Abstract
Obesity-related pathologies, such as nonalcoholic fatty liver disease, are linked to mitochondrial dysfunction and nitric oxide (NO) deficiency. Herein, we tested the hypothesis that a high-fat diet (HFD) modifies the liver mitochondrial proteome and alters proteins involved in NO metabolism, namely arginase 1 and endothelial NO synthase. Male C57BL/6 mice were fed a control or HFD and liver mitochondria were isolated for proteomics and reactive oxygen species measurements. Steatosis and hepatocyte ballooning were present in livers of HFD mice, with no pathology observed in the controls. HFD mice had increased serum glucose and decreased adiponectin. Mitochondrial reactive oxygen species was increased after 8 weeks in the HFD mice, but decreased at 16 weeks compared with the control, which was accompanied by increased uncoupling protein 2. Using proteomics, 22 proteins were altered as a consequence of the HFD. This cohort consists of oxidative phosphorylation, lipid metabolism, sulfur amino acid metabolism, and chaperone proteins. We observed a HFD-dependent increase in arginase 1 and decrease in activated endothelial NO synthase. Serum and liver nitrate + nitrite were decreased by HFD. In summary, these data demonstrate that a HFD causes steatosis, alters NO metabolism, and modifies the liver mitochondrial proteome; thus, NO may play an important role in the processes responsible for nonalcoholic fatty liver disease. Antioxid. Redox Signal. 15, 447–459.
Introduction
Nonalcoholic fatty liver disease (NAFLD), a component of the cardiometabolic syndrome, has emerged as one of the most common etiologies of chronic liver disease. NAFLD can progress from simple fatty liver (i.e., steatosis) to more severe conditions such as nonalcoholic steatohepatitis (NASH), fibrosis, and cirrhosis, if left untreated. NASH is characterized histologically by macrosteatosis, inflammation, hepatocyte ballooning, and pericellular fibrosis in zone 3 of the liver lobule (36). Insulin resistance, disrupted fatty acid metabolism, mitochondrial dysfunction, oxidative stress, and dysregulated cytokine networks are proposed to be critical factors or “hits” responsible for the progression from simple steatosis to NASH (18).
Evidence suggests that mitochondrial dysfunction is a key player and contributor to NAFLD and NASH pathogenesis. Early studies showed that obesity-induced steatosis decreased the capacity for ATP synthesis in liver (6). Similarly, hepatic mitochondria of NASH patients exhibit lower respiratory complex activities (35) and increased mitochondrial DNA mutations (19). Recently, our laboratory reported that chronic consumption of a high-fat diet (HFD) in mice induced NAFLD-like pathology that was accompanied by profound changes in mitochondrial bioenergetics (25). Feeding a HFD for 16 weeks decreased mitochondrial respiration and cytochrome c oxidase activity and increased sensitivity to nitric oxide (NO)-dependent inhibition of mitochondrial respiration when compared with measures in liver mitochondria from the control mice. Similar alterations in mitochondrial bioenergetics and NO–mitochondria signaling occur in alcohol-induced fatty liver disease (47, 48). It is likely that dietary-induced alterations in tissue NO bioavailability underlie these changes in mitochondrial function (7, 17). Although reduced NO concentrations have been shown in cardiovascular and renal tissues of obese and/or diabetic animals (39), the impact of fatty liver (i.e., steatosis) on hepatic levels of NO is not yet known. Understanding the impact of fatty liver on NO metabolism is important, as NO is implicated as a critical physiological regulator for how mitochondria and cells respond to stress in multiple organ systems (16, 17, 42).
In addition to altering NO-dependent mitochondrial respiration, it is hypothesized that chronic exposure to a HFD will modify the liver mitochondrial proteome, which may ultimately compromise mitochondrial function. Identifying NAFLD-dependent alterations to mitochondrial proteins is an important area of research as there are over 600+ proteins thought to comprise this organelle-specific proteome (28). Indeed, studies aimed at determining NAFLD-dependent changes to the mitochondrial proteome represent an attractive approach to better understand the key metabolic and molecular factors that contribute to this important liver disease. Therefore, our next goal was to investigate the liver mitochondrial proteome from wild-type C57BL/6 mice pair-fed a control or HFD for 16 weeks, using two-dimensional isoelectric focusing (2D IEF)/sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). This time point was selected because HFD-dependent impairments in mitochondrial bioenergetics are present at 16 weeks (25). These proteomic studies were complemented by measuring mitochondrial reactive oxygen species (ROS) production and assessing the impact of a HFD on the levels of two key enzymes involved in maintaining tissue NO, namely arginase 1 and endothelial NO synthase (eNOS). To test for a HFD-dependent deficit in NO, we measured serum and liver nitrate + nitrite levels.
Materials and Methods
HFD feeding protocol
Male C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME) at 8 weeks of age and maintained two per cage under a standard 12-h light–dark cycle. Mice were fed a control or HFD for up to 16 weeks before being used in experiments (25). Mice in the HFD group were fed the control diet for days 1–2 before being switched to the HFD. The HFD contains 71% total calories as fat, 11% as carbohydrate, and 18% as protein (24). The control diet is calorically equivalent (1 kcal/ml diet) to the HFD except that the fat content is reduced to 35% total calories, with carbohydrate and protein making up 47% and 18% of total calories, respectively. To perform the pair-feeding protocol, the amount of liquid HFD consumed by each HFD pair of mice was measured daily (i.e., for each 24-h period) and then each corresponding control pair of mice was fed an equal amount of the control diet during the next 24-h time period. Use of a pair-feeding protocol is required for this type of study design so that differences in experimental outcomes between groups (i.e., between the control and the HFD) can be ascribed solely to the presence of higher fat content in the diet and not to increased caloric intake as groups are isocaloric. Animal protocols were approved by the IACUC and mice were treated in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 86-23).
Histology, serum, and liver chemistry measurements
Formalin-fixed liver was processed for hematoxylin–eosin staining and evaluated for the presence of steatosis. Serum levels of adiponectin (B-Bridge International, Mountain View, CA) and insulin (Millipore/Linco, Billerica, MA) and liver levels of tumor necrosis factor α (TNFα; BioLegend, San Diego, CA) were measured by enzyme-linked immunosorbent assay following manufacturer's instructions. Glucose, triglycerides, and alanine aminotransferase (ALT) levels were measured in serum using reagent set kits (Pointe Scientific, Canton, MI). Triglyceride levels were measured in liver cytosolic fractions using a coupled enzymatic-reagent set, triglycerides–GPO (Pointe Scientific). Serum and liver nitrate + nitrite levels were measured using the colorimetric Griess reagent (Cayman Chemical Company, Ann Arbor, MI). Serum and liver samples were filtered to reduce background absorbance per manufacturer's recommendations. Mice were fasted for 4 h before isolation of liver tissue and collection of blood for serum measurements.
Immunoblotting
Total eNOS protein and phosphorylated eNOS (eNOS-PSer1177) protein levels were measured using 1:5000 and 1:1000 dilutions of primary antibodies, respectively (Cell Signaling Technology, Danvers, MA). Levels of arginase 1 protein were measured using a 1:500 dilution of primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Uncoupling protein 2 (UCP2) levels were measured using a 1:1000 dilution of primary antibody (R&D Systems, Minneapolis, MN). Levels of beta-actin protein were measured using a 1:5000 dilution of primary antibody (Sigma-Aldrich, St. Louis, MO). Voltage-dependent anion channel protein was measured using a 1:20,000 dilution of primary antibody (EMD Chemicals, Gibbstown, NJ). After incubation with appropriate horseradish peroxidase-conjugated secondary antibodies, protein bands were visualized by chemiluminescence detection and quantified using Quantity One software (Bio-Rad, Hercules, CA).
Liver mitochondria isolation
Mitochondria were prepared by differential centrifugation of liver homogenates using an ice-cold isolation medium containing 250 mM sucrose, 1 mM EDTA, and 5 mM Tris-HCl (pH 7.5) (21, 25). Protease inhibitors were added to the isolation buffer to prevent protein degradation as required for proteomic analyses. Briefly, liver homogenates were centrifuged at 560 g for 10 min with the resulting postnuclear supernatant centrifuged at 8500 g for 10 min to isolate the mitochondrial fraction. The mitochondrial pellets were subjected to four separate wash cycles (i.e., gentle resuspension of mitochondria followed by centrifugation at 8500 g for 10 min) to obtain the final fraction. All steps in this procedure were conducted at 4°C. Respiration rates and extent of coupling were measured using a Clark-type O2 electrode (25) to ensure high-quality mitochondrial preparation for proteomics and ROS measurements.
Mitochondrial ROS measurements
Mitochondria (0.5 mg/ml) were suspended in HEPES buffer, pH 7.2, which contained 130 mM KCl, 2 mM KH2PO4, 3 mM HEPES, 2 mM MgCl2, and 1 mM EGTA in 50-ml Erlenmeyer flasks. This buffer is identical to that used for respiration measurements. Flasks were sealed with rubber stoppers and continuously gassed with 21% O2:74% N2:5% CO2 throughout the incubation period. Mitochondria were incubated with 2 μM 2′,7′-dichlorodihydrofluorescein diacetate (Invitrogen, Eugene, OR) for 60 min to measure ROS formation as previously described (52). Succinate (0.2 mM) was added as the oxidizable substrate. In a parallel set of experiments, samples were incubated with carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) (1 μM) to dissipate the mitochondrial membrane potential and demonstrate that ROS production was membrane potential dependent. The background fluorescence of the FCCP-treated samples was subtracted from all experimental samples. Select experiments were also performed in the presence of antimycin (15 μM) as a positive control to stimulate mitochondrial superoxide and hydrogen peroxide production. After incubation, the dichlorofluorescein fluorescence of mitochondria suspension was measured with excitation and emission wavelengths set to 488 and 525 nm, respectively.
Proteomic analysis of liver mitochondrial proteins
2D IEF/SDS-PAGE analysis of liver mitochondrial proteins was performed essentially as previously described (2). Briefly, mitochondrial protein (100 μg) was applied to IEF gel strips (ZOOM Strips, pH 3–10; Invitrogen, Carlsbad, CA) and rehydration of IEF strips was done overnight (∼16 h). The following morning, IEF was performed using the following conditions: 175 V for 20 min, ramp to 2000 V for 45 min, hold at 2000 V for 30 min, ramp down to 500 V for 30 min, and then hold at 500 V for 2 h. For SDS-PAGE, IEF gel strips were placed horizontally on top of a 10% resolving gel with 4% stacking gel and sealed into place using warm agarose (1%, w/v), and gels were run at 100 V for 1.5 h. After electrophoresis, gels were stained with Sypro Ruby™ (Invitrogen) for total protein and imaged using a Bio-Rad Fluor-S Imager (Bio-Rad).
Gels were analyzed for differences in protein density using PDQuest software (Bio-Rad) as previously described (2). Individual proteins identified by the software were manually verified to generate a match-set of gels for the control and HFD treatments. The protein density was compared across all gels and a reference gel was selected and served as the master gel image. Automatic matching of proteins in each gel to the corresponding proteins in the master gel was performed and then manually verified to correct for any proteins incorrectly matched to proteins in the reference gel. To correct for intergel protein loading differences, density data for each individual protein in each gel was normalized to the total density of valid proteins for that particular gel. Image analysis revealed that there was no significant difference in total signal density between gels prepared from the control and HFD groups (control: 712,928 ± 15,714; high fat: 698,062 ± 31,545; p-value = 0.78), indicating equal loading and high reproducibility of gels. Statistical analysis was performed as previously described (2).
For protein identification, spots were excised from gels and processed by standard methods used in the UAB mass spectrometry shared facility (www.uab.edu/proteomics) and previously described (2). Samples were analyzed with a Voyager De-Pro mass spectrometer and the obtained peptide masses were submitted to the MASCOT database (www.matrixscience.com) for protein identification. Proteins were classified using the Universal Protein Resource website (www.uniprot.org) maintained by the UniProt Consortium (European Bioinformatics Institute, Swiss Institute of Bioinformatics and Protein Information Resource).
Statistical analysis
Data are reported as the mean ± standard error of the mean. Significant differences (p < 0.05) between the control and HFD groups were obtained using Student's paired t-test where indicated. As a pair feeding study was used on mice from a single batch of animals and mice were paired before starting the experimental liquid diets, differences between means were assessed with Student's t-test for paired samples as described by Snedecor and Cochran (43). Repeated measure analysis of variance was used for analyses of body weight, whereas the difference in liver weight between groups was determined using an analysis of covariance (ANCOVA) as suggested for obesity-related studies (1). Use of ANCOVA is appropriate for this, as ANCOVA revealed that liver weight was highly related to body weight (p < 0.0001) for both feeding time points. In the following section, normalized liver weights are presented as least square means ± standard error of the mean.
Results
Effect of HFD on body weight
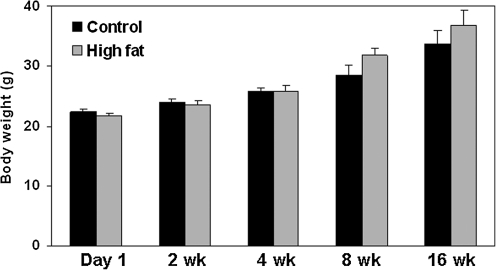
Mice were fed a control or HFD in a pair-fed, isocaloric fashion for up to 16 weeks to determine the impact of chronic exposure to HFD on the liver mitochondrial proteome. As shown in Figure 1, the body weights of the control and HFD mice increased during the 16-week feeding period; however, there was no statistically significant difference in body weight between groups when compared over the entire feeding period (data analyzed by repeated measure analysis of variance).
FIG. 1.
Body weight of mice maintained on control and HFD. Body weights are presented for mice fed the control and HFD for up to 16 weeks. Weights were recorded every Friday afternoon during the 16-week feeding period. Results from repeated measure analysis of variance: diet, p = 0.36; time, p < 0.0001; interaction (diet vs. time), p = 0.26. Data represent the mean ± SEM for n = 8 animals per group. Note that these data are recorded from the same eight mice per treatment during the 16-week feeding protocol. HFD, high-fat diet; SEM, standard error of the mean.
Effect of HFD on liver parameters and circulating metabolic markers
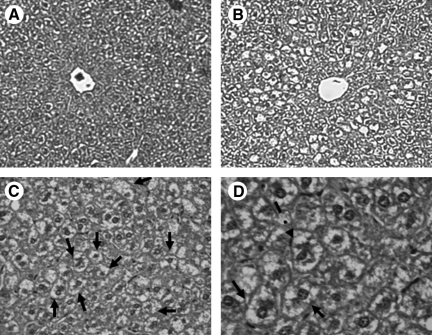
As shown in our previous studies (25), livers from mice fed a HFD for 16 weeks exhibited an accumulation of fat in hepatocytes (Fig. 2B). Moreover, ballooned hepatocytes and Mallory bodies were present in livers of HFD mice (Fig. 2C, D). Steatosis was verified biochemically with increased liver triglycerides present in HFD livers (Table 1). The control livers showed little to no steatosis (Fig. 2A) and had lower triglyceride levels (Table 1). Similar results were found at 8 weeks (data not shown). Serum levels of triglycerides were unaffected by the HFD (Table 1). The difference in liver weights between the control and HFD groups was determined using ANCOVA, with liver weight examined as the dependent variable and body weight as the covariate. At the 8-week time point, the normalized liver weight of the control and HFD groups was 1.43 ± 0.058 and 1.31 ± 0.057 g, respectively (p = 0.0381). At the 16-week time point, the normalized liver weight of the control and HFD groups was 1.46 ± 0.056 and 1.19 ± 0.058 g, respectively (p = 0.008).
FIG. 2.
HFD induces steatosis. Representative hematoxylin/eosin-stained liver sections from mice fed control (A) or HFD (B–D) for 16 weeks. Increased lipid droplets are present within hepatocytes of the HFD mice compared with the control mice. Note the presence of multiple ballooned hepatocytes (solid arrow) and Mallory body (dashed arrow) in the liver from the HFD mice. Magnification: 10 × for panels A and B, 20 × for panel C, and 40 × for panel D.
Table 1.
Effect of a High-Fat Diet on Select Serum and Liver Chemistries
| Control | High fat | |
|---|---|---|
| Serum adiponectin (ng/ml) | 31,962 ± 872 | 22,604 ± 1686a |
| Liver tumor necrosis factor α (pg/mg protein) | 5.95 ± 0.39 | 6.37 ± 0.25 |
| Serum glucose (mg/dl) | 177 ± 25 | 240 ± 21a |
| Serum insulin (ng/ml) | 1.26 ± 0.28 | 1.31 ± 0.26 |
| Serum alanine aminotransferase (IU/l) | 37.4 ± 6.7 | 42.3 ± 8.9 |
| Serum triglycerides (mg/dl) | 74.9 ± 14.5 | 67.5 ± 16.5 |
| Liver triglycerides (mg/mg protein) | 47.3 ± 11 | 109.5 ± 20.2a |
Data represent the mean ± SEM for at least five animals per group for 16-week feeding time point.
p < 0.05, compared with control.
SEM, standard error of the mean.
Accompanying these gross liver parameters, we measured serum ALT levels, an indicator of hepatocyte injury, and saw no difference between the control and HFD groups (Table 1). Liver concentrations of the pro-inflammatory cytokine TNFα were also unchanged between treatment groups (Table 1). Serum TNFα was below the level of detection for the control and HFD animals. In contrast, serum levels of the anti-inflammatory cytokine adiponectin were significantly decreased (30%) in the HFD group compared with the control (Table 1). Fasting levels of serum glucose were significantly increased (35%) in the HFD group compared with the control, with insulin levels unchanged between groups (Table 1).
Effect of HFD on liver mitochondrial proteome
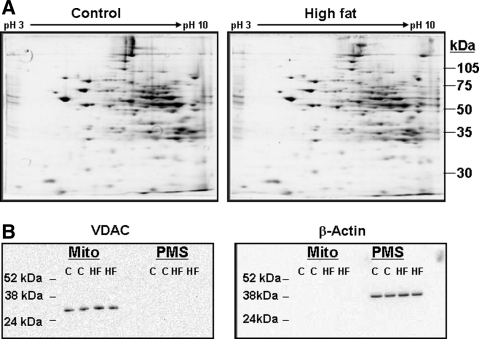
Representative 2D IEF/SDS-PAGE gels from the control and HFD groups are shown in Figure 3A. Using a “mini-2D gel” proteomics system, we typically detect 150–200 mitochondrial proteins (2). It is important to note that the mitochondrial preparations used for these proteomic studies were functionally validated and had robust respiratory capacity (25), which is an absolute criterion for valid proteomic analyses of mitochondria (53). The purity of mitochondrial preparations was also validated by western blot analysis. As shown in Figure 3B, there was an absence of the mitochondrial protein voltage-dependent anion channel in the postmitochondrial fraction and an absence of cytosolic protein beta-actin in the mitochondrial fraction.
FIG. 3.
Representative two-dimensional gel images of liver mitochondrial proteins from the control and HFD groups. (A) Mitochondrial proteins isolated from livers of the control and HFD groups were separated in the first dimension by using pH 3–10 isoelectric focusing gel strips followed by separation in the second dimension on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis acrylamide gels. Please note that proteomic analyses were performed on liver mitochondria from the 16-week feeding time point. (B) Western blot analyses showing purity of mitochondrial preparations. For example, VDAC (outer mitochondrial membrane protein) is not detected in the PMS fraction and beta-actin (cytosolic protein) is not detected in the mitochondrial fraction. VDAC, voltage-dependent anion channel; PMS, postmitochondrial supernatant.
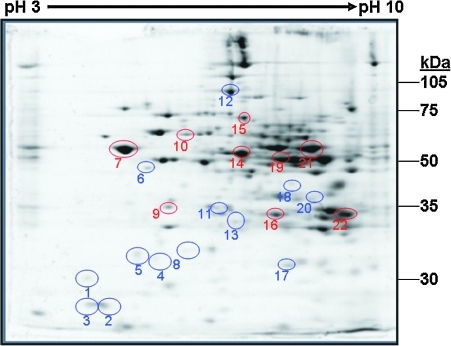
Image analysis of 2D gels detected 171 unique protein “spots” in common across all gels from the control and HFD groups. Densitometry analyses revealed 22 proteins significantly altered in response to the HFD. A 2D gel showing the location of these specific 22 proteins is provided in Figure 4, with a list of their unique proteins IDs and statistical analyses provided in Table 2. In addition to proteins affected by the HFD, we identified 67 proteins that were unchanged between the control and HFD groups. A list of these unaffected proteins with their IDs and location within a representative 2D gel is provided in Supplementary Data (Supplementary Data are available online at www.liebertonline.com/ars).
FIG. 4.
Liver mitochondrial proteins differentially altered by a HFD. Analysis of two-dimensional gels revealed 22 proteins differentially altered in abundance in response to the HFD treatment for 16 weeks. Proteins circled in blue were increased, whereas proteins circled in red were decreased in abundance in response to a HFD. The mass spectrometry identification and biologic functions for these proteins are provided in Tables 2 and 3, respectively. Please note that a separate and different numbering system is used for the global liver mitochondrial proteome map and table provided in Supplementary Data. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Table 2.
Liver Mitochondrial Proteins Altered in Abundance by a High-Fat Diet
| Spot no. | Protein | Control diet (mean ± SEM) | High-fat diet (mean ± SEM) | p-Value | MOWSE score (no. of matched peptides) | Accession no. |
|---|---|---|---|---|---|---|
| 1 | Unidentified | 851 ± 74 | 1121 ± 44 | 0.025 | — | — |
| 2 | Unidentified | 5999 ± 1097 | 8787 ± 1267 | 0.0072 | — | — |
| 3 | uMUP-VIII major urinary protein (18.7 kDa) | 4491 ± 977 | 7958 ± 1958 | 0.0383 | 103 (7) | gi|1839508 |
| 4 | Unidentified | 544 ± 81 | 855 ± 139 | 0.0110 | — | — |
| 5 | Unidentified | 947 ± 186 | 1293 ± 143 | 0.0360 | — | — |
| 6 | Gamma-actin (41 kDa) | 915 ± 118 | 1353 ± 95 | 0.0500 | 121 (7) | gi|809561 |
| 7 | ATP synthase, F1 beta (48 kDa) | 28,095 ± 1601 | 24,030 ± 1441 | 0.0053 | 135 (13) | gi|89574015 |
| 8 | Unidentified | 414 ± 21 | 521 ± 30 | 0.0279 | — | |
| 9 | Pyruvate deHase beta (35 kDa) | 2008 ± 45 | 1842 ± 62 | 0.0148 | 125 (12) | gi|12805431 |
| 10 | Glucose-regulated protein (58 kDa) | 2773 ± 210 | 2401 ± 130 | 0.0500 | 96 (10) | gi|8393322 |
| 11 | Unidentified | 809 ± 169 | 1343 ± 210 | 0.0306 | — | — |
| 12 | Sarcosine deHase (100 kDa) | 5287 ± 396 | 6043 ± 340 | 0.0034 | 158 (20) | gi|20149748 |
| 13 | Nitrilase 2 (30.5 kDa) | 528 ± 51 | 732 ± 72 | 0.0148 | 102 (8) | gi|12963555 |
| 14 | Aldehyde deHase 2 (56.5 kDa) | 16,269 ± 852 | 13,371 ± 754 | 0.0310 | 246 (15) | gi|6753036 |
| 15 | Succinate deHase, subunit a (72.3 kDa) | 5926 ± 89 | 5252 ± 170 | 0.0193 | 124 (17) | gi|15030102 |
| 16 | Electron transfer flavoprotein, subunit alpha (35 kDa) | 6823 ± 747 | 6084 ± 724 | 0.0198 | 112 (11) | gi|21759113 |
| 17 | Ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 (29.3 kDa) | 3150 ± 396 | 3996 ± 309 | 0.0235 | 97 (8) | gi|13385168 |
| 18 | α-methylacyl-CoA racemase (39.6 kDa) | 237 ± 60 | 442 ± 100 | 0.0481 | 106 (7) | gi|2145186 |
| 19 | 3-Hydroxy-3-methylglutaryl-CoA synthase 2 (56.8 kDa) | 13,528 ± 1282 | 6116 ± 713 | 0.0311 | 103 (11) | gi|31560689 |
| 20 | Thiosulfate sulfurtransferase (33.4 kDa) | 424 ± 114 | 908 ± 225 | 0.0204 | 102 (12) | gi|6678449 |
| 21 | ATP synthase, F1 alpha (59.7 kDa) | 17,059 ± 528 | 9282 ± 1199 | 0.0100 | 154 (14) | gi|6680748 |
| 22 | Malate deHase (35.6 kDa) | 16,340 ± 1026 | 9495 ± 1024 | 0.0126 | 109 (11) | gi|126897 |
Liver mitochondrial proteins were identified by two-dimensional isoelectric-focusing/sodium dodecyl sulfate–polyacrylamide gel electrophoresis in combination with mass spectrometry as described in the Methods section. Spot no. is the same as those used to identify proteins shown in the gel provided in Figure 4. Mass is measured in kDa. The MOWSE score is an algorithmic calculation used to assign a statistical weight to each peptide match; therefore, a higher MOWSE score implies higher statistical likelihood of the match being correct. The number of matched peptides (shown in parentheses) is the number of peptides that were matched to allow for protein designation and MOWSE calculation. The accession number is the unique NCBI label given to proteins. Data represent the mean ± SEM for n = 5 animals per treatment group.
Of the 22 proteins altered by chronic exposure to a HFD for 16 weeks, 13 were increased (circled in blue) and 9 were decreased (circled in red) in liver mitochondria (Fig. 4 and Table 2). A brief description of the biological functions of these proteins is provided in Table 3. In general, these proteins fall into the broad categories of energy, amino acid, and lipid metabolism, with some exceptions. Four proteins involved in oxidative phosphorylation were altered by the HFD. The key catalytic subunits responsible for ATP synthesis within complex V, the ATP synthase F1 α and β subunits, were decreased by HFD. Succinate dehydrogenase (i.e., complex II) subunit A (SDH-A) was also decreased, whereas the Rieske Fe-S protein of the ubiquinol-cytochrome bc1 reductase (i.e., complex III) was increased by chronic exposure to HFD. HFD exposure also reduced the levels of malate dehydrogenase and pyruvate dehydrogenase-β, two enzymes involved in mitochondrial energy metabolism. In addition to effects on energy metabolism systems, a HFD exposure also decreased the amount of 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMG-CoA synthase 2) and the electron transfer flavoprotein subunit-α (ETFα). In contrast, the level of α-methylacyl-CoA racemase was significantly increased by the HFD. Other proteins that were significantly increased in response to a HFD were γ-actin, sarcosine dehydrogenase, nitrilase-2, and thiosulfate sulfurtransferase (TSST). Mitochondrial aldehyde dehydrogenase 2 and glucose-regulated protein 58 (GRP58) were decreased by the HFD.
Table 3.
Biological Functions of Mitochondrial Proteins Altered in Abundance by a High-Fat Diet
| Spot no. | Protein | Biologic function | Change with high-fat diet |
|---|---|---|---|
| 3 | uMUP-VIII major urinary protein (18.7 kDa) | Pheromone communication (only in rodents) | ↑ |
| 6 | Gamma-actin (41 kDa) | Cytoskeleton protein; role in liver regeneration | ↑ |
| 7 | ATP synthase, F1 beta (48 kDa) | Catalytic site of ATP synthesis in OxPhos system | ↓ |
| 9 | Pyruvate deHase beta (35 kDa) | Catalyzes the conversion of pyruvate to acetyl CoA | ↓ |
| 10 | Glucose-regulated protein (58 kDa) | Chaperone protein | ↓ |
| 12 | Sarcosine deHase (100 kDa) | Catalyzes the conversion of sarcosine to glycine; last step in choline oxidation pathways | ↑ |
| 13 | Nitrilase 2 (30.5 kDa) | Nitrilase-related metabolism | ↑ |
| 14 | Aldehyde deHase 2 (56.5 kDa) | Acetaldehyde and electrophilic lipid metabolism | ↓ |
| 15 | Succinate deHase, subunit a (72.3 kDa) | Catalyzes the oxidation of succinate to fumarate; flavoprotein | ↓ |
| 16 | Electron transfer flavoprotein, subunit alpha (35 kDa) | Transfers electrons to ubiquinone via electron transfer flavoprotein–ubiquinone oxidoreductase | ↓ |
| 17 | Ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 (29.3 kDa) | Redox activity component of complex III; transfers electrons from ubiquinol to cytochrome c | ↑ |
| 18 | α-methylacyl-CoA racemase (39.6 kDa) | Oxidation of branched chain fatty acids and cholesterol metabolites | ↑ |
| 19 | 3-Hydroxy-3-methylglutaryl-CoA synthase 2 (56.8 kDa) | Catalyzes the condensation of acetoacetyl CoA and acetyl CoA; first step in ketogenesis | ↓ |
| 20 | Thiosulfate sulfurtransferase (33.4 kDa) | Cyanide detoxification; role in iron–sulfur centers; sulfane sulfur metabolism | ↑ |
| 21 | ATP synthase, F1 alpha (59.7 kDa) | Catalytic site of ATP synthesis in OxPhos system | ↓ |
| 22 | Malate deHase (35.6 kDa) | Catalyzes the oxidation of malate to oxaloacetate in Kreb's cycle | ↓ |
Liver mitochondrial proteins were identified by two-dimensional isoelectric-focusing/sodium dodecyl sulfate–polyacrylamide gel electrophoresis in combination with mass spectrometry as described in the Methods section. Major urinary protein-VIII is secreted from liver, filtered into urine, and only used for pheromone communication in mice (9); as such it will not be discussed in this article.
Effect of a HFD on mitochondrial ROS generation
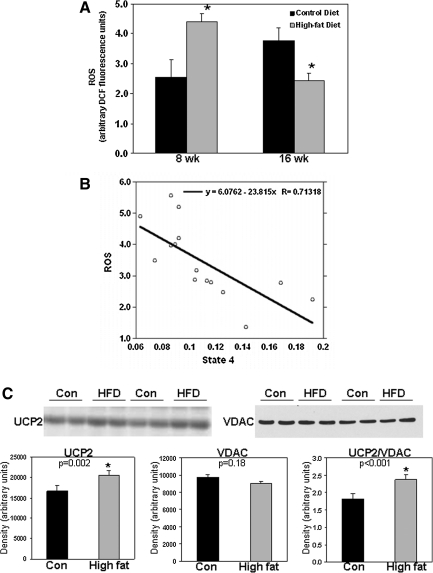
As shown in Figure 5A, chronic feeding of a HFD increased mitochondrial ROS production at 8 weeks, whereas lower rates of production were measured at 16 weeks. Experiments with FCCP (an uncoupling agent) and antimycin (a complex III inhibitor and potent mitochondrial superoxide inducer) show that ROS formation is membrane potential dependent and linked to superoxide and hydrogen peroxide produced from the leakage of electrons from the respiratory chain (see Supplementary Fig. 2). Previously, we reported that mitochondrial state 4 respiration (i.e., ADP independent) was unchanged between the control and HFD groups at early feeding time points (i.e., 3 and 8 weeks), with increased state 4 respiration at the later 16-week time point (25). This result suggests that chronic consumption of a HFD leads to uncoupling of mitochondria, a proposed adaptive condition that can be utilized to dissipate mitochondrial oxidant generation (5). Further analyses support this concept as increased mitochondrial state 4 respiration rates are associated with lower levels of ROS in liver mitochondria (Fig. 5B). Moreover, increased levels of UCP2 protein were observed in livers of the HFD mice, supporting decreased ROS production (Fig. 5C).
FIG. 5.
Effect of a HFD on mitochondrial ROS production and UCP2 in liver. (A) Mitochondrial ROS production was measured using the fluorescent probe 2′,7′-dichlorodihydrofluorescein diacetate as previously described (52). Mitochondria were prepared from livers of mice fed control and HFD for 8 and 16 weeks. (B) Plot of mitochondrial state 4 respiration (microgram atom O/min/mg protein) versus ROS production (fluorescence units). The correlation probability for data shown in panel B is 0.0028, indicating that the two measures are significantly correlated. Note that ROS production was measured under state 4 conditions, that is, in the absence of ADP. (C) Top panels: Representative immunoblots of UCP2 and VDAC protein in liver samples from two separate pairs of control and HFD mice. Results are from groups fed diets for 16 weeks. Bottom panels: Densitometry analysis of UCP2 and VDAC levels along with UCP2 normalized to VDAC. Data shown in panels A and C represent the mean ±SEM for n = 4–7 animals per treatment, with *p < 0.05, compared with control. ROS, reactive oxygen species; UCP, uncoupling protein.
Effect of a HFD on enzymes impacting NO bioavailability
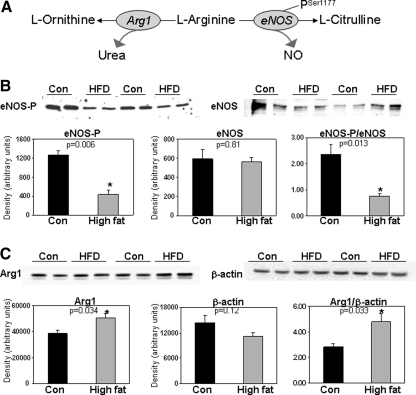
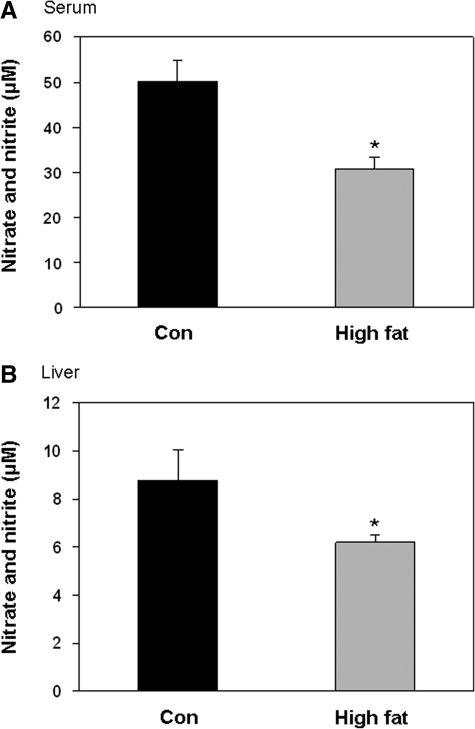
Previous studies suggest that alterations in NO metabolism and bioavailability may contribute to tissue injury from obesity and diabetes (17, 26). The impact of HFD-induced steatosis on two key enzymes involved in the control of NO metabolism in liver, eNOS and arginase 1, are not known (Fig. 6A). Examination of eNOS showed no effect of a HFD on total eNOS protein levels, whereas the extent of Ser1177 phosphorylation of eNOS was significantly decreased by a HFD (Fig. 6B). In contrast, arginase 1 levels were significantly increased by a HFD compared with the controls (Fig. 6C). Accompanying changes in these two key enzymes of NO metabolism, nitrate + nitrite concentrations were decreased in serum and liver of the HFD mice compared with the control mice (Fig. 7A, B). This result supports the concept that chronic consumption of a HFD impairs NO availability in liver.
FIG. 6.
Effect of a HFD on enzymes (arginase 1 and eNOS) affecting NO bioavailability in liver. (A) Simplified schematic showing the role of arginase 1 and eNOS-catalyzed reactions. (B) Top panels: Representative immunoblots of eNOS-PSer1177 and total eNOS in liver samples from two separate pairs of control and HFD mice. Results are from groups fed diets for 16 weeks. Bottom panels: Densitometry analysis of eNOS-PSer1177 and total eNOS levels along with eNOS-P normalized to total eNOS protein. (C) Top panels: Representative immunoblots of arginase 1 and beta-actin protein in liver samples from two separate pairs of control and HFD mice. Results are from groups fed diets for 16 weeks. Bottom panels: Densitometry analysis of arginase 1 and beta-actin levels along with arginase 1 normalized to beta-actin. Data represent the mean ± SEM for n = 4–8 animals per treatment group, with *p < 0.05, compared with control. NO, nitric oxide; eNOS, endothelial NO synthase; eNOS-PSer1177, eNOS phosphorylated at residue Ser 1177; Arg1, arginase 1.
FIG. 7.
Effect of a HFD on serum and liver nitrate + nitrite levels. Consumption of a HFD decreased serum and liver nitrate + nitrite concentrations compared with values measured in the controls. Results are from groups fed diets for 16 weeks. Data represent the mean ± SEM for n = 5 (serum, A) and n = 8 (liver, B) animals per treatment group, with *p < 0.05, compared with control.
Discussion
Defining the molecular mechanisms responsible for NAFLD are important as this disease is the most frequent cause of abnormal liver function tests in the United States, with an estimated prevalence of 15%–30% in the general U.S. population (12, 36). To address this ever-increasing public health problem, numerous experimental animal models have been utilized to understand NAFLD pathobiology. Previously, we adapted a long-term HFD feeding model, originally used in rats (24), to wild-type C57BL/6 mice to study the mitochondrial mechanisms underlying NAFLD (25). Using this approach, mice fed the HFD had higher fasting serum glucose, suggesting hyperglycemia (Table 1). Moreover, mice fed the HFD had decreased serum adiponectin at 16 weeks (Table 1), which is in agreement with studies showing low adiponectin in NAFLD patients (34). Chronic consumption of a HFD in mice also produced macro- and microsteatosis, hepatocyte ballooning, and Mallory bodies (Fig. 2)—some of the hallmark features observed in the liver of NAFLD/NASH patients. However, we did not observe increased serum ALT in the HFD mice (Table 1), which is in agreement with other studies using a HFD in mice (13, 20). Similarly, serum triglycerides were not elevated in the HFD mice (Table 1). It is possible that a HFD-mediated defect in very low-density lipoprotein (VLDL) secretion may explain the absence of increased serum triglycerides in the HFD mice. Studies show that absence of microsomal triglyceride transfer protein, a key protein for VLDL assembly/export, causes hepatic steatosis and reduced serum triglycerides (27). Steatosis was verified because increased triglyceride was present in the livers of the HFD mice (Table 1), although we did not observe a concomitant increase in the liver weight of the HFD mice compared with the controls. This may not be unexpected as fat is less dense than muscle and increased liver weight is typically correlated to water retention and protein accumulation in hepatocytes (11)—processes that may not be altered by a HFD in mice.
Analysis of the mitochondrial proteome revealed changes in two fatty acid metabolism proteins, HMG-CoA synthase 2 and ETFα, which could influence HFD-dependent steatosis. HMG-CoA synthase catalyzes the condensation of acetoacetyl CoA and acetyl CoA to form free CoA and HMG-CoA, which in mitochondria is metabolized to acetoacetate. Important to our findings, HMG-CoA synthase deficiency causes fatty liver (49) and feeding a methionine-choline–deficient diet to mice decreased HMG-CoA synthase (51). Thus, inhibition of ketogenesis linked to HMG-CoA synthase deficiency leads to steatosis. We also observed a significant decrease in ETFα, the electron acceptor for the four chain-length, β-oxidation–specific acyl CoA-dehydrogenases. Reducing equivalents from β-oxidation are transferred sequentially to ETF, ETF-ubiquinone oxidoreductase, and then to ubiquinone, entering the mitochondrial respiratory chain at the level of complex III. A HFD-mediated decrease in this key electron-shuttle protein, coupled with an impaired respiratory chain, is likely to contribute to steatosis because of an inability to metabolize fatty acyl CoAs.
Liver TNFα was unchanged by exposure to a HFD (Table 1), with serum TNFα undetectable in both groups. This observation suggests that a strong pro-inflammatory cytokine response was not activated in liver by this dietary regime at this particular time point. It supports why we did not see elevated serum ALT in this model of HFD-dependent steatosis as serum ALT typically increases in response to cytokine-mediated liver injury (3, 23, 46). Although one might predict increased TNFα in response to a HFD, recent studies show temporal alterations in cytokines and hepatic proteins during chronic feeding studies. For example, robust temporal changes occur in TNFα expression during early and late time points in studies of alcohol-induced fatty liver disease (40). This temporal characteristic is further supported by studies showing a switch from an early inflammatory to a late steatotic transcriptional program during a 16-week exposure to a HFD (38). These studies demonstrate that the impact of a HFD on cytokines, hepatic proteins, and metabolism is dynamic and adaptive. Indeed, several studies show early adaptation to diet and other metabolic stresses, with the eventual failure to adapt to stress leading to disease, such as steatosis (10, 14, 38). Similarly, differential dietary-mediated changes in the liver transcriptome and proteome likely explain why HFD mice in the present study did not have increased body weight when compared with control mice (Fig. 1). Thorens and colleagues showed that C57BL/6 mice have differential metabolic adaptive responses when fed a HFD for up to 9 months, with most mice becoming obese and diabetic; however, a significant percentage remained lean and nondiabetic even when fed the same HFD (10). Specifically, they found that the obese/diabetic phenotype was related to upregulation of lipogenic genes, whereas the lean/nondiabetic phenotype showed decreased lipogenic gene expression and increased eNOS in liver in response to the HFD (10). These studies are important as they indicate that in a genetically homogenous population (e.g., C57BL/6 mice) differential effects on parameters such as body weight can occur even when mice are exposed to the same stressors (e.g., HFD) for equal time periods. These findings support the concept that disease likely occurs in those individuals who are poor adaptors to metabolic stressors, whereas those resistant to disease may be good adaptors. In summary, our results show that feeding a HFD reproduces some, but not all, of the important biochemical and histologic markers observed in NAFLD patients.
Chronic consumption of a HFD also causes defects in hepatic mitochondrial bioenergetics. Previously, we reported that liver mitochondria from mice fed a HFD for 16 weeks have decreased respiration and respiratory complex activities (25). In the present study, we extended our investigations to determine whether changes in oxidative phosphorylation proteins and other mitochondrial proteins occurred in response to a HFD. Herein, we observed a significant decrease in the abundance of SDH-A, as well as decreases in the ATP synthase F1 α and β subunits. SDH-A connects the Kreb's cycle to the respiratory chain through the oxidation of succinate to fumarate with FADH2-linked reduction of the electron carrier ubiquinone. A HFD-mediated deficiency in SDH-A likely contributes to a decrease in respiratory capacity (25). The HFD-mediated decrease in the two ATP synthase subunits (F1 α and β) that catalyze ATP synthesis would also compromise mitochondrial energy conservation. These findings, coupled with a decrease in the content of the other key mitochondrial metabolism enzymes (malate and pyruvate dehydrogenase), provide strong evidence to support the concept that bioenergetic dysfunction occurs in response to chronic exposure to a HFD and can be linked to changes in the proteome.
Accompanying these bioenergetic changes in mitochondria, we observed HFD-dependent alterations in mitochondrial ROS production. Consumption of a HFD for 8 weeks increased ROS, whereas decreased mitochondrial ROS production was observed after 16 weeks of HFD feeding (Fig. 5). Although the underlying molecular mechanism responsible for this result remains undefined, we predict that excess exposure to free fatty acids stimulated mitochondrial ROS production in a short term (i.e., 8 weeks). However, following long-term exposure to fatty acids (i.e., 16 weeks), reactive species-dependent posttranslational modifications to mitochondrial proteins including those of the respiratory chain are predicted to occur. This would most likely impair mitochondrial metabolism, diminish electron transport, and render mitochondria unable to maintain the membrane potential (9, 37). In agreement, we have shown that liver mitochondria from mice fed a HFD for 16 weeks have increased state 4 respiration and a lower mitochondrial membrane potential (25), which is likely due to a HFD-dependent induction of the liver-specific UCP2 (Fig. 5). These events would have the effect of decreasing ROS production, a membrane potential-dependent process (30). In response to increased state 4 respiration at the 16-week time point, mitochondria will be less able to conserve the H+ gradient generated across the inner membrane; thus, mitochondria are less well coupled (i.e., uncoupled). As a consequence, the likelihood of electrons “leaking” to O2 to form superoxide is diminished during increased state 4 respiration because the respiratory complexes involved in superoxide formation are in a less-reduced state during increased rates of electron transfer. Data showing an association between increased state 4 respiration and decreased ROS levels are supportive of this concept (Fig. 5). Although uncoupling may be beneficial in the short term by attenuating ROS production (5), increased state 4 respiration is likely to be highly detrimental in the long term as uncoupling decreases ATP synthesis efficiency. Sustained uncoupling would place hepatocytes under bioenergetic stress and increase susceptibility to cell death, especially when ATP demands are increased acutely in liver. Taken together, these results suggest that although mitochondrial ROS production and oxidative damage may contribute to NAFLD pathobiology early in the disease process, impaired bioenergetics and the inability to preserve hepatocyte viability may be more critical for disease severity as suggested by Diehl and colleagues (18). Future time course studies are needed to evaluate these events in the initiation and progression of NAFLD.
Correspondingly, we have shown that chronic consumption of a HFD increases the sensitivity of mitochondrial respiration to inhibition by NO (25). This observation is important because low, physiologically relevant concentrations of NO can reversibly inhibit mitochondrial respiration at cytochrome c oxidase by competing with O2 at the binuclear center. It is through this interaction that NO may act as a physiologic regulator of mitochondrial respiration, electron transport, and ROS production (8, 16). Although the underlying molecular change within the respiratory chain responsible for increased NO sensitivity is not known, we anticipate that increased sensitivity of mitochondria is related to decreased NO levels (i.e., bioavailability) in steatotic liver (Fig. 7). Specifically, we propose that mitochondria adapt to lower tissue NO concentrations in disease states by increasing their sensitivity to NO (25, 42, 48). In support of this, we observed changes in two key enzymes, arginase 1 and eNOS, which control tissue NO concentrations. First, arginase 1 protein was increased in the liver of the HFD mice compared with the controls (Fig. 6C). Arginase 1 hydrolyzes l-arginine to urea and l-ornithine (Fig. 6A). It is predicted that increased arginase 1 contributes to the decrease in NO metabolites (nitrate + nitrite) in the liver of the HFD mice (Fig. 7) by depleting the eNOS substrate l-arginine. Second, this deficiency in liver NO is compounded as exposure to a HFD also decreased levels of activated eNOS (i.e., decreased eNOS-PSer1177).
A deficiency in eNOS-derived NO may be important in the etiology of NAFLD because NO has beneficial effects in maintaining tissue oxygenation (45), controlling redox-sensitive signaling pathways (8), and supporting several key mitochondrial functions such as energy and lipid metabolism (7). Studies show that eNOS knockout mice have defective beta-oxidation, increased liver and skeletal muscle triglyceride content, and disrupted mitochondria function (22, 41). Moreover, seminal studies by Nisoli, Clementi, and Moncada strengthen this concept as NO is required for functionally active mitochondria via activation of pathways involved in mitochondrial biogenesis (32). As a signaling molecule, NO increases mitochondrial biogenesis by activating soluble guanyl cyclase and generating the second messenger cGMP (31–33). In line with this, NO affects gene expression through regulating the activities of numerous transcription factors (4, 54). Indeed, eNOS-derived NO has been shown to activate the master regulator of mitochondrial biogenesis, peroxisome proliferator-activated receptor coactivator 1α, which stimulates expression of mitochondrial transcription factors initiating expression of genes encoding mitochondrial proteins (31, 33). Although an examination of the effect of NO on the transcriptional networks responsible for the HFD-mediated changes in the mitochondrial proteome is beyond the scope of the present study, it is likely that the HFD-mediated decrease in NO contributes to mitochondrial dysfunction and changes to the mitochondrial proteome. For example, through our proteomic analyses we found a HFD-dependent decrease in mitochondrial GRP58, a protein disulfide isomerase family member that participates in establishing physical associations between the endoplasmic reticulum and mitochondrion and integrates cellular stress responses (15). We predict that the HFD-mediated decrease in GRP58 may be linked to decreased NO as studies show that NO is required for induction of other stress-responsive GRP genes (50). In addition, we observed a HFD-mediated increase in the levels of TSST. TSST is a highly abundant mitochondrial protein responsible for cyanide detoxification. Recently, TSST function has expanded to include sulfane sulfur transfer to other thiol groups, representing a new posttranslational protein thiol modification (29). Although the biological significance of TSST induction in response to HFD remains elusive, pharmacologic NO inhibition increases sulfane sulfur content presumably through modulation of sulfurtransferase expression and/or activity (44). Future studies are warranted to determine the exact underlying mechanisms by which eNOS-derived NO mediates the regulation of these mitochondrial proteins and others identified in this study as being altered by a HFD.
In summary, we provide an analysis of several metabolic changes that occur in liver and mitochondria following chronic exposure to a HFD. A total of 20+ mitochondrial proteins were significantly altered in abundance in response to a HFD, including proteins involved in energy metabolism, chaperone function, fatty acid oxidation, and sulfur amino acid metabolism. These new results are valuable as they highlight key metabolic pathways altered in an experimental model of NAFLD that might be suitable for forthcoming therapeutic intervention studies. Application of more focused proteomic techniques (blue native-PAGE) will likely advance investigations at the subproteome level, for example, the oxidative phosphorylation system, which is a suspected target of NAFLD pathogenesis. We also found a HFD-dependent decrease in serum and liver NO, which is likely related to increased arginase 1 and decreased active eNOS. This NO deficiency is predicted to disrupt mitochondrial function as eNOS-derived NO is critical for proper mitochondrial function and biogenesis. Finally, we propose that decreased NO bioavailability may play a role in the progression of NAFLD to NASH; low NO might be considered as a “second hit” in the disease process. Future studies aimed at using transgenic and/or knockout NOS models are planned to test this concept. Together, our results show that liver NO bioavailability is decreased in response to a HFD and likely contributes to defects in mitochondrial function and liver injury.
Supplementary Material
Abbreviations Used
- 2D IEF/SDS-PAGE
two-dimensional isoelectric focusing/sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- ALT
alanine aminotransferase
- ANCOVA
analysis of covariance
- eNOS
endothelial nitric oxide synthase
- eNOS-PSer1177
endothelial nitric oxide synthase, phosphorylated at Serine 1177
- ETFα
electron transfer flavoprotein-alpha
- FCCP
carbonyl cyanide 4-(trifluoromethoxy)-phenylhydrazone
- GRP58
glucose-regulated protein 58
- HFD
high-fat diet
- HMG-CoA synthase 2
3-hydroxy-3-methylglutaryl-CoA synthase 2
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- NO
nitric oxide
- PMS
postmitochondrial supernatant
- ROS
reactive oxygen species
- SDH-A
succinate dehydrogenase subunit A
- SEM
standard error of the mean
- TNFα
tumor necrosis factor alpha
- TSST
thiolsulfate sulfurtransferase
- UCP2
uncoupling protein 2
- VDAC
voltage-dependent anion channel
Acknowledgments
This work was supported by NIH grants R01AA15172 and R21DK73775 to S.M.B. and by NIH-funded UAB Clinical Nutrition Research Center (P30DK56336, pilot project to S.M.B.). Support for A.L.K. was provided through a Research Supplement to Promote Diversity in Health-Related Research linked to parent grant AA15172. Support for R.N.N. was provided through a summer medical student fellowship funded by the UAB Diabetes Research and Training Center (P60DK79626). The authors thank Dr. S. Barnes and Mr. L. Wilson of the UAB Mass Spectrometry Shared Facility. Mass spectrometers in the Shared Facility came from funds provided by NCRR grants S10RR11329 and S10RR13795 plus UAB HSF General Endowment Fund. Operational funds came from UAB Comprehensive Cancer Center Core Grant (P30CA13148), Purdue-UAB Botanicals Center for Age-Related Disease (P50AT00477), UAB Center for Nutrient-Gene Interaction in Cancer Prevention (U54CA100949), UAB Skin Disease Research Center (P30AR050948), and UAB Polycystic Kidney Disease Center (P30DK74038).
Authors Disclosure Statement
The authors have no conflicts of interest, no disclosures, and no competing financial interests.
References
- 1.Allison DB. Paultre F. Goran MI. Poehlman ET. Heymsfield SB. Statistical considerations regarding the use of ratios to adjust data. Int J Obes Relat Metab Disord. 1995;19:644–652. [PubMed] [Google Scholar]
- 2.Andringa KK. King AL. Eccleston HB. Mantena SK. Landar A. Jhala NC. Dickinson DA. Squadrito GL. Bailey SM. Analysis of the liver mitochondrial proteome in response to ethanol and S-adenosylmethionine treatments: novel molecular targets of disease and hepatoprotection. Am J Physiol Gastrointest Liver Physiol. 2010;298:G732–G745. doi: 10.1152/ajpgi.00332.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumgardner JN. Shankar K. Hennings L. Badger TM. Ronis MJ. A new model for nonalcoholic steatohepatitis in the rat utilizing total enteral nutrition to overfeed a high-polyunsaturated fat diet. Am J Physiol Gastrointest Liver Physiol. 2008;294:G27–G38. doi: 10.1152/ajpgi.00296.2007. [DOI] [PubMed] [Google Scholar]
- 4.Bogdan C. Nitric oxide and the regulation of gene expression. Trends Cell Biol. 2001;11:66–75. doi: 10.1016/s0962-8924(00)01900-0. [DOI] [PubMed] [Google Scholar]
- 5.Brookes PS. Mitochondrial H(+) leak and ROS generation: an odd couple. Free Radic Biol Med. 2005;38:12–23. doi: 10.1016/j.freeradbiomed.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Chavin KD. Yang S. Lin HZ. Chatham J. Chacko VP. Hoek JB. Walajtys-Rode E. Rashid A. Chen CH. Huang CC. Wu TC. Lane MD. Diehl AM. Obesity induces expression of uncoupling protein-2 in hepatocytes and promotes liver ATP depletion. J Biol Chem. 1999;274:5692–5700. doi: 10.1074/jbc.274.9.5692. [DOI] [PubMed] [Google Scholar]
- 7.Clementi E. Nisoli E. Nitric oxide and mitochondrial biogenesis: a key to long-term regulation of cellular metabolism. Comp Biochem Physiol A Mol Integr Physiol. 2005;142:102–110. doi: 10.1016/j.cbpb.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 8.Cooper CE. Giulivi C. Nitric oxide regulation of mitochondrial oxygen consumption II: molecular mechanism and tissue physiology. Am J Physiol Cell Physiol. 2007;292:C1993–C2003. doi: 10.1152/ajpcell.00310.2006. [DOI] [PubMed] [Google Scholar]
- 9.Cooper CE. Patel RP. Brookes PS. Darley-Usmar VM. Nanotransducers in cellular redox signaling: modification of thiols by reactive oxygen and nitrogen species. Trends Biochem Sci. 2002;27:489–492. doi: 10.1016/s0968-0004(02)02191-6. [DOI] [PubMed] [Google Scholar]
- 10.de Fourmestraux V. Neubauer H. Poussin C. Farmer P. Falquet L. Burcelin R. Delorenzi M. Thorens B. Transcript profiling suggests that differential metabolic adaptation of mice to a high fat diet is associated with changes in liver to muscle lipid fluxes. J Biol Chem. 2004;279:50743–50753. doi: 10.1074/jbc.M408014200. [DOI] [PubMed] [Google Scholar]
- 11.Donohue TM., Jr Autophagy and ethanol-induced liver injury. World J Gastroenterol. 2009;15:1178–1185. doi: 10.3748/wjg.15.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabbrini E. Sullivan S. Klein S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology. 2010;51:679–689. doi: 10.1002/hep.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldstein AE. Canbay A. Guicciardi ME. Higuchi H. Bronk SF. Gores GJ. Diet associated hepatic steatosis sensitizes to Fas mediated liver injury in mice. J Hepatol. 2003;39:978–983. doi: 10.1016/s0168-8278(03)00460-4. [DOI] [PubMed] [Google Scholar]
- 14.Hall D. Poussin C. Velagapudi VR. Empsen C. Joffraud M. Beckmann J. Geerts AE. Ravussin Y. Ibberson M. Oresic M. Thorens B. Peroxisomal and microsomal lipid pathways associated with resistance to hepatic steatosis and reduced pro-inflammatory state. J Biol Chem. 2010;285:31011–31023. doi: 10.1074/jbc.M110.127159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi T. Rizzuto R. Hajnoczky G. Su TP. MAM: more than just a housekeeper. Trends Cell Biol. 2009;19:81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill BG. Dranka BP. Bailey SM. Lancaster JR., Jr. Darley-Usmar VM. What part of NO don't you understand? Some answers to the cardinal questions in nitric oxide biology. J Biol Chem. 2010;285:19699–19704. doi: 10.1074/jbc.R110.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang PL. eNOS, metabolic syndrome and cardiovascular disease. Trends Endocrinol Metab. 2009;20:295–302. doi: 10.1016/j.tem.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jou J. Choi SS. Diehl AM. Mechanisms of disease progression in nonalcoholic fatty liver disease. Semin Liver Dis. 2008;28:370–379. doi: 10.1055/s-0028-1091981. [DOI] [PubMed] [Google Scholar]
- 19.Kawahara H. Fukura M. Tsuchishima M. Takase S. Mutation of mitochondrial DNA in livers from patients with alcoholic hepatitis and nonalcoholic steatohepatitis. Alcohol Clin Exp Res. 2007;31:S54–S60. doi: 10.1111/j.1530-0277.2006.00287.x. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy AR. Pissios P. Otu H. Roberson R. Xue B. Asakura K. Furukawa N. Marino FE. Liu FF. Kahn BB. Libermann TA. Maratos-Flier E. A high-fat, ketogenic diet induces a unique metabolic state in mice. Am J Physiol Endocrinol Metab. 2007;292:E1724–E1739. doi: 10.1152/ajpendo.00717.2006. [DOI] [PubMed] [Google Scholar]
- 21.King AL. Bailey SM. Assessment of mitochondrial dysfunction arising from treatment with hepatotoxicants. Curr Protoc Toxicol. 2010;44:14.8.1–14.8.29. doi: 10.1002/0471140856.tx1408s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Gouill E. Jimenez M. Binnert C. Jayet PY. Thalmann S. Nicod P. Scherrer U. Vollenweider P. Endothelial nitric oxide synthase (eNOS) knockout mice have defective mitochondrial beta-oxidation. Diabetes. 2007;56:2690–2696. doi: 10.2337/db06-1228. [DOI] [PubMed] [Google Scholar]
- 23.Li Z. Yang S. Lin H. Huang J. Watkins PA. Moser AB. Desimone C. Song XY. Diehl AM. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology. 2003;37:343–350. doi: 10.1053/jhep.2003.50048. [DOI] [PubMed] [Google Scholar]
- 24.Lieber CS. Leo MA. Mak KM. Xu Y. Cao Q. Ren C. Ponomarenko A. DeCarli LM. Model of nonalcoholic steatohepatitis. Am J Clin Nutr. 2004;79:502–509. doi: 10.1093/ajcn/79.3.502. [DOI] [PubMed] [Google Scholar]
- 25.Mantena SK. Vaughn DP., Jr Andringa KK. Eccleston HB. King AL. Abrams GA. Doeller JE. Kraus DW. Darley-Usmar V. Bailey SM. High fat diet induces dysregulation of hepatic oxygen gradients and mitochondrial function in vivo. Biochem J. 2009;417:183–193. doi: 10.1042/BJ20080868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKnight JR. Satterfield MC. Jobgen WS. Smith SB. Spencer TE. Meininger CJ. McNeal CJ. Wu G. Beneficial effects of L: -arginine on reducing obesity: potential mechanisms and important implications for human health. Amino Acids. 2010;39:349–357. doi: 10.1007/s00726-010-0598-z. [DOI] [PubMed] [Google Scholar]
- 27.Minehira K. Young SG. Villanueva CJ. Yetukuri L. Oresic M. Hellerstein MK. Farese RV., Jr. Horton JD. Preitner F. Thorens B. Tappy L. Blocking VLDL secretion causes hepatic steatosis but does not affect peripheral lipid stores or insulin sensitivity in mice. J Lipid Res. 2008;49:2038–2044. doi: 10.1194/jlr.M800248-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mootha VK. Bunkenborg J. Olsen JV. Hjerrild M. Wisniewski JR. Stahl E. Bolouri MS. Ray HN. Sihag S. Kamal M. Patterson N. Lander ES. Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 29.Mueller EG. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat Chem Biol. 2006;2:185–194. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- 30.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nisoli E. Clementi E. Paolucci C. Cozzi V. Tonello C. Sciorati C. Bracale R. Valerio A. Francolini M. Moncada S. Carruba MO. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 32.Nisoli E. Falcone S. Tonello C. Cozzi V. Palomba L. Fiorani M. Pisconti A. Brunelli S. Cardile A. Francolini M. Cantoni O. Carruba MO. Moncada S. Clementi E. Mitochondrial biogenesis by NO yields functionally active mitochondria in mammals. Proc Natl Acad Sci U S A. 2004;101:16507–16512. doi: 10.1073/pnas.0405432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nisoli E. Tonello C. Cardile A. Cozzi V. Bracale R. Tedesco L. Falcone S. Valerio A. Cantoni O. Clementi E. Moncada S. Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 34.Pagano C. Soardo G. Esposito W. Fallo F. Basan L. Donnini D. Federspil G. Sechi LA. Vettor R. Plasma adiponectin is decreased in nonalcoholic fatty liver disease. Eur J Endocrinol. 2005;152:113–118. doi: 10.1530/eje.1.01821. [DOI] [PubMed] [Google Scholar]
- 35.Perez-Carreras M. Del Hoyo P. Martin MA. Rubio JC. Martin A. Castellano G. Colina F. Arenas J. Solis-Herruzo JA. Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology. 2003;38:999–1007. doi: 10.1053/jhep.2003.50398. [DOI] [PubMed] [Google Scholar]
- 36.Qureshi K. Abrams GA. Metabolic liver disease of obesity and role of adipose tissue in the pathogenesis of nonalcoholic fatty liver disease. World J Gastroenterol. 2007;13:3540–3553. doi: 10.3748/wjg.v13.i26.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci U S A. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radonjic M. de Haan JR. van Erk MJ. van Dijk KW. van den Berg SA. de Groot PJ. Muller M. van Ommen B. Genome-wide mRNA expression analysis of hepatic adaptation to high-fat diets reveals switch from an inflammatory to steatotic transcriptional program. PLoS One. 2009;4:e6646. doi: 10.1371/journal.pone.0006646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajapakse NW. Mattson DL. Role of L-arginine in nitric oxide production in health and hypertension. Clin Exp Pharmacol Physiol. 2009;36:249–255. doi: 10.1111/j.1440-1681.2008.05123.x. [DOI] [PubMed] [Google Scholar]
- 40.Roychowdhury S. McMullen MR. Pritchard MT. Hise AG. van Rooijen N. Medof ME. Stavitsky AB. Nagy LE. An early complement-dependent and TLR-4-independent phase in the pathogenesis of ethanol-induced liver injury in mice. Hepatology. 2009;49:1326–1334. doi: 10.1002/hep.22776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schild L. Dombrowski F. Lendeckel U. Schulz C. Gardemann A. Keilhoff G. Impairment of endothelial nitric oxide synthase causes abnormal fat and glycogen deposition in liver. Biochim Biophys Acta. 2008;1782:180–187. doi: 10.1016/j.bbadis.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 42.Shiva S. Oh JY. Landar AL. Ulasova E. Venkatraman A. Bailey SM. Darley-Usmar VM. Nitroxia: the pathological consequence of dysfunction in the nitric oxide-cytochrome c oxidase signaling pathway. Free Radic Biol Med. 2005;38:297–306. doi: 10.1016/j.freeradbiomed.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 43.Snedecor GW. Cochran WG. Statistical Methods. Ames, IA: The Iowa State University Press; 1967. p. 593. [Google Scholar]
- 44.Sokolowska M. Wlodek L. Srebro Z. Wrobel M. The effect of nitrogen oxide level modulation on the content of thiol compounds and anaerobic sulfur metabolism in mice brains. Neurobiology (Bp) 1999;7:461–477. [PubMed] [Google Scholar]
- 45.Thomas DD. Liu X. Kantrow SP. Lancaster JR., Jr. The biological lifetime of nitric oxide: implications for the perivascular dynamics of NO and O2. Proc Natl Acad Sci U S A. 2001;98:355–360. doi: 10.1073/pnas.011379598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tipoe GL. Ho CT. Liong EC. Leung TM. Lau TY. Fung ML. Nanji AA. Voluntary oral feeding of rats not requiring a very high fat diet is a clinically relevant animal model of non-alcoholic fatty liver disease (NAFLD) Histol Histopathol. 2009;24:1161–1169. doi: 10.14670/HH-24.1161. [DOI] [PubMed] [Google Scholar]
- 47.Venkatraman A. Landar A. Davis AJ. Chamlee L. Sanderson T. Kim H. Page G. Pompilius M. Ballinger S. Darley-Usmar V. Bailey SM. Modification of the mitochondrial proteome in response to the stress of ethanol-dependent hepatotoxicity. J Biol Chem. 2004;279:22092–22101. doi: 10.1074/jbc.M402245200. [DOI] [PubMed] [Google Scholar]
- 48.Venkatraman A. Shiva S. Davis AJ. Bailey SM. Brookes PS. Darley-Usmar V. Chronic alcohol consumption increases the senstivity of rat liver mitochondrial respiration to inhibition by nitric oxide. Hepatology. 2003;38:141–147. doi: 10.1053/jhep.2003.50293. [DOI] [PubMed] [Google Scholar]
- 49.Wolf NI. Rahman S. Clayton PT. Zschocke J. Mitochondrial HMG-CoA synthase deficiency: identification of two further patients carrying two novel mutations. Eur J Pediatr. 2003;162:279–280. doi: 10.1007/s00431-002-1110-x. [DOI] [PubMed] [Google Scholar]
- 50.Xu W. Liu L. Charles IG. Moncada S. Nitric oxide induces coupling of mitochondrial signalling with the endoplasmic reticulum stress response. Nat Cell Biol. 2004;6:1129–1134. doi: 10.1038/ncb1188. [DOI] [PubMed] [Google Scholar]
- 51.Yamazaki Y. Kakizaki S. Takizawa D. Ichikawa T. Sato K. Takagi H. Mori M. Interstrain differences in susceptibility to non-alcoholic steatohepatitis. J Gastroenterol Hepatol. 2008;23:276–282. doi: 10.1111/j.1440-1746.2007.05150.x. [DOI] [PubMed] [Google Scholar]
- 52.Young TA. Cunningham CC. Bailey SM. Reactive oxygen species production by the mitochondrial respiratory chain in isolated rat hepatocytes and liver mitochondria: studies using myxothiazol. Arch Biochem Biophys. 2002;405:65–72. doi: 10.1016/s0003-9861(02)00338-7. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J. Li X. Mueller M. Wang Y. Zong C. Deng N. Vondriska TM. Liem DA. Yang JI. Korge P. Honda H. Weiss JN. Apweiler R. Ping P. Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria. Proteomics. 2008;8:1564–1575. doi: 10.1002/pmic.200700851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou J. Brune B. NO and transcriptional regulation: from signaling to death. Toxicology. 2005;208:223–233. doi: 10.1016/j.tox.2004.11.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.