Abstract
While gliomas are the most common primary brain tumors, their etiology is largely unknown. To identify novel risk loci for glioma, we conducted genome-wide association (GWA) analysis of two case–control series from France and Germany (2269 cases and 2500 controls). Pooling these data with previously reported UK and US GWA studies provided data on 4147 glioma cases and 7435 controls genotyped for 424 460 common tagging single-nucleotide polymorphisms. Using these data, we demonstrate two statistically independent associations between glioma and rs11979158 and rs2252586, at 7p11.2 which encompasses the EGFR gene (population-corrected statistics, Pc = 7.72 × 10−8 and 2.09 × 10−8, respectively). Both associations were independent of tumor subtype, and were independent of EGFR amplification, p16INK4a deletion and IDH1 mutation status in tumors; compatible with driver effects of the variants on glioma development. These findings show that variation in 7p11.2 is a determinant of inherited glioma risk.
INTRODUCTION
Gliomas account for ∼40% of all primary brain tumors and are responsible for around 13 000 deaths in the USA each year. Gliomas are heterogeneous and different tumor subtypes defined in part by malignancy grade [e.g. pilocytic astrocytoma WHO grade I, diffuse ‘low grade’ glioma WHO grade II, anaplastic glioma WHO grade III and glioblastoma (GBM) WHO grade IV] can be distinguished. Most gliomas are associated with a poor prognosis irrespective of clinical care, with the most common type of glioma, GBM, having a median overall survival of 10–15 months (1–3).
While glioma subtypes have distinct molecular profiles resulting from different etiological pathways, no lifestyle exposure have, however, consistently been linked to glioma risk except for ionizing radiation, which only accounts for a very small number of cases (4). Direct evidence for inherited predisposition to glioma is provided by a number of rare inherited cancer syndromes, such as Turcot's and Li–Fraumeni syndromes, and neurofibromatosis (4,5). However, collectively, these diseases account for little of the 2-fold increased risk of glioma seen in first-degree relatives of glioma patients (6), and much of the excess familial risk is likely to be a consequence of the co-inheritance of multiple low-risk variants.
To search for genetic risk variants influencing glioma, we have previously conducted genome-wide association (GWA) studies of UK and US glioma patients with replication of the most significantly associated single-nucleotide polymorphisms (SNPs) in independent case–control series from France, Germany and Sweden. This analysis robustly demonstrated that common variants mapping to 5p15.33 (TERT), 8q24.21 (CCDC26), 9p21.3 (CDKN2A/CDKN2B), 20q13.33 (RTEL1) and 11q23.3 (PHLDB1) contribute to heritable risk of glioma (7). Confirmation of the 9p21.3 and 20q13.33 loci as risk factors for GBM was provided by a contemporaneous study (8).
GWA studies are not contingent on prior information concerning candidate genes or pathways, and thereby have the ability to identify important variants in hitherto unstudied genes. However, the effect sizes of individual variants and the need for stringent thresholds for establishing statistical significance inevitably constrain study power. To increase our power to identify novel genetic risk loci for glioma, we have performed GWA scans on augmented series based on the aforementioned French and German cases and combined these data with our GWA scans of US and UK glioma patients. This analysis identifies genetic variation at 7p11.2 as risk factors for the development of glioma and provides further insight into the biological basis of tumor development.
RESULTS
The four GWA studies collectively provided data on 4147 glioma cases and 7435 controls genotyped for 424 460 common tagging SNPs. As the quantile–quantile (Q–Q) plots for the German and US series showed some evidence of inflation [inflation factor λ = 1.16 and 1.11, respectively, based on the 90% least significant SNPs (9)], we corrected for population substructure using principal-components analyses as implemented in Eigenstrat (Supplementary Material, Fig. S1). This analysis reduced all four inflation factors to <1.05.
Meta-analyses of these data resulted in 37 SNPs showing an association with glioma risk at P< 5.0 × 10−7; 35 of these map to the five loci we have previously shown (7) to influence disease risk namely, 5p15.33, 8q24.21, 9p21.3, 20q13.33 and 11q23.3 (Supplementary Material, Table S1). The strongest evidence for a novel association was provided by two SNPs mapping to 7p11.2.
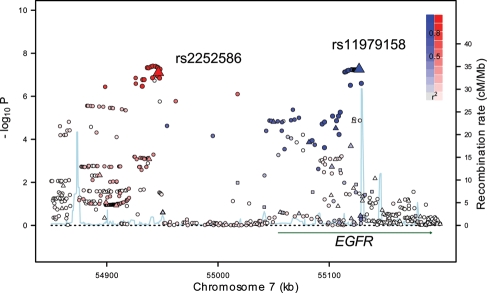
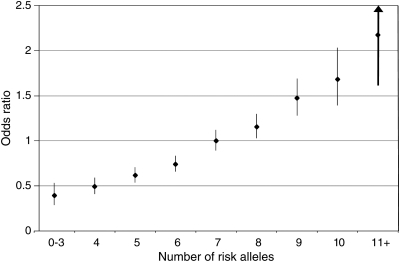
The two SNPs on 7p11.2, rs11979158 and rs2252586, mapping to 55 126 843 and 54 946 418 bp, yielded P-values of 7.03 × 10−8 and 7.89 × 10−8, population-corrected P-values using EIGENSTRAT software, Pc = 7.72 × 10−8 and 2.09 × 10−8, respectively [odds ratio (OR) = 1.23, 95% confidence interval (CI): 1.15–1.35 and OR = 1.18, 95% CI: 1.11–1.25, respectively; Table 1, Fig. 1]. For completeness, we also derived population-corrected P-values using PLINK software (P-values for rs11979158 and rs2252586 were 3.11 × 10−7 and 4.55 × 10−7). Although only one of the associations when corrected for population stratification attained the conventional threshold for genome-wide significance (i.e. P≤5.0 × 10−8), both associations were consistent across the four GWA studies (Phet = 0.44 and 0.27, respectively; Table 1). The SNP rs11979158 maps within intron 1 of the Epidermal Growth Factor Receptor 1 (EGFR) gene and rs2252586 lies 107 kb telomeric from EGFR (Fig. 1). Several lines of evidence support the two SNPs defining independent disease loci. First, there is low linkage disequilibrium (LD) between the two SNPs (D′ = 0.62, r2 = 0.03). Secondly, adjusting rs11979158 through conditional logistic regression for rs2252586 provided evidence of an association (OR = 1.20, 95% CI: 1.10–1.30; P= 1.36 × 10−5). Similarly, adjusting rs2252586 for rs11979158 also provided evidence of an association (OR = 1.14, 95% CI: 1.07–1.22; P= 2.04 × 10−5). Thirdly, there was also an increasing trend in OR with an increasing number of risk alleles (Fig. 2; Supplementary Material, Table S2). To examine the possibility that these SNPs may be correlated with an untyped variant, we made use of HapMap3 and 1000 Genomes data to impute additional variants localizing to 54 850 000–55 200 000 bp region. This analysis provided no SNPs with significantly superior evidence for an association and provides further support for the existence of two independent risk loci at 7p11.2 (Supplementary Material, Fig. S2).
Table 1.
Novel loci with single SNP significance of P < 5.0 × 10−7 for association with glioma on 7p11.2
| Study | Case genotypes |
Control genotypes |
Case MAF | Control MAF | OR (95% CI)* | P-value | Population-corrected P-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Common homozygotes | Heterozygotes | Rare homozygotes | Common homozygotes | Heterozygotes | Rare homozygotes | |||||||
| rs11979158; 55 126 843 bp; minor allele = G; risk allele = A | France | 1091 | 310 | 22 | 858 | 313 | 18 | 0.124 | 0.147 | 1.22 (1.03–1.43) | 0.017 | 6.40 × 10−3 |
| Germany | 635 | 193 | 17 | 893 | 371 | 46 | 0.134 | 0.177 | 1.36 (1.16–1.64) | 2.70 × 10−4 | 7.75 × 10−3 | |
| UK | 471 | 142 | 18 | 1836 | 776 | 85 | 0.141 | 0.175 | 1.28 (1.09–1.54) | 3.75 × 10−3 | 2.75 × 10−3 | |
| USA | 908 | 312 | 27 | 1564 | 601 | 71 | 0.147 | 0.166 | 1.15 (1.01–1.32) | 0.036 | 0.031 | |
| Combined | 1.23 (1.15–1.35) | 7.03 × 10−8 | 7.72 × 10−8 | |||||||||
| rs2252586; 54 946 418 bp; minor allele = T; risk allele = T | France | 601 | 636 | 185 | 580 | 496 | 114 | 0.354 | 0.304 | 1.25 (1.11–1.41) | 1.84 × 10−4 | 7.09 × 10−5 |
| Germany | 366 | 374 | 106 | 652 | 536 | 120 | 0.346 | 0.297 | 1.25 (1.10–1.43) | 7.24 × 10−4 | 8.05 × 10−4 | |
| UK | 285 | 278 | 68 | 1335 | 1131 | 232 | 0.328 | 0.296 | 1.16 (1.02–1.33) | 0.024 | 0.020 | |
| USA | 580 | 551 | 116 | 1091 | 962 | 183 | 0.314 | 0.297 | 1.09 (0.97–1.20) | 0.133 | 0.103 | |
| Combined | 1.18 (1.11–1.25) | 7.89 × 10−8 | 2.09 × 10−8 | |||||||||
*ORs and 95% CI per copy of the risk allele.
Figure 1.
Plot of association results and recombination rates for the 7p11.2 (EGFR) region. –log10P-values (y-axis) of the SNPs are shown according to their chromosomal positions (x-axis). The top two genotyped SNPs, rs11979158 and rs2252586, are labeled. The color intensity of each symbol reflects the extent of LD with the top genotyped SNP; r2 > 0.8 being represented with blue (with rs11979158) and red (with rs2252586) through to white (r2 < 0.2). Genetic recombination rates (cM/Mb), estimated using HapMap CEU samples, are shown with a light blue line. Physical positions are based on build 36 (NCBI) of the human genome. Also shown is the relative position of EGFR.
Figure 2.
Cumulative effect of glioma risk alleles. Plot of increasing ORs for glioma for increasing number of risk alleles. The ORs are relative to the median number of eight risk alleles; vertical bars correspond to 95% CIs. Horizontal line marks the null value (OR = 1.0).
We found no evidence for a pairwise interaction between rs2252586, rs11979158 and the five previously identified risk variants for glioma at 5p15.33 (rs2736100), 8q24.21 (rs4295627), 9p21.3 (rs4977756), 20q13.33 (rs6010620) and 11q23.3 (rs498872) (P> 0.1 after correction for multiple testing; Supplementary Material, Table S3). These data are consistent with each variant having an independent role in defining glioma risk whereby the risk increases with increasing number of variant risk alleles (ORper allele = 1.24, 95% CI: 1.21–1.27, P= 2.89 × 10−72; Supplementary Material, Table S2).
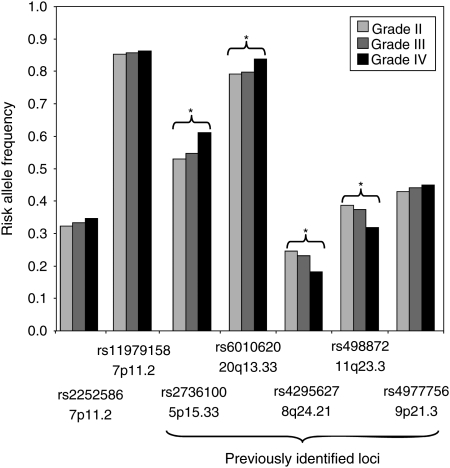
To examine whether the implicated SNPs at 7p11.2 loci were differentially associated with tumor subgroup, we analyzed their prevalence according to histology across all cases where such data were available (n= 4002). The carrier frequencies of the risk alleles for the 7p11.2 SNPs showed no correlation with stratification of tumors by WHO grade in any of the four patient cohorts (Supplementary Material, Table S4). These observations contrast with the strong relationship between genotype and histology seen for 5p15.33 (TERT; rs2736100), 8q24.21 (CCDC26; rs4295627), 20q13.33 (RTEL1; rs6010620) and 11q23.3 (PHLDB1; rs498872) risk variants (Fig. 3).
Figure 3.
Relationship between 7p11.2 genotype and WHO tumor grade. Also shown are the relationships between histology and 5p15.33 (rs2736100), 8q24.21 (rs4295627), 9p21.3 (rs4977756), 20q13.33 (rs6010620) and 11q23.3 (rs498872) genotypes. *Significant association at P< 0.05.
Primary and secondary forms of GBM are recognized; with secondary GBM developing through progression from low-grade diffuse astrocytomas or anaplastic astrocytomas. While usually indistinguishable histologically, distinct molecular pathways characterize the primary and secondary forms. Notably, IDH1 mutations are commonly detectable in low-grade glioma and secondary GBM (>70% of cases) but are rare in primary GBMs (10). In addition, EGFR amplification and p16INK4a deletion are more frequent in primary disease (11). Tumor DNAs were available for a subset of the French cases (n = 761) permitting us to examine the relationship between SNP genotype and EGFR amplification, p16INK4a deletion and IDH1 mutation status in glioma. For both 7p11.2 SNPs, no association was shown with EGFR amplification, p16INK4a deletion or IDH1 mutation (Supplementary Material, Table S5) contrasting with the associations seen between rs2736100, rs4295627, rs498872 and IDH1 status as well as rs4295627and p16INK4a status (Supplementary Material, Table S5).
To explore the possibility that 7p11.2 SNP genotype may influence tumor progression, we examined the relationship between genotype and patient outcome in both the French (n= 1126) and German (n= 614) case series. Survival analysis stratified by histology and adjusting for age at diagnosis, sex, preoperative Karnofsky performance index (KPI), degree of resection, chemotherapy and radiotherapy provided no evidence for an independent relationship between rs11979158 or rs2252586 SNP genotype and overall survival within each of the histological categories in either of patient cohort (Supplementary Material, Table S6).
DISCUSSION
This analysis has shown that the risk of developing glioma is influenced by genetic variation at 7p11.2. Our data support a previous report of an association between rs11979158 and GBM risk based on analysis of a smaller data set (12). These associations are a priori biologically plausible, since EGFR is the prototypical member of the ErbB/EGFR family, which is involved in multiple cellular processes, including cell division, migration, adhesion, differentiation and apoptosis. EGFR is well established to be pivotal in both initiation of primary GBM and progression of lower-grade glioma to grade IV (13); hence genetic variation in EGFR is an attractive basis for predisposition to glioma. Furthermore, the Cancer Genome Atlas (TCGA) consortium reported EGFR as the fourth most highly mutated gene in a compendium of common cancer genes sequenced in GBM (14). While EGFR amplification characterizes 40–70% of primary GBM, it is rarely seen in lower-grade astrocytomas, thereby implicating EGFR activation as a driver of glial tumorigenesis (13). There are multiple mechanisms by which EGFR mediates tumor initiation and progression, all of which are seen in primary glioma. Increased EGFR is common in primary GBM as a consequence of gene amplification and is often associated with exon 2–7 truncation resulting in constitutive receptor activation (EGFRVIII), or less frequently, an activating missense mutation of the EGFR extracellular domain (15–17). While speculative at this juncture, it is entirely plausible that the influence of SNP-mediated risk on glioma development is a consequence of a subtle effect on EGFR expression, thereby having the potential to impact on all histological forms of glioma.
We have previously shown that the TERT and RTEL1 risk variants are primarily associated with high-grade disease and CCDC26 and PHLDB1 with the development of less aggressive glial tumors (18). Moreover, we have now shown that there are differences in the molecular phenotype with respect to p16INK4a and IDH1 status for some of these associations. In contrast, CDKN2A/CDKN2B variation appears independent of the expression of a malignant phenotype, compatible with a driver impact on tumorigenesis (18). Here we have implicated variation at EGFR as risk factors for all forms of glioma which is also reflected in no differences in molecular phenotype as defined by EGFR, p16INK4a or IDH1 status. While these risk variants do not impact on disease outcome, they provide evidence that the glioma subtypes result from different etiological pathways.
As the SNPs genotyped during GWA studies are generally not themselves strong candidates for causality, enumeration of the causal variant at 7p11.2 will involve fine mapping and functional analyses to elucidate the causal basis of the association. While partly speculative as these SNPs are not correlated with known polymorphisms in the coding sequence of EGFR, the glioma association may be mediated through a change in gene expression rather than a sequence change in the expressed protein or through LD with low-frequency variants not catalogued. Although the risk of glioma associated with the 7p11.2 SNPs is modest, the carrier frequency of these risk alleles is high in the European population and therefore, irrespective of the causal basis of the association, these loci make a substantial contribution to the overall development of glioma.
In conclusion, this large study provides unambiguous evidence that common genetic variation in 7p11.2 influences the risk of developing glioma. Furthermore, our findings provide support for the notion that the glioma subtypes result from different etiologic pathways, rather than different stages of tumor evolution within a common carcinogenic pathway. The impact of the currently identified SNPs on glioma risk is small in isolation and hence individually they do not have immediate clinical application. However, the observed differences provide insight into the biological mechanisms that underscore inherited susceptibility to glioma.
MATERIALS AND METHODS
Ethics
Collection of blood samples and clinico-pathological information from patients and controls was undertaken with informed consent and relevant ethical review board approval in accordance with the tenets of the Declaration of Helsinki. Ethical committee approval for this study was obtained from relevant study centers [UK: South East Multicentre Research Ethics Committee (MREC) and the Scottish Multicentre Research Ethics Committee; France: APHP ethical committee-CPP (comité de Protection des Personnes); Germany: Ethics Commission of the Medical Faculty of the University of Bonn and USA: University of Texas MD Anderson Cancer Institutional Review Board].
Subjects
The present study is based on pooling data from GWA studies of UK, US, French and German case–control series. The UK and US GWA studies have been the subject of a previous publication (7). Briefly, the UK study was based on 636 cases (401 male; mean age 46 years) ascertained through the INTERPHONE Study (19). Individuals from the 1958 Birth Cohort served as a source of controls. The US study was based on 1281 cases (786 male; mean age 47 years) ascertained through the MD Anderson Cancer Center, Texas, between 1990 and 2008. Individuals from the Cancer Genetic Markers of Susceptibility (CGEMS) studies served as controls (20,21). The French GWA study comprised 1495 patients with glioma ascertained through the Service de Neurologie Mazarin, Groupe Hospitalier Pitié-Salpêtrière Paris. The controls were ascertained from the SU.VI.MAX (SUpplementation en VItamines et MinerauxAntioXydants) study of 12 735 healthy subjects (women aged 35–60 years; men aged 45–60 years) (22). The German GWA study comprised 880 patients who underwent surgery for a glioma at the Department of Neurosurgery, University of Bonn Medical Center, between 1996 and 2008. Control subjects were taken from three publicly available studies: KORA (Co-operative Health Research in the Region of Augsburg; n = 488) (23,24), POPGEN (Population Genetic Cohort; n = 678) (25) and from the Heinz Nixdorf Recall study (n = 380) (26).
Genotyping and molecular analysis
Genotyping of cases were conducted using Illumina Infinium HD Human610-Quad BeadChips according to the manufacturer's protocols (Illumina, San Diego, CA, USA; Supplementary Material S1). Descriptions of genotyping are included in Supplementary Material S1. To determine amplification, p16INK4a deletion and IDH1 mutation status in gliomas, DNA was extracted from frozen tumors using a standard protocol. CGH-array analysis, EGFR amplification, p16INK4a deletion assessment and sequencing of IDH1 were performed as previously described (2,27).
Statistical analysis
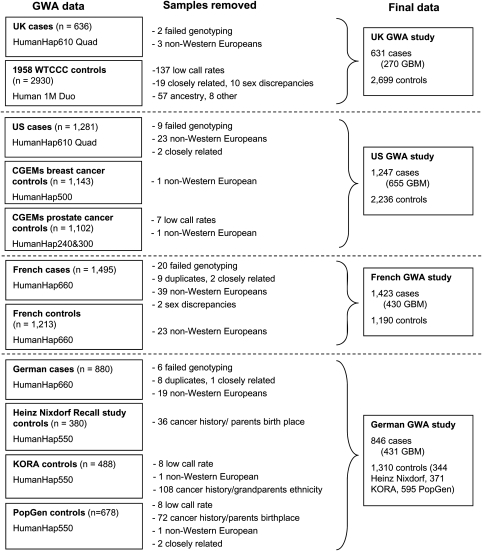
Genotype data from each of the four studies were filtered on the basis of pre-specified quality-control measures (Supplementary Material, Text S1). Individual SNPs were excluded from further analysis if they showed deviation from the Hardy–Weinberg equilibrium with a P-value of <1 × 10−5, an individual SNP genotype yield of <98% or a minor allele frequency of <5%. This filtering resulted in the use of 424 460 SNPs, common to the four case–control series. A total of 363 samples were removed during quality-control steps for reasons including a failure to genotype, unknown duplicates, closely related individuals and sex discrepancies (Fig. 4). Furthermore, for the UK control group, we made the exclusions as proposed by the Wellcome Trust Case Control Consortium (WTCCC; n= 65). For the other controls, we removed people with a history of cancer as well as those whose parents/grandparents were not born in the study regions (n = 216; Fig. 4). We then performed principal-components analyses to identify outlier samples to reduce the effects of population stratification (Supplementary Material, Fig. S1). These analyses resulted in the removal of 644 samples (145 cases and 499 controls), resulting in a total of 4147 patients and 7435 control subjects (Fig. 4).
Figure 4.
Patient exclusion schema for the genome-wide studies.
The association between each SNP and risk of glioma was assessed by the Cochran–Armitage trend test. ORs and associated 95% CIs were calculated by unconditional logistic regression using R software. Relationships between multiple SNPs showing association with glioma risk in the same region were investigated using logistic regression analysis, and the impact of additional SNPs from the same region was assessed by a likelihood ratio test.
To further control for the potentially confounding influence of population stratification, we utilized the program Eigenstrat (28). The Eigenstrat algorithm adjusts genotypes and phenotypes by amounts attributable to ancestry using the calculated principal components of variation. These adjustments were applied to each of the four GWA studies correcting for the top 10 principal components of variation. Figure S3 (Supplementary Material) shows Q–Q plots before and after correction. Adjusted data from each of the four data sets were used to perform a meta-analysis using a modified Cochran–Armitage trend test. The results are reported as Eigenstrat P-values (Pc). The Eigenstrat algorithm involves the adjustment of genotypes and phenotypes by amounts attributable to ancestry using the calculated axes of variation. We also corrected for population structure using PLINK software (http://pngu.mgh.harvard.edu/~purcell/plink/) which implements a logistic regression analysis using 10 principal components as covariates.
Prediction of the untyped SNPs was carried out using IMPUTEv2, based on HapMapIII Release27 (February 2009, NCBI B36, dbSNP26) and the 1000 Genomes Project. Imputed data were analyzed using SNPTESTv2 to account for uncertainties in SNP prediction. LD metrics between HapMap SNPs were based on HapMapIII Release27, viewed using Haploview (v4.2) and plotted using SNAP.
Of the 4147 patients, clinical (age at diagnosis, sex, preoperative KPI , degree of resection, chemotherapy and radiotherapy) and histological covariate data obtained at diagnosis were available for most. Complete survival data were available for 1740 patients (77%) in the French and German cohorts, with a median follow-up interval of 8.9 years for patients without an event. Association analyses of 7p11.2 SNPs with clinical characteristics were performed with the χ2 test on allele and genotype counts. Association with overall survival was performed by comparing the Kaplan–Meier survival curves by means of the log-rank test in a pairwise fashion.
SUPPLEMENTARY MATERIAL
FUNDING
In the UK, funding was provided by the Wellcome Trust and Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund) and the DJ Fielding Medical Research Trust. The UK INTERPHONE study was supported by the European Union Fifth Framework Program ‘Quality of life and Management of Living Resources' (QLK4-CT-1999-01563) and the International Union against Cancer (UICC). The UICC received funds from the Mobile Manufacturers’ Forum and GSM Association. Provision of funds via the UICC was governed by agreements that guaranteed INTERPHONE's scientific independence (http://www.iarc.fr/ENG/Units/RCAd.html). The UK centers were also supported by the Mobile Telecommunications and Health Research (MTHR) Programme and the Northern UK Centre was supported by the Health and Safety Executive, Department of Health and Safety Executive and the UK Network Operators. In the US, funding was provided by NIH grants 5R01(CA119215&5R01 CA070917). Support was also obtained from the American Brain Tumor Association and the National Brain Tumor Society. In France, funding was provided by the Délégation à la Recherche Clinique (MUL03012), the Association pour la Recherche sur les Tumeurs Cérébrales (ARTC), the Institut National du Cancer (INCa;PL046) and the French Ministry of Higher Education and Research. In Germany, funding was provided to M.S. and J.S. by the Deutsche Forschungsgemeinschaft (Si552,Schr285), the Deutsche Krebshilfe (70-2385-Wi2,70-3163-Wi3,10-6262) and BONFOR. Funding for the WTCCC was provided by the Wellcome Trust (076113&085475). The KORA Augsburg studies are supported by grants from the German Federal Ministry of Education and Research (BMBF) and were mainly financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg. This work was financed by the German National Genome Research Network (NGFN) and supported within the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to all the patients and individuals for their participation and we would also like to thank the clinicians and other hospital staff, cancer registries and study staff in respective centers who contributed to the blood sample and data collection. For the UK GWA study, we acknowledge the participation of the clinicians and other hospital staff, cancer registries, study staff and funders who contributed to the blood sample and data collection for this study as listed in Hepworth et al. (BMJ 2006, 332, 883). MD Anderson acknowledges the work on the USA GWA study of Phyllis Adatto, Fabian Morice, Hui Zhang, Victor Levin, Alfred W.K. Yung, Mark Gilbert, Raymond Sawaya, Vinay Puduvalli, Charles Conrad, Fredrick Lang and Jeffrey Weinberg from the Brain and Spine Center. For the German GWA study, we are indebted to B. Harzheim (Bonn) and Dr A. Müller-Erkwoh (Bonn) for help with the acquisition of clinical data and R. Mahlberg (Bonn) who provided technical support. The UK GWA study made use of control genotyping data generated by the Wellcome Trust Case–Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. The US GWA study made use of control genotypes from the CGEMS prostate and breast cancer studies. A full list of the investigators who contributed to the generation of the data is available from http://cgems.cancer.gov/. French controls were taken from the SU.VI.MAX study (22). The German GWA study made use of genotyping data from three population control sources: KORA-gen (23,24), The Heinz-Nixdorf RECALL study (26) and POPGEN (25). We are extremely grateful to all investigators who contributed to the generation of these data sets.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Linnebank M., Semmler A., Moskau S., Smulders Y., Blom H., Simon M. The methylenetetrahydrofolate reductase (MTHFR) variant c.677C>T (A222V) influences overall survival of patients with glioblastoma multiforme. Neuro Oncol. 2008;10:548–552. doi: 10.1215/15228517-2008-020. doi:10.1215/15228517-2008-020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanson M., Marie Y., Paris S., Idbaih A., Laffaire J., Ducray F., El Hallani S., Boisselier B., Mokhtari K., Hoang-Xuan K., et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J. Clin. Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. doi:10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 3.Weller M., Felsberg J., Hartmann C., Berger H., Steinbach J.P., Schramm J., Westphal M., Schackert G., Simon M., Tonn J.C., et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J. Clin. Oncol. 2009;27:5743–5750. doi: 10.1200/JCO.2009.23.0805. doi:10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 4.Bondy M.L., Scheurer M.E., Malmer B., Barnholtz-Sloan J.S., Davis F.G., Il'yasova D., Kruchko C., McCarthy B.J., Rajaraman P., Schwartzbaum J.A., et al. Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008;113:1953–1968. doi: 10.1002/cncr.23741. doi:10.1002/cncr.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodgson S., Maher E. A Practical Guide to Human Cancer Genetics. 2nd edn. Cambridge, England: Cambridge University Press; 1999. pp. 37–38. [Google Scholar]
- 6.Hemminki K., Tretli S., Sundquist J., Johannesen T.B., Granstrom C. Familial risks in nervous-system tumours: a histology-specific analysis from Sweden and Norway. Lancet Oncol. 2009;10:481–488. doi: 10.1016/S1470-2045(09)70076-2. doi:10.1016/S1470-2045(09)70076-2. [DOI] [PubMed] [Google Scholar]
- 7.Shete S., Hosking F.J., Robertson L.B., Dobbins S.E., Sanson M., Malmer B., Simon M., Marie Y., Boisselier B., Delattre J.Y., et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 2009;41:899–904. doi: 10.1038/ng.407. doi:10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrensch M., Jenkins R.B., Chang J.S., Yeh R.F., Xiao Y., Decker P.A., Ballman K.V., Berger M., Buckner J.C., Chang S., et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat. Genet. 2009;41:905–908. doi: 10.1038/ng.408. doi:10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton D.G., Walker N.M., Smyth D.J., Pask R., Cooper J.D., Maier L.M., Smink L.J., Lam A.C., Ovington N.R., Stevens H.E., et al. Population structure, differential bias and genomic control in a large-scale, case–control association study. Nat. Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. doi:10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 10.Nobusawa S., Watanabe T., Kleihues P., Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin. Cancer Res. 2009;15:6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. doi:10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 11.Ohgaki H., Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am. J. Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. doi:10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartzbaum J.A., Xiao Y., Liu Y., Tsavachidis S., Berger M.S., Bondy M.L., Chang J.S., Chang S.M., Decker P.A., Ding B., et al. nherited variation in immune genes and pathways and glioblastoma risk. Carcinogenesis. 2010;31:1770–1777. doi: 10.1093/carcin/bgq152. doi:10.1093/carcin/bgq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang P.H., Xu A.M., White F.M. Oncogenic EGFR signaling networks in glioma. Sci. Signal. 2009;2:re6. doi: 10.1126/scisignal.287re6. doi:10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gan H.K., Kaye A.H., Luwor R.B. The EGFRvIII variant in glioblastoma multiforme. J. Clin. Neurosci. 2009;16:748–754. doi: 10.1016/j.jocn.2008.12.005. doi:10.1016/j.jocn.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Idbaih A., Aimard J., Boisselier B., Marie Y., Paris S., Criniere E., Carvalho Silva R., Laigle-Donadey F., Rousseau A., Mokhtari K., et al. Epidermal growth factor receptor extracellular domain mutations in primary glioblastoma. Neuropathol Appl Neurobiol. 2009;35:208–213. doi: 10.1111/j.1365-2990.2008.00977.x. doi:10.1111/j.1365-2990.2008.00977.x. [DOI] [PubMed] [Google Scholar]
- 17.Lee J.C., Vivanco I., Beroukhim R., Huang J.H., Feng W.L., DeBiasi R.M., Yoshimoto K., King J.C., Nghiemphu P., Yuza Y., et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3:e485. doi: 10.1371/journal.pmed.0030485. doi:10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simon M., Hosking F.J., Marie Y., Gousias K., Boisselier B., Carpentier C., Schramm J., Mokhtari K., Hoang-Xuan K., Idbaih A., et al. Genetic risk profiles identify different molecular etiologies for glioma. Clin. Cancer Res. 2010;16:5252–5259. doi: 10.1158/1078-0432.CCR-10-1502. doi:10.1158/1078-0432.CCR-10-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardis E., Richardson L., Deltour I., Armstrong B., Feychting M., Johansen C., Kilkenny M., McKinney P., Modan B., Sadetzki S., et al. The INTERPHONE study: design, epidemiological methods, and description of the study population. Eur. J. Epidemiol. 2007;22:647–664. doi: 10.1007/s10654-007-9152-z. doi:10.1007/s10654-007-9152-z. [DOI] [PubMed] [Google Scholar]
- 20.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. doi:10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeager M., Orr N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. doi:10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 22.Hercberg S., Galan P., Preziosi P., Bertrais S., Mennen L., Malvy D., Roussel A.M., Favier A., Briancon S. The SU.VI.MAX study: a randomized, placebo-controlled trial of the health effects of antioxidant vitamins and minerals. Arch. Intern. Med. 2004;164:2335–2342. doi: 10.1001/archinte.164.21.2335. doi:10.1001/archinte.164.21.2335. [DOI] [PubMed] [Google Scholar]
- 23.Holle R., Happich M., Lowel H., Wichmann H.E. KORA—a research platform for population based health research. Gesundheitswesen. 2005;67(Suppl. 1):S19–S25. doi: 10.1055/s-2005-858235. doi:10.1055/s-2005-858235. [DOI] [PubMed] [Google Scholar]
- 24.Wichmann H.E., Gieger C., Illig T. KORA-gen—resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl. 1):S26–S30. doi: 10.1055/s-2005-858226. doi:10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 25.Krawczak M., Nikolaus S., von Eberstein H., Croucher P.J., El Mokhtari N.E., Schreiber S. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. doi:10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 26.Schmermund A., Mohlenkamp S., Stang A., Gronemeyer D., Seibel R., Hirche H., Mann K., Siffert W., Lauterbach K., Siegrist J., et al. Assessment of clinically silent atherosclerotic disease and established and novel risk factors for predicting myocardial infarction and cardiac death in healthy middle-aged subjects: rationale and design of the Heinz Nixdorf RECALL Study. Risk Factors, Evaluation of Coronary Calcium and Lifestyle. Am. Heart J. 2002;144:212–218. doi: 10.1067/mhj.2002.123579. doi:10.1067/mhj.2002.123579. [DOI] [PubMed] [Google Scholar]
- 27.Idbaih A., Marie Y., Lucchesi C., Pierron G., Manie E., Raynal V., Mosseri V., Hoang-Xuan K., Kujas M., Brito I., et al. BAC array CGH distinguishes mutually exclusive alterations that define clinicogenetic subtypes of gliomas. Int. J. Cancer. 2008;122:1778–1786. doi: 10.1002/ijc.23270. doi:10.1002/ijc.23270. [DOI] [PubMed] [Google Scholar]
- 28.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi:10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.