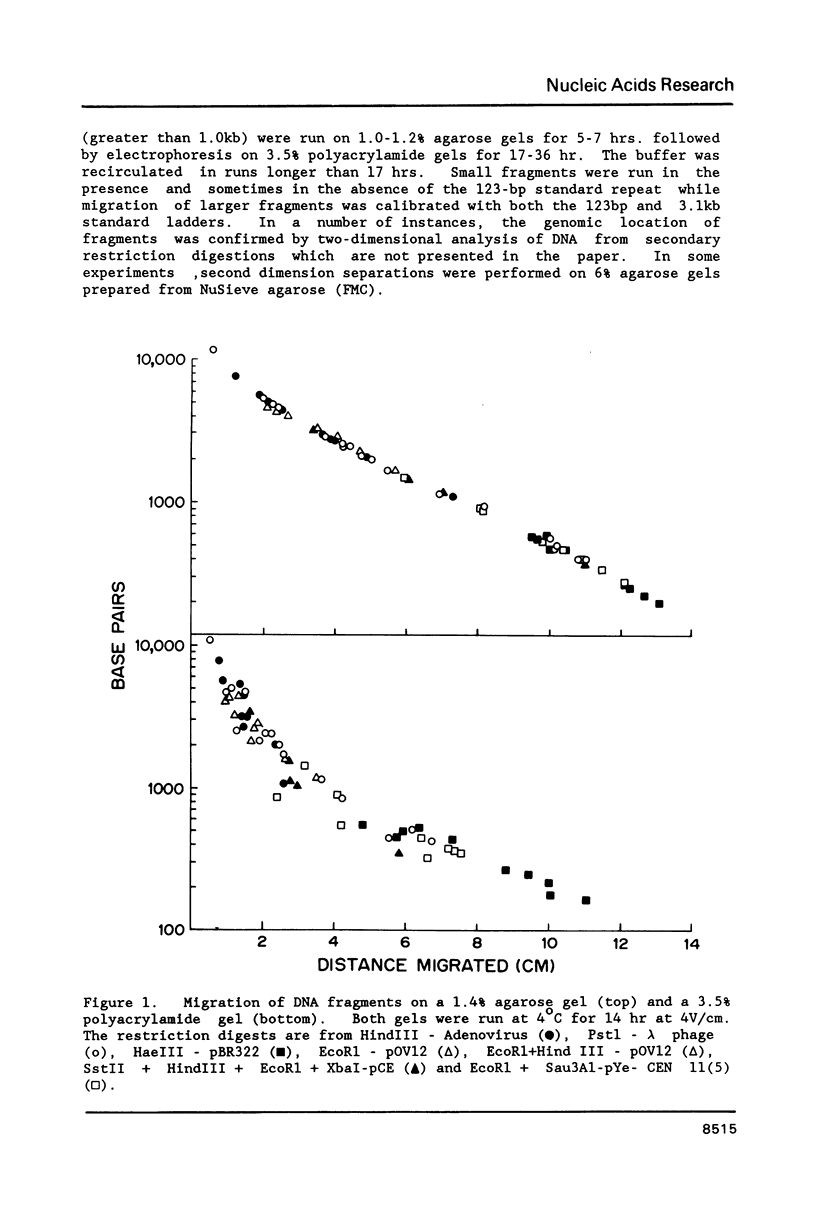
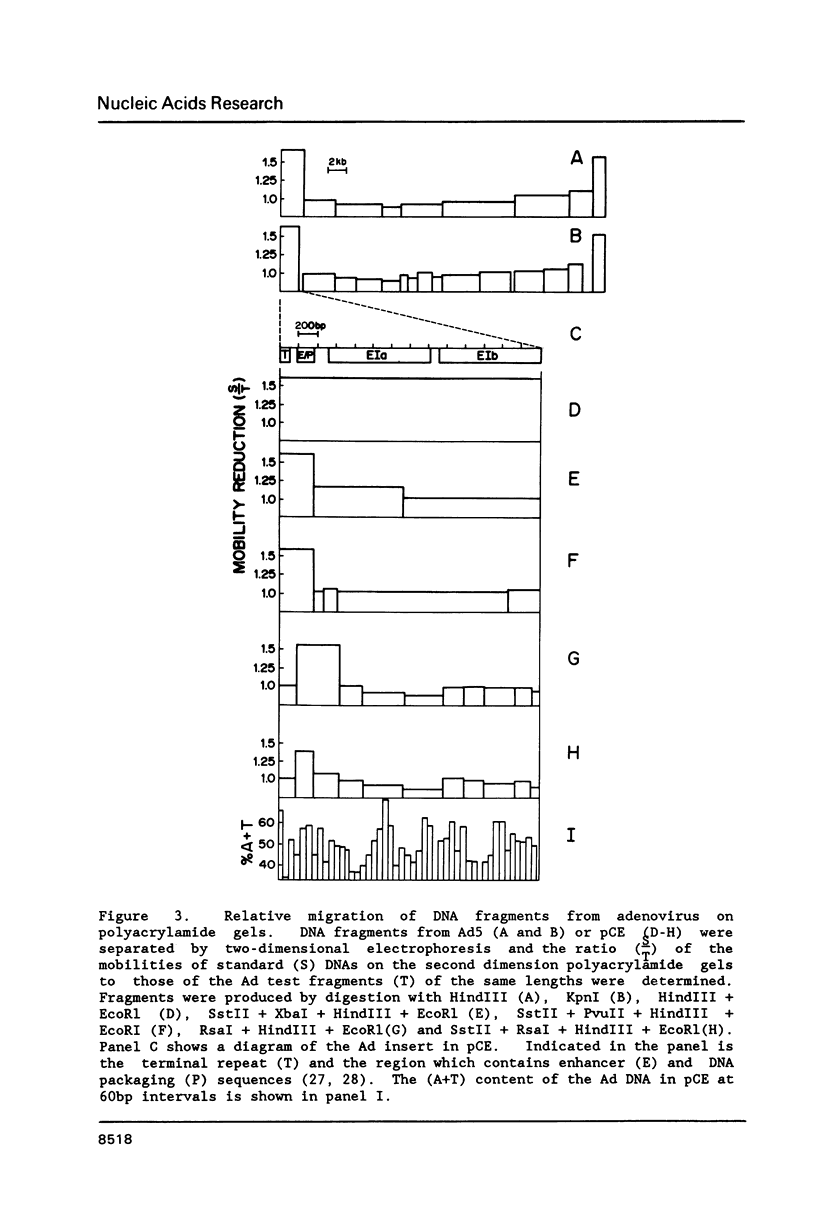
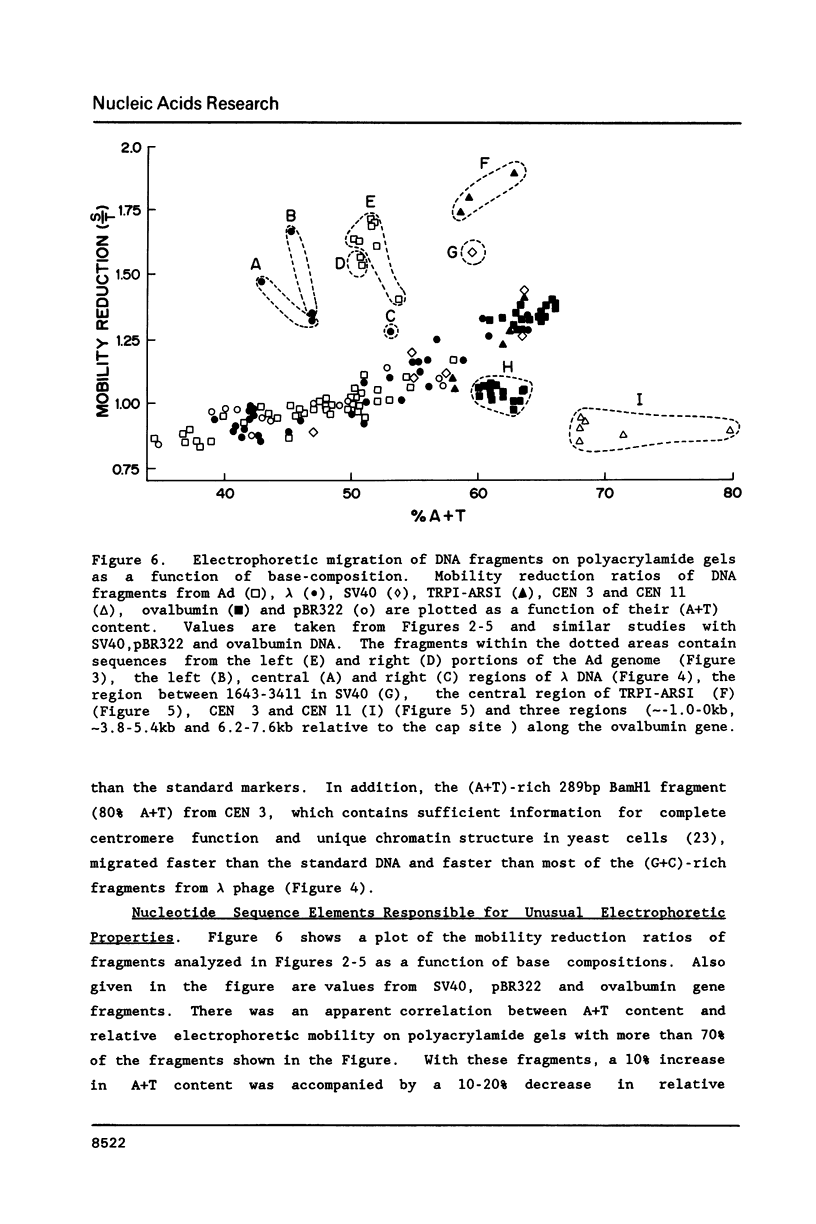
Abstract
Unusual DNA structures were detected by an electrophoretic procedure in which DNA fragments were separated according to size on agarose gels and then by shape on polyacrylamide gels. Fragments from yeast centromeres migrated faster in polyacrylamide than predicted from their base composition and size and this property was attributed to a nonrandom distribution of oligomeric A tracts that exhibited minima at 10-11 base intervals. Fragments from seven loci in 107 kb of DNA migrated anomalously slow and these fragments contained blocks of A2-6 in a 10-11 base periodicity which is indicative of bent DNA. The most pronounced bent sequences were found within yeast ARS1 and centered at 245 and 240 bp from the left and right ends of the adenovirus genome. Each sequence is approximately 150 bp away from a replication origin and the adenovirus sequences are within 50 bp of enhancers. Nuclear matrix attachment sites, which are also adjacent to enhancers, contain sequences characteristic of bent DNA. These results suggest that bent structures reside at the base of DNA loops in chromosomes.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bloom K. S., Amaya E., Carbon J., Clarke L., Hill A., Yeh E. Chromatin conformation of yeast centromeres. J Cell Biol. 1984 Nov;99(5):1559–1568. doi: 10.1083/jcb.99.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli E., Franzini C., Leoni L., Savino M. Nucleosome "phasing" and cruciform structures in circular supercoiled pBR322 DNA. Cell Biophys. 1984 Mar;6(1):23–31. doi: 10.1007/BF02788578. [DOI] [PubMed] [Google Scholar]
- Carrì M. T., Micheli G., Graziano E., Pace T., Buongiorno-Nardelli M. The relationship between chromosomal origins of replication and the nuclear matrix during the cell cycle. Exp Cell Res. 1986 Jun;164(2):426–436. doi: 10.1016/0014-4827(86)90041-8. [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Sweder K., Srienc F., Bailey J. E., Campbell J. L. Deletion mutations affecting autonomously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2455–2466. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg S. S., Englund P. T. Heterogeneity of minicircles in kinetoplast DNA of Leishmania tarentolae. J Mol Biol. 1980 Apr 15;138(3):447–472. doi: 10.1016/s0022-2836(80)80012-x. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. The structure and function of yeast centromeres. Annu Rev Genet. 1985;19:29–55. doi: 10.1146/annurev.ge.19.120185.000333. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Colbert D. A., Knoll B. J., Woo S. L., Mace M. L., Tsai M. J., O'Malley B. W. Differential hormonal responsiveness of the ovalbumin gene and its pseudogenes in the chick oviduct. Biochemistry. 1980 Nov 25;19(24):5586–5592. doi: 10.1021/bi00565a020. [DOI] [PubMed] [Google Scholar]
- Diekmann S., Wang J. C. On the sequence determinants and flexibility of the kinetoplast DNA fragment with abnormal gel electrophoretic mobilities. J Mol Biol. 1985 Nov 5;186(1):1–11. doi: 10.1016/0022-2836(85)90251-7. [DOI] [PubMed] [Google Scholar]
- Dijkwel P. A., Wenink P. W., Poddighe J. Permanent attachment of replication origins to the nuclear matrix in BHK-cells. Nucleic Acids Res. 1986 Apr 25;14(8):3241–3249. doi: 10.1093/nar/14.8.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. Anatomy of hypersensitive sites. Nature. 1984 May 17;309(5965):213–214. doi: 10.1038/309213a0. [DOI] [PubMed] [Google Scholar]
- Fisher M. P., Dingman C. W. Role of molecular conformation in determining the electrophoretic properties of polynucleotides in agarose-acrylamide composite gels. Biochemistry. 1971 May 11;10(10):1895–1899. doi: 10.1021/bi00786a026. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Fütterer J., Winnacker E. L. Adenovirus DNA replication. Curr Top Microbiol Immunol. 1984;111:41–64. doi: 10.1007/978-3-642-69549-0_2. [DOI] [PubMed] [Google Scholar]
- Gough G. W., Lilley D. M. DNA bending induced by cruciform formation. Nature. 1985 Jan 10;313(5998):154–156. doi: 10.1038/313154a0. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Evidence for the existence of stable curvature of DNA in solution. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4632–4636. doi: 10.1073/pnas.81.15.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence dependence of the curvature of DNA: a test of the phasing hypothesis. Biochemistry. 1985 Dec 3;24(25):7033–7037. doi: 10.1021/bi00346a001. [DOI] [PubMed] [Google Scholar]
- Hammarskjöld M. L., Winberg G. Encapsidation of adenovirus 16 DNA is directed by a small DNA sequence at the left end of the genome. Cell. 1980 Jul;20(3):787–795. doi: 10.1016/0092-8674(80)90325-6. [DOI] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A enhancer contains two functionally distinct domains: one is specific for E1A and the other modulates all early units in cis. Cell. 1986 Apr 25;45(2):229–236. doi: 10.1016/0092-8674(86)90387-9. [DOI] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Hiwasa T., Segawa M., Yamaguchi N., Oda K. Phasing of nucleosomes in SV40 chromatin reconstituted in vitro. J Biochem. 1981 May;89(5):1375–1389. doi: 10.1093/oxfordjournals.jbchem.a133329. [DOI] [PubMed] [Google Scholar]
- Kearsey S. Structural requirements for the function of a yeast chromosomal replicator. Cell. 1984 May;37(1):299–307. doi: 10.1016/0092-8674(84)90326-x. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kunkel G. R., Martinson H. G. Nucleosomes will not form on double-stranded RNa or over poly(dA).poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 1981 Dec 21;9(24):6869–6888. doi: 10.1093/nar/9.24.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linxweller W., Hörz W. Reconstitution experiments show that sequence-specific histone-DNA interactions are the basis for nucleosome phasing on mouse satellite DNA. Cell. 1985 Aug;42(1):281–290. doi: 10.1016/s0092-8674(85)80123-9. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Effron P. N., Goodman T. C., Singleton C. K., Wells R. D., Wartell R. M., Englund P. T. Physical characterization of a kinetoplast DNA fragment with unusual properties. J Biol Chem. 1984 Jul 25;259(14):8974–8979. [PubMed] [Google Scholar]
- Mason P. J., Török I., Kiss I., Karch F., Udvardy A. Evolutionary implications of a complex pattern of DNA sequence homology extending far upstream of the hsp70 genes at loci 87A7 and 87C1 in Drosophila melanogaster. J Mol Biol. 1982 Mar 25;156(1):21–35. doi: 10.1016/0022-2836(82)90456-9. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Montiel J. F., Norbury C. J., Tuite M. F., Dobson M. J., Mills J. S., Kingsman A. J., Kingsman S. M. Characterization of human chromosomal DNA sequences which replicate autonomously in Saccharomyces cerevisiae. Nucleic Acids Res. 1984 Jan 25;12(2):1049–1068. doi: 10.1093/nar/12.2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R. Structural wrinkles and the genomic regulatory sites of eukaryotes. J Mol Evol. 1985;22(2):150–159. doi: 10.1007/BF02101693. [DOI] [PubMed] [Google Scholar]
- Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA) . poly(dT). EMBO J. 1982;1(2):173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. Nucleosome cores reconstituted from poly (dA-dT) and the octamer of histones. Nucleic Acids Res. 1979;6(5):1805–1816. doi: 10.1093/nar/6.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder K., Silver S., DeLucia A. L., Fanning E., Tegtmeyer P. An altered DNA conformation in origin region I is a determinant for the binding of SV40 large T antigen. Cell. 1986 Mar 14;44(5):719–725. doi: 10.1016/0092-8674(86)90838-x. [DOI] [PubMed] [Google Scholar]
- Simpson L. Isolation of maxicircle component of kinetoplast DNA from hemoflagellate protozoa. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1585–1588. doi: 10.1073/pnas.76.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Künzler P. Cromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 1979 Apr;6(4):1387–1415. doi: 10.1093/nar/6.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Stellwagen N. C. Anomalous electrophoresis of deoxyribonucleic acid restriction fragments on polyacrylamide gels. Biochemistry. 1983 Dec 20;22(26):6186–6193. doi: 10.1021/bi00295a023. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Struhl K., Davis R. W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979 Nov 1;282(5734):39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Thomas T. J., Bloomfield V. A. Chain flexibility and hydrodynamics of the B and Z forms of poly(dG-dC).poly(dG-dC). Nucleic Acids Res. 1983 Mar 25;11(6):1919–1930. doi: 10.1093/nar/11.6.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts C. Viral DNA sequences from incomplete particles of human adenovirus type 7. Cell. 1977 Sep;12(1):243–249. doi: 10.1016/0092-8674(77)90202-1. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Curved DNA. CRC Crit Rev Biochem. 1985;19(2):89–106. doi: 10.3109/10409238509082540. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N., Sussman J. L. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- Wang J. N., Hogan M. An equilibrium between distorted and undistorted DNA in the adult chicken beta A-globin gene. J Biol Chem. 1985 Jul 5;260(13):8194–8202. [PubMed] [Google Scholar]
- Wasylyk B., Oudet P., Chambon P. Preferential in vitro assembly of nucleosome cores on some AT-rich regions of SV40 DNA. Nucleic Acids Res. 1979 Oct 10;7(3):705–713. doi: 10.1093/nar/7.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985 Oct 3;317(6036):451–453. doi: 10.1038/317451a0. [DOI] [PubMed] [Google Scholar]
- Zeiger R. S., Salomon R., Dingman C. W., Peacock A. C. Role of base composition in the electrophoresis of microbial and crab DNA in polyacrylamide gels. Nat New Biol. 1972 Jul 19;238(81):65–69. doi: 10.1038/newbio238065a0. [DOI] [PubMed] [Google Scholar]
- Zhurkin V. B. Specific alignment of nucleosomes on DNA correlates with periodic distribution of purine-pyrimidine and pyrimidine-purine dimers. FEBS Lett. 1983 Jul 25;158(2):293–297. doi: 10.1016/0014-5793(83)80598-5. [DOI] [PubMed] [Google Scholar]






