Abstract
The developing retina is an excellent model to study cellular fate determination and differentiation in the context of a complex tissue. Over the last decade, many basic principles and key genes that underlie these processes have been experimentally identified. In this review, we construct network models to summarize known gene interactions that underlie determination and fundamentally affect differentiation of each retinal cell type. These networks can act as a scaffold to assemble subsequent discoveries. In addition, these summary networks provide a rational segue to systems biology approaches necessary to understand the many events leading to appropriate cellular determination and differentiation in the developing retina and other complex tissues.
Keywords: retina, cell fate determination, network, systems biology
Introduction
Multicellular organisms are made of tissues with multiple specialized cell types. Understanding the determination and differentiation of heterogeneous cell types within the context of complex tissues is fundamental to many areas of biology. This knowledge will have widespread application in treatment of developmental disorders and disease states such as cancer and will be critical for successful bioengineering and transplantation of tissue types to replace damaged or degenerate structures. The determination and differentiation of a given cell within a tissue is the culmination of the expression of many gene products and their subsequent intra- and intercellular signaling events. To address the challenge of understanding cell fate determination and differentiation we must adopt a broad systems biology approach to adequately take into account the activities of large numbers of genes and signaling pathways.
One emerging systems-based strategy to analyze and integrate large datasets is to generate network models, in which genes or proteins are represented by nodes and their relationships by edges in the graph (network). However, most large expression datasets are too sparse to infer high statistical confidence gene relationships which are based on the estimate of a covariance matrix.1 In addition, the networks generated de novo are often large, and do not facilitate prioritization of candidate genes and gene relationships for hypothesis based validation. To address this problem, we have previously described a heuristic approach that uses a seed network to summarize prior knowledge of a small part of the gene network involved in cellular development.2,3 The seed network can then be used to query large datasets in order to identify additional molecules with putative relationships to seed genes. These candidate molecules can then be used to expand the network and are the basis for generating testable hypotheses to validate their functional role.
Cell fate determination and differentiation in the vertebrate retina provides many opportunities to generate and utilize systems-based tools and approaches to understand development of cells within complex tissues. First, development of the retina is well-characterized4–6 and the sequence of cell genesis and differentiation is well-documented and largely conserved among vertebrates.7–11 Thus, activity of gene networks that underlie the fate determination and differentiation in a particular retinal cell type will take place in known cells with known birthdates and known locations within the tissue. Second, the retina is highly accessible and is very amenable to in vivo hypothesis testing,12 thus the role of hypothesized gene candidates and network interactions in cell fate determination and differentiation can be readily assessed. Third, we can build on the foundational system-based approaches developed through the study of single cell organisms like yeast,13 diffuse systems like the immune system,14 or cultured tissue systems,15 and extend these methods to examine the development of more complex tissues that comprise living organisms.
Here we review what is presently known about the genetic networks that underlie cell fate determination and differentiation in the developing retina and present the seed networks that we have constructed based on our examination of published literature. The developing retina is an extensively reviewed16–20 system regarding cell fate determination during retinogenesis, but a summary of literature-curated gene networks underlying differentiation of each retinal cell type has not been previously presented. In order to demonstrate its potential as a model to study determination and differentiation of multiple cell types within the context of a complex tissue, we have assembled seed networks to summarize what is known about the genes and their relationships that underlie cell fate determination and largely influence the differentiation of each of the basic retinal cell types. Finally, we demonstrate that the experimentally-based summary network for photoreceptors can be extracted from an independent gene expression data set.
Retinal Cell Types
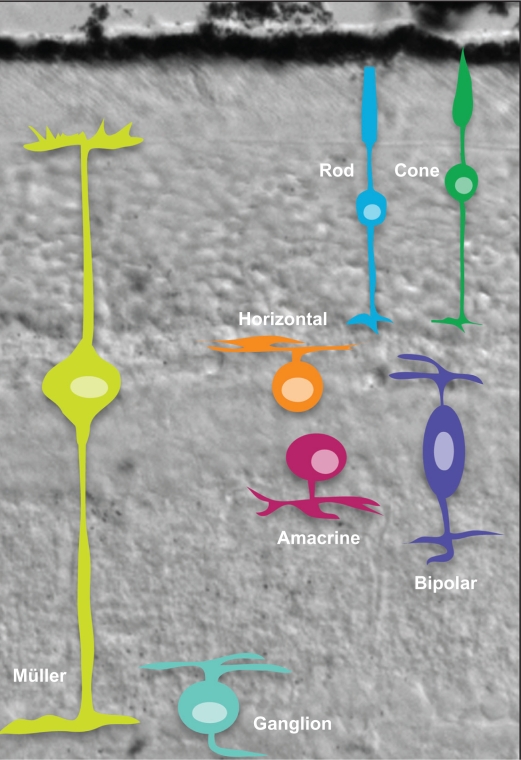
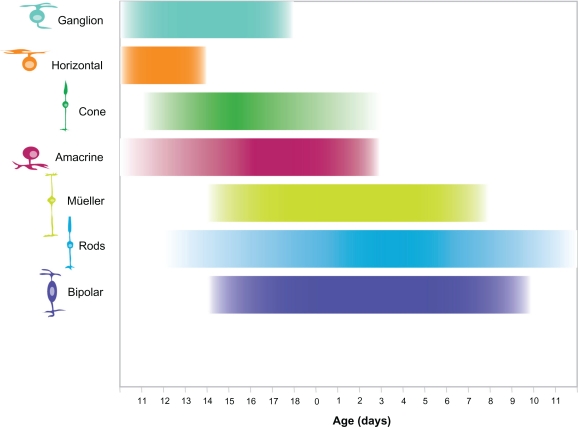
The mature mouse retina is composed of seven basic cell types, six neuronal and one glial (Fig. 1). While this review focuses on only the differentiation of the basic cell types, many retinal cells can be further subdivided morphologically, biochemically and functionally.21–31 Photoreceptors (rods and cones) reside in the outer nuclear layer (ONL) and are responsible for phototransduction and necessary for vision.32 Photoreceptors synapse with bipolar cells, neurons that reside in the inner nuclear layer (INL). Bipolar cells relay visual stimulus to retinal ganglion cells in the ganglion cell layer either directly or indirectly via amacrine cells, which also reside in the INL. Other cells present in the INL are horizontal cells, which mediate lateral interactions between photoreceptors and Müller glia that play a critical role in retinal homeostasis.33 Axons of the retinal ganglion cells project into the visual centers in the brain, thereby relaying the visual information detected by the retina. While appropriate processing of visual stimuli requires the function of all retinal cell types, most blinding retinal diseases are the result of the degeneration of photoreceptors or ganglion cells.34,35 Interestingly, the seven cell types that comprise the retina are derived from a common pool of retinal progenitor cells.17 Thus, the developing retina provides a relatively simple, yet elegant system to study the generation and maturation of a complex tissue. We know that the cell fate decisions made by retinal progenitor cells are governed by an intrinsic genetic program that determines their response to extrinsic cues from their environment.16,17 The sequence of retinal cell genesis is highly conserved in vertebrates,8,11,36–39 following a general progression of retinal ganglion cells (RGCs), horizontal cells (HCs), cone photoreceptors followed by amacrine cells (ACs), and subsequently bipolar cells (BCs), rod photoreceptors and Müller glial cells (MCs) (Fig. 2). Based on this general progression of birth order, retinal cell types can be divided into cohorts of early-born cells which include ganglion cells and cone photoreceptors, and late-born cells which include rod photoreceptors, bipolar cells and Müller glia.8
Figure 1.
The retinal cell types in the adult mouse retina.
The adult mouse retina is comprised of three cellular layers separated by two synaptic layers. Rod and cone photoreceptors reside in the outer nuclear layer (ONL), and form synaptic contacts in the outer plexiform layer (OPL) with horizontal cells and bipolar cells, both of which reside in the inner nuclear layer (INL). In addition, amacrine cells and the cell bodies of Müller glia are found in the INL. Synaptic contacts between bipolar cells, amacrine cells and ganglion cells are present in the inner plexiform layer (IPL) and ganglion cells reside in the innermost cellular layer, the ganglion cell layer.
Figure 2.
Time course of cell genesis in the developing mouse retina. Retinal cell types are listed on the Y-axis, developmental time on the X-axis. Birth of the animal is indicated as 0, embryonic development is left of 0, postnatal development to the right. The approximate time course of cell genesis is indicated by the bar adjacent each cell type. This figure is based on the work reported by Young.8
Gene Families that Underlie the Specification of Retinal Cell Types
There are a number of genes that are well known to act in the specification of and/or largely influence the differentiation of retinal cells. They compose a regulatory network that can integrate extrinsic information through signaling pathways like Notch, as well as implement intrinsic programming via transcription factors, many of which can be grouped into the basic helix loop helix (bHLH) gene family and the homeobox gene family.
The family of basic helix loop helix (bHLH) genes is characterized by an α helix-loop-α helix structural motif. The bHLH genes Mash1, Math3, NeuroD, Math5 and Ngn2 cross-regulate each other to specify neuronal types in developing retina,19,40 while Ptf1a,41,42 Bhlhb443 and Bhlhb544 have roles in the development of more specific retinal cell types or subtypes. Other family members such as Hes1 and Hes5 work as effectors of Notch signaling. These bHLH genes interact with members of homeobox gene subfamilies including Pax, CVC, POU, Lim and Sox. The Pax gene subfamily has critical roles in embryogenesis45 and Pax6 functions as an early regulatory gene in the development of eye.46 In contrast, the CVC homeodomain subfamily members Vsx1 and Chx10 have more specific roles in retinogenesis across vertebrate species like mouse,47,48 chicken49 and fish.50,51 The POU homeodomain subfamily members have a variety of functions related to neural development,52 and genes Brn3b, Brn3c53 and Brn3a54 are all involved in mouse retinal development. The LIM homeodomain gene subfamily are involved in neural patterning55 with Isll and Lim1 playing crucial roles in retinal development. The Sox subfamily genes56 are indispensible in many aspects of development including neurogenesis, and Sox8, Sox9 are implicated in retinogenesis. Like the bHLH genes, some homeobox genes like Vsx1,57,58 Barhl259 and Irx560 appear to specify retinal cell subtypes. Together these genes (see Supplementary Information) work in concert to specify cell fate in the developing retina.
Using a scaffold of bHLH and homeobox gene family members, we developed a seed network to summarize key gene relationships that govern the development of each of the retinal cell types in mouse retina. These seed networks are based on published studies that have demonstrated a role for the seed genes in the determination and differentiation of retinal cell types via either loss of function experiments41,61–68 gain of function experiments69,70 or transcriptional regulation experiments.71,72 Genes involved in the specification of multiple retinal cell types (see Supplementary Information) are not always included, in a given seed network due to the lack of strong evidence they interact with other essential genes in the seed network specifying a particular cell type. These seed networks can be used in two complimentary ways: 1) to design database queries to identify additional key molecules for cell-specific development, 2) to assemble a comprehensive summary of known gene relationships and identify key decision points in cell-specific specification that may be important regulatory targets for future application.
Müller Glial Cells
The gene relationships that underlie Müller glia determination and differentiation are summarized in the seed network in Figure 3. Müller glia are the only glial cells to arise from the retinal progenitor cell population. Thus, the factors which influence the progenitor cell choice between gliogenesis and neurogensis are critical for the creation of these cells. Previous work has demonstrated that Notch signaling plays a major role in the choice between neural and glial cell fate.73 Notch is a transmembrane receptor that functions at the cell surface to both receive extracellular signals and to regulate gene expression in the nucleus. Notch signaling is widely used to control developmental processes in many animal species. 74 In the developing retina, the Notch pathway is implicated in the control of progenitor cell proliferation and apoptosis, as well as the multipotency of progenitor cells.75 In addition to its role in maintaining the undifferentiated and proliferative state of retinal progenitor cells (RPCs), Notch also seems to regulate the neuronal versus glial cell fate choice by inhibiting the photoreceptor cell fate in mouse retina.76,77
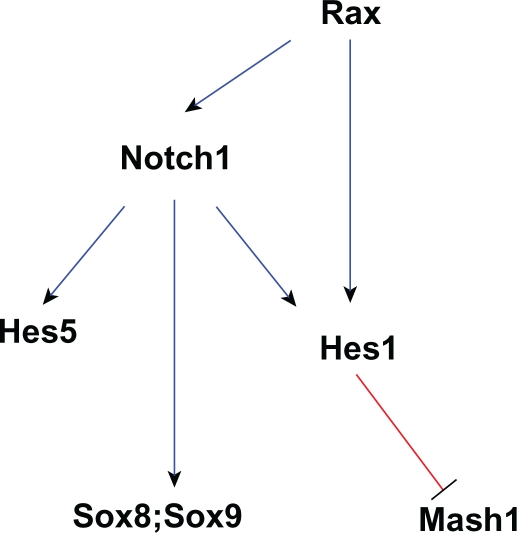
Figure 3.
A network of genes essential for Müller glia development. Edges in this graph are based on evidence that Rax promotes notch1 and Hes1 transcription,73 Notch signaling positively regulates expression of hes1, hes5,78 sox8 and sox9,71,80 and Hes1 suppresses the proneuronal gene Mash1.79,162 Blue edges between genes indicate activation, while red edges indicate repression.
As essential effectors of Notch signaling,78 bHLH genes Hes1 and Hes5 have partly overlapping but distinct roles in Müller cell determination and differentiation. Both Hes1 and Hes5 are thought to repress expression of neuronal bHLH genes.19 However, their specific target genes appear to be different since Hes1 maintains the progenitors and inhibits both neuronal and glial differentiation, whereas Hes5 cooperatively regulates maintenance of progenitors but promotes the glial cell fate.61,62 Specifically, Hes1 is known to inhibit the proneuronal gene Mash179 and thus promotes glial cell determination. Consistent with their different effects, both Hes1 and Hes5 are expressed in undifferentiated cells while Hes5 is also expressed in differentiating Müller glial cells.
The homeobox gene, Rax, promotes the glial cell fate choice, potentially via activation of promoters of Notch1 and Hes1.73 The homeobox genes Sox8 and Sox9 have also been implicated in the specification of Müller glial cells,71,80 though neither of them alone is sufficient to induce Müller glial cell differentiation. Notch signaling regulates Sox8 and Sox9 transcription, though it does not appear to be through its activation of Hes1 and Hes5.71
Retinal Ganglion Cells
The gene relationships that underlie retinal ganglion cell determination and differentiation are summarized in the seed network in Figure 4. The bHLH gene Math5 plays a critical role in retinal ganglion cell (RGC) development. The targeted deletion of Math5 results in the loss of more than 80% of RGCs,64 and a cell fate shift to other retinal cell types.64,81,82 It seems that Math5 underlies RGC differentiation in two ways. First, Math5 activates a downstream transcriptional network that controls ganglion cell differentiation and development.83,84 Second, Math5 suppresses other bHLH proneuronal genes such as Math3, NeuroD and Ngn2 that are involved in the adoption of other retinal cell fates.82,84 The available evidence suggests that Math5 is directly regulated by Pax6.85,86 Downstream of Math5, Brn3b and lsl-1 are known to play critical roles in ganglion cell differentiation.64,87,88 Brn3b, a POU subfamily gene, while not required for the initial commitment of RGC fate, is essential for early retinal ganglion cell differentiation.63,89 Homozygous disruption of Brn3b leads to a selective loss of 70% RGCs63 suggesting not all RGC differentiation is dependent on Brn3b.90 Consistent with this, it is hypothesized that Brn3b regulates genes important for formation of RGC axons and axon path-finding.89,91 In addition to loss of Brn3b, deletion of the Lim family gene lsl-1 also causes a marked reduction in the number of ganglion cells.65 Recent studies indicate that both lsl-1 and Brn3b regulate genes such as Eomes and Shh.87,88 Eomes is a T-box transcription factor, now known as a direct target of Brn3b and required for RGCs and optic nerve development.92 Other Brn3b-related genes are also found to contribute to ganglion cell development. For example, the zinc finger protein Wt1, acts upstream of Brn3b, and plays a role in the development of RGCs.93,94 Barhl2 functions downstream of Brn3b to regulate the maturation and survival of RGCs.59 Math5 and Brn3b are essential for ganglion cell determination. In addition here are other Brn3b dependent-genes,72 Math5-dependent genes,84 and genes identified in RGC single cell expression studies. 95 However, the relationships of these genes to the network described here are not yet understood and were not included in our seed network.
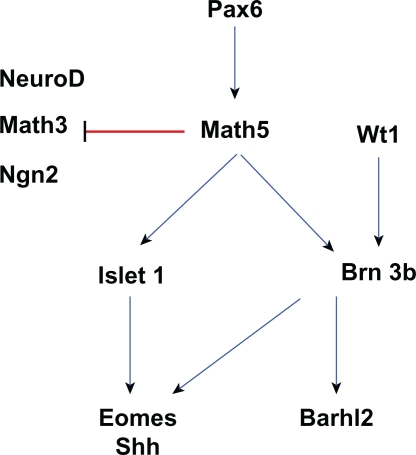
Figure 4.
A network of genes essential for ganglion cell development. Edges in this graph are based on evidence that Pax6 actives Math5 expression163 and Math5 suppresses Math3 and NeuroD to promote ganglion cell fate.82,84 In addition, Math5 promotes Brn3b and Isletl expression,64,87,88 which in turn positively regulate genes like Eomes,92 Shh164 and Barhl2.59 Brn3b is also activated by Wt1.94 Blue edges between genes indicate activation, while red edges indicate repression.
Bipolar Cells
Compared to other retinal cell types, data supporting the relationships among genes essential for bipolar cells specification and differentiation are relatively sparse; however, the genes with key regulatory roles in bipolar cells determination and differentiation are summarized in Figure 5. The bHLH gene Mash 1 plays a pivotal role in bipolar cell differentiation. In both rat and mouse, the onset of Mash1 expression (about E15) most closely correlates with the appearance of bipolar cells and Müller glia.96,97 In Mash1 –/– retinal explants, the differentiation of all late born retinal cells (bipolar cells, rod photoreceptors and Müller glia) was delayed, and the number of the mature bipolar cells was significantly reduced, though the number of vimentin-positive cells (likely Müller glial cells) was increased.98 Additionally, Mash1 is expressed by a subset (10%–30%, depending on age) of the total proliferating progenitor cells, providing a molecular marker of heterogeneity among retinal progenitor cells (RPCs).97 Together, this evidence suggests that Mash1 plays a role in the commitment and/or differentiation of late born retinal cells, particularly bipolar cells.
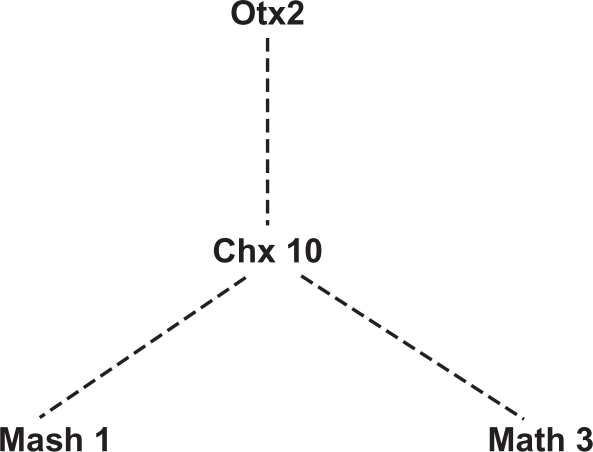
Figure 5.
A network of genes essential for bipolar cell development. The edges in this graph are based on evidence that Otx2 may affect the competence of progenitor cells to adopt a bipolar vs. rod photoreceptor cell fate,103 that Chx10 is hypothesized to work downstream of Otx2 to promote bipolar cell fate,102 and that Chx10, together with Mash1 and Math3, specify bipolar cell fate.67,100 Dotted edges indicate indirect or poorly characterized gene relationships.
Mash1 and Math3 are co-expressed in various regions of CNS suggesting these genes may have some functional redundancy.67 Interestingly, the Xenopus homolog of Math3, Ath3, was shown to directly convert non-neuronal or undifferentiated cells to a neural fate,99 though the phenotype of Math3 (–/–) mice suggests Math3 is not essential for neuronal commitment.67 However, in Math3 (–/–)-Mash1 (–/–) mice, in regions where the two genes are normally co-expressed, neuronal fate is blocked at the neural precursor stage and cells that normally differentiate into neurons adopted the glial fate. The retinas in these animals lack bipolar cells and have a significantly increased population of Müller glia.67 It has been shown that Math3 and Mash1 are expressed by differentiating bipolar cells in the retina.97,99 However, misexpression of Mash1 or Math3 does not promote bipolar cell generation, rather it inhibits Müller gliogenesis. 100 Taken together these studies suggest that Mash1, with the cooperation of Math3, prevents gliogenesis in the developing retina and contributes significantly, but not entirely, to the specification of the bipolar cell fate.
The expression of the homeobox gene Chx10 is also integral to bipolar cell fate. Chx10 is restricted to the inner nuclear layer (INL) in the mature retina, though in the developing mouse eye, the Chx10 transcript is confined to the anterior optic vesicle and all neuroblasts of the optic cup.101 Loss of Chx10 results in reduced proliferation of retinal progenitors and a specific absence of differentiated bipolar cells.68 Misexpression of Chx10 induces generation of inner nuclear layer cells,100 while misexpression of Mash1 or Math3 together with Chx10 increases the number of mature bipolar cells while decreasing the mature Müller glial cell number.100 Thus, it is proposed that Chx10 confers the specific inner nuclear layer identity to retinal neurons while bHLH genes such as Mash1 and Math3 subsequently specify the bipolar cell fate.100 In addition, Chx10 promotes bipolar cell fate determination by inhibiting photoreceptor specification, presumably by acting downstream of Otx2 or other Otx genes.102 Otx2 subcellular localization is hypothesized to play a role in the rod versus bipolar cell fate choice.103 In the retina of a postnatal, bipolar-cell-specific-Otx2 conditional knockout mouse the expression of mature bipolar cell markers is significantly down-regulated,104 demonstrating its importance in bipolar cell differentiation.
Amacrine Cells
The gene relationships that underlie amacrine cell determination and differentiation are summarized in the seed network in Figure 6. For amacrine cell specification, the bHLH gene Math3 cooperates with another bHLH gene, NeuroD, and amacrine cells are completely missing in Math3-NeuroD double mutant retinas. The cells in the double knockout retinas that fail to differentiate into amacrine cells adopt both ganglion and Müller glial cell fates. However, while these genes are necessary for amacrine cell fate determination, they are not sufficient; misexpression of either Math3 or NeuroD alone cannot induce amacrine cell genesis.69
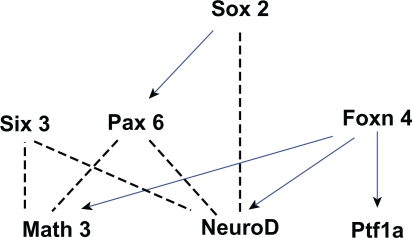
Figure 6.
A network of genes essential for amacrine cell development. The edges in this graph are based on evidence that Sox2 activates Pax6 and NeuroD to promote amacrine cell fate,70,85 that Pax6 and Six3, with the cooperation of Math3 and NeuroD, specify amacrine cell fate,69 and that Foxn4 positively regulates Ptf1a, Math3 and NeuroD expression.41,42,66 Blue edges between genes indicate activation while dotted edges indicate indirect or poorly characterized relationships between genes.
In the Pax6-knockout mouse retina, the retinal progenitor cells become totally restricted to an amacrine cell fate.85 While misexpression of Pax6, Math3 or NeuroD alone does not induce amacrine genesis, the misexpression of a combination of bHLH genes Math3 or NeuroD with homeobox genes Pax6 or Six3 (the transcription of which is independent of Pax6)105 does promote amacrine cell genesis.69 Furthermore, misexpression of Pax6 with only Math3 results in the production of amacrine cells and horizontal cells, while the combination of Pax6 and NeuroD predominantly increases only the number of amacrine cells, suggesting that when expressed with Pax6, NeuroD is more specific for amacrine cell differentiation than Math3.69 The homeobox gene, Sox2, is expressed in a subset of amacrine cells and misexpression of Sox2 results in a dramatic increase of amacrine cells in INL. Experimental evidence indicates that Sox2 transcriptionally induces Pax6 and may also induce NeuroD.70 Taking all these data into account, it appears that Sox2 functions upstream of Pax6 and NeuroD to affect/promote amacrine cell fate.
The expression of the forkhead gene family member Foxn4 in mouse retina correlates closely with the birth date of amacrine cells and misexpression of Foxn4 promotes amacrine cell genesis.66 Further, Foxn4-null mice exhibit a significant decrease in amacrine cells and a complete loss of horizontal cells.66 The effect of Foxn4 on amacrine cell differentiation may be via activation upstream of NeuroD and Math3 signaling, since in Foxn4–/– retinas there is a marked downregulation of NeuroD and Math3 with no observable alteration in Math5, Ngn2, Chx10 or Pax6 expression. 66 Downstream of Foxn4 is Ptf1a.106 Lineage tracing reveals that Ptf1a expression in the developing mouse retina marks the horizontal and amacrine cell precursors. 42 Loss of Ptf1a affects the differentiation of a small population of amacrine cells and the entire population of horizontal cells. While Foxn4 may influence amacrine cell differentiation via NeuroD and Math3, Ptf1a does not appear to work in this way as in the Ptf1a-null retina expression of the two genes was unaffected.41,42
Horizontal Cells
The gene relationships that underlie horizontal cell determination and differentiation are summarized in the seed network in Figure 7. It appears that amacrine (Fig. 6) and horizontal (Fig. 7) cell fates are linked as they share several key regulatory genes including Foxn4, Ptf1a, Math3, and Pax6 (Figs. 6 and 7). As previously mentioned, misexpression of Pax6 with Math3 results in an increase of both horizontal cells and amacrine cells, though the effect on horizontal cell genesis is greater (14% increase) than the effect on amacrine cell genesis (7% increase).69 At the same time, deletion of Foxn4 results in complete loss of horizontal cells, presumably via the downregulation of Math3.66
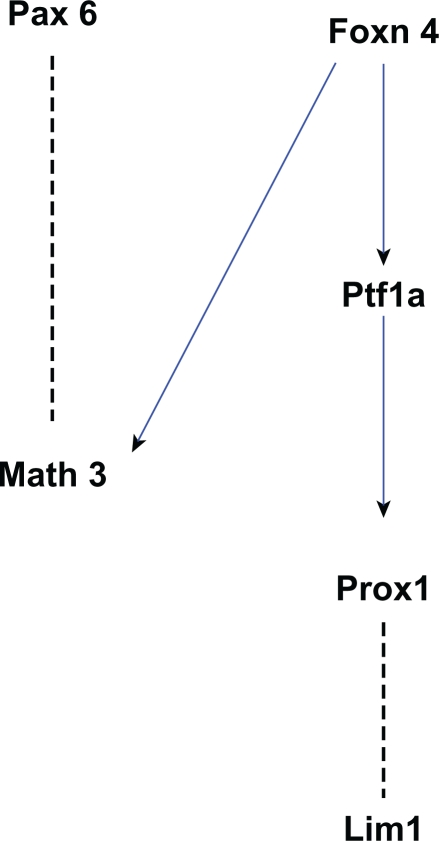
Figure 7.
A network of genes essential for horizontal cell development. The edges in this graph are based on evidence that Foxn4 positively regulates Math3 and Ptf1a expression,66 that coexpression of Pax6 and Math3 promotes horizontal cell fate,69 and that Ptf1a positively regulates Prox1 expression41,66,107 which in turn affects Lim1 expression.106
Prox1, the Prospero-related homeobox 1, is also important for horizontal cell differentiation. Prox1 is expressed in, and is required for efficient cell cycle exit for, early RPCs (but not in late RPCs).107 Prox1-null retinas exhibit a complete loss of horizontal cells and the misexpression of Prox1 results in the production of horizontal cells.107,108 Considering the fact that there is a lack of Prox1 expression in Foxn4-null retina and a downregulation of Prox1 in Ptf1a-null retina,41,66 Prox1 seems to promote horizontal cell fate by acting downstream of the Foxn4-Ptf1a axis. Downstream of Foxn4-Ptf1a-Prox1 is another essential gene, Lim1.106 Lim1 is required for specific morphogenesis of horizontal cells in chick retina.109 In mouse retina, Lim1 is essential to instruct the differentiation and migration of horizontal cells to the correct laminar position.110,111
Cone and Rod Photoreceptors
Both cones and rods employ phototransduction, a process that captures and converts photons of light to an electrical signal; however, each cell type expresses a particular visual protein (opsin) to absorb a specific portion of the light spectrum. In mice, cones express either a S-opsin (short wavelength sensitive) or a M-opsin (middle wavelength sensitive) while rods express rhodopsin. Interestingly, both rod and cone photoreceptors share several key genes essential for cell fate specification and differentiation. Thus, the relationships of genes underlying the differentiation of cones and rods are shown together in a single network (Fig. 8).
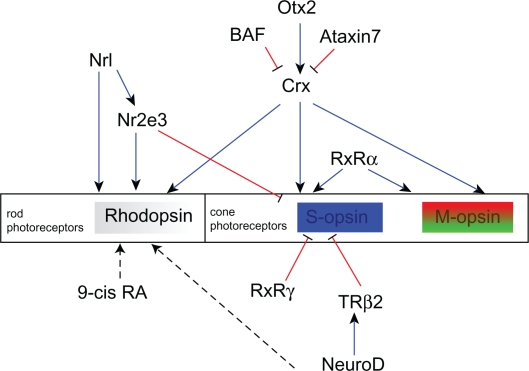
Figure 8.
A network of genes essential for rod and cone photoreceptor cell development.
The edges in this graph are based on evidence that Otx2 activates Crx12 while Ataxin-7118 and BAF119 repress Crx transactivation. Crx and Nrl synergically activate the rod specific pigment rhodopsin,114 while Crx promotes expression of M and S cone-specific opsins.115,116 Nr2e3, which is activated by Nrl represses expression of both S- and M-opsin.127 NeuroD is necessary for sustained expression of TRβ2,113 which inhibits S-opsin and activates M-opsin expression.142 RXRα146 promotes cone-specific gene expression while RXRγ145 represses S-opsin expression. Blue edges between genes indicate activation, while red edges indicate repression.
NeuroD is the only bHLH gene known to be essential for photoreceptor differentiation. NeuroD is expressed in developing photoreceptors and is maintained in a subset of mature photoreceptors in the adult mouse retina.112,113 In the NeuroD-null retina, the number of rods is reduced, while the number of the bipolar cells is increased in a dose-dependent fashion112 Misexpression of NeuroD not only blocks gliogenesis, but also favors rod photoreceptor differentiation while reducing bipolar cell differenation.112 NeuroD is also necessary for sustained expression of TRβ2, an essential gene for cone photoreceptor development.113
Photoreceptor cell types are generated by common activity of genes like Crx (Cone rod homeobox), Nrl (neural retina leucine zipper), and Nr2e3. For example, Crx is expressed early in the developing retina, and is predominantly expressed in photoreceptors in mature retina.114 Crx transactivates the Rhodopsin promoter and acts synergistically with Nrl to drive rhodopsin expression in rods.114 Crx also activates cone opsins.115,116 Two genes are known to suppress Crx function Ataxin-7117,118 and BAF,119 both contribute to photoreceptor degenerative disease. Otx2, a member of Otx homeobox gene family, transactivates Crx120 and misexpression of Otx2 directs retinal progenitor cells towards photoreceptor fate.120
Nrl is a basic motif–leucine zipper transcription factor preferentially expressed in rod photoreceptors,121,122 which positively regulates rhodopsin.123,124 In the Nrl−/− mouse retina, cone-like photoreceptor cells are clearly different from WT rods and cones, revealing a functional transformation from rods to S-cones.125,126 From these results, it is inferred that Nrl modulates rod-specific genes as well as inhibits S-cone differentiation through the activation of Nr2e3.126,127 Nr2e3 expression is restricted to photoreceptor cells. It is a ligand-dependent transcription factor that requires itself for the repression of its own transcription.128,129 Mutation of Nr2e3 causes enhanced S cone syndrome (ESCS),130 a retinal degenerative disease in humans that results in an abundance of short-wavelength sensitively cones (S cones) at the expense of rod photoreceptors.131 It is hypothesized that when photoreceptors are first generated the defective Nr2e3 cannot prevent a ‘default’ shift of rod progenitors to an S-cone fate, producing a large number of S-cones and an absence of rods.132 This is supported by the fact that Nr2e3 acts as a repressor of cone-specific genes in rods,133 and directly interacts with Crx to enhance rhodopsin and repress cone opsins.134
In addition to upstream genes including Otx2, Crx, Nrl and Nr2e3 as well as photoreceptor-specific genes like rhodopsin, S-opsin and M-opsin, retinoid receptors are indispensible for appropriate photoreceptor differentiation. Retinoid receptors belong to a steroid receptor superfamily of proteins that serve as ligand-dependent transcription factors. Retinoic acid (RA) plays its role in transcription through retinoic acid receptors (RARs) and retinoid X receptors (RXRs). 9-cis RA binds to and transactivates both RXRs and RARs.135 In addition, 9-cis RA directs progenitor cells to the rod cell fate through activation of members of the steroid/thyroid superfamily of receptors.136 Another effector of this family, thyroid hormone (TH), is found to induce progenitor cells to differentiate into cones in embryonic rat retinal cultures.137 Many effects of TH are mediated by TH receptors (TRs).138 The most important TR in retina development is TRPβ. TRPβ is expressed in the outer nuclear layer of the embryonic retina.139,140 The mouse retina has an opposing S-cone (greater expression ventrally) and M-cone (expressed more dorsally) distribution. Deletion of TRβ2 in mice causes the selective loss of M-cones and a concomitant increase in S-opsin immunoreactive cones, disturbing the gradient of an opposing S- (ventral) and M-cone (dorsal) distribution.141 TH is also required to inhibit S-opsin and activate M-opsin expression.142 Other studies confirm that thyroid hormone action is required for normal cone opsin expression during mouse retinal development.143,144 RXRγ cooperates with TRβ2 to suppress S-opsin in all immature cones and in dorsal cones of the mature retina though it is not necessary for M-opsin regulation. 145 Finally, RXRα acts in synergy with Crx to activate many cone-specific genes.146
Identification of Experimentally-Determined Gene Relationships in a High Throughput Gene Expression Dataset
The gene relationships in the seed networks described above are supported by experimental evidence and thus have been validated in the narrow sense by identifying direct or indirect interactions between two genes under particular experimental conditions. The next step to identify the ‘system’ of genes that work together to influence cell-specific determination and differentiation will require the use of large gene expression datasets and potentially additional dataset types such as protein-protein interaction datasets, ChIP-chip datasets, datasets from animals with specific mutations, etc. We have previously demonstrated the successful application of literature-derived seed-networks to query high-throughput gene expression datasets.2,3 One motivation for this review article was to assemble the available experimental evidence in a way that it might be readily applied to future studies of other cell types, and perhaps to even guide the experimental design processes that underlie the generation of new datasets.
An implicit assumption when using large gene expression datasets is that legitimate gene relationships will be discoverable by identifying a correlation of expression between them. An important question, then is, are known experimentally-determined gene relationships identifiable in large gene expression datasets as high correlation coefficients? We used the seed-network that describes photoreceptor differentiation (Fig. 8) to address this question.
Using previously published data collected from developing rod photoreceptors isolated from the retina at E16, P0, P2, P6 and P10,147 we calculated the correlation coefficients between all pairs of genes (edges) present in the seed network (Table 1). In the photoreceptor seed-network, there were 13 genes and 17 edges (relationships) between them. Two genes (BAF and 9-cis-RA) were not present in the dataset, which left 15 edges to identify. Seven of the 15 edges were recognized as high correlation coefficients (>|0.85|) and an additional three of the 15 edges were supported with weaker correlation coefficients (>|0.45|).
Table 1.
Pairwise correlation coefficients between genes of the photoreceptor-specific seed network. Pearson correlation coefficients were calculated based on the developmental gene expression in rod photoreceptors isolated from retina at ages E16, P2, P6 and P10.147 Two genes, BAF and 9-cis RA were not present in the expression dataset and therefore no correlation coefficient could be calculated (NO DATA). Seed network is shown in Figure 8.
| Gene | Gene | Correlation |
|---|---|---|
| BAF | crx | NO DATA |
| ataxin7 | crx | 0.658655867 |
| crx | rhodopsin | 0.596816525 |
| crx | s-opsin | –0.287648519 |
| crx | m-opsin | 0.072625073 |
| nrl | nr2e3 | 0.995021406 |
| nrl | rhodopsin | 0.910738221 |
| nr2e3 | rhodopsin | 0.867910201 |
| nr2e3 | s-opsin | –0.983763422 |
| neurod | trb2 | –0.27275455 |
| neurod | rhodopsin | 0.097272022 |
| rxrg | s-opsin | 0.940893433 |
| trb2 | s-opsin | –0.919529756 |
| rxra | s-opsin | –0.59339804 |
| rxra | m-opsin | 0.455414641 |
| 9-cis RA | rhodopsin | NO DATA |
Thus, two-thirds of the seed-network relationships are present in the dataset and nearly half of the seed-network relationships are strongly correlated. Encouragingly, our result suggests that a significant number of legitimate gene relationships can be discovered using gene expression data. Previously, we have used seed networks to discover new candidate genes by focusing on genes that were correlated with multiple seed-network genes.2,3 Ultimately, it appears that it will require a combination of datasets and approaches to describe the entire gene network that underlies cell fate determination and differentiation.
Summary
The seed networks presented here can be the basis for queries of high throughput datasets to identify larger, more comprehensive networks that participate in cellular fate specification and differentiation in the developing mouse retina. In addition to summarizing prior knowledge of these processes, seed networks can also be the basis for comparative studies between tissue types within a species or between diverged organisms in order to identify genetic pathways that are conserved through development and evolution.3,148–150 while a more generalized gene-by-gene comparative approach has been effective in identifying orthologs that may play a role in a complex process or a disease state in different organisms,151–154 it is the conservation of not only the gene, but of its relationships to other genes in a network, that dramatically increases the likelihood that the gene, in fact, functions in similar way. Being able to include relational data is one advantage of the seed network approach over more generalized comparative studies. The effectiveness of a cross-species seed network approach has been demonstrated elsewhere.3,149
These seed networks were constructed to help demonstrate the potential of the developing vertebrate retina as a model system for the development and evaluation of systems based approaches. In addition to its characteristic organization and developmental time course, there is a significant amount of high throughput data that has been collected from the developing retina,155–159 and single cells from the developing retina.95,147,160,161 Because of its characteristic organization during development, candidate molecules that are generated using systems based approaches can be rapidly, albeit cursorily, evaluated based on in situ spatial and temporal expression.3,160 Finally, due to its accessibility, candidates can be functionally evaluated in developing retinas using in vivo electroporation to either drive overexpression or knockdown expression of candidate molecules.161
Networks and network representation of processes have an important role in the implementation of systems based approaches and the analysis of large datasets and complex processes. Demonstrating the ability of these seed networks to effectively focus the generation of hypotheses from high throughput data sets would significantly advance the discoveries that depend upon this type of data. In addition, we have also demonstrated that seed networks are an effective way to do comparative analysis of retinal development and use knowledge of one model system to drive discovery in another.3 The use of seed networks to identify conserved networks that act in similar ways (as opposed to conserved genes) will be tremendously useful in the extrapolation of discovery in one model system to another. Thus, development of systems based approaches to investigate cell fate determination in the developing mouse retina will not only lead to important discoveries in the developing retina, but strategies that can be broadly generalized to address many biological questions.
Supplementary Materials
Footnotes
Disclosure
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material
References
- 1.Johnson RA, Wichern DW. Applied multivariate statistical analysis. 5 ed. Prentice Hall; 2002. [Google Scholar]
- 2.Hecker LA, Alcon TC, Honavar VG, Greenlee MH. Using a seed-network to query multiple large-scale gene expression datasets from the developing retina in order to identify and prioritize experimental targets. Bioinform Biol Insights. 2008;2:401–12. doi: 10.4137/bbi.s417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serb JM, Orr MC, West Greenlee MH. Using evolutionary conserved modules in gene networks as a strategy to leverage high throughput gene expression queries. PLoS One. S(9) [DOI] [PMC free article] [PubMed]
- 4.Sernagor E, Eglen S, Harris B, Wong R. Retinal Development. Cambridge University Press; 2006. [Google Scholar]
- 5.Dowling JE. THE RETINA AN APPROACHABLE PART OF THE BRAIN. The Belknap Press of Harvard University Press; 1987. [Google Scholar]
- 6.Chalupa LM, Williams RW. Eye, Retina, and visual system of the mouse: The MIT Press; 2008. [Google Scholar]
- 7.Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J Comp Neurol. 1979 Nov 15;188(2):263–72. doi: 10.1002/cne.901880205. [DOI] [PubMed] [Google Scholar]
- 8.Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985 Jun;212(2):199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- 9.LaVail MM, Rapaport DH, Rakic P. Cytogenesis in the monkey retina. J Comp Anat. 1991;309:86–114. doi: 10.1002/cne.903090107. [DOI] [PubMed] [Google Scholar]
- 10.Stiemke MM, Hollyfield JG. Cell birthdays in Xenopus laevis retina. Differentiation. 1995 Feb;58(3):189–93. doi: 10.1046/j.1432-0436.1995.5830189.x. [DOI] [PubMed] [Google Scholar]
- 11.Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM. Timing and topography of cell genesis in the rat retina. J Comp Neurol. 2004 Jun 21;474(2):304–24. doi: 10.1002/cne.20134. [DOI] [PubMed] [Google Scholar]
- 12.Song N, Lang RA. Animal Models in Eye Research. In: Tsonis PA, editor. Animal Models in Eye Research. Elsevier; 2008. pp. 120–33. [Google Scholar]
- 13.Mustacchi R, Hohmann S, Nielsen J. Yeast systems biology to unravel the network of life. Yeast. 2006 Feb;23(3):227–38. doi: 10.1002/yea.1357. [DOI] [PubMed] [Google Scholar]
- 14.Polpitiya AD, McDunn JE, Burykin A, Ghosh BK, Cobb JP. Using systems biology to simplify complex disease: immune cartography. Crit Care Med. 2009 Jan;37(l Suppl):S16–21. doi: 10.1097/CCM.0b013e3181920cb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirouac DC, Ito C, Csaszar E, et al. Dynamic interaction networks in a hierarchically organized tissue. Mol Syst Biol. Oct 5;6:417. doi: 10.1038/msb.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001 Feb;2(2):109–18. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 17.Marquardt T, Gruss P. Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci. 2002 Jan;25(1):32–8. doi: 10.1016/s0166-2236(00)02028-2. [DOI] [PubMed] [Google Scholar]
- 18.Marquardt T. Transcriptional control of neuronal diversification in the retina. Prog Retin Eye Res. 2003 Sep;22(5):567–77. doi: 10.1016/s1350-9462(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 19.Hatakeyama J, Kageyama R. Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol. 2004 Feb;15(1):83–9. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Res. 2008 Feb 4;1192:90–8. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 21.Szel A, Rohlich P, Caffe AR, Juliusson B, Aguirre G, Van Veen T. Unique topographic separation of two spectral classes of cones in the mouse retina. J Comp Neurol. 1992 Nov 15;325(3):327–42. doi: 10.1002/cne.903250302. [DOI] [PubMed] [Google Scholar]
- 22.Applebury ML, Antoch MP, Baxter LC, et al. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000 Sep;27(3):513–23. doi: 10.1016/s0896-6273(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh KK, Bujan S, Haverkamp S, Feigenspan A, Wassle H. Types of bipolar cells in the mouse retina. J Comp Neurol. 2004 Jan 26;469(1):70–82. doi: 10.1002/cne.10985. [DOI] [PubMed] [Google Scholar]
- 24.Wassle H, Puller C, Muller F, Haverkamp S. Cone contacts, mosaics, and territories of bipolar cells in the mouse retina. J Neurosci. 2009 Jan 7;29(1):106–17. doi: 10.1523/JNEUROSCI.4442-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharpe LT, Stockman A. Rod pathways: the importance of seeing nothing. Trends Neurosci. 1999 Nov;22(11):497–504. doi: 10.1016/s0166-2236(99)01458-7. [DOI] [PubMed] [Google Scholar]
- 26.Chalupa LM, Gunhan E. Development of On and Off retinal pathways and retinogeniculate projections. Prog Retin Eye Res. 2004 Jan;23(1):31–51. doi: 10.1016/j.preteyeres.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Brooks DE, Komaromy AM, Kallberg ME. Comparative retinal ganglion cell and optic nerve morphology. Vet Ophthalmol. 1999;2(1):3–11. doi: 10.1046/j.1463-5224.1999.00047.x. [DOI] [PubMed] [Google Scholar]
- 28.MacNeil MA, Masland RH. Extreme diversity among amacrine cells: implications for function. Neuron. 1998 May;20(5):971–82. doi: 10.1016/s0896-6273(00)80478-x. [DOI] [PubMed] [Google Scholar]
- 29.Perez De Sevilla Muller L, Shelley J, Weiler R. Displaced amacrine cells of the mouse retina. J Comp Neurol. 2007 Nov 10;505(2):177–89. doi: 10.1002/cne.21487. [DOI] [PubMed] [Google Scholar]
- 30.Kolb H, Fernandez E, Schouten J, Ahnelt P, Linberg KA, Fisher SK. Are there three types of horizontal cell in the human retina. J Comp Neurol. 1994 May 15;343(3):370–86. doi: 10.1002/cne.903430304. [DOI] [PubMed] [Google Scholar]
- 31.Masland RH. The fundamental plan of the retina. Nat Neurosci. 2001 Sep;4(9):877–86. doi: 10.1038/nn0901-877. [DOI] [PubMed] [Google Scholar]
- 32.Smith WC. Phototransduction and Photoreceptor Physiology. In: Heckenlively JR, Arden GB, editors. Principles and Practice of Clinical Electrophysiology of Vision. The MIT Press; 2006. [Google Scholar]
- 33.Bringmann A, Pannicke T, Biedermann B, et al. Role of retinal glial cells in neurotransmitter uptake and metabolism. Neurochem Int. 2009 Mar-Apr;54(3–4):143–60. doi: 10.1016/j.neuint.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 34.Dejneka NS, Bennett J. Gene therapy and retinitis pigmentosa: advances and future challenges. Bioessays. 2001 Jul;23(7):662–8. doi: 10.1002/bies.1092. [DOI] [PubMed] [Google Scholar]
- 35.Parameswaran S, Balasubramanian S, Babai N, et al. Induced pluripotent stem cells generate both retinal ganglion cells and photoreceptors: therapeutic implications in degenerative changes in glaucoma and age-related macular degeneration. Stem Cells. Apr;28(4):695–703. doi: 10.1002/stem.320. [DOI] [PubMed] [Google Scholar]
- 36.Edqvist PH, Hallbook F. Newborn horizontal cells migrate bi-directionally across the neuroepithelium during retinal development. Development. 2004 Mar;131(6):1343–51. doi: 10.1242/dev.01018. [DOI] [PubMed] [Google Scholar]
- 37.Morrow EM, Chen CM, Cepko CL. Temporal order of bipolar cell genesis in the neural retina. Neural Dev. 2008;3:2. doi: 10.1186/1749-8104-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drager UC. Birth dates of retinal ganglion cells giving rise to the crossed and uncrossed optic projections in the mouse. Proc R Soc Lond B Biol Sci. 1985 Mar 22;224(1234):57–77. doi: 10.1098/rspb.1985.0021. [DOI] [PubMed] [Google Scholar]
- 39.Farah MH, Easter SS., Jr Cell birth and death in the mouse retinal ganglion cell layer. J Comp Neurol. 2005 Aug 15;489(1):120–34. doi: 10.1002/cne.20615. [DOI] [PubMed] [Google Scholar]
- 40.Akagi T, Inoue T, Miyoshi G, et al. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J Biol Chem. 2004 Jul 2;279(27):28492–8. doi: 10.1074/jbc.M400871200. [DOI] [PubMed] [Google Scholar]
- 41.Fujitani Y, Fujitani S, Luo H, et al. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006 Nov;133(22):4439–50. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- 42.Nakhai H, Sel S, Favor J, et al. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development. 2007 Mar;134(6):1151–60. doi: 10.1242/dev.02781. [DOI] [PubMed] [Google Scholar]
- 43.Bramblett DE, Pennesi ME, Wu SM, Tsai MJ. The transcription factor Bhlhb4 is required for rod bipolar cell maturation. Neuron. 2004 Sep 16;43(6):779–93. doi: 10.1016/j.neuron.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 44.Feng L, Xie X, Joshi PS, et al. Requirement for Bhlhb5 in the specification of amacrine and cone bipolar subtypes in mouse retina. Development. 2006 Dec;133(24):4815–25. doi: 10.1242/dev.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balczarek KA, Lai ZC, Kumar S. Evolution of functional diversification of the paired box (Pax) DNA-binding domains. Mol Biol Evol. 1997 Aug;14(8):829–42. doi: 10.1093/oxfordjournals.molbev.a025824. [DOI] [PubMed] [Google Scholar]
- 46.Glardon S, Holland LZ, Gehring WJ, Holland ND. Isolation and developmental expression of the amphioxus Pax-6 gene (AmphiPax-6): insights into eye and photoreceptor evolution. Development. 1998 Jul;125(14):2701–10. doi: 10.1242/dev.125.14.2701. [DOI] [PubMed] [Google Scholar]
- 47.Chow RL, Snow B, Novak J, et al. Vsx1, a rapidly evolving paired-like homeobox gene expressed in cone bipolar cells. Mech Dev. 2001 Dec;109(2):315–22. doi: 10.1016/s0925-4773(01)00585-8. [DOI] [PubMed] [Google Scholar]
- 48.Clark AM, Yun S, Veien ES, et al. Negative regulation of Vsx1 by its paralog Chx10/Vsx2 is conserved in the vertebrate retina. Brain Res. 2008 Feb 4;1192:99–113. doi: 10.1016/j.brainres.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorval KM, Bobechko BP, Ahmad KF, Bremner R. Transcriptional activity of the paired-like homeodomain proteins CHX10 and Vsx1. J Biol Chem. 2005 Mar 18;280(11):10100–8. doi: 10.1074/jbc.M412676200. [DOI] [PubMed] [Google Scholar]
- 50.Levine EM, Passini M, Hitchcock PF, Glasgow E, Schechter N. Vsx-1 and Vsx-2: two Chx10-like homeobox genes expressed in overlapping domains in the adult goldfish retina. J Comp Neurol. 1997 Oct 27;387(3):439–48. [PubMed] [Google Scholar]
- 51.Passini MA, Levine EM, Canger AK, Raymond PA, Schechter N. Vsx-1 and Vsx-2: differential expression of two paired-like homeobox genes during zebrafish and goldfish retinogenesis. J Comp Neurol. 1997 Nov 24;388(3):495–505. doi: 10.1002/(sici)1096-9861(19971124)388:3<495::aid-cne11>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 52.Schonemann MD, Ryan AK, Erkman L, McEvilly RJ, Bermingham J, Rosenfeld MG. POU domain factors in neural development. Adv Exp Med Biol. 1998;449:39–53. doi: 10.1007/978-1-4615-4871-3_4. [DOI] [PubMed] [Google Scholar]
- 53.Wang SW, Mu X, Bowers WJ, et al. Brn3b/Brn3c double knockout mice reveal an unsuspected role for Brn3c in retinal ganglion cell axon outgrowth. Development. 2002 Jan;129(2):467–77. doi: 10.1242/dev.129.2.467. [DOI] [PubMed] [Google Scholar]
- 54.Badea TC, Cahill H, Ecker J, Hattar S, Nathans J. Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron. 2009 Mar 26;61(6):852–64. doi: 10.1016/j.neuron.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000 Feb;16(2):75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- 56.Kiefer JC. Back to basics: Sox genes. Dev Dyn. 2007 Aug;236(8):2356–66. doi: 10.1002/dvdy.21218. [DOI] [PubMed] [Google Scholar]
- 57.Chow RL, Volgyi B, Szilard RK, et al. Control of late off-center cone bipolar cell differentiation and visual signaling by the homeobox gene Vsx1. Proc Natl Acad Sci U S A. 2004 Feb 10;101(6):1754–9. doi: 10.1073/pnas.0306520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohtoshi A, Wang SW, Maeda H, et al. Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr Biol. 2004 Mar 23;14(6):530–6. doi: 10.1016/j.cub.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 59.Ding Q, Chen H, Xie X, Libby RT, Tian N, Gan L. BARHL2 differentially regulates the development of retinal amacrine and ganglion neurons. JNeurosci. 2009 Apr 1;29(13):3992–4003. doi: 10.1523/JNEUROSCI.5237-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng CW, Chow RL, Lebel M, et al. The Iroquois homeobox gene, Irx5, is required for retinal cone bipolar cell development. Dev Biol. 2005 Nov 1;287(1):48–60. doi: 10.1016/j.ydbio.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 61.Hojo M, Ohtsuka T, Hashimoto N, Gradwohl G, Guillemot F, Kageyama R. Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development. 2000 Jun;127(12):2515–22. doi: 10.1242/dev.127.12.2515. [DOI] [PubMed] [Google Scholar]
- 62.Takatsuka K, Hatakeyama J, Bessho Y, Kageyama R. Roles of the bHLH gene Hes1 in retinal morphogenesis. Brain Res. 2004 Apr 9;1004(1–2):148–55. doi: 10.1016/j.brainres.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 63.Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci U S A. 1996 Apr 30;93(9):3920–5. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang SW, Kim BS, Ding K, et al. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001 Jan 1;15(1):24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elshatory Y, Everhart D, Deng M, Xie X, Barlow RB, Gan L. lslet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. J Neurosci. 2007 Nov 14;27(46):12707–20. doi: 10.1523/JNEUROSCI.3951-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004 Sep 16;43(6):795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 67.Tomita K, Moriyoshi K, Nakanishi S, Guillemot F, Kageyama R. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 2000 Oct 16;19(20):5460–72. doi: 10.1093/emboj/19.20.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burmeister M, Novak J, Liang MY, et al. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996 Apr;12(4):376–84. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- 69.Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE, Kageyama R. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002 Feb;129(4):831–42. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- 70.Lin YP, Ouchi Y, Satoh S, Watanabe S. Sox2 plays a role in the induction of amacrine and Muller glial cells in mouse retinal progenitor cells. Invest Ophthalmol Vis Sci. 2009 Jan;50(1):68–74. doi: 10.1167/iovs.07-1619. [DOI] [PubMed] [Google Scholar]
- 71.Muto A, lida A, Satoh S, Watanabe S. The group E Sox genes Sox8 and Sox9 are regulated by Notch signaling and are required for Muller glial cell development in mouse retina. Exp Eye Res. 2009 May 31; doi: 10.1016/j.exer.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 72.Mu X, Beremand PD, Zhao S, et al. Discrete gene sets depend on POU domain transcription factor Brn3b/Brn-3.2/POU4f2 for their expression in the mouse embryonic retina. Development. 2004 Mar;131(6):1197–210. doi: 10.1242/dev.01010. [DOI] [PubMed] [Google Scholar]
- 73.Furukawa T, Mukherjee S, Bao ZZ, Morrow EM, Cepko CL. rax, Hes1, and Notch1 promote the formation of Muller glia by postnatal retinal progenitor cells. Neuron. 2000 May;26(2):383–94. doi: 10.1016/s0896-6273(00)81171-x. [DOI] [PubMed] [Google Scholar]
- 74.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004 Mar;131(5):965–73. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 75.Jadhav AP, Cho SH, Cepko CL. Notch activity permits retinal cells to progress through multiple progenitor states and acquire a stem cell property. Proc Natl Acad Sci U S A. 2006 Dec 12;103(50):18998–9003. doi: 10.1073/pnas.0608155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jadhav AP, Mason HA, Cepko CL. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development. 2006 Mar;133(5):913–23. doi: 10.1242/dev.02245. [DOI] [PubMed] [Google Scholar]
- 77.Yaron O, Farhy C, Marquardt T, Applebury M, Ashery-Padan R. Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development. 2006 Apr;133(7):1367–78. doi: 10.1242/dev.02311. [DOI] [PubMed] [Google Scholar]
- 78.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999 Apr 15;18(8):2196–207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bae S, Bessho Y, Hojo M, Kageyama R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development. 2000 Jul;127(13):2933–43. doi: 10.1242/dev.127.13.2933. [DOI] [PubMed] [Google Scholar]
- 80.Poche RA, Furuta Y, Chaboissier MC, Schedl A, Behringer RR. Sox9 is expressed in mouse multipotent retinal progenitor cells and functions in Muller glial cell development. J Comp Neurol. 2008 Sep 20;510(3):237–50. doi: 10.1002/cne.21746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001 Jul;128(13):2497–508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL. Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev Biol. 2006 Jul 15;295(2):764–78. doi: 10.1016/j.ydbio.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 83.Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003 Dec 1;264(1):240–54. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 84.Mu X, Fu X, Sun H, Beremand PD, Thomas TL, Klein WH. A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Dev Biol. 2005 Apr 15;280(2):467–81. doi: 10.1016/j.ydbio.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 85.Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001 Apr 6;105(1):43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 86.Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998 Dec;125(23):4821–33. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- 87.Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008 Jun;135(11):1981–90. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mu X, Fu X, Beremand PD, Thomas TL, Klein WH. Gene regulation logic in retinal ganglion cell development: Isll defines a critical branch distinct from but overlapping with Pou4f2. Proc Natl Acad Sci U S A. 2008 May 13;105(19):6942–7. doi: 10.1073/pnas.0802627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gan L, Wang SW, Huang Z, Klein WH. POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev Biol. 1999 Jun 15;210(2):469–80. doi: 10.1006/dbio.1999.9280. [DOI] [PubMed] [Google Scholar]
- 90.Xiang M. Requirement for Brn-3b in early differentiation of postmitotic retinal ganglion cell precursors. Dev Biol. 1998 May 15;197(2):155–69. doi: 10.1006/dbio.1998.8868. [DOI] [PubMed] [Google Scholar]
- 91.Erkman L, Yates PA, McLaughlin T, et al. A POU domain transcription factor-dependent program regulates axon pathfinding in the vertebrate visual system. Neuron. 2000 Dec;28(3):779–92. doi: 10.1016/s0896-6273(00)00153-7. [DOI] [PubMed] [Google Scholar]
- 92.Mao CA, Kiyama T, Pan P, Furuta Y, Hadjantonakis AK, Klein WH. Eomesodermin, a target gene of Pou4f2, is required for retinal ganglion cell and optic nerve development in the mouse. Development. 2008 Jan;135(2):271–80. doi: 10.1242/dev.009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wagner KD, Wagner N, Vidal VP, et al. The Wilms’ tumor gene Wt1 is required for normal development of the retina. EMBO J. 2002 Mar 15;21(6):1398–405. doi: 10.1093/emboj/21.6.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wagner KD, Wagner N, Schley G, Theres H, Scholz H. The Wilms’ tumor suppressor Wt1 encodes a transcriptional activator of the class IV POU-domain factor Pou4f2 (Brn-3b) Gene. 2003 Feb 27;305(2):217–23. doi: 10.1016/s0378-1119(02)01231-3. [DOI] [PubMed] [Google Scholar]
- 95.Trimarchi JM, Stadler MB, Roska B, et al. Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J Comp Neurol. 2007 Jun 20;502(6):1047–65. doi: 10.1002/cne.21368. [DOI] [PubMed] [Google Scholar]
- 96.Guillemot F, Joyner AL. Dynamic expression of the murine Achaete-Scute homologue Mash-1 in the developing nervous system. Mech Dev. 1993 Aug;42(3):171–85. doi: 10.1016/0925-4773(93)90006-j. [DOI] [PubMed] [Google Scholar]
- 97.Jasoni CL, Reh TA. Temporal and spatial pattern of MASH-1 expression in the developing rat retina demonstrates progenitor cell heterogeneity. J Comp Neurol. 1996 May 27;369(2):319–27. doi: 10.1002/(SICI)1096-9861(19960527)369:2<319::AID-CNE11>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 98.Tomita K, Nakanishi S, Guillemot F, Kageyama R. Mash1 promotes neuronal differentiation in the retina. Genes Cells. 1996 Aug;l(8):765–74. doi: 10.1111/j.1365-2443.1996.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 99.Takebayashi K, Takahashi S, Yokota C, et al. Conversion of ectoderm into a neural fate by ATH-3, a vertebrate basic helix-loop-helix gene homologous to Drosophila proneural gene atonal. EMBO J. 1997 Jan 15;16(2):384–95. doi: 10.1093/emboj/16.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hatakeyama J, Tomita K, Inoue T, Kageyama R. Roles of homeobox and bHLH genes in specification of a retinal cell type. Development. 2001 Apr;128(8):1313–22. doi: 10.1242/dev.128.8.1313. [DOI] [PubMed] [Google Scholar]
- 101.Liu IS, Chen JD, Ploder L, et al. Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron. 1994 Aug;13(2):377–93. doi: 10.1016/0896-6273(94)90354-9. [DOI] [PubMed] [Google Scholar]
- 102.Livne-Bar I, Pacal M, Cheung MC, et al. Chx10 is required to block photoreceptor differentiation but is dispensable for progenitor proliferation in the postnatal retina. Proc Natl Acad Sci U S A. 2006 Mar 28;103(13):4988–93. doi: 10.1073/pnas.0600083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baas D, Bumsted KM, Martinez JA, Vaccarino FM, Wikler KC, Barnstable CJ. The subcellular localization of Otx2 is cell-type specific and developmentally regulated in the mouse retina. Brain Res Mol Brain Res. 2000 May 31;78(1–2):26–37. doi: 10.1016/s0169-328x(00)00060-7. [DOI] [PubMed] [Google Scholar]
- 104.Koike C, Nishida A, Ueno S, et al. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol. 2007 Dec;27(23):8318–29. doi: 10.1128/MCB.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oliver G, Mailhos A, Wehr R, Copeland NG, Jenkins NA, Gruss P. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995 Dec;121(12):4045–55. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 106.Boije H, Edqvist PH, Hallbook F. Temporal and spatial expression of transcription factors FoxN4, Ptf1a, Prox1, Isll and Lim1 mRNA in the developing chick retina. Gene Expr Patterns. 2008 Jan;8(2):117–23. doi: 10.1016/j.modgep.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 107.Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003 May;34(1):53–8. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- 108.Cook T. Cell diversity in the retina: more than meets the eye. Bioessays. 2003 Oct;25(10):921–5. doi: 10.1002/bies.10356. [DOI] [PubMed] [Google Scholar]
- 109.Suga A, Taira M, Nakagawa S. LIM family transcription factors regulate the subtype-specific morphogenesis of retinal horizontal cells at post-migratory stages. Dev Biol. 2009 Jun 15;330(2):318–28. doi: 10.1016/j.ydbio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 110.Liu W, Wang JH, Xiang M. Specific expression of the LIM/homeodomain protein Lim-1 in horizontal cells during retinogenesis. Dev Dyn. 2000 Mar;217(3):320–5. doi: 10.1002/(SICI)1097-0177(200003)217:3<320::AID-DVDY10>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 111.Poche RA, Kwan KM, Raven MA, Furuta Y, Reese BE, Behringer RR. Lim1 is essential for the correct laminar positioning of retinal horizontal cells. J Neurosci. 2007 Dec 19;27(51):14099–107. doi: 10.1523/JNEUROSCI.4046-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Morrow EM, Furukawa T, Lee JE, Cepko CL. NeuroD regulates multiple functions in the developing neural retina in rodent. Development. 1999 Jan;126(1):23–36. doi: 10.1242/dev.126.1.23. [DOI] [PubMed] [Google Scholar]
- 113.Liu H, Etter P, Hayes S, et al. NeuroDl regulates expression of thyroid hormone receptor 2 and cone opsins in the developing mouse retina. J Neurosci. 2008 Jan 16;28(3):749–56. doi: 10.1523/JNEUROSCI.4832-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen S, Wang QL, Nie Z, et al. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997 Nov;19(5):1017–30. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 115.Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999 Dec;23(4):466–70. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- 116.Peng GH, Chen S. Crx activates opsin transcription by recruiting HAT-containing co-activators and promoting histone acetylation. Hum Mol Genet. 2007 Oct 15;16(20):2433–52. doi: 10.1093/hmg/ddm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.La Spada AR, Fu YH, Sopher BL, et al. Polyglutamine-expanded ataxin-7 antagonizes CRX function and induces cone-rod dystrophy in a mouse model of SCA7. Neuron. 2001 Sep 27;31(6):913–27. doi: 10.1016/s0896-6273(01)00422-6. [DOI] [PubMed] [Google Scholar]
- 118.Chen S, Peng GH, Wang X, et al. Interference of Crx-dependent transcription by ataxin-7 involves interaction between the glutamine regions and requires the ataxin-7 carboxy-terminal region for nuclear localization. Hum Mol Genet. 2004 Jan 1;13(1):53–67. doi: 10.1093/hmg/ddh005. [DOI] [PubMed] [Google Scholar]
- 119.Wang X, Xu S, Rivolta C, et al. Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J Biol Chem. 2002 Nov 8;277(45):43288–300. doi: 10.1074/jbc.M207952200. [DOI] [PubMed] [Google Scholar]
- 120.Nishida A, Furukawa A, Koike C, et al. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003 Dec;6(12):1255–63. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- 121.Swain PK, Hicks D, Mears AJ, et al. Multiple phosphorylated isoforms of NRL are expressed in rod photoreceptors. J Biol Chem. 2001 Sep 28;276(39):36824–30. doi: 10.1074/jbc.M105855200. [DOI] [PubMed] [Google Scholar]
- 122.Swaroop A, Xu JZ, Pawar H, Jackson A, Skolnick C, Agarwal N. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):266–70. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kumar R, Chen S, Scheurer D, et al. The bZIP transcription factor Nrl stimulates rhodopsin promoter activity in primary retinal cell cultures. J Biol Chem. 1996 Nov 22;271(47):29612–8. doi: 10.1074/jbc.271.47.29612. [DOI] [PubMed] [Google Scholar]
- 124.Rehemtulla A, Warwar R, Kumar R, Ji X, Zack DJ, Swaroop A. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc Natl Acad Sci U S A. 1996 Jan 9;93(1):191–5. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Daniele LL, Lillo C, Lyubarsky AL, et al. Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest Ophthalmol Vis Sci. 2005 Jun;46(6):2156–67. doi: 10.1167/iovs.04-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mears AJ, Kondo M, Swain PK, et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001 Dec;29(4):447–52. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 127.Oh EC, Cheng H, Hao H, Jia L, Khan NW, Swaroop A. Rod differentiation factor NRL activates the expression of nuclear receptor NR2E3 to suppress the development of cone photoreceptors. Brain Res. 2008 Oct 21;1236:16–29. doi: 10.1016/j.brainres.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kobayashi M, Takezawa S, Hara K, et al. Identification of a photoreceptor cell-specific nuclear receptor. Proc Natl Acad Sci U S A. 1999 Apr 27;96(9):4814–9. doi: 10.1073/pnas.96.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Corbo JC, Cepko CL. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 2005 Aug;1(2):ell. doi: 10.1371/journal.pgen.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Haider NB, Jacobson SG, Cideciyan AV, et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000 Feb;24(2):127–31. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- 131.Hood DC, Cideciyan AV, Roman AJ, Jacobson SG. Enhanced S cone syndrome: evidence for an abnormally large number of S cones. Vision Res. 1995 May;35(10):1473–81. doi: 10.1016/0042-6989(95)98727-q. [DOI] [PubMed] [Google Scholar]
- 132.Cornish EE, Xiao M, Yang Z, Provis JM, Hendrickson AE. The role of opsin expression and apoptosis in determination of cone types in human retina. Exp Eye Res. 2004 Jun;78(6):1143–54. doi: 10.1016/j.exer.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 133.Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005 Jan 5;25(1):118–29. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005 Mar 15;14(6):747–64. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- 135.Heyman RA, Mangelsdorf DJ, Dyck JA, et al. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992 Jan 24;68(2):397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 136.Kelley MW, Turner JK, Reh TA. Retinoic acid promotes differentiation of photoreceptors in vitro. Development. 1994 Aug;120(8):2091–102. doi: 10.1242/dev.120.8.2091. [DOI] [PubMed] [Google Scholar]
- 137.Kelley MW, Turner JK, Reh TA. Ligands of steroid/thyroid receptors induce cone photoreceptors in vertebrate retina. Development. 1995 Nov;121(11):3777–85. doi: 10.1242/dev.121.11.3777. [DOI] [PubMed] [Google Scholar]
- 138.Lazar MA. Thyroid hormone action: a binding contract. J Clin Invest. 2003 Aug;112(4):497–9. doi: 10.1172/JCI19479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hodin RA, Lazar MA, Wintman Bl, et al. Identification of a thyroid hormone receptor that is pituitary-specific. Science. 1989 Apr 7;244(4900):76–9. doi: 10.1126/science.2539642. [DOI] [PubMed] [Google Scholar]
- 140.Sjoberg M, Vennstrom B, Forrest D. Thyroid hormone receptors in chick retinal development: differential expression of mRNAs for alpha and N-terminal variant beta receptors. Development. 1992 Jan;114(1):39–47. doi: 10.1242/dev.114.1.39. [DOI] [PubMed] [Google Scholar]
- 141.Ng L, Hurley JB, Dierks B, et al. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001 Jan;27(1):94–8. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- 142.Roberts MR, Srinivas M, Forrest D, Morreale de Escobar G, Reh TA. Making the gradient: thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc Natl Acad Sci U S A. 2006 Apr 18;103(16):6218–23. doi: 10.1073/pnas.0509981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Applebury ML, Farhangfar F, Glosmann M, et al. Transient expression of thyroid hormone nuclear receptor TRbeta2 sets S opsin patterning during cone photoreceptor genesis. Dev Dyn. 2007 May;236(5):1203–12. doi: 10.1002/dvdy.21155. [DOI] [PubMed] [Google Scholar]
- 144.Pessoa CN, Santiago LA, Santiago DA, et al. Thyroid hormone action is required for normal cone opsin expression during mouse retinal development. Invest Ophthalmol Vis Sci. 2008 May;49(5):2039–45. doi: 10.1167/iovs.07-0908. [DOI] [PubMed] [Google Scholar]
- 145.Roberts MR, Hendrickson A, McGuire CR, Reh TA. Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci. 2005 Aug;46(8):2897–904. doi: 10.1167/iovs.05-0093. [DOI] [PubMed] [Google Scholar]
- 146.Fujieda H, Bremner R, Mears AJ, Sasaki H. Retinoic acid receptor-related orphan receptor alpha regulates a subset of cone genes during mouse retinal development. J Neurochem. 2009 Jan;108(1):91–101. doi: 10.1111/j.1471-4159.2008.05739.x. [DOI] [PubMed] [Google Scholar]
- 147.Akimoto M, Cheng H, Zhu D, et al. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc Natl Acad Sci U S A. 2006 Mar 7;103(10):3890–5. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kim SK. Common aging pathways in worms, flies, mice and humans. J Exp Biol. 2007 May;210(Pt 9):1607–12. doi: 10.1242/jeb.004887. [DOI] [PubMed] [Google Scholar]
- 149.Bell R, Hubbard A, Chettier R, et al. A human protein interaction network shows conservation of aging processes between human and invertebrate species. PLoS Genet. 2009 Mar;5(3):el000414. doi: 10.1371/journal.pgen.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dimitriadi M, Sleigh JN, Walker A, et al. Conserved genes act as modifiers of invertebrate SMN loss of function defects. PLoS Genet. 6(10):el001172. doi: 10.1371/journal.pgen.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994 Aug 5;265(5173):785–9. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 152.Liu YH, Jakobsen JS, Valentin G, Amarantos I, Gilmour DT, Furlong EE. A systematic analysis of Tinman function reveals Eya and JAK-STAT signaling as essential regulators of muscle development. Dev Cell. 2009 Feb;16(2):280–91. doi: 10.1016/j.devcel.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 153.Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left-right asymmetry. Nat Rev Genet. 2002 Feb;3(2):103–13. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- 154.Sinclair A, Smith C, Western P, McClive P. A comparative analysis of vertebrate sex determination. Novartis FoundSymp. 2002;244:102–111. discussion 111–104, 203–106, 253–107. [PubMed] [Google Scholar]
- 155.Yoshida S, Mears AJ, Friedman JS, et al. Expression profiling of the developing and mature Nrl−/− mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum Mol Genet. 2004 Jul 15;13(14):1487–503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- 156.Barnhill AE, Hecker LA, Kohutyuk O, Buss JE, Honavar VG, Greenlee HW. Characterization of the retinal proteome during rod photoreceptor genesis. BMC Res Notes. 3:2S. doi: 10.1186/1756-0500-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001 Nov 30;107(5):579–89. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 158.Liu J, Wang J, Huang Q, et al. Gene expression profiles of mouse retinas during the second and third postnatal weeks. Brain Res. 2006 Jul 7;1098(1):113–25. doi: 10.1016/j.brainres.2006.04.086. [DOI] [PubMed] [Google Scholar]
- 159.Dorrell Ml, Aguilar E, Weber C, Friedlander M. Global gene expression analysis of the developing postnatal mouse retina. Invest Ophthalmol VisSci. 2004 Mar;45(3):1009–19. doi: 10.1167/iovs.03-0806. [DOI] [PubMed] [Google Scholar]
- 160.Trimarchi JM, Cho SH, Cepko CL. Identification of genes expressed preferentially in the developing peripheral margin of the optic cup. Dev Dyn. 2009 Sep;238(9):2327–9. doi: 10.1002/dvdy.21973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004 Jan 6;101(1):16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995 Dec 15;9(24):3136–48. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 163.Mu X, Klein WH. A gene regulatory hierarchy for retinal ganglion cell specification and differentiation. Semin Cell Dev Biol. 2004 Feb;15(1):115–23. doi: 10.1016/j.semcdb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 164.Wang Y, Dakubo GD, Thurig S, Mazerolle CJ, Wallace VA. Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development. 2005 Nov;132(22):5103–13. doi: 10.1242/dev.02096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.