Abstract
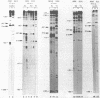
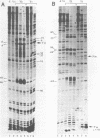
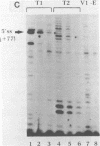
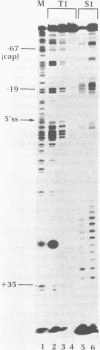
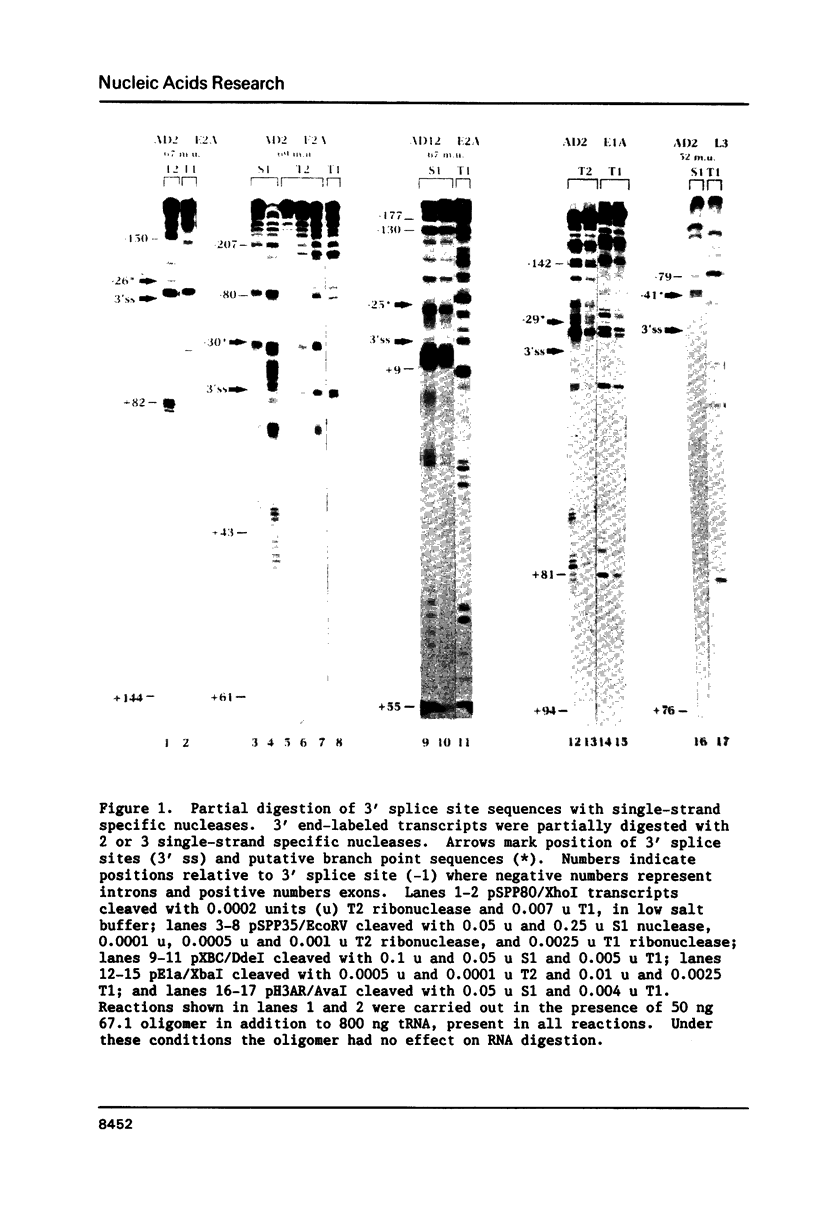
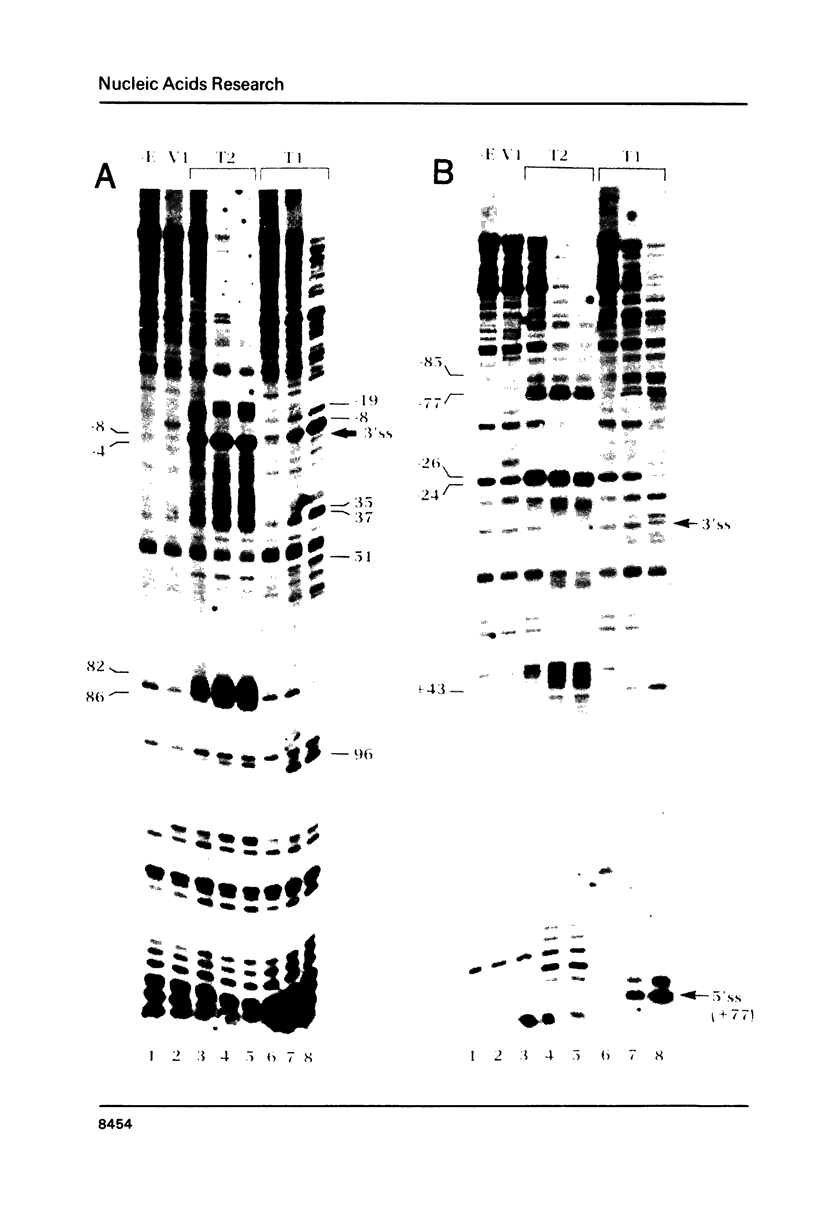
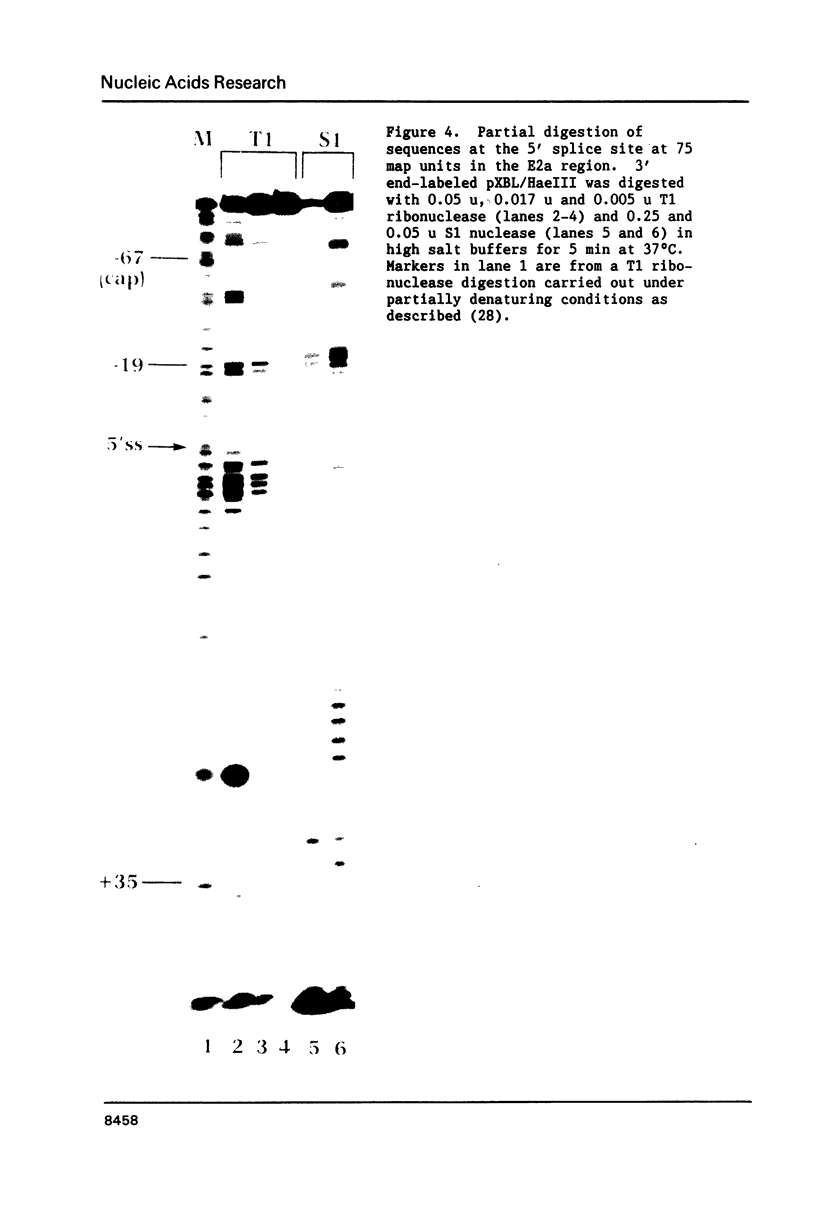
The conformation of RNA sequences spanning five 3' splice sites and two 5' splice sites in adenovirus mRNA was probed by partial digestion with single-strand specific nucleases. Although cleavage of nucleotides near both 3' and 5' splice sites was observed, most striking was the preferential digestion of sequences near the 3' splice site. At each 3' splice site a region of very strong cleavage is observed at low concentrations of enzyme near the splice site consensus sequence or the upstream branch point consensus sequence. Additional sites of moderately strong cutting near the branch point consensus sequence were observed in those sequences where the splice site was the preferred target. Since recognition of the 3' splice site and branch site appear to be early events in mRNA splicing these observations may indicate that the local conformation of the splice site sequences may play a direct or indirect role in enhancing the accessibility of sequences important for splicing.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldi M. I., Mattoccia E., Tocchini-Valentini G. P. Role of RNA structure in splicing: excision of the intervening sequence in yeast tRNA3leu is dependent on the formation of a D stem. Cell. 1983 Nov;35(1):109–115. doi: 10.1016/0092-8674(83)90213-1. [DOI] [PubMed] [Google Scholar]
- Beyer A. L., Christensen M. E., Walker B. W., LeStourgeon W. M. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977 May;11(1):127–138. doi: 10.1016/0092-8674(77)90323-3. [DOI] [PubMed] [Google Scholar]
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Brody E., Abelson J. The "spliceosome": yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985 May 24;228(4702):963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986 Jan 31;44(2):207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- Chabot B., Black D. L., LeMaster D. M., Steitz J. A. The 3' splice site of pre-messenger RNA is recognized by a small nuclear ribonucleoprotein. Science. 1985 Dec 20;230(4732):1344–1349. doi: 10.1126/science.2933810. [DOI] [PubMed] [Google Scholar]
- Chu G., Sharp P. A. A gene chimaera of SV40 and mouse beta-globin is transcribed and properly spliced. Nature. 1981 Jan 29;289(5796):378–382. doi: 10.1038/289378a0. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Seiler S. R., Sharp P. A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985 Aug;42(1):345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- Green M., Mackey J. K., Wold W. S., Rigden P. Thirty-one human adenovirus serotypes (Ad1-Ad31) form five groups (A-E) based upon DNA genome homologies. Virology. 1979 Mar;93(2):481–492. doi: 10.1016/0042-6822(79)90251-4. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Rodriguez J. R., Rosbash M. A quantitative analysis of the effects of 5' junction and TACTAAC box mutants and mutant combinations on yeast mRNA splicing. Cell. 1985 Dec;43(2 Pt 1):423–430. doi: 10.1016/0092-8674(85)90172-2. [DOI] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohavong P., Gattoni R., Schmitt P., Stévenin J. The different intron 2 species excised in vivo from the E2A premRNA of adenovirus-2: an approach to analyse alternative splicing. Nucleic Acids Res. 1986 Jul 11;14(13):5207–5227. doi: 10.1093/nar/14.13.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchingman G. R. Sequence of the DNA-binding protein of a human subgroup E adenovirus (type 4): comparisons with subgroup A (type 12), subgroup B (type 7), and subgroup C (type 5). Virology. 1985 Oct 15;146(1):90–101. doi: 10.1016/0042-6822(85)90055-8. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Grabowski P. J., Padgett R. A., Sharp P. A. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature. 1985 Feb 14;313(6003):552–557. doi: 10.1038/313552a0. [DOI] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Kruijer W., van Schaik F. M., Speijer J. G., Sussenbach J. S. Structure and function of adenovirus DNA binding protein: comparison of the amino acid sequences of the Ad5 and Ad12 proteins derived from the nucleotide sequence of the corresponding genes. Virology. 1983 Jul 15;128(1):140–153. doi: 10.1016/0042-6822(83)90325-2. [DOI] [PubMed] [Google Scholar]
- Krämer A., Keller W., Appel B., Lührmann R. The 5' terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984 Aug;38(1):299–307. doi: 10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
- Lockard R. E., Kumar A. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 1981 Oct 10;9(19):5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrand S., Setyono B., Greenberg J. R., Pederson T. Structure of nuclear ribonucleoprotein: identification of proteins in contact with poly(A)+ heterogeneous nuclear RNA in living HeLa cells. J Cell Biol. 1981 Aug;90(2):380–384. doi: 10.1083/jcb.90.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe S. H. A large inverted repeat sequence overlaps two acceptor splice sites in adenovirus. Nucleic Acids Res. 1983 Dec 20;11(24):8891–8900. doi: 10.1093/nar/11.24.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe S. H. Ribonucleoprotein structure of adenovirus nuclear RNA probed by nuclease digestion. J Mol Biol. 1982 Dec 15;162(3):585–606. doi: 10.1016/0022-2836(82)90390-4. [DOI] [PubMed] [Google Scholar]
- Munroe S. H. Secondary structure of splice sites in adenovirus mRNA precursors. Nucleic Acids Res. 1984 Nov 26;12(22):8437–8456. doi: 10.1093/nar/12.22.8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Peebles C. L., Perlman P. S., Mecklenburg K. L., Petrillo M. L., Tabor J. H., Jarrell K. A., Cheng H. L. A self-splicing RNA excises an intron lariat. Cell. 1986 Jan 31;44(2):213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Teem J. L., Rosbash M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: implications for U1 RNA and metazoan mRNA splicing. Cell. 1983 Sep;34(2):395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Specific and stable intron-factor interactions are established early during in vitro pre-mRNA splicing. Cell. 1985 Nov;43(1):131–142. doi: 10.1016/0092-8674(85)90018-2. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Schmelzer C., Schweyen R. J. Self-splicing of group II introns in vitro: mapping of the branch point and mutational inhibition of lariat formation. Cell. 1986 Aug 15;46(4):557–565. doi: 10.1016/0092-8674(86)90881-0. [DOI] [PubMed] [Google Scholar]
- Schneuwly S., Kuroiwa A., Baumgartner P., Gehring W. J. Structural organization and sequence of the homeotic gene Antennapedia of Drosophila melanogaster. EMBO J. 1986 Apr;5(4):733–739. doi: 10.1002/j.1460-2075.1986.tb04275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelness G. S., Williams D. L. Secondary structure analysis of apolipoprotein II mRNA using enzymatic probes and reverse transcriptase. Evaluation of primer extension for high resolution structure mapping of mRNA. J Biol Chem. 1985 Jul 15;260(14):8637–8646. [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Tonegawa S., Maxam A. M., Tizard R., Bernard O., Gilbert W. Sequence of a mouse germ-line gene for a variable region of an immunoglobulin light chain. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1485–1489. doi: 10.1073/pnas.75.3.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell B. C., Tolstoshev P., Crystal R. G. Secondary structures for splice junctions in eukaryotic and viral messenger RNA precursors. Nucleic Acids Res. 1980 Aug 25;8(16):3659–3672. doi: 10.1093/nar/8.16.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary C. P., Vournakis J. N. RNA structure analysis using T2 ribonuclease: detection of pH and metal ion induced conformational changes in yeast tRNAPhe. Nucleic Acids Res. 1984 Sep 11;12(17):6763–6778. doi: 10.1093/nar/12.17.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel S., Brötz M., Kruczek I., Neumann R., Eick D., Winterhoff U., Doerfler W. Cloned fragments of human adenovirus type-12 DNA. Gene. 1981 Nov;15(2-3):273–278. doi: 10.1016/0378-1119(81)90136-0. [DOI] [PubMed] [Google Scholar]
- Vournakis J. N., Celantano J., Finn M., Lockard R. E., Mitra T., Pavlakis G., Troutt A., van den Berg M., Wurst R. M. Sequence and structure analysis of end-labeled RNA with nucleases. Gene Amplif Anal. 1981;2:267–298. [PubMed] [Google Scholar]
- Waring R. B., Ray J. A., Edwards S. W., Scazzocchio C., Davies R. W. The Tetrahymena rRNA intron self-splices in E. coli: in vivo evidence for the importance of key base-paired regions of RNA for RNA enzyme function. Cell. 1985 Feb;40(2):371–380. doi: 10.1016/0092-8674(85)90151-5. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Liau G., Mudryj M., Obici S., de Crombrugghe B. Conservation of the sizes for one but not another class of exons in two chick collagen genes. 1984 Jul 26-Aug 1Nature. 310(5975):333–337. doi: 10.1038/310333a0. [DOI] [PubMed] [Google Scholar]
- van der Veen R., Arnberg A. C., van der Horst G., Bonen L., Tabak H. F., Grivell L. A. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell. 1986 Jan 31;44(2):225–234. doi: 10.1016/0092-8674(86)90756-7. [DOI] [PubMed] [Google Scholar]