Abstract
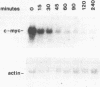
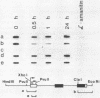
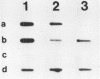
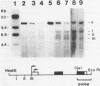
DMSO (dimethylsulfoxide), a potent inducer of granulocytic differentiation in HL60 cells, causes a rapid decrease of cytoplasmic steady state c-myc RNA. This decrease is regulated mainly at the level of transcript elongation. Elongation is blocked within the untranslated c-myc leader. Twelve hours after transcriptional shut off of c-myc, DNAase I hypersensitive site II was still detectable, indicating that closing of this site upstream of the gene does not correlate with reduction in the steady state level of c-myc RNA.
Full text
PDF




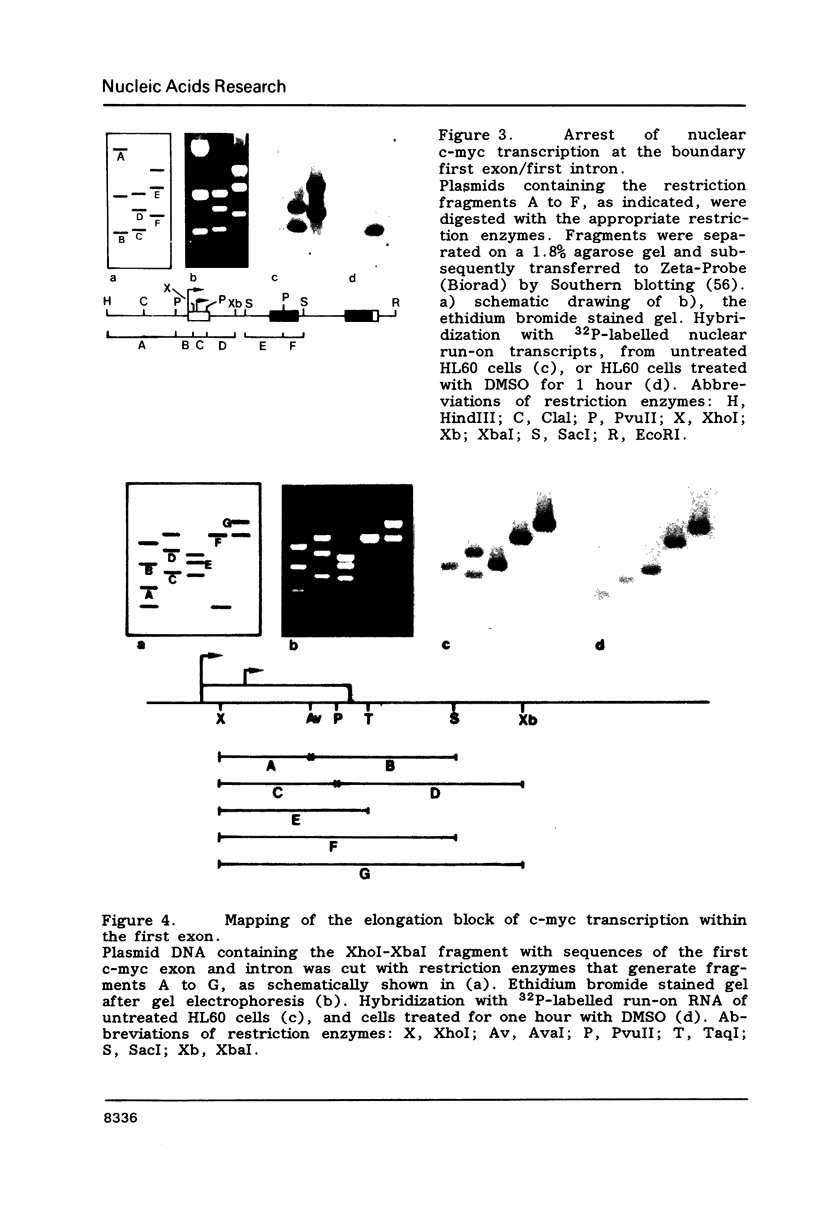

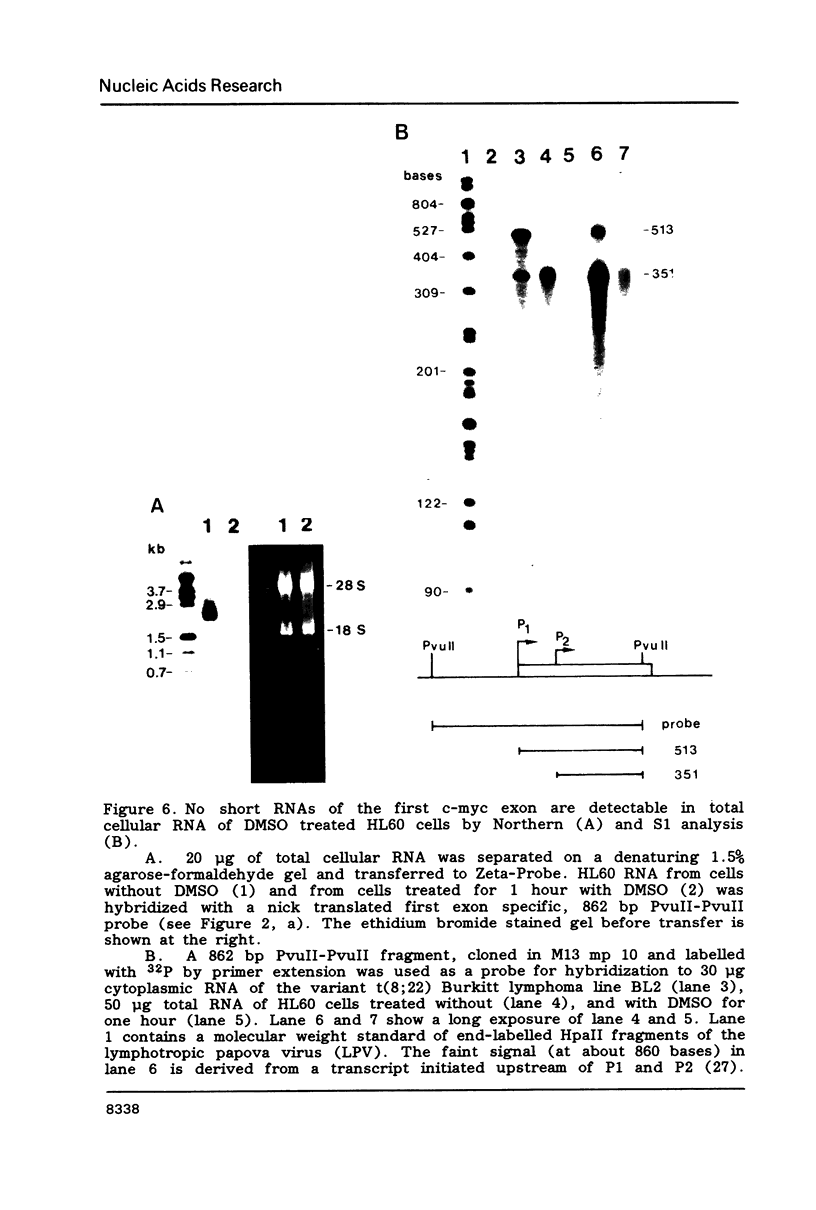
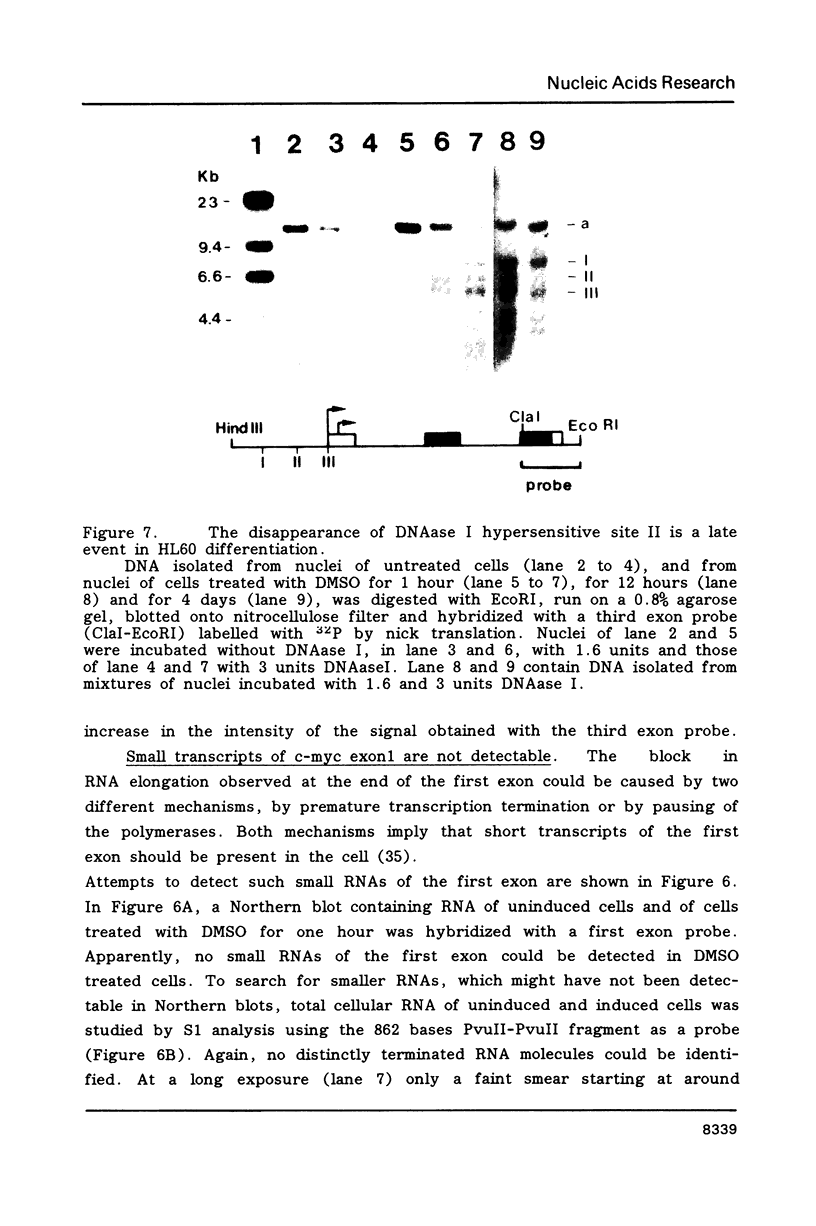


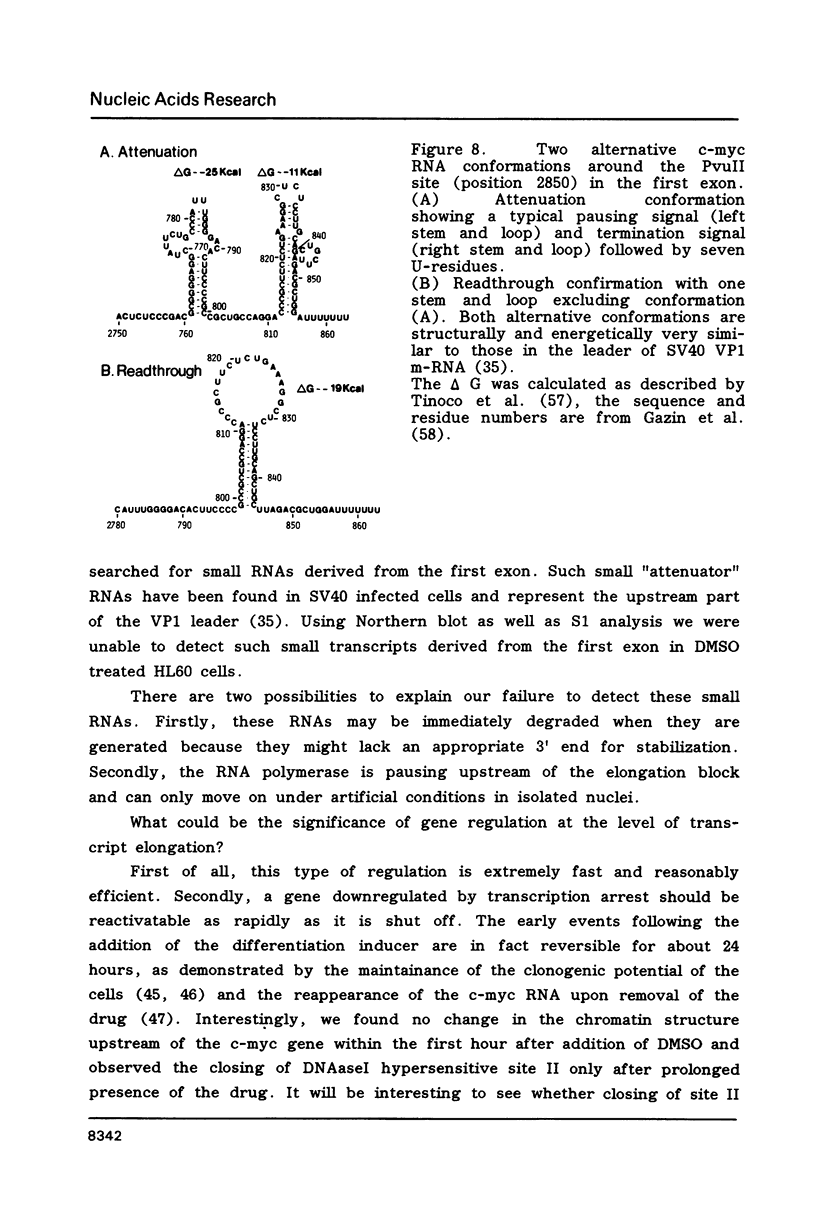




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Harris A. W., Pinkert C. A., Corcoran L. M., Alexander W. S., Cory S., Palmiter R. D., Brinster R. L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985 Dec 12;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Boyd A. W., Metcalf D. Induction of differentiation in HL60 leukaemic cells: a cell cycle dependent all-or-none event. Leuk Res. 1984;8(1):27–43. doi: 10.1016/0145-2126(84)90029-8. [DOI] [PubMed] [Google Scholar]
- Campisi J., Gray H. E., Pardee A. B., Dean M., Sonenshein G. E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984 Feb;36(2):241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- Chiu N. H., Bruszewski W. B., Salzman N. P. Evidence for the role of double-helical structures in the maturation of simian virus-40 messenger RNA. Nucleic Acids Res. 1980 Jan 11;8(1):153–168. doi: 10.1093/nar/8.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. J., Ruscetti F. W., Gallagher R. E., Gallo R. C. Terminal differentiation of human promyelocytic leukemia cells induced by dimethyl sulfoxide and other polar compounds. Proc Natl Acad Sci U S A. 1978 May;75(5):2458–2462. doi: 10.1073/pnas.75.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S., Groudine M. Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature. 1982 Aug 12;298(5875):679–681. doi: 10.1038/298679a0. [DOI] [PubMed] [Google Scholar]
- Cory S., Graham M., Webb E., Corcoran L., Adams J. M. Variant (6;15) translocations in murine plasmacytomas involve a chromosome 15 locus at least 72 kb from the c-myc oncogene. EMBO J. 1985 Mar;4(3):675–681. doi: 10.1002/j.1460-2075.1985.tb03682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce C. M., Thierfelder W., Erikson J., Nishikura K., Finan J., Lenoir G. M., Nowell P. C. Transcriptional activation of an unrearranged and untranslocated c-myc oncogene by translocation of a C lambda locus in Burkitt. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6922–6926. doi: 10.1073/pnas.80.22.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla-Favera R., Wong-Staal F., Gallo R. C. Onc gene amplification in promyelocytic leukaemia cell line HL-60 and primary leukaemic cells of the same patient. Nature. 1982 Sep 2;299(5878):61–63. doi: 10.1038/299061a0. [DOI] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani C., Mechti N., Piechaczyk M., Lebleu B., Jeanteur P., Blanchard J. M. Increased rate of degradation of c-myc mRNA in interferon-treated Daudi cells. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4896–4899. doi: 10.1073/pnas.82.15.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M., Malcolm S., Rabbitts T. H. Chromosome translocation can occur on either side of the c-myc oncogene in Burkitt lymphoma cells. Nature. 1984 Mar 15;308(5956):286–288. doi: 10.1038/308286a0. [DOI] [PubMed] [Google Scholar]
- Denny C. T., Hollis G. F., Magrath I. T., Kirsch I. R. Burkitt lymphoma cell line carrying a variant translocation creates new DNA at the breakpoint and violates the hierarchy of immunoglobulin gene rearrangement. Mol Cell Biol. 1985 Nov;5(11):3199–3207. doi: 10.1128/mcb.5.11.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dony C., Kessel M., Gruss P. Post-transcriptional control of myc and p53 expression during differentiation of the embryonal carcinoma cell line F9. Nature. 1985 Oct 17;317(6038):636–639. doi: 10.1038/317636a0. [DOI] [PubMed] [Google Scholar]
- Dunnick W., Shell B. E., Dery C. DNA sequences near the site of reciprocal recombination between a c-myc oncogene and an immunoglobulin switch region. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7269–7273. doi: 10.1073/pnas.80.23.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson P. J., Littlewood T. D., Forster A., Rabbitts T. H. Chromatin structure of transcriptionally active and inactive human c-myc alleles. EMBO J. 1985 Nov;4(11):2885–2891. doi: 10.1002/j.1460-2075.1985.tb04018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Piechaczyk M., Henglein B., Blanchard J. M., Traub B., Kofler E., Wiest S., Lenoir G. M., Bornkamm G. W. Aberrant c-myc RNAs of Burkitt's lymphoma cells have longer half-lives. EMBO J. 1985 Dec 30;4(13B):3717–3725. doi: 10.1002/j.1460-2075.1985.tb04140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einat M., Resnitzky D., Kimchi A. Close link between reduction of c-myc expression by interferon and, G0/G1 arrest. Nature. 1985 Feb 14;313(6003):597–600. doi: 10.1038/313597a0. [DOI] [PubMed] [Google Scholar]
- Erikson J., ar-Rushdi A., Drwinga H. L., Nowell P. C., Croce C. M. Transcriptional activation of the translocated c-myc oncogene in burkitt lymphoma. Proc Natl Acad Sci U S A. 1983 Feb;80(3):820–824. doi: 10.1073/pnas.80.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrlander P. D., Piechaczyk M., Marcu K. B. Chromatin structure of the murine c-myc locus: implications for the regulation of normal and chromosomally translocated genes. EMBO J. 1985 Dec 1;4(12):3195–3202. doi: 10.1002/j.1460-2075.1985.tb04065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazin C., Dupont de Dinechin S., Hampe A., Masson J. M., Martin P., Stehelin D., Galibert F. Nucleotide sequence of the human c-myc locus: provocative open reading frame within the first exon. EMBO J. 1984 Feb;3(2):383–387. doi: 10.1002/j.1460-2075.1984.tb01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M., Adams J. M., Cory S. Murine T lymphomas with retroviral inserts in the chromosomal 15 locus for plasmacytoma variant translocations. 1985 Apr 25-May 1Nature. 314(6013):740–743. doi: 10.1038/314740a0. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Grosso L. E., Pitot H. C. Transcriptional regulation of c-myc during chemically induced differentiation of HL-60 cultures. Cancer Res. 1985 Feb;45(2):847–850. [PubMed] [Google Scholar]
- Hay N., Aloni Y. Attenuation of late simian virus 40 mRNA synthesis is enhanced by the agnoprotein and is temporally regulated in isolated nuclear systems. Mol Cell Biol. 1985 Jun;5(6):1327–1334. doi: 10.1128/mcb.5.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Skolnik-David H., Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982 May;29(1):183–193. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Kelly K., Cochran B. H., Stiles C. D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983 Dec;35(3 Pt 2):603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Klein G., Klein E. Evolution of tumours and the impact of molecular oncology. Nature. 1985 May 16;315(6016):190–195. doi: 10.1038/315190a0. [DOI] [PubMed] [Google Scholar]
- Klein G. Specific chromosomal translocations and the genesis of B-cell-derived tumors in mice and men. Cell. 1983 Feb;32(2):311–315. doi: 10.1016/0092-8674(83)90449-x. [DOI] [PubMed] [Google Scholar]
- Knight E., Jr, Anton E. D., Fahey D., Friedland B. K., Jonak G. J. Interferon regulates c-myc gene expression in Daudi cells at the post-transcriptional level. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1151–1154. doi: 10.1073/pnas.82.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachman H. M., Skoultchi A. I. Expression of c-myc changes during differentiation of mouse erythroleukaemia cells. Nature. 1984 Aug 16;310(5978):592–594. doi: 10.1038/310592a0. [DOI] [PubMed] [Google Scholar]
- Leder P., Battey J., Lenoir G., Moulding C., Murphy W., Potter H., Stewart T., Taub R. Translocations among antibody genes in human cancer. Science. 1983 Nov 18;222(4625):765–771. doi: 10.1126/science.6356357. [DOI] [PubMed] [Google Scholar]
- Ley T. J., Anagnou N. P., Pepe G., Nienhuis A. W. RNA processing errors in patients with beta-thalassemia. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4775–4779. doi: 10.1073/pnas.79.15.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little C. D., Nau M. M., Carney D. N., Gazdar A. F., Minna J. D. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983 Nov 10;306(5939):194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Bishop J. M., Varmus H. E. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982 Jan 21;295(5846):209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Courtneidge S. A., Crittenden L. B., Fadly A. M., Bishop J. M., Varmus H. E. Analysis of avian leukosis virus DNA and RNA in bursal tumours: viral gene expression is not required for maintenance of the tumor state. Cell. 1981 Feb;23(2):311–322. doi: 10.1016/0092-8674(81)90127-6. [DOI] [PubMed] [Google Scholar]
- Pelicci P. G., Knowles D. M., 2nd, Magrath I., Dalla-Favera R. Chromosomal breakpoints and structural alterations of the c-myc locus differ in endemic and sporadic forms of Burkitt lymphoma. Proc Natl Acad Sci U S A. 1986 May;83(9):2984–2988. doi: 10.1073/pnas.83.9.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. P. Consequences of myc invasion of immunoglobulin loci: facts and speculation. Cell. 1983 Jul;33(3):647–649. doi: 10.1016/0092-8674(83)90006-5. [DOI] [PubMed] [Google Scholar]
- Pragnell I. B., Arndt-Jovin D. J., Jovin T. M., Fagg B., Ostertag W. Commitment to differentiation in Friend cells and initiation of globin mRNA synthesis occurs during the G1 phase of the cell cycle. Exp Cell Res. 1980 Feb;125(2):459–470. doi: 10.1016/0014-4827(80)90140-8. [DOI] [PubMed] [Google Scholar]
- Rabbitts T. H., Forster A., Hamlyn P., Baer R. Effect of somatic mutation within translocated c-myc genes in Burkitt's lymphoma. Nature. 1984 Jun 14;309(5969):592–597. doi: 10.1038/309592a0. [DOI] [PubMed] [Google Scholar]
- Rabbitts T. H., Hamlyn P. H., Baer R. Altered nucleotide sequences of a translocated c-myc gene in Burkitt lymphoma. Nature. 1983 Dec 22;306(5945):760–765. doi: 10.1038/306760a0. [DOI] [PubMed] [Google Scholar]
- Rapp U. R., Cleveland J. L., Brightman K., Scott A., Ihle J. N. Abrogation of IL-3 and IL-2 dependence by recombinant murine retroviruses expressing v-myc oncogenes. Nature. 1985 Oct 3;317(6036):434–438. doi: 10.1038/317434a0. [DOI] [PubMed] [Google Scholar]
- Rappold G. A., Hameister H., Cremer T., Adolph S., Henglein B., Freese U. K., Lenoire G. M., Bornkamm G. W. c-myc and immunoglobulin kappa light chain constant genes are on the 8q+ chromosome of three Burkitt lymphoma lines with t(2;8) translocations. EMBO J. 1984 Dec 1;3(12):2951–2955. doi: 10.1002/j.1460-2075.1984.tb02239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stewart T. A., Pattengale P. K., Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984 Oct;38(3):627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- Sun L. K., Showe L. C., Croce C. M. Analysis of the 3' flanking region of the human c-myc gene in lymphomas with the t(8;22) and t(2;8) chromosomal translocations. Nucleic Acids Res. 1986 May 27;14(10):4037–4050. doi: 10.1093/nar/14.10.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R., Kelly K., Battey J., Latt S., Lenoir G. M., Tantravahi U., Tu Z., Leder P. A novel alteration in the structure of an activated c-myc gene in a variant t(2;8) Burkitt lymphoma. Cell. 1984 Jun;37(2):511–520. doi: 10.1016/0092-8674(84)90381-7. [DOI] [PubMed] [Google Scholar]
- Taub R., Moulding C., Battey J., Murphy W., Vasicek T., Lenoir G. M., Leder P. Activation and somatic mutation of the translocated c-myc gene in burkitt lymphoma cells. Cell. 1984 Feb;36(2):339–348. doi: 10.1016/0092-8674(84)90227-7. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Webb E., Adams J. M., Cory S. Variant (6 ; 15) translocation in a murine plasmacytoma occurs near an immunoglobulin kappa gene but far from the myc oncogene. Nature. 1984 Dec 20;312(5996):777–779. doi: 10.1038/312777a0. [DOI] [PubMed] [Google Scholar]
- Westergaard O., Gocke E., Nielsen O. F., Leer J. C. Effect of lucanthone (miracil D) on transcription of ribosomal RNA genes from Tetrahymena in vivo and in vitro. Nucleic Acids Res. 1979 Jun 11;6(7):2391–2402. doi: 10.1093/nar/6.7.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin E. H., Wong-Staal F., Gelmann E. P., Dalla-Favera R., Papas T. S., Lautenberger J. A., Eva A., Reddy E. P., Tronick S. R., Aaronson S. A. Expression of cellular homologues of retroviral onc genes in human hematopoietic cells. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2490–2494. doi: 10.1073/pnas.79.8.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannarell A., Niemann M., Schumm D. E., Webb T. E. Proflavine sensitivity of RNA processing in isolated nuclei. Nucleic Acids Res. 1977 Mar;4(3):503–511. doi: 10.1093/nar/4.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]