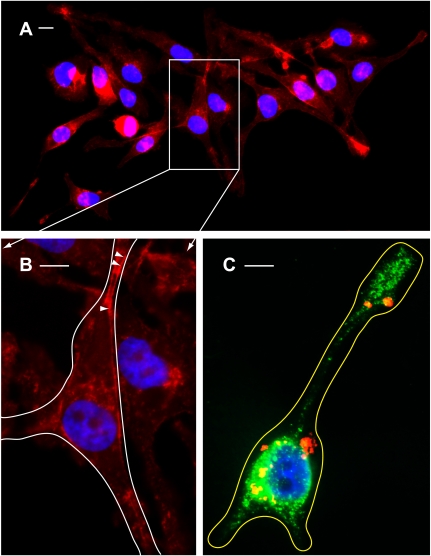
Having established an efficient method of aldolase downregulation, we next studied its effect on actin-dependent cell morphology. The actin cytoskeleton was labeled in normal and aldolase knockdown cells using Alexa 647-conjugated phalloidin. Knockdown of aldolase using either siRNA-1 (Fig. 7B) or siRNA-2 (Fig. 7C) leads to dramatic phenotypic changes in cell morphology characterized by the formation of elongated protrusions (Fig. 7, B3 and C3), which are absent in cells treated with control siRNA (Fig. 7A3). Importantly, staining of the actin cytoskeleton with Alexa 647-conjugated phalloidin revealed its role in the formation of long protrusions and short filopodia, which are involved in contacts between cells and give rise to the formation of an intensive cellular network in aldolase knockdown cells (Fig. 7C5; see also Supplemental Fig. S2B, showing an enlarged version of Fig. 7C5) but not in control cells (Fig. 7A5; see also Supplemental Fig. S2A, showing an enlarged version of Fig. 7A5). Finally, a detailed epifluorescence microscopy analysis revealed the involvement of the cortical actin cytoskeleton in the formation of cell protrusions (Fig. 8B, arrowheads).
Fig. 7.
Aldolase knockdown induces the formation of cell protrusions loaded with V-ATPase-dependent acidic vesicles. After transfection with negative Alexa 555-conjugated control siRNA (A1–A4), aldolase siRNA-1 (B1–B4), or aldolase siRNA-2 (C1–C4), HeLa cells were analyzed by quadruple epifluorescence microscopy. A1, B1, C1: cells transfected with Alexa 555-conjugated siRNA (red) and nuclei stained with 4,6-diamidino-2-phenylindole (DAPI; blue). Scale bars correspond to 5 μm for all images from first to fourth rows. A2, B2, C2: cells with acidic vesicles loaded with DAMP and stained with Alexa 488-conjugated dinitrophenyl-KLH (anti-DNP) antibodies (green). A3, B3, C3: merged images of A1–C1 and A2–C2. The borders of representative cells are outlined in white. A4, B4, C4: actin staining with Alexa 647-conjugated phalloidin (purple). A5 and C5: enlarged black and white images of boxes outlined in A4 and C4. These 2 enlarged images clearly show the borders of the cells and are also depicted in Supplemental Fig. S2. White and yellow lines outlining the borders of representative cells show the formation of cell protrusions and digitations in aldolase knockdown but not in control siRNA-transfected cells. Scale bar corresponds to 1 μm in A5 and 3 μm in C5.
Fig. 8.
Morphological changes characterized by the formation of cell protrusions and redistribution of acidic vesicles as a result of aldolase knockdown. A: wide-field view of cells transfected with Alexa 555-conjugated siRNA-1 (red) followed by staining of actin cytoskeleton with Alexa 647-conjugated phalloidin (red) and nuclei with DAPI (blue). Long cellular protrusions (up to 20–40 μm) were clearly detected in all cells. B: enlarged image of box outlined in A. Arrowheads show cortical actin filaments. The border of a representative cell is outlined in white. C: epifluorescence microscopy merged image showing the aldolase knockdown phenotype characterized by cell shape remodeling accompanied by formation of long protrusions loaded with acidic vesicles. This representative cell was transfected with Alexa 555-conjugated siRNA-1 (red) followed by loading of acidic vesicles with DAMP and staining with Alexa 488-conjugated anti-DNP antibodies (green) and nuclei staining with DAPI (blue). The border of the cell is outlined in yellow. Scale bars all correspond to 5 μM.
Aldolase knockdown causes subcellular redistribution of V-ATPase-driven acidic vesicles but does not change acidification capacity of endosomal/lysosomal compartments.
To study the effect of aldolase knockdown on the distribution of acidic vesicles, we used a DAMP assay. It consists of loading cells in vivo with the acidic pH probe DAMP, followed by cell fixation and detection with Alexa 488-conjugated anti-DNP antibodies and visualization of vesicles by epifluorescence microscopy. In these experiments we could not detect any difference in DAMP labeling of acidic vesicles between control siRNA (Fig. 7, A2 and A3) and aldolase siRNA-treated HeLa cells (Fig. 7, B2, B3, C2, and C3). However, it is clear that morphological changes of the cell shape caused by aldolase knockdown are accompanied by a major redistribution and clustering of acidic vesicles in the apex of newly formed protrusions and in the perinuclear region (Fig. 7, A3, B3, and C3, and Fig. 8C). To study the effect of aldolase knockdown on the acidification capacity of endosomal/lysosomal compartments, we implemented two additional quantitative assays using LysoTracker red DND-99 fluorometry and FITC-dextran ratiometric fluorometry (48). Experiments with LysoTracker demonstrated no changes in vesicular acidification in cells transfected with negative control siRNA, aldolase siRNA-1, or aldolase siRNA-2, as well as in mock-transfected cells. Bafilomycin used as a positive control decreased LysoTracker uptake and its fluorescence up to 90% of control, nontreated cells (not shown). Since no differences in the acidification of endosomes/lysosomes between control and aldolase siRNA-treated cells were detected using the LysoTracker approach, we then used a more sensitive and quantitative FITC-dextran ratiometric fluorometry method (48). The calculated pH range using this approach for both control and aldolase knockdown cells was 5.4–5.7, whereas treatment with bafilomycin increased intravesicular pH to about 6.5.
Aldolase expression level is inversely correlated with gelsolin expression levels.
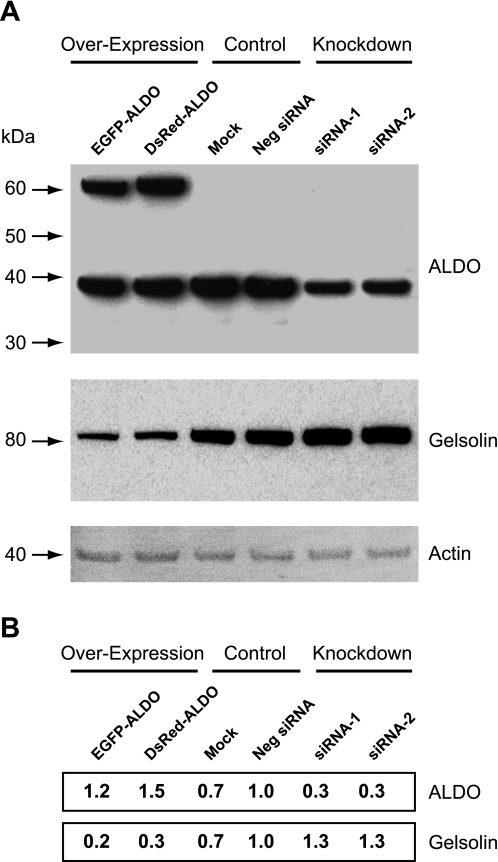
In addition to a prominent role of aldolase (73, 74) and ARNO/Arf6 (11, 56) in actin cytoskeleton remodeling, gelsolin is considered as the prototype of a family of actin binding proteins that also control actin cytoskeleton organization. Gelsolin is a well-studied protein that can bind, sever, and cap actin filaments and is involved in the control of cell morphology (67). Therefore, we next studied whether morphological changes observed in aldolase siRNA-treated HeLa cells are accompanied by changes in gelsolin expression. Indeed, we found that in aldolase knockdown cells, gelsolin expression was increased by about 30% compared with negative control siRNA-treated cells (Fig. 9, A and B). In contrast, overexpression of either EGFP-aldolase or DsRed-aldolase recombinant proteins to up to 150% of its endogenous level reduced gelsolin expression to about 20–30% of mock/negative siRNA-treated cells. Thus there is a clear reverse correlation between the expression levels of aldolase and gelsolin in HeLa cells.
Fig. 9.
Effect of ALDO-A knockdown or overexpression on the level of gelsolin expression in HeLa cells. A: cells were transfected with aldolase-specific siRNA or aldolase cDNA to knockdown or overexpress aldolase. Levels of protein expression were analyzed by Western blotting. The same blot was probed with antibodies for ALDO-A (top), gelsolin (middle), and actin as a loading control (bottom). B: densitometric quantification of the levels of expression of ALDO-A and gelsolin protein in HeLa cells. Numbers correspond to the intensities of the bands relative to the negative control siRNA-transfected cells (Neg siRNA). EGFP, enhanced green fluorescent protein; DsRed, monomeric red fluorescent protein. Values are shown in the same order as the bands presented in A.
DISCUSSION
Previously, we demonstrated that in kidney proximal tubule epithelial cells, ARNO and Arf6 are targeted to endosomal compartments of the protein degradative pathway, where they colocalize with endosomal V-ATPase in situ as well as in purified early endosomes in vitro (23, 40). We also showed that V-ATPase-dependent intraendosomal acidification modulates the direct interaction of ARNO with the a2-isoform of V-ATPase and that these interactions play an important role in the regulation of the endosomal/lysosomal pathway (29, 40). We recently mapped the binding sites of interaction between these two proteins (43). In the current study, we have demonstrated that ARNO not only directly interacts with the a2-subunit isoform but also directly interacts with the a1-, a3-, and a4-isoforms of the V-ATPase. Importantly, the apparent differential binding capacity of a-isoforms with ARNO in pull-down experiments may reflect their differential interaction affinities, which need to be further studied using the SPR assay. This approach was successfully used in our recent study to demonstrate and estimate the high affinity of interaction between a2N of V-ATPase and ARNO (43). Because of the widespread and differential cellular distribution of a-subunit isoforms (24, 42), these results indicate that the association of membrane-bound V-ATPases with cytosolic small GTPases is a general phenomenon that may take place on a variety of cell and organelle membranes, and thus this event may regulate a variety of cell biological processes.
Next, we aimed to search for other proteins that could directly interact with both V-ATPase and ARNO and might be involved in the formation and/or modulation of V-ATPase/small GTPase complex function. We focused on aldolase as a potential candidate based on current experimental evidence linking aldolase with regulation of V-ATPase function (32, 35, 50). An emerging important regulatory mechanism of V-ATPase function is the reversible assembly/disassembly of the V0 and V1 sectors, which, for instance, plays a crucial role in the regulation of the endosomal/lysosomal protein degradative pathway in dendritic cells (70). In yeast, the assembly mechanism is regulated by a-isoforms (VPH1/STV1) of the V-ATPase and is dependent on the cellular and membrane environment of the vacuole or Golgi compartments in which these two isoforms are located (8, 24, 53, 61). Thus in this study we investigated the potential interaction of ARNO with aldolase. Using a combination of protein-protein interaction techniques, we demonstrated that ARNO indeed directly and specifically interacts with both recombinant and native aldolases. Moreover, our real-time binding and kinetic analysis using SPR (BIAcore) revealed a high-avidity interaction (KD = 2.84 × 10−10 M) between ARNO and aldolase. Recently, using peptides and recombinant proteins pull-down experiments, we identified the catalytic Sec7 domain of ARNO as the major binding site with the V-ATPase a2-subunit isoform (43). In the present study, we used a similar approach to identify the regulatory lipid-binding PH domain of ARNO as the unique binding site with aldolase. Importantly, the PH domain of ARNO was also shown to be involved in interaction with Arf6 (15) and Arl4 proteins (27), which are involved in modulation of its autoinhibited state and regulation of its enzymatic and cell biological activities (66).
Using an iodixanol gradient-based cell fractionation approach, in the present study we demonstrated for the first time the association of aldolase with early endosomal membranes containing the megalin/albumin-FITC receptor/ligand complex (29). Separation of endosomal fractions using iodixanol gradients has been previously used for kidney-derived Madin-Darby canine kidney (MDKC) (62) and baby hamster kidney (BHK) cells (25). It is noteworthy that in our fractionation experiments with MTCs, aldolase was found associated with the light (∼10% iodixanol) membrane endosomal fractions. Similarly, ARNO was also found associated with the light ∼10% iodixanol membrane fraction in BHK cells (25). Consistent with these findings, our previous studies demonstrated that ARNO interacts with the a2-isoform of V-ATPase in proximal tubule endosomes (29). In summary, our data demonstrate that detectable levels of aldolase, which is generally considered a predominantly cytosolic protein, are associated with the membranes of purified early endosomes, where it could directly interact with ARNO and V-ATPase, and thus it may modulate their respective functions.
Previously, the role of aldolase in the regulation of V-ATPase and the actin cytoskeleton was examined in yeast and mammalian cells. Since our findings suggested that aldolase could modulate the interactions and regulate activities of both V-ATPase and ARNO, we next studied the effect of aldolase knockdown on actin cytoskeleton-dependent cell morphology and distribution of endosomal/lysosomal compartments. We used two specific siRNAs capable of downregulating by ∼50% the levels of the predominant HeLa cell aldolase-A. These experiments demonstrated that this level of aldolase knockdown was sufficient to induce dramatic morphological changes characterized by the formation of cell protrusions and the redistribution of V-ATPase-containing acidic vesicles. Since a regulatory role of aldolase in assembly/disassembly of V-ATPase has been previously suggested (6, 35–37), we next investigated whether aldolase knockdown could lead to changes in the V-ATPase-driven acidification of endosomal/lysosomal compartments. However, using three independent vesicle acidification assays, we demonstrated that 50% aldolase knockdown does not alter the acidification capacity of endosomal/lysosomal compartments in these cells. We suggest the following explanations for this finding, which could be tested in future studies. First, we hypothesized that although a 50% aldolase knockdown is sufficient for inducing generalized changes in cell morphology, a more efficient knockdown of aldolase may be required to affect other more localized and specific events regulated by the V-ATPase/ARNO/aldolase complex on endosomal compartments. Second, it is well known that glucose/aldolase-dependent modulation of V-ATPase in yeast strongly depends on the origin of the vesicular compartments examined (vacuoles vs. Golgi) and their cellular microenvironment (53). Similar mechanisms could also exist in mammalian cells. Therefore, the use of more specific and sensitive techniques to measure exclusively endosomal acidification vs. total endosomal/lysosomal acidification may be required to test this possibility.
To explain the dramatic effect of aldolase knockdown on cell morphology and acidic vesicle redistribution, we also propose the following three cell biological mechanisms. The first is a direct effect of aldolase on actin cytoskeleton rearrangement. The direct interaction of aldolase with F-actin in vitro (73) and its important role in regulation of the actin cytoskeleton in vivo have been previously examined (74). It was proposed that aldolase may play a direct structural and/or regulatory role in actin cytoskeletal function, cell compartmentalization, and mobility (73, 74). Therefore, the results obtained in our aldolase knockdown experiments on cell morphology could be explained by a direct role of aldolase in actin cytoskeleton-dependent cell shape remodeling. The second mechanism is a novel role of aldolase as a regulator of ARNO/Arf6 signaling during actin cytoskeleton rearrangement. Previously, both ARNO and Arf6 have been implicated as important regulators of actin cytoskeleton remodeling (11, 56). Therefore, the high affinity and specific interaction of aldolase with the regulatory PH domain of ARNO uncovered in this study could also be involved in the modulation of ARNO and Arf6 function on actin cytoskeleton via either 1) a generalized action, e.g., modulation of the actin cytoskeleton induces general cell shape remodeling and protrusion formation; and/or 2) a localized action, e.g., modulation of the V-ATPase/Arf6/ARNO/aldolase complex on endosomes followed by a local rearrangement of the actin cytoskeleton near the endosomal membranes, eliciting a redistribution of the acidic vesicles. The third proposed mechanism is an unexpected role of aldolase as a potential regulator of the ARNO-dependent gene expression signaling cascade that is modulated during actin cytoskeleton rearrangement events. Rearrangement of the actin cytoskeleton and modulation of cell morphology involve a variety of actin binding proteins, including gelsolin (67). Interestingly, modulation of the actin cytoskeleton via gelsolin could also regulate V-ATPase recycling (4). Importantly, previous studies revealed that gelsolin/actin-dependent epithelial cell invasion during tumorigenesis is a highly regulated process dependent on signaling of Ras, Rac, and Arf small GTPases (19). In particular, it has been shown that in HeLa cells, Arf6-dependent formation of cell protrusions was accompanied by simultaneous redistribution and colocalization of gelsolin, indicating cross talk between the ARNO/Arf6 and actin/gelsolin pathways (54). On the other hand, it is noteworthy that in addition to their role in modulation of Arf small GTPase activity, proteins of the cytohesin family have recently emerged as important transcriptional regulators. In particular, cytohesin-2 (ARNO) has been shown to participate in regulation of gene expression via the MAPK signaling pathway during serum-mediated transcriptional activation in nonimmune cells (68), whereas cytohesin-1 has been shown to be involved in activating the IL-2 gene promoter in T cells (51). Moreover, the scaffolding protein specifically interacting with cytohesin-1 in T cells, called cytohesin binder and regulator (Cybr), also has been shown to be involved in MAPK-mediated and T cell antigen receptor-dependent gene transcription (12). These studies indicate that transcriptional regulation by cytohesins may be a widespread phenomenon among the small GEFs, and particular members of the cytohesin family could be regulated by cell type-specific interacting molecules. Thus our data suggest that aldolase could be one of these cytohesin-2 (ARNO)-interacting molecules that may be involved in modulation of ARNO transcriptional activity via its interaction with the regulatory PH domain of ARNO described in our study. This novel and exciting hypothesis awaits experimental testing and confirmation in future studies.
In summary, this study uncovers a novel interaction between ARNO, a crucial modulator of Arf-small GTPase signaling, and aldolase, an “old” glycolytic enzyme with emerging novel “moonlighting” functions as a scaffolder and modulator of protein complexes, including the actin cytoskeleton and the V-ATPase nanomotor, among others. By employing a variety of protein-protein interaction techniques, we identified aldolase as a specific and high-affinity interactor of ARNO, which, in turn, interacts with all four a-subunit isoforms of the V-ATPase. Thus these data suggest that the interaction of ARNO with aldolase and V-ATPases, which are specifically targeted to different subcellular compartments by the a-subunit isoforms, is a widespread phenomenon that may take place on a variety of cell membranes and organelles. In searching for the functional significance of this interaction, we uncovered a critical role of aldolase in actin cytoskeleton remodeling that results in the formation of cell protrusions and acidic vesicle redistribution. Thus we propose that aldolase is a novel modulator of V-ATPase/Arf6/ARNO and actin/gelsolin signaling pathways and is involved in the coordination of actin-dependent cell shape remodeling and acidic vesicle redistribution. However, the detailed interplay between these pathways and the intertwined role of aldolase in regulation of both vesicular trafficking and the cellular cytoskeleton remain to be tested in further studies.
GRANTS
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant DK038452 (to V. Marshansky, D. Brown, and D. A. Ausiello) and a pilot and feasibility study supported by a Boston Area Diabetes Research Center NIDDK Grant DK057521-08 (to V. Marshansky).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Masamitsu Futai for providing cDNAs of the full-length wild-type mouse V-ATPase a1-, a2-, a3-, and a4-isoforms. We are also grateful to Dr. Jim Casanova and Dr. Sylvain Bourgoin for providing cDNA of triglycine variant of human wild-type ARNO. We thank Dr. Dean Tolan and Carolyn Ritterson Lew (Boston University) for providing reagents, equipment, and training to perform aldolase activity assay and also for constructive suggestions and discussions during preparation of aldolase-A siRNA knockdown experiments.
REFERENCES
- 1. Amsellem S, Gburek J, Hamard G, Nielsen R, Willnow TE, Devuyst O, Nexo E, Verroust PJ, Christensen EI, Kozyraki R. Cubilin is essential for albumin reabsorption in the renal proximal tubule. J Am Soc Nephrol 21: 1859–1867, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arakaki TL, Pezza JA, Cronin MA, Hopkins CE, Zimmer DB, Tolan DR, Allen KN. Structure of human brain fructose 1,6-(bis)phosphate aldolase: linking isozyme structure with function. Protein Sci 13: 3077–3084, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balasubramanian N, Scott DW, Castle JD, Casanova JE, Schwartz MA. Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nat Cell Biol 9: 1381–1391, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beaulieu V, Da Silva N, Pastor-Soler N, Brown CR, Smith PJ, Brown D, Breton S. Modulation of the actin cytoskeleton via gelsolin regulates vacuolar H+-ATPase recycling. J Biol Chem 280: 8452–8463, 2005 [DOI] [PubMed] [Google Scholar]
- 5. Birn H, Christensen EI. Renal albumin absorption in physiology and pathology. Kidney Int 69: 440–449, 2006 [DOI] [PubMed] [Google Scholar]
- 6. Bond S, Forgac M. The Ras/cAMP/protein kinase A pathway regulates glucose-dependent assembly of the vacuolar (H+)-ATPase in yeast. J Biol Chem 283: 36513–36521, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348: 125–132, 1990 [DOI] [PubMed] [Google Scholar]
- 8. Brown D, Breton S, Ausiello DA, Marshansky V. Sensing, signaling and sorting events in kidney epithelial cell physiology. Traffic 10: 275–284, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown D, Marshansky V. Renal V-ATPase: physiology and pathophysiology. In: Handbook of ATPases, edited by Futai M, Wada Y, Kaplan JH. Weinheim, Germany: Wiley-VCH, 2004, p. 414–442 [Google Scholar]
- 10. Buscaglia CA, Penesetti D, Tao M, Nussenzweig V. Characterization of an aldolase-binding site in the Wiskott-Aldrich syndrome protein. J Biol Chem 281: 1324–1331, 2006 [DOI] [PubMed] [Google Scholar]
- 11. Casanova JE. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic 8: 1476–1485, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Chen Q, Coffey A, Bourgoin SG, Gadina M. Cytohesin binder and regulator augments T cell receptor-induced nuclear factor of activated T cells AP-1 activation through regulation of the JNK pathway. J Biol Chem 281: 19985–19994, 2006 [DOI] [PubMed] [Google Scholar]
- 13. Choukroun GJ, Marshansky V, Gustafson CE, McKee M, Hajjar RJ, Rosenzweig A, Brown D, Bonventre JV. Cytosolic phospholipase A2 regulates golgi structure and modulates intracellular trafficking of membrane proteins. J Clin Invest 106: 983–993, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chun J, Shapovalova Z, Dejgaard SY, Presley JF, Melancon P. Characterization of class I and II ADP-ribosylation factors (Arfs) in live cells: GDP-bound class II Arfs associate with the ER-Golgi intermediate compartment independently of GBF1. Mol Biol Cell 19: 3488–3500, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell 18: 2244–2253, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 7: 347–358, 2006 [DOI] [PubMed] [Google Scholar]
- 17. Dalby A, Dauter Z, Littlechild JA. Crystal structure of human muscle aldolase complexed with fructose 1,6-bisphosphate: mechanistic implications. Protein Sci 8: 291–297, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dalby AR, Tolan DR, Littlechild JA. The structure of human liver fructose-1,6-bisphosphate aldolase. Acta Crystallogr D Biol Crystallogr 57: 1526–1533, 2001 [DOI] [PubMed] [Google Scholar]
- 19. De Corte V, Bruyneel E, Boucherie C, Mareel M, Vandekerckhove J, Gettemans J. Gelsolin-induced epithelial cell invasion is dependent on Ras-Rac signaling. EMBO J 21: 6781–6790, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diepholz M, Venzke D, Prinz S, Batisse C, Florchinger B, Rossle M, Svergun DI, Bottcher B, Fethiere J. A different conformation for EGC stator subcomplex in solution and in the assembled yeast V-ATPase: possible implications for regulatory disassembly. Structure 16: 1789–1798, 2008 [DOI] [PubMed] [Google Scholar]
- 21. Donaldson JG, Jackson CL. Regulators and effectors of the ARF GTPases. Curr Opin Cell Biol 12: 475–482, 2000 [DOI] [PubMed] [Google Scholar]
- 22. Donaldson JG, Klausner RD. ARF: a key regulatory switch in membrane traffic and organelle structure. Curr Opin Cell Biol 6: 527–532, 1994 [DOI] [PubMed] [Google Scholar]
- 23. El-Annan J, Brown D, Breton S, Bourgoin S, Ausiello DA, Marshansky V. Differential expression and targeting of endogenous Arf1 and Arf6 small GTPases in kidney epithelial cells in situ. Am J Physiol Cell Physiol 286: C768–C778, 2004 [DOI] [PubMed] [Google Scholar]
- 24. Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol 8: 917–929, 2007 [DOI] [PubMed] [Google Scholar]
- 25. Frank S, Upender S, Hansen SH, Casanova JE. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J Biol Chem 273: 23–27, 1998 [DOI] [PubMed] [Google Scholar]
- 26. Garceau V, Houle MG, Chouinard F, Gagnon S, Harbour D, Naccache PH, Bourgoin SG. Characterization of cytohesin-1 monoclonal antibodies: expression in neutrophils and during granulocytic maturation of HL-60 cells. J Immunol Methods 249: 121–136, 2001 [DOI] [PubMed] [Google Scholar]
- 27. Hofmann I, Thompson A, Sanderson CM, Munro S. The Arl4 family of small G proteins can recruit the cytohesin Arf6 exchange factors to the plasma membrane. Curr Biol 17: 711–716, 2007 [DOI] [PubMed] [Google Scholar]
- 28. Hosokawa H, Merkulova M, Zhuang JZ, Dip PV, Xiaoying J, Randazzo PA, Grüber G, Ausiello DA, Marshansky V. V-ATPase modulate enzymatic activity of Arf-GEF ARNO and controls protein degradative pathway. 50th Annual Meeting of American Society for Cell Biology (Abstract of Poster Presentation), 2010 [Google Scholar]
- 29. Hurtado-Lorenzo A, Skinner M, El Annan J, Futai M, Sun-Wada GH, Bourgoin S, Casanova J, Wildeman A, Bechoua S, Ausiello DA, Brown D, Marshansky V. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat Cell Biol 8: 124–136, 2006 [DOI] [PubMed] [Google Scholar]
- 30. Jackson CL, Casanova JE. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol 10: 60–67, 2000 [DOI] [PubMed] [Google Scholar]
- 31. Kane PM. Regulation of V-ATPases by reversible disassembly. FEBS Lett 469: 137–141, 2000 [DOI] [PubMed] [Google Scholar]
- 32. Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev 70: 177–191, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karlsson R, Mo JA, Holmdahl R. Binding of autoreactive mouse anti-type II collagen antibodies derived from the primary and the secondary immune response investigated with the biosensor technique. J Immunol Methods 188: 63–71, 1995 [DOI] [PubMed] [Google Scholar]
- 34. Kim JH, Lee S, Lee TG, Hirata M, Suh PG, Ryu SH. Phospholipase D2 directly interacts with aldolase via its PH domain. Biochemistry 41: 3414–3421, 2002 [DOI] [PubMed] [Google Scholar]
- 35. Lu M, Ammar D, Ives H, Albrecht F, Gluck SL. Physical interaction between aldolase and vacuolar H+-ATPase is essential for the assembly and activity of the proton pump. J Biol Chem 282: 24495–24503, 2007 [DOI] [PubMed] [Google Scholar]
- 36. Lu M, Holliday LS, Zhang L, Dunn WA, Jr, Gluck SL. Interaction between aldolase and vacuolar H+-ATPase: evidence for direct coupling of glycolysis to the ATP-hydrolyzing proton pump. J Biol Chem 276: 30407–30413, 2001 [DOI] [PubMed] [Google Scholar]
- 37. Lu M, Sautin YY, Holliday LS, Gluck SL. The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J Biol Chem 279: 8732–8739, 2004 [DOI] [PubMed] [Google Scholar]
- 38. Malay AD, Allen KN, Tolan DR. Structure of the thermolabile mutant aldolase B, A149P: molecular basis of hereditary fructose intolerance. J Mol Biol 347: 135–144, 2005 [DOI] [PubMed] [Google Scholar]
- 39. Mansour M, Lee SY, Pohajdak B. The N-terminal coiled coil domain of the cytohesin/ARNO family of guanine nucleotide exchange factors interacts with the scaffolding protein CASP. J Biol Chem 277: 32302–32309, 2002 [DOI] [PubMed] [Google Scholar]
- 40. Maranda B, Brown D, Bourgoin S, Casanova JE, Vinay P, Ausiello DA, Marshansky V. Intra-endosomal pH-sensitive recruitment of the Arf-nucleotide exchange factor ARNO and Arf6 from cytoplasm to proximal tubule endosomes. J Biol Chem 276: 18540–18550, 2001 [DOI] [PubMed] [Google Scholar]
- 41. Marshansky V. The V-ATPase a2-subunit as a putative endosomal pH-sensor. Biochem Soc Trans 35: 1092–1099, 2007 [DOI] [PubMed] [Google Scholar]
- 42. Marshansky V, Futai M. The V-type-ATPase in vesicular trafficking: targeting, regulation and function. Curr Opin Cell Biol 20: 415–426, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Merkulova M, Bakulina A, Thaker YR, Grüber G, Marshansky V. Specific motifs of the V-ATPase a2-subunit isoform interact with catalytic and regulatory domains of ARNO. Biochim Biophys Acta 1797: 1398–1409, 2010 [DOI] [PubMed] [Google Scholar]
- 44. Miranda KC, Karet FE, Brown D. An extended nomenclature for mammalian V-ATPase subunit genes and splice variants. PLoS One 5: e9531, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Muench SP, Huss M, Song CF, Phillips C, Wieczorek H, Trinick J, Harrison MA. Cryo-electron microscopy of the vacuolar ATPase motor reveals its mechanical and regulatory complexity. J Mol Biol 386: 989–999, 2009 [DOI] [PubMed] [Google Scholar]
- 46. Myers KR, Casanova JE. Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol 18: 184–192, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nishi T, Forgac M. The vacuolar-ATPases—nature's most versatile proton pumps. Nat Rev Mol Cell Biol 3: 94–103, 2002 [DOI] [PubMed] [Google Scholar]
- 48. Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci USA 75: 3327–3331, 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oka T, Murata Y, Namba M, Yoshimizu T, Toyomura T, Yamamoto A, Sun-Wada GH, Hamasaki N, Wada Y, Futai M. a4, a unique kidney-specific isoform of mouse vacuolar H+-ATPase subunit a. J Biol Chem 276: 40050–40054, 2001 [DOI] [PubMed] [Google Scholar]
- 50. Parra KJ, Kane PM. Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol Cell Biol 18: 7064–7074, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perez OD, Mitchell D, Jager GC, South S, Murriel C, McBride J, Herzenberg LA, Kinoshita S, Nolan GP. Leukocyte functional antigen 1 lowers T cell activation thresholds and signaling through cytohesin-1 and Jun-activating binding protein 1. Nat Immunol 4: 1083–1092, 2003 [DOI] [PubMed] [Google Scholar]
- 52. Pezza JA, Stopa JD, Brunyak EM, Allen KN, Tolan DR. Thermodynamic analysis shows conformational coupling and dynamics confer substrate specificity in fructose-1,6-bisphosphate aldolase. Biochemistry 46: 13010–13018, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Qi J, Forgac M. Cellular environment is important in controlling V-ATPase dissociation and its dependence on activity. J Biol Chem 282: 24743–24751, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Radhakrishna H, Klausner RD, Donaldson JG. Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. J Cell Biol 134: 935–947, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rangarajan ES, Park H, Fortin E, Sygusch J, Izard T. Mechanism of aldolase control of sorting nexin 9 function in endocytosis. J Biol Chem 285: 11983–11990, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Santy LC, Casanova JE. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J Cell Biol 154: 599–610, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Santy LC, Frank SR, Hatfield JC, Casanova JE. Regulation of ARNO nucleotide exchange by a PH domain electrostatic switch. Curr Biol 9: 1173–1176, 1999 [DOI] [PubMed] [Google Scholar]
- 58. Sautin YY, Lu M, Gaugler A, Zhang L, Gluck SL. Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol Cell Biol 25: 575–589, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schafer DA, D'Souza-Schorey C, Cooper JA. Actin assembly at membranes controlled by ARF6. Traffic 1: 892–903, 2000 [DOI] [PubMed] [Google Scholar]
- 60. Schekman R, Orci L. Coat proteins and vesicle budding. Science 271: 1526–1533, 1996 [DOI] [PubMed] [Google Scholar]
- 61. Shao E, Forgac M. Involvement of the nonhomologous region of subunit A of the yeast V-ATPase in coupling and in vivo dissociation. J Biol Chem 279: 48663–48670, 2004 [DOI] [PubMed] [Google Scholar]
- 62. Sheff DR, Daro EA, Hull M, Mellman I. The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J Cell Biol 145: 123–139, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Smith AN, Finberg KE, Wagner CA, Lifton RP, Devonald MA, Su Y, Karet FE. Molecular cloning and characterization of Atp6n1b: a novel fourth murine vacuolar H+-ATPase a-subunit gene. J Biol Chem 276: 42382–42388, 2001 [DOI] [PubMed] [Google Scholar]
- 64. Smith AN, Lovering RC, Futai M, Takeda J, Brown D, Karet FE. Revised nomenclature for mammalian vacuolar-type H+-ATPase subunit genes. Mol Cell 12: 801–803, 2003 [DOI] [PubMed] [Google Scholar]
- 65. Smith AN, Skaug J, Choate KA, Nayir A, Bakkaloglu A, Ozen S, Hulton SA, Sanjad SA, Al-Sabban EA, Lifton RP, Scherer SW, Karet FE. Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat Genet 26: 71–75, 2000 [DOI] [PubMed] [Google Scholar]
- 66. Stalder D, Barelli H, Gautier R, Macia E, Jackson CL, Antonny B. Kinetic studies of the Arf activator Arno on model membranes in the presence of Arf effectors suggest control by a positive feedback loop. J Biol Chem 286: 3873–3883, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem 274: 33179–33182, 1999 [DOI] [PubMed] [Google Scholar]
- 68. Theis MG, Knorre A, Kellersch B, Moelleken J, Wieland F, Kolanus W, Famulok M. Discriminatory aptamer reveals serum response element transcription regulated by cytohesin-2. Proc Natl Acad Sci USA 101: 11221–11226, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Toyomura T, Oka T, Yamaguchi C, Wada Y, Futai M. Three subunit a isoforms of mouse vacuolar H+-ATPase. Preferential expression of the a3 isoform during osteoclast differentiation. J Biol Chem 275: 8760–8765, 2000 [DOI] [PubMed] [Google Scholar]
- 70. Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science 299: 1400–1403, 2003 [DOI] [PubMed] [Google Scholar]
- 71. Volker KW, Knull H. A glycolytic enzyme binding domain on tubulin. Arch Biochem Biophys 338: 237–243, 1997 [DOI] [PubMed] [Google Scholar]
- 72. Wagner CA, Finberg KE, Breton S, Marshansky V, Brown D, Geibel JP. Renal vacuolar-ATPase. Physiol Rev 84: 1263–1314, 2004 [DOI] [PubMed] [Google Scholar]
- 73. Wang J, Morris AJ, Tolan DR, Pagliaro L. The molecular nature of the F-actin binding activity of aldolase revealed with site-directed mutants. J Biol Chem 271: 6861–6865, 1996 [PubMed] [Google Scholar]
- 74. Wang J, Tolan DR, Pagliaro L. Metabolic compartmentation in living cells: structural association of aldolase. Exp Cell Res 237: 445–451, 1997 [DOI] [PubMed] [Google Scholar]
- 75. Zhang Z, Zheng Y, Mazon H, Milgrom E, Kitagawa N, Kish-Trier E, Heck AJ, Kane PM, Wilkens S. Structure of the yeast vacuolar ATPase. J Biol Chem 283: 35983–35995, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.