Summary
Rapid regulation of oxidative phosphorylation is crucial for mitochondrial adaptation to swift changes in fuels availability and energy demands. An intra-mitochondrial signaling pathway regulates cytochrome oxidase (COX), the terminal enzyme of the respiratory chain, through reversible phosphorylation. We find that PKA-mediated phosphorylation of a COX subunit dictates mammalian mitochondrial energy fluxes, and identify the specific residue (S58) of COX subunit IV-1 (COXIV-1) that is involved in this mechanism of metabolic regulation. Using protein mutagenesis, molecular dynamics simulations, and induced fit docking, we show that mitochondrial energy metabolism regulation by phosphorylation of COXIV-1 is coupled with prevention of COX allosteric inhibition by ATP. This regulatory mechanism is essential for efficient oxidative metabolism and cell survival. We propose that S58 COXIV-1 phosphorylation has evolved as a metabolic switch that allows mammalian mitochondria to rapidly toggle between energy utilization and energy storage.
Keywords: mitochondria, oxidative phosphorylation, PKA, ATP, allosteric inhibition, cytochrome oxidase, COXIV-1, protein phosphorylation
Introduction
To maintain cellular energy homeostasis mitochondria adapt to changes in substrate availability and metabolic demands by modulating oxidative phosphorylation (OXPHOS). Reversible phosphorylation of mitochondrial proteins is an important player in OXPHOS modulation (Hopper et al., 2006; Pagliarini and Dixon, 2006), but only recently the molecular mechanisms have started to emerge. We described a metabolic-sensing signaling pathway in the mitochondrial matrix, involving mitochondrial soluble adenylyl cyclase (sAC), which generates cAMP and activates PKA in response to mitochondrial CO2 generated in the Krebs cycle. This pathway modulates cytochrome oxidase (COX), the terminal enzyme of the electron transfer chain (ETC), as one of its targets (Acin-Perez et al., 2009a; Acin-Perez et al., 2009b).
COX translocates protons across the inner mitochondrial membrane, contributing to the proton electrochemical gradient used by the ATP synthase to generate ATP. In living cells, COX is a pacemaker of ETC function (D’Aurelio et al., 2001; Dalmonte et al., 2009; Villani et al., 1998), and its activity is modulated by phosphorylation of its subunits (Helling et al., 2008; Lee et al., 2005; Miyazaki et al., 2003; Samavati et al., 2008; Steenaart and Shore, 1997) and by allosteric ATP inhibition (Bender and Kadenbach, 2000; Kadenbach, 2003). However, the molecular mechanisms of COX regulation in response to changes of substrate availability have been elusive.
The nuclear-encoded subunits of COX have tissue specific isoforms (Lomax and Grossman, 1989) regulated during development (Bonne et al., 1993). Subunit IV (COXIV) has two isoforms, COXIV-1 and COXIV-2, which evolutionarily diverged about 320My ago. COXIV-1 is expressed ubiquitously, while COXIV-2 is highly expressed in adult lung and only at low levels in brain and heart (Huttemann et al., 2001).
Here, we identify phosphorylation of amino acid residue S58 in the matrix loop of COXIV-1 as a mechanism for PKA-dependent regulation of COX. We show, by computational means, that S58 is involved in binding ATP at the allosteric site and that its phosphorylation prevents this interaction. S58 phosphorylation modulates the kinetics of the enzyme by controlling ATP allosteric inhibition. We propose that this is a regulatory mechanism of oxidative phosphorylation, because it allows cells to match mitochondrial ETC activity with the availability of metabolic substrates and energy consumption requirements.
Results
Serine 58 is a candidate target for intramitochondrial PKA phosphorylation of COXIV-1
We had identified COXIV as a putative PKA target (Acin-Perez et al., 2009b). Since most mammalian tissues express COXIV-1 and not COXIV-2, we narrowed our study to the former. Serines and threonines of murine COXIV-1 were analyzed for probability of phosphorylation. Six candidates were identified (Table 1), four of which were predicted to be PKA targets. Three were excluded: S16 is in the cleaved signaling peptide; S157 is in the COXII/IV interface in the inner membrane; T142 is in the intermembrane space. The fourth, S58, is localized on the matrix side of COXIV-1, has a high PKA prediction score and is predicted to be a PKA target by Kinase Phos 2.0 and pka Prediction Site (not shown).
Table 1.
Prediction of S/T phosphorylation in mouse COXIV-1 (NP_034071.1). NetPhos 2.0 predicts 6 putative phosphoresidues with different probability scores. The amino acid positions refer to the precursor protein, including the signal peptide. Putative kinases for S/T residues were identified by PhosphoMotif Finder. PKA, protein kinase A; PKC, protein kinase C; CK II, casein kinase II. The putative function of the protein domain containing phosphorylated amino acids is indicated (NCBI Reference Sequence features).
| Net Phos 2.O Prediction | PhosphoMotif Finder | |||
|---|---|---|---|---|
| Position | Sequence | Score | Kinase | localization |
| S16 | KRAISTSVC | 0.930 | PKA/PKC/Calmodulin K | signal peptide |
| S58 | MLSASQKAL | 0.992 | PKA/PKC | matrix |
| S74 | WSSLSRDEK | 0.966 | CK II | matrix |
| S157 | IQGFSAKWD | 0.830 | PKA/PKC | Subunit IV/II interface |
| T17 | RAISTSVCL | 0.721 | CK II | signal peptide |
| T142 | VAIETKRML | 0.949 | PKA/PKC | IMS |
Newly generated polyclonal antibodies against a mouse S58phospho-COXIV-1 peptide were used to show that S58phospho-COXIV-1 signal in mouse liver mitochondria was increased by 8Br-cAMP (PKA activator) and decreased by H89, PKI 14-22 (PKA inhibitors) and KH7 (sAC inhibitor) (Supplementary Fig. S1), thus confirming S58 as a PKA phosphorylation target.
Mutagenesis of S58 of COXIV-1
To prove that S58 phosphorylation is involved in COX regulation we used mouse fibroblasts (C1), where COXIV-1 is silenced (Li et al., 2006), resulting in marked decrease of COXIV-1 expression (Supplementary Fig. S2A), COX activity (Supplementary Fig. S2B,C), and cell respiration (Supplementary Fig. S2D).
Transient expressed rat COXIV-1 (NP_058898.1) that has 92% identity with mouse, but contains one mismatch in the RNAi target region (Supplementary Fig. S3A) that prevents its silencing, partially complemented COX activity in C1 cells (Supplementary Fig. S3B-C).
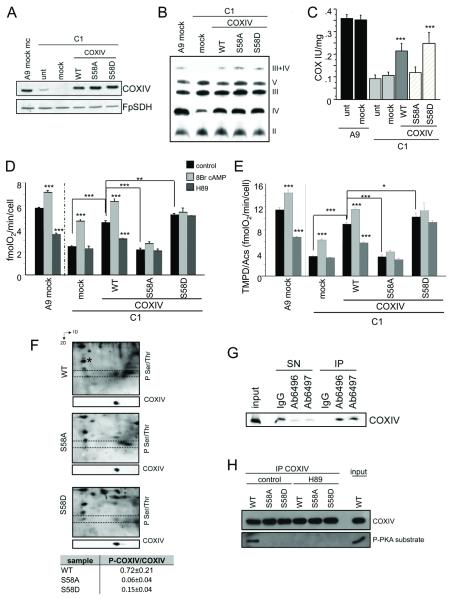
Next, we generated C1 cells stably expressing wild type (WT), or a non-phosphorylatable S58A, or a phospho-mimetic S58D COXIV-1. The levels of COXIV-1 in mass cultures (Fig. 1A) and 6 clones (Supplementary Fig. S3D) were recovered to normal levels. Blue-Native Gel Electrophoresis (BNGE) both in first (Fig. 1B) and second (Supplementary Fig. S3E) dimensions showed restoration of COX assembly. COX activity was partially rescued, but did not reach the values of A9 controls (Fig. 1C; Supplementary Fig. S3F), possibly because assembly of rat COXIV-1 in the mouse enzyme results in a mildly defective activity (Dey et al., 2000). We also excluded a compensatory increase in the expression of endogenous COXIV-1 or COXIV-2 (Supplementary Fig. S3G,H). Importantly, expression of S58A COXIV-1 resulted in reduced COX activity (Fig. 1C and Supplementary Fig. S3F), intact cell respiration (Fig. 1D), and COX-driven respiration (Fig. 1E), as compared to WT and S58D cells, indicating that loss of COXIV-1 S58 phosphorylation results in COX deficiency.
Figure 1. C1 cells expressing WT and mutant COXIV-1.
A) Western Blot of C1 cells expressing WT, S58A, and S58D COXIV-1 using anti COXIV anitbodies. FpSDH is a loading control. B) First dimension BNGE to detect COX assembly using anti-COXI antibodies. βATPase is a loading control. C) COX activity measured spectrophotometrically in C1 cells expressing COXIV-1. D) Intact cell respiration upon PKA activation or inhibition by 8Br cAMP (1mM, for 2 hrs) or H89 (1μM, for 2 hrs), respectively. E) Modulation assessed by TMPD/ascorbate driven respiration. Bars are averages of mass cultures and 2 clones (in triplicate). Statistical differences are relative to mock C1 cells (transfected with empty vector). A9 mock cells are included as reference. *, p<0.01; ***, p<0.0001. Unt, untransfected cells. F) 2D-BNGE and Western Blot of mitochondria from transfected cells treated with 8Br cAMP (1mM, for 2 hrs), using an anti-phospho Ser/Thr antibody. The dashed box indicates the region of COXIV migration (inset below each panel), when the membrane was re-probed with anti-COXIV antibody. P-COXIV/COXIV ratios are shown (average of three independent gels ± SD). G) Immunoprecipitation of phosphorylated COXIV-1 in mitochondrial lysates from A9 cells with S58phospho-COXIV-1 (Ab6496 or Ab6497) and detection with COXIV antibodies. Rabbit IgG were used as negative control; IP, immunoprecipitation; SN, supernatant. H) Immunoprecipitation of COXIV from untreated and H89 treated (1μM, for 2 hrs) cells expressing WT, S58A, and S58D COXIV-1.
In WT COXIV-1 cells, PKA stimulation with membrane permeant 8Br-cAMP or its inhibition with H89, resulted in increased and decreased respiration and COX activity, respectively (Fig. 1D,E). C1 cells displayed a greater OXPHOS increase and a lesser decrease than controls, because the enzyme is constitutively de-phosphorylated (Acin-Perez et al., 2009a). However, S58A and S58D cells, which cannot be phosphorylated, had blunted responses to PKA modulation, indicating loss of sensitivity to PKA modulation.
Protein phosphorylation is reduced in COXIV-1 S58 mutants
COXIV-1 phosphorylation was investigated by 2D-BNGE, in cells treated with 8Br-cAMP. The intensity of phosphorylated COXIV-1 detected using a phospho Ser/Thr antibody was normalized by an unknown phosphoprotein (asterisk in Fig. 1F), which was unchanged in all samples. This ratio was then normalized by total COXIV determined with anti-COXIV antibody. S58A and S58D COXIV-1 were less phosphorylated than WT COXIV-1 (Fig. 1F), indicating that S58 accounts for most of PKA-dependent COXIV-1 phosphorylation. Most COXIV-1 appeared phosphorylated in A9 control cells, as shown by almost complete immunodepletion from the supernatant after immunoprecipitation with S58phospho-COXIV-1 antibodies (Fig.1G). These antibodies did not detect the rat protein, due to sequence differences in the epitope. Nevertheless, by immunoprecipitation with COXIV antibody and detection with an antibody that recognizes PKA phosphorylated peptides, in C1 cells expressing the WT, but not the mutant rat COXIV-1, the protein was phosphorylated, and H89 decreased its phosphorylation (Fig. 1H).
Molecular dynamics simulations of COX containing WT or mutant COXIV-1
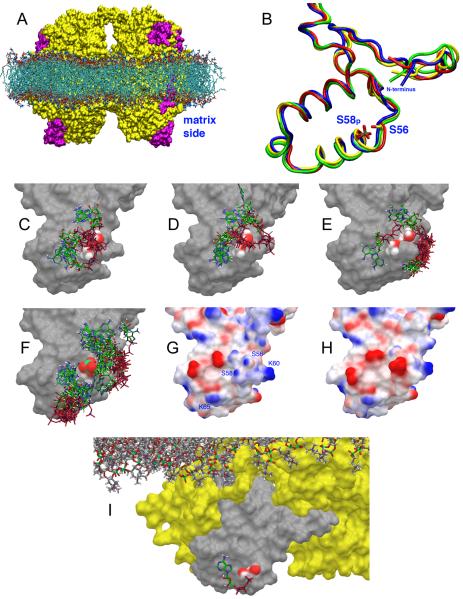
Dimeric WT (Fig. 2A) and mutant COX (COXIV-1 S58A, S58D, and S58phospho) were simulated in the membrane by molecular dynamics (MD). The mean backbone (bb) root mean square deviations (RMSD) from the initial structure, and the mean backbone fluctuations (RMSF) around the average structure of each subunit (Supplementary Table 1) suggest that neither mutation of COXIV-1 S58 to A or D nor its phosphorylation change COX conformation significantly. A superposition of the most prevalent conformations of the matrix domain of WT and mutant COXIV-1 (Fig. 2B) did not show major structural changes (bb-RMSDs between each superimposed structure and the average structure of 0.753, 0.732, 0.702, and 0.629 Å for WT, S58A, S58D, and S58phospho, respectively). Thus, the effects of S58A and S58D mutations reflect the functional role played by S58 in COX regulation.
Fig. 2. Structural models of COX and ATP binding site on the matrix side of COX.
A) Molecular surface of membrane (teal) embedded dimeric WT COX. The enzyme is in yellow with the two COXIV subunits in magenta. B) Superposition of the matrix domain of COXIV in WT (blue), S58A (red), S58D (green), and S58phopsho (yellow), as derived from the most prevalent conformations in the MD. C) Binding poses of ATP on the surface of WT COXIV-1 in the most prevalent conformation (Cluster 1). S56 and S58 side chains atoms are color-coded (carbon, grey; oxygen, red; hydrogen, white). D,E) Binding poses of ATP on the surface of WT COXIV-1 in the conformations of Cluster 2 and 3, respectively. F) Binding poses of ATP on the surface of S58phospho COXIV-1 derived from the entire MD simulation superimposed on the conformation of Cluster 1. The phosphate moiety of S58phospho is color-coded (phosphorus, green; carbon, grey; oxygen, red; hydrogen, white). G, H) Molecular surfaces of WT COXIV-1 and S58phospho COXIV-1 colored according to electrostatic potentials (+1 kT/e, blue; −1 kT/e, red). I) Matrix side of COX (yellow surface) embedded in the membrane (shown as sticks) with ATP bound to subunit IV-1 (grey surface).
ATP binding site in the matrix domain of COXIV-1
At high ATP:ADP ratios, ATP binds COXIV and other enzyme subunits and reversibly inhibits COX activity (Arnold and Kadenbach, 1997; Bender and Kadenbach, 2000; Napiwotzki et al., 1997). Two ATP binding sites have been proposed in COXIV, one facing the intermembrane space and regulated by cytosolic ATP (Napiwotzki and Kadenbach, 1998), another facing the matrix and regulated by intramitochondrial ATP (Arnold and Kadenbach, 1997). We hypothesized that phosphorylation of COXIV-1 S58 regulates allosteric inhibition of COX by matrix ATP. The binding site for ATP in the matrix domain of COXIV-1 was studied using a combination of Induced Fit Docking (IFD) and QM-Polarized Ligand Docking (QPLD). Five clusters of conformations were identified in the MD simulations of WT and S58phospho COXIV-1, and ranked from 1 to 5 based on the number of frames contained (more frames reflect more likely conformations). Charges on the ligand (ATP) and protein were assigned based on a mitochondrial matrix pH of 8.0±0.2 (Metoki and Hommes, 1984). In Cluster 1 from WT COXIV-1 there were two groups of ATP binding poses (Fig. 2C) characterized by an interaction of the γ- and/or β-phosphate of ATP with the hydroxyl moieties of the side chains of S56 and S58. In Cluster 2 (Fig. 2D) there was binding of ATP at S56/S58, but a minor site near the K60 side chain (Fig. 2G) started being occupied. This site was fully occupied in the remaining conformations (~20% of all frames, Fig. 2E). S56 and S58 are situated in the 1st helix of the matrix domain of COXIV-1 (Fig. 2B), and the dipole moment of the helix provides 0.5-0.7 units of positive charge that stabilizes the binding of the γ- and β-phosphates of ATP (Fig. 2G). The phosphate group of S58phospho obliterates the positive electrostatic potential (Fig. 2H), thereby preventing ATP binding. This is shown by failure of the IFD to identify a pose corresponding to the binding of ATP to the side chains of S56 and S58 of COXIV-1 S58phospho (Fig. 2F). Low affinity binding sites for ATP were recognized near the side chains of K60 and K65. The pose with the best IFD score was refined by QPLD (Fig. 2I). The phosphate of ATP wraps around the side chain of S58, while the sugar and base are contained in nearby pockets. When COXIV-1 is phosphorylated, the phosphate of S58phospho replaces the γ-phosphate of ATP, significantly weakening its binding. The pose in Fig. 2I was used to calculate by free-energy perturbation (FEP) changes in the binding energy of ATP, when S58 is replaced by A or D or is phosphorylated. ΔΔGbind values for ATP with respect to WT were 0.82±2.9 kcal/mol for S58A, 7.15±3.5 kcal/mol for S58D, and 8.6±3.5 kcal/mol for S58phospho (more positive values indicate weaker binding), corresponding respectively to a ~4-fold, ~175,000-fold, and ~2,100,000-fold increase in Kd.
ATP allosteric inhibition of COX is regulated by COXIV-1 phosphorylation
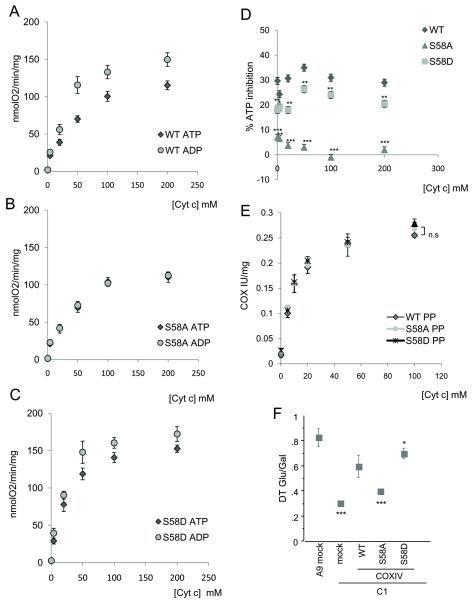
Since S58 of COXIV-1 is a PKA target and S58phospho prevents ATP binding, we investigated the functional consequences of these molecular interactions. COX activity was measured in permeabilized cells in an ATP regenerating system with increasing concentrations of reduced cytochrome c, in the presence of either ATP or ADP. WT COXIV-1 cells had ATP-induced inhibition at all cytochrome c concentrations, while S58A cells were insensitive to ATP, and S58D cells had a lesser degree of ATP inhibition (Fig. 3A-D).
Figure 3. COX allosteric inhibition by ATP in cells expressing COXIV-1 and its role in OXPHOS.
Oxygen consumption using increasing concentrations of cytochrome c in the presence of ATP (5mM) or ADP (5mM) measured in WT (A), S58A (B), and S58D (C) permeabilized COXIV-1 expressing cells (n=3). D) Percentage of ATP inhibition at different cytochrome c concentrations from values from A-C (n=3). E) COX activity measured spectrophotometrically in sonicated mitochondria after pyrophosphatase (PP) treatment (n=6 for each point). F) Doubling time ratio (n=3) of cells grown in glucose versus galactose medium (DT Glu/Gal). Statistically significant differences are relative to WT COXIV-1 cells. *, p<0.01; **, p<0.001; ***, p<0.0001.
Next, mitochondria were sonicated and treated with pyrophosphatase to hydrolyze ATP and ADP. The removal of ATP from all binding sites eliminated the differences in COX activity between WT and S58 mutant cells (Fig. 3E), indicating that ATP is responsible for the differences.
COXIV-1 S58 phosphorylation is necessary for efficient OXPHOS function
To study the physiological significance of S58 phosphorylation cell growth was assessed in WT and S58 mutant cells in medium containing galactose as the main carbon source, where cells are forced to utilize OXPHOS to generate ATP (Acin-Perez et al., 2009a). C1 cells had a severe growth defect in galactose (relative to glucose, Fig. 3F). Cells expressing WT or S58D COXIV-1, but not S58A COXIV-1, had a significant improvement of growth in galactose, suggesting that loss of S58 COXIV-1 phosphorylation results in COX inhibition and a failure to maintain ATP synthesis, when cells need to upregulate oxidative metabolism.
Discussion
The discovery of a signaling system through sAC and PKA (Acin-Perez et al., 2009b) contributed to our understanding of the regulation of mitochondrial metabolism. However, the molecular mechanisms underlying this regulation had not been defined.
Many proteins involved in OXPHOS function are targets of phosphorylation (Balaban; Deng et al.; Hopper et al., 2006). In this study, we focused on COX, but it is likely that other OXPHOS components may be involved in PKA-mediated metabolic regulation (Papa et al., 2008). COX contains 13 subunits, 10 of which are phosphorylated at various sites (Thomson, 2002). With a combined proteomic (Acin-Perez et al., 2009b) and informatics strategy we determined that S58 in COXIV-1 fulfilled the criteria for being accessible to matrix PKA and part of a PKA target motif. Furthermore, phosphorylation of this residue had been shown earlier in bovine heart mitochondria (Helling et al., 2008). We pinpointed S58 of COXIV-1, as a player in COX regulation by reversible phosphorylation and identified S58 as part of the allosteric binding site for ATP on the matrix side of COXIV-1, showing that phosphorylation of S58 dramatically weakens the interaction with ATP. Furthermore, we have shown that lack of COXIV-1 phosphorylation in S58A mutant cells results in reduced COX activity and defective growth under oxidative conditions. These results suggest a link between activation of the intramitochondrial CO2-cAMP-PKA pathway and phosphorylation of COXIV-1 S58, which regulates COX allosteric inhibition by matrix ATP. This interpretation does not exclude that extramitochondrial ATP may bind other sites in COX and regulate its activity differently.
Our findings suggest that mitochondria can switch from a “storage mode” to a “consumption mode”. When ATP builds up in mitochondria, because of reduced requirements, enzymes are inhibited and fuel utilization diminishes, resulting in low intramitochondrial CO2 levels, low sAC activity and cAMP, and low PKA activity. COXIV-1 is dephosphorylated and COX activity inhibited by ATP, shunting substrates towards fat and glycogen accumulation. On the other hand, when cellular ATP consumption is high, large amounts of substrates are oxidized, CO2 stimulates COXIV-1 phosphorylation, transiently preventing ATP inhibition and allowing for maximal electron flux through COX and high ATP production. Since the phospho-mimetic S58D results in a small increase in COX activity and the S58phospho-COXIV-1 antibodies immuno-capture most COXIV-1, we inferred that normally the majority of COXIV-1 is phosphorylated. Studies in more complex organisms will assess whether this applies to organs and tissues in vivo.
The S58 residue is conserved among mammals (Supplementary Table 2), but not in non-mammalian species, suggesting that S58 has coevolved with mammals. Furthermore, two different isoforms of COXIV are found in many species. Similarly to other nuclear-encoded COX subunits, COXIV plays a regulatory role, in part through differential temporal and spatial expression of its isoforms. COXIV-2 does not have a PKA phosphorylation site corresponding to S58. Instead, it has three glutamic acids, carrying negative charges that may impede ATP binding. COXIV-2 confers higher activity to COX than COXIV-1, thereby allowing for a fast electron flux and possibly reducing the risk of forming reactive oxygen species in highly oxygenated tissues (Huttemann et al., 2007). Under oxygen deprivation COXIV-2 is induced in brain, where it abolishes sensitivity to ATP inhibition (Horvat et al., 2006). Furthermore, an increase of COXIV-2 expression regulated by hypoxia-inducible factor 1 occurs under hypoxia, in association with COXIV-1 degradation (Fukuda et al., 2007). These responses could be part of a hypoxia-preconditioning program to prevent the wave of free radical production, following reperfusion.
The presence of S58 in COXIV-1 of mammalian species could be related to a specific need for metabolic regulation in animals exposed to variations in ambient temperature and food availability. In modern days, metabolic regulation through reversible phosphorylation of respiratory chain subunits may represent a trigger for metabolic diseases, such as obesity and diabetes, or a potential target for therapy with approaches to modulate consumption and storage of metabolic supplies.
Experimental Procedures
Cell lines and treatments
The A9 cells and COXIV-1 silenced cells (C1) were reported elsewhere (Li et al., 2006). Cell replication in glucose and galactose medium was measured as described (Acin-Perez et al., 2009a). 8Br-cAMP 1mM (Sigma-Aldrich) and H89 1μM (Calbiochem) for 2 hrs were used to stimulate or inhibit PKA, respectively.
Mitochondrial isolations
Mitochondria-enriched fractions were obtained from 20×106 cells, as described elsewhere (Birch-Machin and Turnbull, 2001). Mitochondria isolation from mouse liver and pharmacological treatments were performed as described (Acin-Perez et al., 2009b).
Cloning and transfection of rat COXIV-1
Rat COXIV-1 cDNA was obtained by reverse transcription (Superscript reverse transcriptase®, Invitrogen) of rat PC12 cells total RNA using appropriate primers (Supplementary Methods) and cloned in pcDNA3.1 (Invitrogen).
S58A and S58D mutants of COXIV-1 were generated by in situ mutagenesis using the QuickChange® Site-Directed Mutagenesis kit (Stratagene) with appropriate primers (Supplementary Methods).
Stable cell lines expressing WT, S58A, and S58D COXIV-1 were generated by transfection with FuGene6 (Roche), followed by selection in 250μg/ml G418 for 3 weeks.
Oxygen consumption and enzymatic assays
Oxygen consumption determinations in intact cells were carried out as described (Hofhaus et al., 1996). COX activity was measured spectrophotometrically on mitochondrial fractions (2-5μg of protein) or in cell lysates (30-50μg of protein) as described (Birch-Machin and Turnbull, 2001). COX allosteric inhibitory effects of ATP were measured as described (Bender and Kadenbach, 2000) in the presence of ATP or ADP (5mM). Cell respiration was measured in 50μg of Tween-permeabilized cells with increasing concentrations of reduced cytochrome c (0.25-200μM).
For pyrophosphatase treatments, mitochondria were sonicated as described (Acin-Perez et al., 2009b), centrifuged at 20,000g for 5 min at 4°C, and resuspended in 10mM Tris–HCl (pH 7.4), 25mM sucrose, 75mM sorbitol, 100mM KCl, 10mM K2HPO4, 0.05mM EDTA, 5mM MgCl2, 1mg/ml BSA. Pyrophosphatase (2 Units, Sigma-Aldrich) was added for 10 min at 25°C. COX activity was measured (Birch-Machin and Turnbull, 2001) with increasing concentrations of reduced cytochrome c (0.1-100μM).
mRNA content measurements by RT-PCR
mRNA levels of mouse COXIV-1 and 2; rat COXIV-1 and β-actin were measured using a LightCycler® RNA Master SYBR Green I kit (Roche), in a LightCycler capillary RT-PCR instrument (Roche) using appropriate primers (Supplementary Methods).
Immunoprecipitation of COXIV
Mitochondrial lysates (dodecyl maltoside; 1.6g/g protein) (Schagger and von Jagow, 1991) were incubated overnight with 1-3μg of antibody (COXIV-1 or Ab6496 or Ab6497) or control rabbit IgG bound to Protein G Sepharose beads (Invitrogen). Beads were pulled down and washed twice with PBS containing 0.5% dodecyl maltoside. Proteins were eluted in 50-100μl of 0.5M Tris-HCl pH 6.8, 5% glycerol, 2% SDS and 100mM DTT and used for Western blot analyses.
Western Blot analyses
For denaturing SDS-PAGE proteins (10-25μg from cell lysates and 25μl from the immunoprecipitated) were electrophoresed in a 12.5% acrylamide denaturing gel and electroblotted onto PVDF (BioRad) filters.
For 1D-BNGE and 2D-BNGE, 50-75μg of mitochondrial protein were applied on a 5-13% gradient BN gel followed by separation in a 12.5% denaturing gel (Schagger and von Jagow, 1991). After electrophoresis, proteins were electroblotted onto PVDF filters.
For protein detection the following antibodies were used: FpSDH, Core2, COXIV, and βATPase (all from Invitrogen); cyt c (BD Pharmingen); GAPDH (Abcam); phospho Ser/Thr (BD Biosciences), PKA-P-substrate (Cell Signaling). Rabbit polyclonal antibodies against the mouse COXIV-1 S58phospho-peptide VAHVTMLSApSQKALKEKE (Ab6496 and Ab6497) were generated (Phosphosolutions).
Informatics analyses
Phosphorylation prediction was by NetPhos 2.0 software (http://www.cbs.dtu.dk/services/NetPhos/) with a threshold score > 0.5. Kinase prediction was with PhosphoMotif Finder (http://www.hprd.org/PhosphoMotif_finder). PKA-dependent phosphorylation prediction was by Kinase Phos 2.0 (http://kinasephos2.mbc.nctu.edu.tw/). Potential PKA phosphorylation sites were predicted by PKA Prediction Sites (pkaPS) (http://mendel.imp.ac.at/sat/pkaPS/). COXIV-1 cDNA sequence alignment was by Nucleotide BLAST (http://blast.ncbi.nlm.nih.gov/). Protein sequence alignments were by ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
Molecular Dynamics simulations
Models of membrane embedded COX were built from the X-ray structure of dimeric bovine COX (PDB entry 1V55) with Desmond (D.E. Shaw Research) (Bowers et al., 2006), by adding an explicit bilayer of 1-palmitoyl-2-oleoyl-phosphatidylcholine around COX and solvating the enzyme and both sides of the membrane with SPC water (Berendsen et al., 1981): a minimum distance of 12Å was left between any protein atom and the edge of the simulation box. The ensemble was energy minimized under periodic boundaries condition using the 2005 OPLS-AA force-field (Jorgensen et al., 1996). 1.2ns MD simulations were carried out with Desmond in the NPT ensemble at 298.15K (25°C), using the Nose-Hoover thermostat method (Evans and Holian, 1985) and the Martyna-Tobias-Klein barostat method (Martyna et al., 1994). SHAKE constraints (Ryckaert et al., 1977) were imposed on all the heavy-atom-hydrogen covalent bonds. Coordinates were saved every 4.8ps.
Identification of the binding site for ATP in the matrix domain of COXIV
Identification of the ATP binding site in the matrix domain of COXIV-1 was by IFD (Sherman et al., 2006). The QPLD protocol (Cho et al., 2005) was used to refine selected poses to improve the partial charges on the ligand atoms in a docking run, by replacing them with charges derived from quantum mechanical calculations on the ligand in the field of the receptor. IFD and QPLD were implemented in the Schrödinger Suite 2010 (Schrödinger LLC.). FEP (Zwanzig, 1954) were calculated with Desmond in the NPT ensemble at 298.15K in the protein:ATP complex+solvent and the protein alone+solvent environments. Simulations ran for 0.6ns for each λ window (12 windows for each perturbation). Results were collected and analyzed by the Bennett method (Bennet, 1976; Shirts et al., 2003). ΔΔGbind values were calculated by subtracting the protein+solvent ΔGFEP from the complex+solvent ΔGFEP value. Kd changes for ATP binding to COXIV-1 were calculated from the relationship
Electrostatic potentials at the molecular surface were determined with the Poisson-Boltzmann solver of QSite (Schrödinger LLC), using a continuum model with a solvent dielectric constant εs=78, and a protein dielectric constant εp=2.
Statistical analyses
Comparisons among groups were made using one-way ANOVA. Pair wise comparisons were made by post hoc Fisher PLSD test using StatView (Adept Scientific). Differences were considered statistically significant at p<0.05. In all graphs, error bars indicate standard deviations.
Supplementary Material
Acknowledgements
This work was supported by grants from NIH R01 GM088999 (GM), GM69840 (DLG), the Muscular Dystrophy Association (GM), and the United Mitochondrial Disease Foundation (RAP).
We thank Dr. Anatoly Starkov (Weill Medical College of Cornell University) for discussion and helpful advice.
Footnotes
The authors have no conflicts of interest with the work presented here.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acin-Perez R, Salazar E, Brosel S, Yang H, Schon EA, Manfredi G. Modulation of mitochondrial protein phosphorylation by soluble adenylyl cyclase ameliorates cytochrome oxidase defects. EMBO Mol Med. 2009a;1:392–406. doi: 10.1002/emmm.200900046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acin-Perez R, Salazar E, Kamenetsky M, Buck J, Levin LR, Manfredi G. Cyclic AMP produced inside mitochondria regulates oxidative phosphorylation. Cell Metab. 2009b;9:265–276. doi: 10.1016/j.cmet.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S, Kadenbach B. Cell respiration is controlled by ATP, an allosteric inhibitor of cytochrome-c oxidase. Eur J Biochem. 1997;249:350–354. doi: 10.1111/j.1432-1033.1997.t01-1-00350.x. [DOI] [PubMed] [Google Scholar]
- Balaban RS. The mitochondrial proteome: a dynamic functional program in tissues and disease states. Environ Mol Mutagen. 2010;51:352–359. doi: 10.1002/em.20574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender E, Kadenbach B. The allosteric ATP-inhibition of cytochrome c oxidase activity is reversibly switched on by cAMP-dependent phosphorylation. FEBS Lett. 2000;466:130–134. doi: 10.1016/s0014-5793(99)01773-1. [DOI] [PubMed] [Google Scholar]
- Bennet CH. Efficient estimation of free energy differences from Monte Carlo data. J. Comp. Phys. 1976;22:245–268. [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B, editor. Intermolecular Forces. Reidel Publishing Company; Dordrecht: 1981. pp. 331–342. [Google Scholar]
- Birch-Machin MA, Turnbull DM. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- Bonne G, Seibel P, Possekel S, Marsac C, Kadenbach B. Expression of human cytochrome c oxidase subunits during fetal development. Eur J Biochem. 1993;217:1099–1107. doi: 10.1111/j.1432-1033.1993.tb18342.x. [DOI] [PubMed] [Google Scholar]
- Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregerson BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DEN- ScalableAlgorithmsforMolecularDynamicsSimulationsonCommodity Clusters; ACM/IEEE Conference on Supercomputing(SC06); Tampa, Florida. IEEE Computer Society; 2006. p. 43. 2006. [Google Scholar]
- Cho AE, Guallar V, Berne BJ, Friesner R. Importance of accurate charges in molecular docking: quantum mechanical/molecular mechanical (QM/MM) approach. J Comput Chem. 2005;26:915–931. doi: 10.1002/jcc.20222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aurelio M, Pallotti F, Barrientos A, Gajewski CD, Kwong JQ, Bruno C, Beal MF, Manfredi G. In vivo regulation of oxidative phosphorylation in cells harboring a stop-codon mutation in mitochondrial DNA-encoded cytochrome c oxidase subunit I. J Biol Chem. 2001;276:46925–46932. doi: 10.1074/jbc.M106429200. [DOI] [PubMed] [Google Scholar]
- Dalmonte ME, Forte E, Genova ML, Giuffre A, Sarti P, Lenaz G. Control of respiration by cytochrome c oxidase in intact cells: role of the membrane potential. J Biol Chem. 2009;284:32331–32335. doi: 10.1074/jbc.M109.050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng N, Zhang J, Zong C, Wang Y, Lu H, Yang P, Wang W, Young GW, Korge P, Lotz C, Doran P, Liem DA, Apweiler R, Weiss JN, Duan H, Ping P. Phosphoproteome analysis reveals regulatory sites in major pathways of cardiac mitochondria. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M110.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey R, Barrientos A, Moraes CT. Functional constraints of nuclear-mitochondrial DNA interactions in xenomitochondrial rodent cell lines. J Biol Chem. 2000;275:31520–31527. doi: 10.1074/jbc.M004053200. [DOI] [PubMed] [Google Scholar]
- Evans DJ, Holian BL. The Nose--Hoover thermostat. The Journal of Chemical Physics. 1985;83:4069–4074. [Google Scholar]
- Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- Helling S, Vogt S, Rhiel A, Ramzan R, Wen L, Marcus K, Kadenbach B. Phosphorylation and kinetics of mammalian cytochrome c oxidase. Mol Cell Proteomics. 2008 doi: 10.1074/mcp.M800137-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofhaus G, Shakeley RM, Attardi G. Use of polarography to detect respiration defects in cell cultures. Methods Enzymol. 1996;264:476–483. doi: 10.1016/s0076-6879(96)64043-9. [DOI] [PubMed] [Google Scholar]
- Hopper RK, Carroll S, Aponte AM, Johnson DT, French S, Shen RF, Witzmann FA, Harris RA, Balaban RS. Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium. Biochemistry. 2006;45:2524–2536. doi: 10.1021/bi052475e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvat S, Beyer C, Arnold S. Effect of hypoxia on the transcription pattern of subunit isoforms and the kinetics of cytochrome c oxidase in cortical astrocytes and cerebellar neurons. J Neurochem. 2006;99:937–951. doi: 10.1111/j.1471-4159.2006.04134.x. [DOI] [PubMed] [Google Scholar]
- Huttemann M, Kadenbach B, Grossman LI. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene. 2001;267:111–123. doi: 10.1016/s0378-1119(01)00385-7. [DOI] [PubMed] [Google Scholar]
- Huttemann M, Lee I, Liu J, Grossman LI. Transcription of mammalian cytochrome c oxidase subunit IV-2 is controlled by a novel conserved oxygen responsive element. FEBS J. 2007;274:5737–5748. doi: 10.1111/j.1742-4658.2007.06093.x. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. Journal of the American Chemical Society. 1996;118:11225–11236. [Google Scholar]
- Kadenbach B. Intrinsic and extrinsic uncoupling of oxidative phosphorylation. Biochim Biophys Acta. 2003;1604:77–94. doi: 10.1016/s0005-2728(03)00027-6. [DOI] [PubMed] [Google Scholar]
- Lee I, Salomon AR, Ficarro S, Mathes I, Lottspeich F, Grossman LI, Huttemann M. cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity. J Biol Chem. 2005;280:6094–6100. doi: 10.1074/jbc.M411335200. [DOI] [PubMed] [Google Scholar]
- Li Y, Park JS, Deng JH, Bai Y. Cytochrome c oxidase subunit IV is essential for assembly and respiratory function of the enzyme complex. J Bioenerg Biomembr. 2006;38:283–291. doi: 10.1007/s10863-006-9052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax MI, Grossman LI. Tissue-specific genes for respiratory proteins. Trends Biochem Sci. 1989;14:501–503. doi: 10.1016/0968-0004(89)90185-0. [DOI] [PubMed] [Google Scholar]
- Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. The Journal of Chemical Physics. 1994;101:4177–4189. [Google Scholar]
- Metoki K, Hommes FA. The pH of mitochondria of fibroblasts from a hyperornithinaemia, hyperammonaemia, homocitrullinuria-syndrome patient. J Inherit Metab Dis. 1984;7:9–11. doi: 10.1007/BF01805611. [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Neff L, Tanaka S, Horne WC, Baron R. Regulation of cytochrome c oxidase activity by c-Src in osteoclasts. J Cell Biol. 2003;160:709–718. doi: 10.1083/jcb.200209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napiwotzki J, Kadenbach B. Extramitochondrial ATP/ADP-ratios regulate cytochrome c oxidase activity via binding to the cytosolic domain of subunit IV. Biol Chem. 1998;379:335–339. doi: 10.1515/bchm.1998.379.3.335. [DOI] [PubMed] [Google Scholar]
- Napiwotzki J, Shinzawa-Itoh K, Yoshikawa S, Kadenbach B. ATP and ADP bind to cytochrome c oxidase and regulate its activity. Biol Chem. 1997;378:1013–1021. doi: 10.1515/bchm.1997.378.9.1013. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Dixon JE. Mitochondrial modulation: reversible phosphorylation takes center stage? Trends Biochem Sci. 2006;31:26–34. doi: 10.1016/j.tibs.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Papa S, De Rasmo D, Scacco S, Signorile A, Technikova-Dobrova Z, Palmisano G, Sardanelli AM, Papa F, Panelli D, Scaringi R, Santeramo A. Mammalian complex I: a regulable and vulnerable pacemaker in mitochondrial respiratory function. Biochim Biophys Acta. 2008;1777:719–728. doi: 10.1016/j.bbabio.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- Samavati L, Lee I, Mathes I, Lottspeich F, Huttemann M. TNFalpha inhibits oxidative phosphorylation through tyrosine phosphorylation at subunit I of cytochrome c oxidase. J Biol Chem. 2008 doi: 10.1074/jbc.M801954200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- Shirts MR, Bair E, Hooker G, Pande VS. Equilibrium Free Energies from Nonequilibrium Measurements Using Maximum-Likelihood Methods. Physical Review Letters. 2003;91:140601. doi: 10.1103/PhysRevLett.91.140601. [DOI] [PubMed] [Google Scholar]
- Steenaart NA, Shore GC. Mitochondrial cytochrome c oxidase subunit IV is phosphorylated by an endogenous kinase. FEBS Lett. 1997;415:294–298. doi: 10.1016/s0014-5793(97)01145-9. [DOI] [PubMed] [Google Scholar]
- Thomson M. Evidence of undiscovered cell regulatory mechanisms: phosphoproteins and protein kinases in mitochondria. Cell Mol Life Sci. 2002;59:213–219. doi: 10.1007/s00018-002-8417-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani G, Greco M, Papa S, Attardi G. Low reserve of cytochrome c oxidase capacity in vivo in the respiratory chain of a variety of human cell types. J Biol Chem. 1998;273:31829–31836. doi: 10.1074/jbc.273.48.31829. [DOI] [PubMed] [Google Scholar]
- Zwanzig RW. High-Temperature Equation of State by a Perturbation Method. I. Nonpolar Gases. J. Chem. Phys. 1954;22:1420–1426. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.