Abstract
We have previously identified several colorectal cancer (CRC)-associated polymorphisms using genome-wide association (GWA) analysis. We sought to fine-map the location of the functional variants for three of these regions at 8q23.3 (EIF3H), 16q22.1 (CDH1/CDH3) and 19q13.11 (RHPN2). We genotyped two case–control sets at high density in the selected regions and used existing data from four other case–control sets, comprising a total of 9328 CRC cases and 10 480 controls. To improve marker density, we imputed genotypes from the 1000 Genomes Project and Hapmap3 data sets. All three regions contained smaller areas in which a cluster of single nucleotide polymorphisms (SNPs) showed clearly stronger association signals than surrounding SNPs, allowing us to assign those areas as the most likely location of the disease-associated functional variant. Further fine-mapping within those areas was generally unhelpful in identifying the functional variation based on strengths of association. However, functional annotation suggested a relatively small number of functional SNPs, including some with potential regulatory function at 8q23.3 and 16q22.1 and a non-synonymous SNP in RPHN2. Interestingly, the expression quantitative trait locus browser showed a number of highly associated SNP alleles correlated with mRNA expression levels not of EIF3H and CDH1 or CDH3, but of UTP23 and ZFP90, respectively. In contrast, none of the top SNPs within these regions was associated with transcript levels at EIF3H, CDH1 or CDH3. Our post-GWA study highlights benefits of fine-mapping of common disease variants in combination with publicly available data sets. In addition, caution should be exercised when assigning functionality to candidate genes in regions discovered through GWA analysis.
INTRODUCTION
In recent genome-wide association (GWA) studies, we have identified 14 common, low-penetrance colorectal cancer (CRC) susceptibility alleles within regions at chromosomes 1q41, 3q26.2, 8q23.3, 8q24.21, 10p14, 11q23.1, 12q13.13, 14q22.2, 15q13.3, 16q22.1, 18q21.1, 19q13.11, 20p12.3 and 20q13.33 (1–8). These loci were indicated by tagSNPs that are unlikely to be the functional variant associated with the disease. The discovery of functional variants may be aided by a deep examination of genetic variation in the linkage disequilibrium (LD) blocks in which the tagSNPs reside. Such discovery is likely to benefit from recent efforts such as the 1000 Genomes Project, where a comprehensive discovery of novel variants is being carried out in several populations. In two previous studies, we have used a combination of sequencing, genotyping, bioinformatics and functional experiments to fine-map and identify the genetic variation associated with CRC risk at 8q24 and 18q21 (9,10). At 8q24, the original tagSNP rs6983267 was associated with differential response to Wnt signalling (10), while a single nucleotide polymorphism (SNP) in LD with the tagSNP (denoted as ‘novel 1') in the 18q21 region was associated with differential expression of the SMAD7 gene (9).
In this study, we had three aims. First, we wished to refine the most likely location of the ‘disease-causing/functional' variant based on association testing at genotyped and imputed SNPs. Secondly, we wished to determine the SNP(s) most likely to be functional within the fine-mapped regions based on effect sizes and strengths of association. Thirdly, we wished to identify potentially functional variants by annotating SNPs in high LD with the original tagSNP. To achieve these goals, we used data from four different GWA studies and from two replication sample sets (N = 19 808 samples in total) to fine-map the genetic variation associated with CRC tagSNPs at chromosomes 8q23.3, 16q22.1 and 19q13.11. In addition to the genotyping of a large number of polymorphisms in these three regions, we used data from the 1000 Genomes Project and HapMap3 data sets to generate in silico genotypes for a large number of additional SNPs. This combination of high-density genotyping and imputation allowed a wide and deep examination of SNPs with minor allele frequency (MAF) > 0.01 in these regions. We have also carried out a comprehensive annotation of the most strongly associated variants using the expression quantitative trait locus (eQTL) browser (http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/) and Galaxy (http://main.g2.bx.psu.edu/). We detect SNPs more strongly associated with disease than the original tagSNPs and/or find evidence of functional variation at each locus.
RESULTS
We studied six non-overlapping CRC case–control series of Northern European ancestry. For fine-mapping, the NSCCG and Scotland2 CRC case–control series were genotyped with custom Illumina arrays that had a high SNP density within the three regions targeted in this study. To select the additional SNPs for fine mapping in these regions, we used Haploview (11) and SequenceLDHot (12) to define the haplotype blocks and recombination hotspots containing the tagSNP previously found to be associated with CRC in each region: rs16892766 (8q23.3), rs9929218 (16q22.1) and rs10411210 (19q13.11). (4,7). We determined the haplotype block in which the tagSNP lay and identified from dbSNP (build 128) all SNPs with MAF > 0.05 between the recombination hotspots flanking the haplotype block, irrespective of their relationship to the original tagSNP. If there was evidence of long-range LD outside the haplotype block, additional SNPs within such regions were also identified. All these SNPs were submitted to Illumina for assay design and those with a design score > 0.8 were genotyped. In the other sample series (see Materials and Methods section)—CORGI, Scotland1, CFR, VQ58—a small number of SNPs in each region had been genotyped using proprietary SNP arrays. Additional SNPs were imputed in each sample set using HapMap3 and 1000 Genomes Project as reference data sets.
8q23.3/EIF3H region
The original CRC-associated tagSNP, rs16892766 (chr8: 117 699 864), lies within a region of extended but irregular patterns of LD (chr8:117 650 000–117 895 000, Fig. 1). We analysed data for 456 genotyped or imputed SNPs in the 8q23.3 region (Supplementary Material, Tables S1 and S2) around rs16892766. No evidence of additional, independent association signals was found (details not shown). Four markers close to rs16892766 reached genome-wide significance (rs28535528, rs11986063, rs16888589, rs16888611, P < 5 × 10−8; see Supplementary Material, Table S2) in the meta-analyses of the six case–control sets examined in the study.
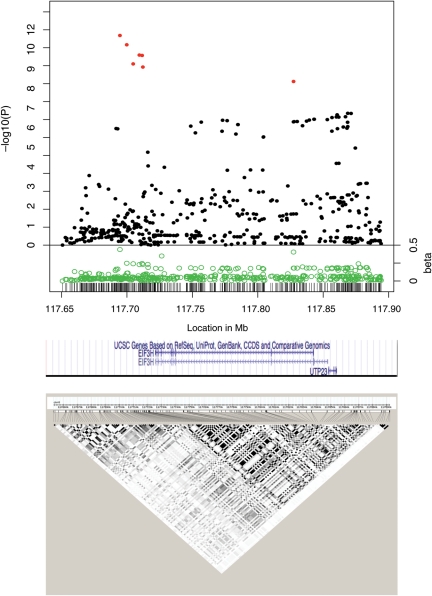
Figure 1.
Associations between 8q23.3 SNPs and CRC risk in six case–control studies of European descent. The top part of the figure shows the meta-analysis P-values and their associated beta coefficients. The P-values for rs16892766, the regions’ original tagSNP, and for 8-117694643, rs11986063, rs28535528, rs16888589, rs16888611 and 8-117827346, are shown in red. The x-axis indicates the SNP location (in Mb) based on the human genome build 36. The y-axis indicates the −log10 of the P-value (top) and absolute value of the beta coefficients (bottom) for each SNP. The middle part of the figure shows the approximate locations of the EIF3H and UTP23 genes. The lower part of the figure shows the patterns of LD (r2) in the CEPH population.
The association signals in the region of these five SNPs (chr8:117 694 643–117 712 171) were notably stronger than those for surrounding SNPs, suggesting that this region had a higher likelihood for being the location of the functional variation within the region (Fig. 1). One of these five SNPs, 8-117694643, imputed using the 1000 Genomes Project data, was more strongly associated with CRC (P = 2.07 × 10−12, β = 0.438, data not shown) than the original tagSNP (rs16892766, P = 6.81 × 10−12, β = 0.247, Fig. 1, Supplementary Material, Table S2). To evaluate the accuracy of the 8-117694643 imputation, we sequenced 170 CORGI samples and found several differences, including four heterozygous predicted as homozygous by the imputed data of this rare SNP (MAF = 2%), suggesting that this was a spurious association caused by inaccurate imputation of rare genotypes. rs16892766 thus remained the most strongly associated SNP in the 8q23.3 region. A further SNP (8-117827346), which lies in an EIF3H intron outside the chr8:117 694 643–117 712 171 region, had an association signal that reached genome-wide significance. The effect size of this SNP was relatively strong (β = 0.403, Fig. 1). However, the level of significance was lower than that of the SNPs in the chr8:117 694 643–117 712 171 region and levels of association at surrounding SNPs were unremarkable.
We assessed the location of each of the seven most strongly associated SNPs with regard to the ENCODE functional annotation (http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=173506627&c=chr8&g=wgEncodeReg). Most SNPs were in unremarkable locations according to the ENCODE data and none was associated with histone methylation or transcription factor binding, although rs11986063 maps to a DNAse I hypersensitive site (Table 1).
Table 1.
A summary of the association of this study's most significantly associated SNPs and genomic features identified at the UCSC genome browser using Galaxy
| Region | SNP | UCSC Genome Browser Track | Location of the feature (Build 36 coordinates) | Observations |
|---|---|---|---|---|
| 8q23.3 | rs16892766 | 7X Regulatory Potential | 8:117 699 862–117 699 866 | High regulatory potential |
| 8q23.3 | rs16892766 | 28X + 17X Conservation | 8:117 699 862–117 699 866 | Highly conserved |
| 8q23.3 | rs11986063 | ENCODE DNAse I hypersensitivity | 8:117 709 321–117 709 510 | Hypersensitivity cluster detected in 4/71 cell lines, score = 286/1000 |
| 16q22.1 | rs35158985 | Aceview genes | 16:67 353 526–67 353 862 | Overlaps with rary.aApr07-unspliced |
| 16q22.1 | rs35158985 | Burge RNA-seq | 16:67 354 245–67 354 249 | Overlaps with novel colonic mRNA |
| 16q22.1 | rs58548890 | ENCODE ChIP-seq | 16:67 347 448–67 349 230 | IRF4 TF binding, score = 1000/1000 |
| 16q22.1 | rs13339591 | ENCODE DNAse I hypersensitivity | 16:67 366 661–67 366 890 | Hypersensitivity cluster detected in 5/71 cell lines, score = 118/1000 |
| 16q22.1 | rs2961 | ENCODE DNAse I hypersensitivity | 16:67 376 241–67 376 410 | Hypersensitivity cluster detected in 2/71 cell lines, score = 144/1000 |
| 16q22.1 | rs9929218, rs9929239 | ENCODE histone mark H3K4Me3 | 16:67 378 336–67 378 812 | H3K4Me3 mark in 1/9 cell lines |
| 16q22.1 | rs9929218 | ENCODE DNAse I hypersensitivity | 16:67 378 081–67 378 590 | Hypersensitivity cluster detected in 39/71 cell lines, score = 1000/1000 |
| 16q22.1 | rs9929218, rs9929239 | ENCODE ChIP-seq | 16:67 377 982–67 378 706 | FOSL2 TF binding, score = 126/1000 |
| 16q22.1 | rs9929218 | ENCODE ChIP-seq | 16:67 378 047–67 378 501 | HNF4A TF binding, score = 686 1000 |
| 16q22.1 | rs9929218 | ENCODE ChIP-seq | 16:67 378 198–67 378 511, | HEY TF binding, score = 62/1000 |
| 16q22.1 | rs9929239 | ENCODE ChIP-seq | 16:67 378 523–67 378 728 | NRSF TF binding, score = 76/1000 |
| 19q13.11 | 19-38209763 | Alternative splicing | 19:38 209 378–38 210 017 | Overlaps with bleeding exon |
| 19q13.11 | 19-38209763 | Aceview genes | 19:38 161 332–38 227 053 | Overlaps with RHPN2.bApr07 |
| 19q13.11 | rs10411210 | 7X Regulatory Potential | 19:38 224 138–38 224 142 | High regulatory potential |
| 19q13.11 | rs10411210 | 28X +17X Conservation | 19:38 224 138–38 224 138 | Highly conserved |
| 19q13.11 | rs28626308 | 28X +17X Conservation | 19:38 209 353–38 209 357 | Highly conserved |
| 19q13.11 | rs28570619, rs17841839, rs28626308 | ENCODE histone mark H3K4Me3 | 19:38 208 858–38 209 668 | H3K4Me3 mark in 1/9 cell lines |
| 19q13.11 | rs73039426 rs73039428 | ENCODE ChIP-seq | 19:38 212 501–38 213 096 | GR TF binding, score = 359/1000 |
| 19q13.11 | rs10411210 | ENCODE DNAse I hypersensitivity | 19:38 224 001–38 224 390 | Hypersensitivity cluster detected in 38/71 cell lines, score = 1000/1000 |
| 19q13.11 | rs10411210 | ENCODE ChIP-seq | 19:38 223 760–38 224 730 | HEY1 TF binding, score = 752/1000 |
| 19q13.11 | rs10411210 | ENCODE ChIP-seq | 19:38 223 809–38 224 543 | USF-1 TF binding, score = 199/1000 |
| 19q13.11 | rs10411210 | ENCODE ChIP-seq | 19:38 223 841–38 224 677 | RXRA TF binding, score = 1000/1000 |
| 19q13.11 | rs73039426, rs73039428 | ENCODE ChIP-seq | 19:38 223 868–38 224 590 | FOSL2 TF binding, score = 1000/1000 |
| 19q13.11 | rs10411210 | ENCODE ChIP-seq | 19:38 223 870–38 224 607 | GR TF binding, score = 448/1000 |
| 19q13.11 | rs10411210 | ENCODE ChIP-seq | 19:38 223 926–38 224 530 | JunD TF , score = 1000/1000 |
| 19q13.11 | rs10411210 | ENCODE ChIP-seq | 19:38 223 934–38 224 387 | CEBPB TF binding, score = 97/1000 |
| 19q13.11 | rs10411210 | ENCODE ChIP-seq | 19:38 223 953–38 224 519 | P300 binding, score = 1000/1000 |
| 19q13.11 | rs73039426, rs73039428 | ENCODE ChIP-seq | 19:38 223 970–38 224 639 | HNF4A TF binding, score = 446/1000 |
| 19q13.11 | rs10411210 | ENCODE ChIP-seq | 19:38 224 009–38 224 531 | SREBPl TF binding, score = 11/1000, |
| 19q13.11 | rs10411210 | ENCODE ChIP-seq | 19:38 224 040–38 224 204 | BAF155 TF binding, score = 624/1000 |
| 19q13.11 | rs10411210 | ENCODE ChIP-seq | 19:38 224 057–38 224 230 | NFKB TF binding, score = 58/1000 |
We then assessed eQTLs in the region around the tagSNP rs16892766. Transcript levels in monocytes of the UTP23 [small subunit (SSU) processome component, homologue (yeast)] gene were positively associated with genotypes at three 8q23.3 SNPs (rs11986063, rs16888589 and rs7837208) that had the third, fourth and ninth strongest associations with CRC (3.61 × 10−11 < P < 8.89 × 10−8; see Table 2, Supplementary Material, Tables S2 and S5). Although rs16892766 was not present in the eQTL browser data, the three SNPs associated with UTP23 expression were in high LD with rs16892766 (0.73 < D′ < 0.82, 0.49 < r2 < 0.68; Supplementary Material, Fig. S1).
Table 2.
Most significant cis eQTLs at 8q23.3 and 16q22.1
| Region | eQTL SNP | Target gene | Association P-value (rank)* |
|---|---|---|---|
| 8q23.3 | rs11986063 | UTP23 | 2.5 × 10−11 (3) |
| 8q23.3 | rs16888589 | UTP23 | 7.9 × 10−11 (4) |
| 8q23.3 | rs7837208 | UTP23 | 7.0 × 10−8 (9) |
| 16q22.1 | rs2059254 | ZFP90 | 1.7 × 10−7 (6) |
| 16q22.1 | rs8056538 | ZFP90 | 2.7 × 10−7 (19) |
| 16q22.1 | rs9925923 | ZFP90 | 2.8 × 10−7 (20) |
| 16q22.1 | rs2113200 | ZFP90 | 4.6 × 10−7 (43) |
All these associations between SNPs and gene expression levels have been reported in monocytes (22) and were compiled from the eQTL genome browser (http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/). The full list of eQTLs in the three regions investigated in the study is shown in Supplementary Material, Table S5.
*P-value between the eQTL SNP and CRC risk obtained in the association meta-analyses. In parenthesis the rank of the eQTL SNP in the association testing is shown. For details see Supplementary Material, Tables S2 and S3.
Interestingly, of the 21 cis eQTLs around rs16892766 that have been reported in the eQTL browser, 16 involved UTP23 (associations with CRC risk, 2.55 × 10−11 < P < 0.043; Table 2 and Supplementary Material, Tables S2 and S5) and three, RAD21 (for all, P > 0.08). Only one eQTL, rs12675038, involved EIF3H—and this SNP showed no association with CRC (P = 0.97; see Supplementary Material, Tables S2 and S5). This analysis suggests that UTP23, rather than EIF3H, is the target of the genetic variation associated with CRC in the 8q23.3 region.
Pittman et al. (13) had previously undertaken an independent assessment of the 8q23.3 region associated with CRC risk, using re-sequencing, direct genotyping of 154 SNPs in 1964 CRC cases and 2081 controls, and imputation of an additional 112 SNPs from HapMap2. Pittman et al. had also found the same five SNPs in the region to be highly associated with CRC. They also identified a SNP, ‘Novel 28', that was not present in public databases, that was in nearly complete LD with rs16892766 and that was more strongly associated with CRC than any other SNP in the region. Pittman et al. undertook detailed functional studies of the region and found, in contrast to the absence of functional features in the ENCODE data, that a 750 bp region around rs16888589 acted as a repressor of gene expression and interacted with the EIF3H promoter in CRC cell lines.
16q22.1/CDH1 locus
The CRC-associated tagSNP, rs9929218 (chr16:67 378 447), lies within a haplotype block located at ∼chr16:67 100 000–67 400 000, which contains many strongly associated SNPs (Fig. 2). We evaluated data for 676 SNPs in and around this region (Supplementary Material, Tables S1 and S3). Eight markers (rs7199991, rs2961, rs35158985, rs9929239, rs9923610, rs2059254, rs1981871 and rs13339591; Supplementary Material, Table S3), four of which were genotyped in the two largest case–control sets, had P-values lower than that obtained for rs9929218 (P = 1.97 × 10−7, β = −0.118), with rs7199991 showing the lowest P-value (P = 1.31 × 10−7, β = −0.120; Supplementary Material, Table S3). The association signals suggested that the most likely location of the functional SNP was chr16:67 350 000–67 380 000. The levels of LD seen between these top 16q22.1 markers were very high (0.72 < r2 < 1; Supplementary Material, Fig. S2). We failed to detect, using logistic regression, additional risk loci in the region (details not shown). Thus, we believe that there is only one common 16q22.1 CRC risk allele.
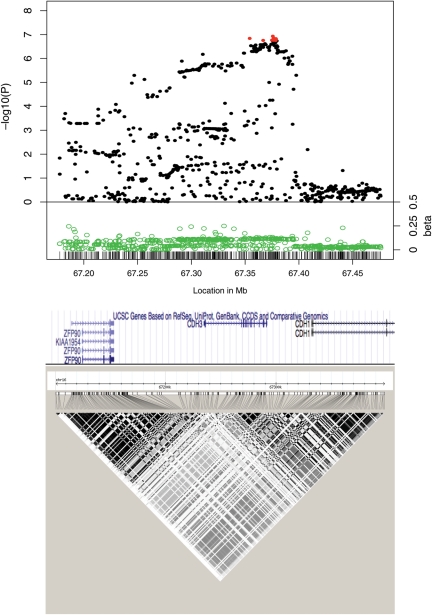
Figure 2.
Associations between 16q22.1 SNPs and CRC risk in six case–control studies of European descent. The top part of the figure shows the meta-analysis P-values and their associated beta coefficients. The P-values for rs9929218, the regions’ original tagSNP, and for rs7199991, rs2961, rs35158985, rs9929239, rs9923610, rs2059254, rs1981871 and rs13339591, the most strongly associated SNPs, are shown in red. The x-axis indicates the SNP location (in Mb) based on the human genome build 36. The y-axis indicates the −log10 of the P-value (top) and absolute value of the beta coefficients (bottom) for each SNP. The middle part of the figure shows the approximate locations of the ZPF90, CDH3 and CDH1 genes. The lower part of the figure shows the patterns of LD (r2) in the CEPH population.
The chr16:67 350 000–67 380 000 region lies within intron 2 of CDH1 and contains at least four sites with strong evidence of regulatory activity from the ENCODE project. Our annotation of the genomic regions where the most highly associated 16q22.1 SNPs map identified the best evidence of regulatory features close to rs9929239 (chr16:67 378 627) and rs9929218 (chr16: 67 378 447). These two highly associated SNPs are separated by 192 bp and map to a region with a strong H3K4Me3 mark, suggestive of enhancer activity, a DNAse I hypersensitivity, and predicted binding sites for the FOSL2, NRFS and HNF4A transcription factors (Table 1). rs2961 and rs13339591, two other highly associated SNPs in the region, map to DNAse I hypersensitivity segments. A full list of the genomic features associated with highly associated SNPs at 16q22.1 is shown in Table 1.
eQTL database interrogation showed that genotypes at rs2059254, the marker with the sixth lowest P-value in the region (P = 1.91 × 10−7, β = −0.118, Supplementary Material, Table S3), were associated with transcript levels in monocytes of ZFP90 (Zinc finger protein 90, chr16:67 130 617–67 158 540), a gene that maps to the proximal end of the haplotype block (Fig. 2). In all, 23 out of the 29 SNPs associated with ZFP90 transcript levels were associated with CRC at P < 0.05 (Supplementary Material, Table S5). Four of these associations with ZFP90 transcript levels involved SNPs (rs2059254, rs8056538, rs9925923 and rs2113200), genotyped in the NSSCG and Scotland studies, that were associated with CRC risk at P < 5 × 10−7 (Table 2, Supplementary Material, Tables S3 and S5). In contrast, none of the 22 eQTLs involving CDH1, the strongest CRC candidate gene at 16q22.1, showed strong associations with CRC risk. For example, the SNP most strongly associated with CDH1 expression, rs4783566, had the 160th lowest association P-value with CRC in our study (P = 0.0004, Supplementary Material, Tables S3 and S5). These eQTL findings suggest that ZFP90, rather than CDH1 or CDH3, is the most likely target of the 16q22.1 genetic variation associated with increased CRC risk. This was a somewhat surprising result giving that CDH1 and CDH3, the two genes that map to this haplotype block, represented very strong candidates given their known involvement in colorectal tumorigenesis (14,15). Previous reports have suggested that the functional variation at 16q22.1 is rs16260 (chr16:67 328 535), a SNP in strong LD (r2 = 0.91) with rs9929218 that lies within the CDH1 promoter (16). However, rs16260 lies outside the region with the strongest association with CRC—it had the 83rd lowest P-value in our study (P = 1.88 × 10−6; Supplementary Material, Table S3)—and the evidence that it affects CDH1 transcription is mixed (17,18).
19q13.11/RHPN2 locus
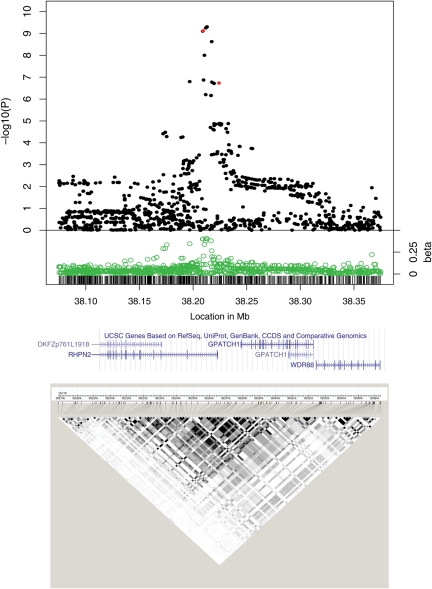
The CRC-associated tagSNP, rs10411210 (chr19: 38 224 140), lies within an intron of RHPN2. It is in a haplotype block at ∼chr19:38 168 000–38 364 000. We evaluated 865 polymorphisms in and around this region (Supplementary Material, Tables S1 and S4). Eleven markers reached genome-wide significance (Fig. 3, Supplementary Material, Table S3 and Fig. S3) and lay within a region (∼chr19: 38 208 992–38 222 500) of clearly stronger association than the surrounding SNPs. Most of the top-ranking SNPs at 19q13 had P-values that were nearly three orders of magnitude lower than that of rs10411210, and were in modest LD with this tagSNP (r2 < 0.21, D′ < 0.71; Supplementary Material, Table S4 and Fig. S3). One of these top-ranking SNPs, rs28626308, was a non-synonymous change in the RhoGTP-binding domain of RHPN2 (R70Q, P = 7.49 × 10−10, β = −0.46, 28X conservation = 85%, 17X conservation = 86%; Table 1 and Supplementary Material, Table S4). rs28626308 is in moderate LD with rs10411210 (D′ = 0.71, r2 = 0.21). The risk (minor) rs28626308 allele was predicted as damaging by SIFT (19) and as probably damaging by Poly-Phen2 (20). Owing to the potential importance of rs28626308, we sequenced 192 samples to investigate the imputation accuracy of this SNP. Although the concordance between imputed and genotyped genotypes was high (>95%), we found lower, suboptimal concordance for the heterozygous genotypes (∼70%, data not shown). Despite this, rs28626308 remains a very good functional candidate.
Figure 3.
Associations between 19q13.11 SNPs and CRC risk in six case–control studies of European descent. The top part of the figure shows the meta-analysis P-values and their associated beta coefficients. The P-values for rs10411210, the regions’ original tagSNP, and for rs28628368, a RPHN2 nsSNP are shown in red. The x-axis indicates the SNP location (in Mb) based on the human genome build 36. The y-axis indicates the −log10 of the P-value (top) and absolute value of the beta coefficients (bottom) for each SNP. The middle part of the figure shows the approximate location of the RPHN2 gene. The lower part of the figure shows the patterns of LD (r2) in the CEPH population.
Our search for association between 19q13.11 SNPs and transcript levels at genes in the region identified 44 cis eQTLs reported in monocytes, liver and lymphoblastoid cell lines (Supplementary Material, Table S5). Only nine of these associations involved SNPs that were nominally associated (at P < 0.05) with CRC risk. The SNP with the strongest eQTL signal (rs7253595, P = 0.006) had the 112th lowest P-value in our study and was associated with transcript levels at FLJ14640 and SLC7A9 (Supplementary Material, Tables S4 and S5). Four SNPs—rs7253624, rs16967665, rs16967578 and rs12983019—were associated with RHPN2 transcript levels but not with CRC risk, and showed the 650th, 693th, 843th and 862th lowest P-values in our study (P > 0.26 for all four SNPs; see Supplementary Material, Tables S4 and S5). This provides further support for the proposal that the missense variant in RHPN2, rs28626308, is the functional variation associated with CRC risk at 19q13.11. However, we must be cautious, since rs10411210, the original 19q13.11 tag SNP, lies in a region that was recently identified as highly regulatory by the ENCODE project: it overlaps with H3K4Me1 methylation marks and DNAse I hypersensitivity signals in 38 out of the 77 cell lines examined by the ENCODE project. Furthermore, several important transcription factors, including HEY1, RXRA, FOSL2, JunD, P300 and BAF155 bind strongly to the region encompassing rs10411210 (Table 1).
DISCUSSION
We have carried out a comprehensive evaluation of common SNPs in three genomic regions that harbour CRC risk loci discovered by GWA studies. We genotyped between 22 and 156 SNPs in each region in up to 9328 cases and 10 480 controls. In all samples, we also imputed in each region between 324 and 1025 SNPs that had recently been identified or validated by the 1000 Genomes Project or HapMap3.
Genotype imputation led to a large increase in informative markers. For example, we, respectively, obtained a 20-, 12- and 7-fold increase in the number of informative markers when our own data from the Illumina Hap300, Hap550 and Hap1M arrays were used. Even for the two data sets with the highest genotyping density (NSCCG and Scotland2), the use of imputation increased the number of markers by nearly 5-fold and led to a marker density of ∼1 SNP/kb. The recently released 1000 Genomes Project data particularly provided extra information. For example, ∼6% of the top 25 markers were not present in dbSNP and ∼60% were not characterized by the HapMap project (data not shown). Therefore, the imputation to the 1000 Genomes Project was, in principle, capable of providing more accurate and comprehensive information about local patterns of LD.
It is not necessarily the case that SNPs with the strongest evidence of association with disease are those that are truly functional and, as a result, some caution must be exercised in fine-mapping signals around tagSNPs identified in GWA studies. However, our study has led to a considerable reduction in the size of the genomic region most likely to contain the functional variants (for example see Figs 1–3). In all cases, some of the SNPs in the most strongly associated region were genotyped, at least in some studies, and some were imputed. In general, the imputed SNPs provided very similar results to the genotyped SNPs and provided good supporting evidence.
Imputation was less successful in identifying the most likely functional SNPs within the region of strongest association based on strengths of association. We discovered, in two of the three regions (16q22.1 and 19q13.11), SNPs showing stronger associations than those found for the original tagSNP. In total, there were 19 markers with P-values that were lower than that of the reported tagSNPs at 16q22.1 and 19q13.11; many of these SNPs had low minor allele frequencies (MAFs) (for example at 19q13.11) and most of them were either fully imputed (16q22.1: 2 SNPs, 19q13.11: 12 SNPs) or genotyped only in the NSCCG/Scotland2 studies and imputed in the other case–control sets (16q22.1: 5 SNPs, 19q13.11: 1 SNP). Only one marker was fully genotyped in all six case–control sets (rs7199991, the top SNP at 16q22.1) and had a P-value lower that the original tagSNP. Although imputation suggested SNPs, such as 8-117694643 and rs28626308, with stronger associations with CRC than the original tagSNPs, assessment of the accuracy of imputation using direct sequencing showed small, but important, errors. At least for uncommon genotypes imputed to the 1000 Genomes Project data, it seems unlikely that imputation can truly provide definitive evidence of SNP functionality.
In contrast with imputation, annotation of eQTLs and other genomic features within the regions of the top CRC-associated SNPs revealed variants that may be functional. Interestingly, and against previous expectations, our study did not support a role of a number of candidate genes in the haplotype blocks containing these risk alleles. For example, in the past, we proposed that EIF3H, CDH1 and RHPN2 were the clearly the best candidates in these blocks because they were the nearest genes to the tagSNPs and because there was some evidence of their involvement in tumorigenesis (4,7). Such latter evidence was particularly compelling for CDH1, a gene that is mutated or epigenetically silenced in colorectal tumours (15), and for EIF3H. RHPN2 remains the most likely candidate gene, but through the effect of coding rather than regulatory variation. However, with the important caveat that colorectal eQTL data sets are not available and that these associations were detected in blood cells, CDH1 does not seem to be the most likely target of the genetic variation associated with CRC risk at 16q22.1. Instead, ZFP90 emerged as the best candidates in this region. Interestingly, rs1728785, a SNP recently associated with ulcerative colitis risk (21), lies within an intron of ZFP90 and is associated with its transcript levels at this gene (22), suggesting that the gene may be involved in predisposition to both CRC and ulcerative colitis.
At 8q23.3, our analysis suggested that UTP23 was the most likely target of the functional variation. Although our fine mapping results were very similar to those of Pittman et al. (13), that study found good evidence that EIF3H was the target of the functional variation in the region. It is, however, entirely conceivable that both genes are coordinately regulated, given that they have related roles in mRNA translation.
In conclusion, we have carried out a large fine-mapping and annotational study of three CRC risk loci. This study is the fourth one to fine-map regions and to propose functional variants at CRC loci discovered by GWA studies (9,10,13); the remaining regions are under investigation, but show more complex patterns of LD or have evidence for multiple independent variants that require further analysis (23). We have refined the size of the disease-associated regions on 8q23.3, 16q22.1 and 19q13.11, and identified a number of candidate SNPs that might include functional alleles at these regions. We identified an RHPN2 non-synonymous polymorphism as a strong functional candidate, and discovered evidence suggesting that the risk alleles near CDH1 and EIF3H affect the expression of unexpected candidate genes. Our investigation establishes a foundation for future work, including expression studies in colorectal mucosa, to assign disease functionality to these SNPs using experimental and laboratory-based assays.
MATERIALS AND METHODS
Samples
We used six non-overlapping case–control series of Northern European ancestry that included the following: (i) 931 familial colorectal tumour cases and 929 cancer-free controls of white British origin from the COloRectal Gene Identification (CORGI) consortium; (ii) 1003 early-onset Scottish CRC cases (<55 years) and 979 Scottish population controls (Scotland1); (iii) 2860 incident CRC cases and 2849 population controls from the United Kingdom National Study of Colorectal Cancer Genetics (NSCCG); (iv) 2006 sporadic CRC cases and 2057 population controls of Scottish origin (Scotland2); (v) 1186 familial cases and 998 controls from Australia and North America of the Colon Cancer Family Registry (Colon CFR) study (24); and (vi) 1425 British Dukes stage B and C CRC patients from the VICTOR (http://www.octo-oxford.org.uk/alltrials/infollowup/vic.html, N = 923) and QUASAR2 (http://www.octo-oxford.org.uk/alltrials/trials/q2.html, N = 502) clinical trials, together with publicly available data from 2690 population controls from the UK 1958 Birth Cohort (VQ58).
Two of these GWA studies (CORGI and Scotland1) were originally used in the discovery of the four loci fine-mapped in the present study (see below). While we acknowledge that this could introduce minor bias into the fine-mapping, selection for the validation phases was non-stringent (at P < 0.05) and we considered that the gain in power from including the discovery phase samples would outweigh any such bias.
Collection of blood samples from patients and controls was undertaken with informed consent and ethical review board approval in accordance with the tenets of the Declaration of Helsinki.
Genotyping
The GWA study samples were genotyped using proprietary Illumina SNP arrays: CORGI on Hap550; Scotland1 on Hap300 + Hap240S; CFR and 1958 Birth Cohort on Hap1M; and VQ on Hap300, Hap370 or Hap660.
Details about the SNPs genotyped using the proprietary and custom arrays can be seen in Supplementary Material, Tables S2–S4.
General genotyping quality control assessment was as previously described and all SNPs presented in this study passed the required thresholds (1,4). Duplicate samples were used to check genotyping quality. SNPs and samples with <95% SNPs genotyped were eliminated from the analyses. Genotype frequencies at each SNP were tested for deviation from the Hardy–Weinberg equilibrium and rejected at P < 10−4.
We examined 192 samples with Kaspar genotyping or direct Sanger sequencing to verify the imputation accuracy of selected SNPs at 8q23.3 (8-117694643) and 19q13.11 (rs28626308). Primers used to type these polymorphisms are shown in Supplementary Material, Table S6.
Statistical analysis
Association statistics, using an additive model, were obtained with Snptest (www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html). We used genotype data from the 1000 Genomes (CEPH) and HapMap3 (CEPH and TSI) samples and the Imputev2 software (25) to generate in silico genotypes at additional SNPs in all these regions. Imputed genotypes were only called if they had a probability >0.90. Genotypes from the 1000 Genome CEPH VCF files were extracted with the VCFtools program (http://vcftools.sourceforge.net/) and LD patterns were visualized with Haploview (11). Association meta-analyses only included markers with proper_info scores >0.5, imputed call rates/SNP > 0.9 and MAFs > 0.01. Meta-analyses were carried out with Meta (http://www.stats.ox.ac.uk/~jsliu/meta.html) using the genotype probabilities from Imputev2, where a SNP was not directly typed. To test for the presence of additional independent risk alleles in each region, we carried out logistic regression analysis that included all SNPs with evidence of association in the meta-analysis at P < 5 × 10−4. To combine data from genotyped and imputed SNPs in logistic regression analyses, we used the genotyped probabilities obtained after imputation for each of the three genotypes using the following formula:
where pAA, pAB and pBB are the probabilities of having AA, AB and BB genotypes, respectively.
SNP annotation
We annotated all SNPs with association signals similar to or stronger than that of the region's original tag SNP. We used Galaxy (26) to carry out a comprehensive examination of genomic features associated with SNPs in the UCSC genome browser.
The genomic features investigated with Galaxy included overlaps with (i) highly conserved regions (conservation >40%) in alignments of 17 (17X) and 28 (28X) vertebrate genomes; (ii) regions with high regulatory potential scores derived from the alignment of seven genomes (7X Reg Potential); (iii) known Ensembl, UCSC or Encode genes; (iv) predicted transcripts or genes; (v) human mRNAs; (vi) RNA and tRNA genes; (vii) miRNAs; (viii) alternative splicing events; (ix) alternative promoters; (x) VISTA enhancers; (xi) CpG islands; (xii) predicted and conserved transcription binding sites; (xiii) predicted targetScan miRNA binding sites; (xiv) novel transcripts discovered by RNA-seq of colon tissue; (xv) regions of low nucleosome occupancy; and (xvi) repeated regions and copy number variants. We also searched for overlaps between the most strongly associated SNPs and the recently released Encode data on H3K4Me1, H3K4Me3 and H3K27Ac Histone marks, DNAaseI hypersensitivity clusters and Transcription Factor Chip-Seq. We noted features with intensity signals higher than 200/1000 (for details on these scores, see the Encode Regulation Super-track at http://genome.ucsc.edu/cgi-bin/hgTrackUi?g=wgEncodeReg). We also identified cis eQTLs in all four regions using the eQTL browser (http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/), a database that includes eQTLs identified in the liver (27), brain (28), fibroblasts (29), T-cells (29), monocytes (22) and lymphoblastoid cell lines (29–33).
SUPPLEMENTARY MATERIAL
FUNDING
Cancer Research UK provided principal funding for this study. The Oxford Comprehensive Biomedical Research Centre (I.T.) and the EU FP7 CHIBCHA grant (to L.G.C.-C. and I.T.) also provided funding. Core infrastructure support to the Wellcome Trust Centre for Human Genetics, Oxford, was provided by grant 075491/Z/04. The Medical Research Council (G0000657-53203), CORE and Scottish Executive Chief Scientist's Office (K/OPR/2/2/D333, CZB/4/449) provided additional funding (to M.D.). The Colon Cancer Family Registry was supported by the National Cancer Institute and National Institutes of Health under Request for Application #CA-95-011, and through cooperative agreements with the Australian Colorectal Cancer Family Registry (UO1 CA097735), the USC Familial Colorectal Neoplasia Collaborative Group (UO1 CA074799), Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (UO1 CA074800), Ontario Registry for Studies of Familial Colorectal Cancer (UO1 CA074783), Seattle Colorectal Cancer Family Registry (UO1 CA074794) and The University of Hawaii Colorectal Cancer Family Registry (UO1 CA074806).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to many colleagues within UK Clinical Genetics Departments (for CORGI) and to many collaborators who participated in the VICTOR and QUASAR2 trials. We would also like to thank colleagues from the UK National Cancer Research Network (for NSCCG). We are grateful to the COGS and SOCCS administrative teams, Roseanne Cetnarskyj and the research nurse teams, for recruiting individuals to the studies; the Wellcome Trust Clinical Research Facility for sample preparation; and all clinicians and pathologists throughout Scotland at collaborating centres. The study used the biological and data resource of Generation Scotland. This study made use of genotyping data on the 1958 Birth Cohort and NBS samples, kindly made available by the Wellcome Trust Case–Control Consortium 2. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk/. We are grateful to Jake Byrnes and Adam Auton for their help with Galaxy and VCFtools. Finally, we would like to thank all individuals who participated in the study.
Conflicts of Interest statement. None declared.
REFERENCES
- 1.Tomlinson I., Webb E., Carvajal-Carmona L., Broderick P., Kemp Z., Spain S., Penegar S., Chandler I., Gorman M., Wood W., et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 2007;39:984–988. doi: 10.1038/ng2085. doi:10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 2.Broderick P., Carvajal-Carmona L., Pittman A.M., Webb E., Howarth K., Rowan A., Lubbe S., Spain S., Sullivan K., Fielding S., et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat. Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. doi:10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 3.Jaeger E., Webb E., Howarth K., Carvajal-Carmona L., Rowan A., Broderick P., Walther A., Spain S., Pittman A., Kemp Z., et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat. Genet. 2008;40:26–28. doi: 10.1038/ng.2007.41. doi:10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 4.Tomlinson I.P., Webb E., Carvajal-Carmona L., Broderick P., Howarth K., Pittman A.M., Spain S., Lubbe S., Walther A., Sullivan K., et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet. 2008;40:623–630. doi: 10.1038/ng.111. doi:10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 5.Pittman A.M., Webb E., Carvajal-Carmona L., Howarth K., Di Bernardo M.C., Broderick P., Spain S., Walther A., Price A., Sullivan K., et al. Refinement of the basis and impact of common 11q23.1 variation to the risk of developing colorectal cancer. Hum. Mol. Genet. 2008;17:3720–3727. doi: 10.1093/hmg/ddn267. doi:10.1093/hmg/ddn267. [DOI] [PubMed] [Google Scholar]
- 6.Tenesa A., Farrington S.M., Prendergast J.G., Porteous M.E., Walker M., Haq N., Barnetson R.A., Theodoratou E., Cetnarskyj R., Cartwright N., et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet. 2008;40:631–637. doi: 10.1038/ng.133. doi:10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houlston R.S., Webb E., Broderick P., Pittman A.M., Di Bernardo M.C., Lubbe S., Chandler I., Vijayakrishnan J., Sullivan K., Penegar S., et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. doi:10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houlston R.S., Cheadle J., Dobbins S.E., Tenesa A., Jones A.M., Howarth K., Spain S.L., Broderick P., Domingo E., Farrington S., et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet. 2010;42:973–977. doi: 10.1038/ng.670. doi:10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pittman A.M., Naranjo S., Webb E., Broderick P., Lips E.H., van Wezel T., Morreau H., Sullivan K., Fielding S., Twiss P., et al. The colorectal cancer risk at 18q21 is caused by a novel variant altering SMAD7 expression. Genome Res. 2009;19:987–993. doi: 10.1101/gr.092668.109. doi:10.1101/gr.092668.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuupanen S., Turunen M., Lehtonen R., Hallikas O., Vanharanta S., Kivioja T., Bjorklund M., Wei G., Yan J., Niittymaki I., et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 2009;41:885–890. doi: 10.1038/ng.406. doi:10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 11.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview-analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. doi:10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 12.Fearnhead P. SequenceLDhot-detecting recombination hotspots. Bioinformatics. 2006;22:3061–3066. doi: 10.1093/bioinformatics/btl540. doi:10.1093/bioinformatics/btl540. [DOI] [PubMed] [Google Scholar]
- 13.Pittman A.M., Naranjo S., Jalava S.E., Twiss P., Ma Y., Olver B., Lloyd A., Vijayakrishnan J., Qureshi M., Broderick P., et al. Allelic variation at the 8q23.3 colorectal cancer risk locus functions as a cis-acting regulator of EIF3H. PLoS. Genet. 2010;6:e1001126. doi: 10.1371/journal.pgen.1001126. doi:10.1371/journal.pgen.1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behrens J. The role of the Wnt signalling pathway in colorectal tumorigenesis. Biochem. Soc. Trans. 2005;33:672–675. doi: 10.1042/BST0330672. doi:10.1042/BST0330672. [DOI] [PubMed] [Google Scholar]
- 15.Wheeler J.M., Kim H.C., Efstathiou J.A., Ilyas M., Mortensen N.J., Bodmer W.F. Hypermethylation of the promoter region of the E-cadherin gene (CDH1) in sporadic and ulcerative colitis associated colorectal cancer. Gut. 2001;48:367–371. doi: 10.1136/gut.48.3.367. doi:10.1136/gut.48.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pittman A.M., Twiss P., Broderick P., Lubbe S., Chandler I., Penegar S., Houlston R.S. The CDH1–160C>A polymorphism is a risk factor for colorectal cancer. Int. J. Cancer. 2009;125:1622–1625. doi: 10.1002/ijc.24542. doi:10.1002/ijc.24542. [DOI] [PubMed] [Google Scholar]
- 17.Li L.C., Chui R.M., Sasaki M., Nakajima K., Perinchery G., Au H.C., Nojima D., Carroll P., Dahiya R. A single nucleotide polymorphism in the E-cadherin gene promoter alters transcriptional activities. Cancer Res. 2000;60:873–876. [PubMed] [Google Scholar]
- 18.Lei H., Sjoberg-Margolin S., Salahshor S., Werelius B., Jandakova E., Hemminki K., Lindblom A., Vorechovsky I. CDH1 mutations are present in both ductal and lobular breast cancer, but promoter allelic variants show no detectable breast cancer risk. Int. J. Cancer. 2002;98:199–204. doi: 10.1002/ijc.10176. doi:10.1002/ijc.10176. [DOI] [PubMed] [Google Scholar]
- 19.Ng P.C., Henikoff S. SIFT- Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. doi:10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. doi:10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett J.C., Lee J.C., Lees C.W., Prescott N.J., Anderson C.A., Phillips A., Wesley E., Parnell K., Zhang H., Drummond H., et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H., et al. Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS. One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. doi:10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomlinson I.P.M., Carvajal-Carmona L.G., Dobbins S.E., Tenesa A., Jones A., Howarth K., Palles C., Broderick P., Jaeger E., Farrington S., et al. The GREM1, BMP4 loci harbor multiple common susceptibility variants for colorectal cancer. PLoS. Gene. 2011 doi: 10.1371/journal.pgen.1002105. doi:10.1371/journal.pgen.1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newcomb P.A., Baron J., Cotterchio M., Gallinger S., Grove J., Haile R., Hall D., Hopper J.L., Jass J., Le Marchand L., et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol. Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. doi:10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 25.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS. Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. doi:10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blankenberg D., Taylor J., Schenck I., He J., Zhang Y., Ghent M., Veeraraghavan N., Albert I., Miller W., Makova K.D., et al. A framework for collaborative analysis of ENCODE data: making large-scale analyses biologist-friendly. Genome Res. 2007;17:960–964. doi: 10.1101/gr.5578007. doi:10.1101/gr.5578007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. Mapping the genetic architecture of gene expression in human liver. PLoS. Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. doi:10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers A.J., Gibbs J.R., Webster J.A., Rohrer K., Zhao A., Marlowe L., Kaleem M., Leung D., Bryden L., Nath P., et al. A survey of genetic human cortical gene expression. Nat. Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. doi:10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 29.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M., et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. doi:10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D., et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. doi:10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veyrieras J.B., Kudaravalli S., Kim S.Y., Dermitzakis E.T., Gilad Y., Stephens M., Pritchard J.K. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS. Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. doi:10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. doi:10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montgomery S.B., Sammeth M., Gutierrez-Arcelus M., Lach R.P., Ingle C., Nisbett J., Guigo R., Dermitzakis E.T. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. doi:10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.