Abstract
Huntington's disease is initiated by the expression of a CAG repeat-encoded polyglutamine region in full-length huntingtin, with dominant effects that vary continuously with CAG size. The mechanism could involve a simple gain of function or a more complex gain of function coupled to a loss of function (e.g. dominant negative-graded loss of function). To distinguish these alternatives, we compared genome-wide gene expression changes correlated with CAG size across an allelic series of heterozygous CAG knock-in mouse embryonic stem (ES) cell lines (HdhQ20/7, HdhQ50/7, HdhQ91/7, HdhQ111/7), to genes differentially expressed between Hdhex4/5/ex4/5 huntingtin null and wild-type (HdhQ7/7) parental ES cells. The set of 73 genes whose expression varied continuously with CAG length had minimal overlap with the 754-member huntingtin-null gene set but the two were not completely unconnected. Rather, the 172 CAG length-correlated pathways and 238 huntingtin-null significant pathways clustered into 13 shared categories at the network level. A closer examination of the energy metabolism and the lipid/sterol/lipoprotein metabolism categories revealed that CAG length-correlated genes and huntingtin-null-altered genes either were different members of the same pathways or were in unique, but interconnected pathways. Thus, varying the polyglutamine size in full-length huntingtin produced gene expression changes that were distinct from, but related to, the effects of lack of huntingtin. These findings support a simple gain-of-function mechanism acting through a property of the full-length huntingtin protein and point to CAG-correlative approaches to discover its effects. Moreover, for therapeutic strategies based on huntingtin suppression, our data highlight processes that may be more sensitive to the disease trigger than to decreased huntingtin levels.
INTRODUCTION
Huntington's disease (HD) features degeneration, especially of the striatum, along with motor, cognitive and psychiatric symptoms (1). All cases of HD are caused by an expanded CAG trinucleotide repeat in one copy of the 4p16.3 HD gene (2). In humans, the CAG alleles span a continuum from fully penetrant HD alleles (≥40 CAGs), which lead to HD pathology and clinical symptoms, through alleles with progressively reduced penetrance that can often (36–39 CAGs) (2–4), or only very rarely (27–35 CAGs), result in overt HD (5), to the shorter, more frequent, ‘normal’ alleles at the lower end of the distribution (6–26 CAGs), for which no HD case has been reported (6).
The dominant effects of endogenous CAG repeats are graded with the allele size. In the fully penetrant range, CAG length is inversely correlated with the age at onset of overt HD symptoms in humans (2,7,8) and with the onset of striatal phenotypes in heterozygote Huntington's disease homolog (Hdh) CAG repeat knock-in mice (made to replicate the human CAG spectrum) (9–16). Graded effects of the polymorphic CAG stretch are also evident through the reduced penetrance HD range, for example, in sensitizing major depressive disorder (17), as well as in the normal range, as part of a continuous relationship of CAG size with ATP/ADP levels in human lymphoblastoid cells (18). Thus, the CAG repeat is a functional polymorphism, specifying a polyglutamine region in the full-length 3144 amino acid huntingtin protein (2), such that most humans express full-length huntingtins with polyglutamine repeats of different sizes. This large alpha-helical HEAT domain protein (19–22) is thought to act as a mechanical facilitator of multi-subunit complexes (21), as suggested for other predominantly HEAT proteins (23).
The preponderance of evidence argues that the polyglutamine segment modulates the full-length huntingtin in a manner most consistent with a simple gain of a novel function, rather than either a simple or a dominant-negative loss of function. Chiefly, huntingtin deficiency fails to mimic CAG expansion. Individuals with a balanced chromosomal translocation inactivating one HD allele express 50% of normal levels of huntingtin without exhibiting HD symptoms (24). In mice, more severe decreases in full-length huntingtin levels, below 50%, produce developmental defects and embryonic lethality (9,25–28). Yet HD homozygote humans (29,30) and knock-in mice (9,13,15,16,31) do exist, despite expressing no normal-range full-length huntingtin. There is mounting evidence that the polyglutamine repeat, though not required for basal activity (32), enhances, rather than impairs, the full-length huntingtin function in repressing energy metabolism (18,32) and in stimulating the multi-subunit enzyme, polycomb repressive complex 2 (21).
However, some findings from studies on CAG knock-in and full-length HD transgenic mouse systems seem more consistent with a dominant-negative effect or a combination of gain of function and loss of function. These include the failure of the full-length mutant huntingtin to exhibit functional activities attributed to the normal protein, such as in NRSE/REST target gene transcription (33), as well as SREBP (34–36) and LXR (37) regulated transcription of genes involved in cholesterol and lipid metabolism that lead to decreased biosynthetic intermediates. The alternate to the simple gain-of-function hypothesis is that the polyglutamine repeat confers a novel property on the full-length huntingtin that produces, or combines with, a loss of huntingtin function (37–40).
To more fully understand the molecular response to the impact of the polyglutamine region on the full-length huntingtin, and to reconcile seemingly conflicting evidence, we have created an isogenic allelic panel of heterozygous CAG knock-in mouse embryonic stem (ES) cells, augmenting an existing wild-type and huntingtin-null knock-out ES cell panel. With this new resource, we have conducted an unbiased genome-wide search for dominant CAG length-dependent gene expression changes and tested the validity of predictions that distinguish the alternate hypotheses: CAG-length-dependent gene changes would either be novel (simple gain of function) or overlap with the molecular effects of absent huntingtin (dominant-negative loss of function or combined gain and loss of function). The results of our analyses distinguish these alternatives and offer insights into the biology of the full-length huntingtin, as revealed by structure–function and deficiency analyses.
RESULTS
An isogenic panel of heterozygous CAG knock-in ES cell lines
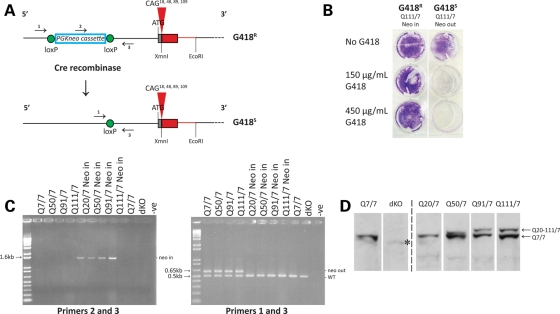
To enable a discovery strategy based on the continuous relationship between CAG size and phenotype, we generated an allelic series of heterozygous Hdh CAG knock-in mouse ES cell lines, which would appropriately express full-length huntingtins from endogenous alleles. This series was produced from a matching series of targeted ‘PGKneo-in’ CAG exon 1 knock-in (CAG 18, CAG 48, CAG 89 and CAG 109) ES cell lines (14,27). As the ‘neo-in’ alleles express decreased levels of full-length huntingtin, due to the floxed PGKneo cassette in the promotor region (9,27), the selection cassette (conferring G418 resistance) was removed by Cre-recombinase-mediated excision (Fig. 1A) and multiple G418-sensitive ES subclones were identified for each line (Fig. 1B) (Materials and Methods). The results of polymerase chain reaction (PCR) amplification assays with genomic DNAs confirmed proper PGKneo excision (Fig. 1C), and DNA sequence analysis confirmed the size of the CAG repeat (data not shown). Immunoblot analysis revealed the ∼350 kDa band of the wild-type full-length huntingtin with seven glutamines and, with progressively decreased mobility, a band of full-length huntingtin with a polyglutamine region from the normal human range (Q20), the adult-onset (Q50) and the juvenile-onset (Q91 and Q111) HD range (Fig. 1D). As expected, the wild-type parental ES cells expressed only the 7-glutamine full-length huntingtin and no huntingtin band was detected in the extract of double knock-out (null) Hdhex4/5/ex4/5 ES cells (25) (Fig. 1D).
Figure 1.
Generation of an allelic panel of heterozygous CAG knock-in ES cell lines. (A) The upper schematic depicts the targeted Hdh CAG knock-in allele in heterozygous Hdhneo20/7, Hdhneo50/7, Hdhneo91/7 and Hdhneo111/7 ES cell lines, with the location of the loxP-flanked PGKneo selection cassette in the promotor region upstream of the chimeric mouse (gray)/human (red) exon 1 with different CAG repeat sizes (CAG 18, CAG 48, CAG 89 and CAG 109), encoding adjacent CAA, CAG codons, the polyglutamine repeat in the full-length endogenous huntingtin. The corresponding PGKneo-out-targeted allele in the cognate Hdh20/7, Hdh50/7, Hdh91/7 and Hdh111/7 ES cell subclones created by Cre-recombinase-mediated excision is depicted below. Arrows denote the locations of primer sets for ‘neo-in’ (2 and 3) and ‘neo-out’ (1 and 3) PCR amplification assays. The schematic is not drawn to scale. The wild-type allele is not shown. (B) Trypan blue staining reveals the growth of G418-resistant (G418R) Hdhneo111/7 (Q111 neo-in) ES cells and the lack of growth of a subclone of G418-sensitive (G418S) Hdh111/7 (Q111 neo-out) ES cells, derived by Cre-recombinase excision of the pGKneo selection cassette. (C) Agarose gel analysis of PCR amplification products generated by specific PGKneo-in (left gel) and PGKneo-out (right gel) assays. The expected bands confirmed the presence of the PGKneo cassette in the parental Hdhneo20/7, Hdhneo50/7, Hdhneo91/7and Hdhneo111/7 ES cell genomic DNAs (lanes left gel) and the proper removal of the PGKneo cassette from the targeted allele in the cognate neo-out Hdh20/7, Hdh50/7, Hdh91/7 and Hdh111/7 ES cell DNAs (lanes on the left gel). The latter assay also amplifies the expected product from the wild-type allele present in all of the ES cell lines, including the Hdhex4/5/ex4/5 ES cell genomic DNA (dKO), which harbor alleles with a targeted inactivating mutation replacing/deleting exons 4 and 5 (data not shown). PCR products were not detected in genomic DNA minus lanes (−ve) for each assay. (D) The immunoblot, detected with mAb2166, confirms proper expression of both full-length wild-type huntingtin (7 glutamines) and, migrating more slowly, huntingtins from the targeted allele with distinct polyglutamine tracts (Q20, Q50, Q91 and Q111) in protein extracts of Hdh20/7, Hdh50/7, Hdh91/7 and Hdh111/7 ES cells, respectively. Wild-type ES cells (Q7/7) express full-length 7-glutamine huntingtin from both alleles and Hdhex4/5/Hdhex4/5-null ES cells (dKO) express no full-length huntingtin. mAb2166 and other anti-huntingtin antibodies differentially detect the huntingtin with the shorter polyglutamine tract, compared with those with the longer tracts, for reasons that are not yet understood. Bands are ∼350 kDa (the position of the 250 kDa marker is not depicted on this immunoblot). An asterisk indicates a cross-reacting band, previously noted (25).
The entire allelic panel comprises three to four heterozygous CAG knock-in ES cell subclones for each repeat length, and subclones of wild-type ES cells and double knock-out Hdhex4/5/ex4/5 ES cells, that were chosen based upon morphological, developmental and molecular criteria. The chosen subclones displayed an appropriate stem cell morphology (Supplementary Material, Fig. S1A), the expression of stem cell marker mRNAs (Supplementary Material, Fig. S1B), the ability to develop into embryoid bodies (Supplementary Material, Fig. S1C) and the expression of germ-layer marker mRNAs (Supplementary Material, Fig. S1D).
The members of this heterozygous Hdh CAG knock-in ES cell panel, which share an 129Sv genetic background with the wild-type HdhQ7/7 and huntingtin-null Hdhex4/5/ex4/5 ES cell lines, are named HdhQ20/7, HdhQ50/7, HdhQ91/7 and HdhQ111/7 to denote the polyglutamine region, encoded by the pure CAG tract and adjacent CAA, CAG codons, in the endogenous full-length huntingtin proteins expressed from the wild-type and targeted CAG alleles.
ATP/ADP ratio decreased with increased CAG length
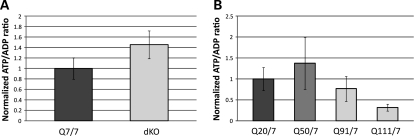
The ATP/ADP ratio, a dominant quantitative phenotype known to vary with CAG size in human lymphoblastoid cells (18), was measured in all members of the ES cell panel, to assess the utility of this new resource for evaluating dominant CAG-correlated effects. Results of the HPLC analysis demonstrated that huntingtin-null Hdhex4/5/ex4/5 cells showed a trend toward higher ATP/ADP ratios than the wild-type HdhQ7/7 ES cells (Fig. 2A), consistent with the proposal that the full-length huntingtin negatively regulates this energy measure (18,32). In contrast, across the HdhQ20/7, HdhQ50/7, HdhQ91/7 and HdhQ111/7 ES cell panel, ATP/ADP ratios decreased concomitantly (Pearson's correlation test, P = 0.018) with the increased size of the longer CAG allele (Fig. 2B), confirming that lengthening the polyglutamine region in full-length huntingtin expressed from one allele enhanced the negative regulation of this energy measure, as reported previously (18,32).
Figure 2.
ATP/ADP ratio across the members of the ES cell panel. (A) The bar plot summarizes the results of HPLC determination of ATP/ADP ratio for replicates of Hdhex4/5/Hdhex4/5-null ES cells (dKO), expressed as a percentage of the parental wild-type ES cell ATP/ADP ratio, showing a trend (P = 0.057) toward increased ATP/ADP ratio in the absence of full-length huntingtin. All data points are included. (B) The bar plot summarizes the results of HPLC determination of ATP/ADP ratio for two biological replicates performed in duplicate for each member of the Hdh20/7, Hdh50/7, Hdh91/7 and Hdh111/7 knock-in ES series, expressed as a percentage of the Hdh20/7 ATP/ADP ratio, demonstrating a decrease with increasing CAG repeat length (Pearson's correlation coefficient P = 0.018). All data points are included.
In demonstrating the utility of the allelic ES cell panel for assessing a candidate CAG length-dependent phenotype, these results also implied that comprehensive phenotyping across the members of the panel would permit the discovery of new dominant CAG length-dependent responses.
Continuous analysis effectively identified changes correlated with CAG size
As an unbiased discovery approach, we utilized Affymetrix Mouse 430 2.0 gene chips to generate genome-wide gene expression datasets from RNA isolated from three independent subclones for each of the HdhQ20/7, HdhQ50/7, HdhQ91/7 and HdhQ111/7 ES cell genotypes (12 samples), as well as six wild-type HdhQ7/7 and six null Hdhex4/5/ex4/5 ES cell replicates (12 samples). The raw data sets were subject to standard quality control filtering steps (Materials and Methods).
The 12 CAG knock-in ES cell data sets were then analyzed to identify probes/genes with expression altered progressively with the CAG repeat size. As the CAG length is a single source of variation expected to produce subtle incremental changes, we reasoned that a continuous analytic strategy should effectively identify such alterations from among the gene expression changes due to all other factors (technical, biological and chance), whereas the standard dichotomous comparison (normal range versus HD range) would be expected to highlight any probes with altered expression, regardless of the cause.
Continuous analysis to uncover graded differences in signal intensity, with increased CAG size, was conducted by comparing the probe intensities from the HdhQ20/7, HdhQ50/7, HdhQ91/7 and HdhQ111/7 data sets with the CAG length of the corresponding cell lines using Pearson's correlation analysis. A total of 73 probes (Supplementary Material, Table S1) passed a rigorous statistical cut-off (absolute correlation coefficient > 0.8; P< 0.001). Of these, 48 probes were negatively correlated and 25 probes were positively correlated with the length of the CAG allele. Validation of a subset of these genes, by RT Q-PCR assay, verified that the RNA levels of the majority were significantly altered in the predicted direction (Supplementary Material, Fig. S2), strongly implying that the RNA levels of most of the genes in the set identified by this continuous analysis approach were likely to truly vary in a continuous fashion with CAG length.
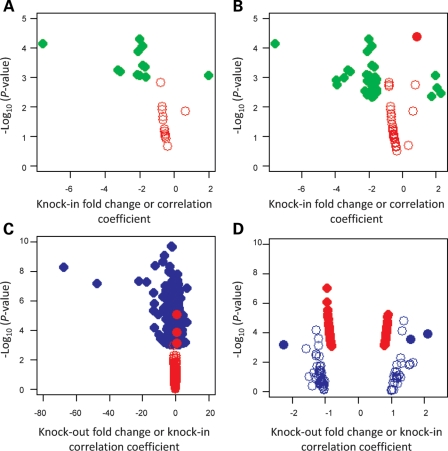
The dichotomous normal-range versus HD-range allele analysis was conducted by comparing the normalized average signal intensity for each probe from the combined expanded CAG data sets (HdhQ50/7, HdhQ91/7 and HdhQ111/7) with the average signal intensity in the normal human CAG allele datasets (HdhQ20/7). Thirteen probes met stringent statistical criteria (Student's t-test, P< 0.001; absolute fold-change > 1.5) and 37 probes passed a less stringent cut-off (P< 0.005; absolute fold-change > 1.5), providing a set of probes whose expression differed significantly between ES cells with short and long CAGs (Supplementary Material, Table S2). However, as shown in the volcano plots presented in Figure 3, none of the 13 high-stringency probes (Fig. 3A) and only one of the 37 relaxed-stringency probes (Fig. 3B, filled red circle) exhibited a signal intensity that correlated with the CAG size. These results indicated that the traditional dichotomous analytical approach, most often used in the HD field, disclosed variation due to non-CAG repeat factors and did not efficiently uncover gene expression changes continuously correlated with the CAG length.
Figure 3.
Comparisons of CAG-correlated and huntingtin-null gene sets. (A) Volcano plot showing −log10(P) (Y-axis) for the 13 probes discovered by the dichotomous analysis of the CAG knock-in ES cell data sets at high statistical stringency (P< 0.001) relative to fold-change (X-axis) (green symbols) and the −log10(P) (Y-axis) for each of these probes in the continuous analysis across the CAG knock-in ES cell data sets relative to Pearson's correlation coefficient (X-axis) (red symbols), demonstrating that none of the significant probes in the dichotomous analysis of the CAG knock-in ES cells was significantly correlated with the CAG repeat size. (B) Volcano plot showing −log10(P) (Y-axis) for the 37 probes discovered by the dichotomous analysis of the CAG knock-in ES cell data sets at more relaxed statistical stringency (P< 0.005) relative to fold-change (X-axis) (green symbols) and the −log10(P) (Y-axis) for each of these probes in the continuous analysis across the CAG knock-in ES cell data sets relative to Pearson's correlation coefficient (X-axis) (red symbols), demonstrating that only one probe from the less-stringent dichotomous analysis of the CAG knock-in ES cells was significantly correlated with the CAG repeat size (filled red circle). (C) Volcano plot showing −log10(P) (Y-axis) for the 754 probes significantly (P< 0.001) different in huntingtin-null ES cell data sets, compared with wild-type parental ES cell data sets (P< 0.001) relative to fold-change (X-axis) (blue symbols) and the −log10(P) (Y-axis) for each of these probes in the continuous analysis across the CAG knock-in ES cell data sets relative to Pearson's correlation coefficient (X-axis) (red symbols), revealing that few of the significant probes in the huntingtin-null ES cells were significantly correlated with the CAG repeat size. Mest, an outlier, was excluded from this analysis. (D) Volcano plot showing −log10(P) (Y-axis) for the 73 probes with expression significantly (P< 0.001) correlated with CAG repeat length across the CAG knock-in ES cell data sets relative to Pearson's correlation coefficient > 0.8 (X-axis) (red symbols) and the −log10(P) (Y-axis) for each of these probes in the huntingtin-null versus wild-type ES cell data set comparison relative to fold-change (X-axis) (blue symbols), demonstrating that most of the CAG-correlated probes were not significantly changed in the absence of huntingtin. The X-axis corresponds to fold-change for the dichotomous analysis and Pearson's correlation coefficient for continuous analysis. Open and filled circles represent non-significant and significant probes, respectively.
Genes altered with CAG size are distinct from genes altered with lack of huntingtin
Identification of a set of probes/genes whose expression conformed to the HD trigger mechanism (dominant, continuous with CAG length) allowed an empirical test to distinguish the main alternative hypotheses for the effects of the impact of the polyglutamine region on the full-length huntingtin: a simple gain of a novel function versus a more complex gain-of-function/loss-of-function scenario, including a dominant-negative loss of function. We determined whether the probes altered in a dominant CAG-continuous fashion, as outcomes of the full-length huntingtin gain of function might also be prominent in the ES cell response to the absence of huntingtin. Any model in which the expanded CAG contributes to a loss of huntingtin function would predict an extensive overlap in the molecular responses observed. However, the simple gain of a novel function hypothesis does not require any overlap.
First, the set of probes/genes with expression that differed between the Hdhex4/5/ex4/5 and wild-type ES cell data sets was identified by Student's t-test (Material and Methods). This comparison disclosed 754 probes with significantly different signal intensities (absolute fold-change > 1.5; P< 0.001): 398 increased and 356 decreased in the huntingtin-null ES data sets (Supplementary Material, Table S3). RT Q-PCR analysis of a subset of these genes generally confirmed the microarray data (Supplementary Material, Fig. S2A), though one gene (Mest), altered 20-fold in the RT Q-PCR analysis, was an outlier on the microarray platform (200-fold change).
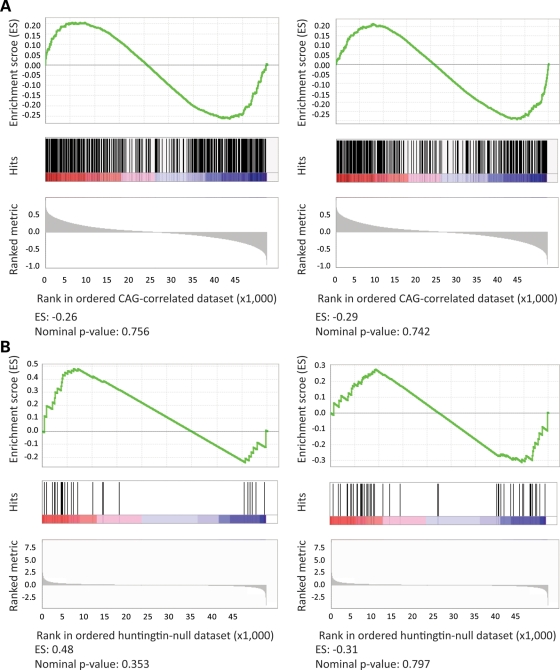
Comparison of the ‘huntingtin-null’ probe set with the set of probes whose expression was continuously altered with CAG length revealed virtually no overlap; 99.6% (751 of 754) of the probes altered in cells lacking full-length huntingtin did not vary with CAG size (Fig. 3C) and 96% (70 of 73) of the CAG-correlated probes did not distinguish wild-type and full-length huntingtin-null ES cells (Fig. 3D). Probes for only three genes (Mapt, Erdr1 and Mll5) were altered in both genetic paradigms. A second more global analytical approach, gene set enrichment analysis, also failed to disclose a significant overlap. The rank order of the probes significant in one paradigm, when assessed in the entire ∼45 000 probe data set from the other paradigm, did not differ from what might be observed by chance, with low enrichment scores and high nominal P-values (Fig. 4).
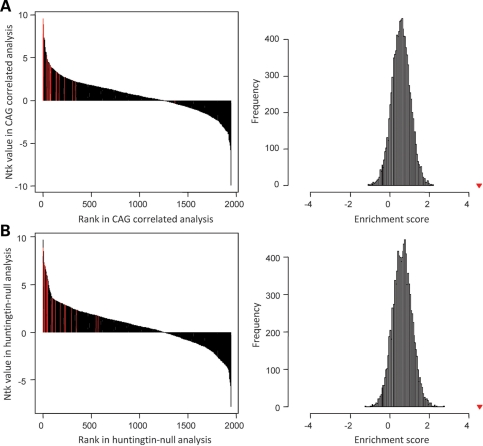
Figure 4.
Tests of gene set enrichment by phenotype permutation analysis. The results of GSEA, using phenotype permutation (1000 iterations), are presented. Top: enrichment score (ES) representing the degree to which the test gene set is overrepresented at the top or bottom of the ranked genes from the other data set; middle: position of the test set probes in the other dataset; bottom: ranked metric (Pearson's correlation coefficient for the CAG-correlated data set or t-statistic for the huntingtin-null data set). The enrichment score (ES) and nominal P-value are shown at the bottom. (A) The probes with significantly increased (left panel) or decreased (right panel) expression in the huntingtin-null ES cell comparison were not significantly enriched in the CAG-correlated knock-in ES cell data set. (B) The probes with expression significantly positively correlated (left) or negatively correlated (right) with the CAG length in the knock-in ES cell data sets were not significantly enriched in the huntingtin-null ES cell data set.
The dominant molecular effects of extending the polyglutamine region in the full-length huntingtin, therefore, did not mimic the responses to lack of huntingtin function, contradicting dominant-negative loss of function or mixed gain-of-function/loss-of-function mechanisms, but consistent with a simple gain of a novel function hypothesis.
Related huntingtin pathways
If extending the polyglutamine region in the full-length huntingtin confers a simple gain of function, how then to explain reports of phenotypes implying loss of function? We reasoned that the response to the polyglutamine region in the full-length huntingtin (a structural alteration with a dominant impact, potentially acting through an intrinsic activity of huntingtin) and the response to lack of huntingtin (classic deficiency) might be expected to engage related pathways, predicting that the probe sets from the two genetic paradigms, though almost completely distinct, might actually be connected via common pathways.
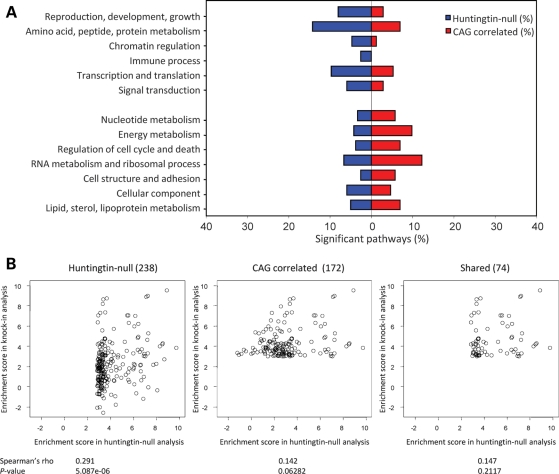
sigPathway analysis identified 238 pathways significantly (gene set permutation q < 0.01 and phenotype permutation q < 0.01) altered between wild-type and huntingtin-null ES cell data sets (Supplementary Material, Table S4) and 172 pathways that correlated with CAG size across the knock-in ES cell panel data sets (Supplementary Material, Table S5). In support of overlap at the pathway, if not the probe level, 74 pathways were significant in both genetic paradigms (Supplementary Material, Table S6), and the gene set enrichment analysis (GSEA) with the top 20 ranked pathways from each paradigm disclosed significant enrichment in the entire 1947 pathways for the other paradigm (Fig. 5A and B). These results were unlikely to have been produced by chance because the true enrichment score (i.e. average rank of top 20 pathways in the other paradigm) significantly deviated from the majority of the enrichment scores obtained from simulations for each paradigm, with 10 000 iterations of permuted data sets. In addition, the majority of these pathways could be assigned to 13 categories at the network level, though, as shown in Figure 6A, six clusters were more prominently altered in the huntingtin-null ES cells (reproduction/development/growth, amino acid/peptide/protein metabolism, chromatin regulation, immune process, transcription/translation and signal transduction) and seven clusters were more prominently altered by extending the polyglutamine region in the full-length huntingtin (nucleotide metabolism, energy metabolism, regulation of cell cycle/death, RNA metabolism/ribosomal process, cell structure/adhesion, cellular component and lipid/sterol/lipoprotein metabolism).
Figure 5.
Tests of enrichment of significant pathways by permutation analysis. The results of permutation-based enrichment analysis to test whether a sort of the top 20 significant pathways in one paradigm was also significantly enriched in the other paradigm. Left: original sigPathway results (black bars) representing rank (X-axis) and NTk value from gene set permutations (Y-axis), with the rank of test pathways highlighted (red bars). Right: average ranks of test pathways (enrichment score) compared with the distribution obtained from a random sampling of 20 pathways from the compared data set (10 000 permutations). (A) Top 20 huntingtin-null significant pathways in CAG-correlated knock-in ES cell pathways indicated significant enrichment, with an enrichment score of 4.51, compared with a distribution of enrichment scores obtained from a random selection of 20 pathways in knock-in pathway results. (B) Top 20 CAG-correlated pathways in huntingtin-null pathways indicated significant enrichment, with an enrichment score of 4.55, compared with a distribution of enrichment scores obtained from a random selection of 20 pathways in knock-out pathway results. Red triangles in the right panels represent true enrichment scores.
Figure 6.
CAG-correlated and huntingtin-null pathway categories. A summary of sigPathway analysis results, with significant pathways for each paradigm grouped by category is presented (A), with results of correlation analysis testing the relationship between huntingtin-null significant pathways (KO) and the CAG-correlated pathways (KI) (B–D). (A) Bar plot of the proportion (X-axis) of the 238 pathways significantly enriched in the huntingtin-null ES cell comparison (blue) and the 172 significantly CAG-correlated pathways across the knock-in ES cell data sets (red) within 13 shared categories (Y-axis). (B) Left scatter plot displays the relationship of the enrichment scores of 238 huntingtin-null significant pathways (X-axis; NTk values) with the enrichment score for these pathways in the continuous CAG ES cell knock-in pathways analysis (Y-axis; NTk values). The Spearman's rank correlation statistic (rho) and the significant P-value indicate that the huntingtin-null pathways capture to some extent the CAG-correlated pathway set. Middle scatter plot displays the relationship of the enrichment scores of the 172 continuous CAG-correlated pathways (X-axis) with the enrichment score for these pathways from the huntingtin-null analysis (Y-axis). The Spearman's rank correlation statistic (rho) and the non-significant P-value indicate that the pathways significantly altered with the CAG size are not altered by the absence of huntingtin. The right scatter plot shows the relationship between the enrichment scores of 74 pathways significant in both the huntingtin-null ES cell analysis (X-axis) and the CAG continuous ES cell data analysis (Y-axis). The Spearman's rank correlation statistic (rho) and the non-significant P-value indicate that the enrichment score in one analysis does not predict the enrichment score in the other paradigm, indicating that these pathways are not consistently altered in the different paradigms.
Despite these general features of commonality and interconnection of the pathways altered in the two genetic paradigms, statistical analysis continued to highlight the lack of one-to-one correspondence between the specific alterations detected in these pathway data sets. The alterations of the 238 significant pathways in huntingtin-null were mildly predictive of a significant effect on the same pathways in the continuous CAG comparison (Spearman's rho 0.291; P = 5.08E−6) (Fig. 6B). However, alterations of the 172 full-length huntingtin polyglutamine region-correlated pathways were not at all predictive of a significant effect in the wild-type/huntingtin-null data set comparison (Spearman's rho 0.142; P = 0.06282) (Fig. 6C). Nor did the alterations observed in the 74 common pathways predict statistical significance from one genetic comparison to the other (Spearman's rho 0.147; P = 0.2117) (Fig. 6D). Therefore, while the huntingtin-null pathways captured a portion of the dominant response to increase the full-length huntingtin polyglutamine region, the conversing was not true.
Taken together, these findings implied that the probes/genes contributing to the CAG-correlation genetic paradigm and those contributing to the huntingtin-null genetic paradigm represent either different members of the same pathways or members of distinct but interconnected pathways within the same category networks.
Interconnectedness of CAG-correlated and huntingtin-null pathways
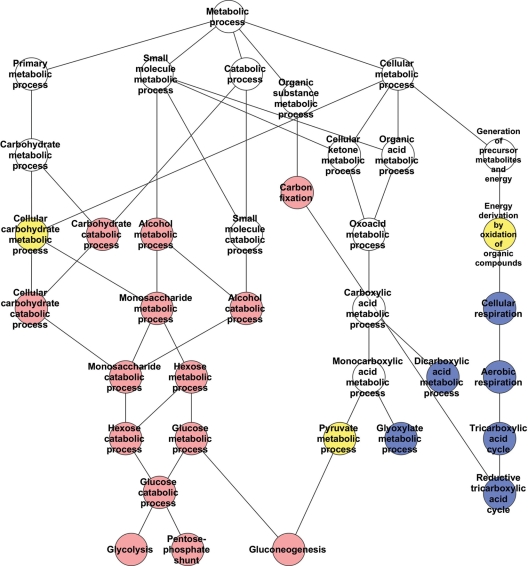
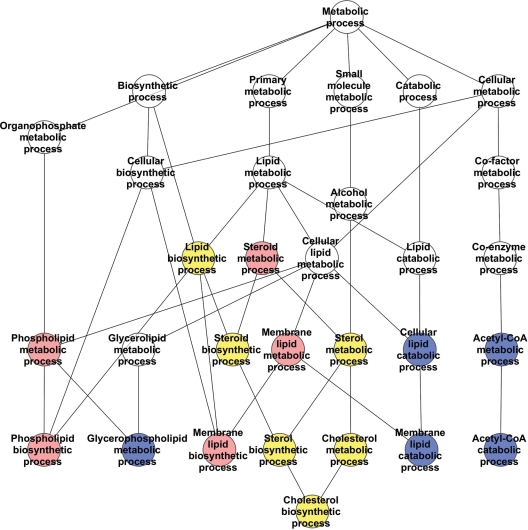
To explore this possibility, we examined the hierarchical relationships of the pathways in the energy and lipid/cholesterol/sterol categories. The energy network (24 pathways) (Fig. 7 and Supplementary Material, Fig. S3) comprised pathways altered with the size of the full-length huntingtin polyglutamine region (carbohydrate metabolism, glycolysis) and other pathways prominent in huntingtin deficiency (mitochondrial, TCA cycle, oxidative respiration) with only a few pathways common to both. In contrast, the lipid/sterol/lipoprotein network (17 pathways) (Fig. 8 and Supplementary Material, Fig. S4) comprised nearly equal proportions of processes prominent in the response to extending the full-length huntingtin polyglutamine region (phospholipid metabolism), pathways prominent in the wild-type/huntingtin-null comparison (acetyl-CoA, glycerophospholipid metabolism) and shared pathways common to both genetic paradigms (cholesterol, sterol).
Figure 7.
Energy category pathways network. A schematic demonstrating the hierarchy of relationships between the processes/pathways significantly correlated with the CAG repeat length (red), significantly altered in the absence of huntingtin (blue) or significantly altered in both full-length huntingtin genetic paradigms (yellow). The cellular component ‘Mitochondrial matrix’, significantly altered only in the huntingtin-null ES cell analysis, is not shown. Only directly related pathways are illustrated. The KEGG/GO annotations for the pathways are provided in Supplementary Material, Figure S3.
Figure 8.
Lipid, sterol and lipoprotein metabolism category pathways network. A schematic demonstrating the hierarchy of relationships between the processes/pathways significantly correlated with the CAG repeat length (red), significantly altered in the absence of huntingtin (blue) or significantly altered in both full-length huntingtin genetic paradigms (yellow). The catalytic activity ‘phospholipase A2’, which is significantly changed only in the absence of huntingtin, is not depicted on the pathway annotation diagram. Only directly related pathways are illustrated. The KEGG/GO annotations for the pathways are provided in Supplementary Material, Figure S4.
These hierarchies demonstrated that some of the genes that were uniquely changed with increasing CAG repeat length were different members of the same pathways as genes that were altered by the complete absence of huntingtin, while other CAG-repeat-altered genes were members of distinct pathways that were not significantly affected by the lack of huntingtin. However, even the latter CAG-correlated pathways, though distinct, were often closely connected in the network to the pathways changed in the absence of huntingtin. The highly interconnected character of these networks, which are impacted by altering both the full-length huntingtin polyglutamine repeat and the dose of the full-length huntingtin, predicts that, integrating across the network, both genetic paradigms might in some cases be expected to produce similar phenotypic outcomes despite emanating from different initial insults, whereas in other cases the phenotypic outcomes may be distinct.
DISCUSSION
The accumulated evidence indicates that the HD CAG repeat acts as a functional DNA polymorphism, encoding a polyglutamine region in the full-length huntingtin that exerts dominant effects that are graduated with CAG length across an allelic continuum. Yet HD homozygotes exist, despite expressing no normal-range full-length huntingtin, and conversely, huntingtin deficiency does not mimic HD. A parsimonious explanation is that the polyglutamine repeat confers a simple gain of a novel function, such that the full-length huntingtin is modulated, though not impaired, in a graded manner that yields dominant effects that are continuous with polyglutamine length.
Investigations of this hypothesis and alternative proposals, such as a gain of a dominant-negative loss of full-length huntingtin function, have been hampered by the lack of an appropriate series of CAG alleles in a cell culture system, for which there are also HD knock-out alleles. The isogenic allelic ES cell panel that we have generated now provides a suitable cell culture system. It comprises a series of heterozygous CAG knock-in ES cell lines, appropriately expressing full-length huntingtins with 20, 50, 91 or 111 polyglutamine repeats (from endogenous alleles), augmenting the wild-type and huntingtin-null ES cell lines, previously reported. The members of this ES cell panel can undergo development, allowing studies in differentiated cell types, such as neuronal cells.
The allelic ES cell panel allows investigations to determine whether candidate phenotypes may (or may not) be dominant and progressive with CAG size. ATP/ADP ratios, continuously decreased with CAG size in human lymphoblastoid cell lines (18), were decreased with CAG length across the knock-in ES cell members, and ATP/ADP ratio was increased in the absence of full-length huntingtin, in knock-out ES cells, consistent with the proposal that the polyglutamine repeat enhances an as-yet uncharacterized role of full-length huntingtin in negatively regulating this energetic measure (18,32). The allelic CAG ES cell series, therefore, offers a resource with which to further assess purported HD-associated differences from studies in human and non-human systems to determine whether they conform to the dominant continuous effects of the HD CAG repeat, rather than emanating from other sources of variation that may distinguish HD and non-HD samples.
The discovery of new dominant CAG length-dependent effects can also be efficiently accomplished by phenotyping across the members of a CAG allelic panel. Continuous analysis, though not dichotomous analysis, efficiently uncovered genes with CAG-correlated expression from genome-wide data sets. The success of the continuous analytical approach, with four different CAG allele lengths on an isogenic genetic background, predicts that this strategy can be effective in identifying the dominant effects of the CAG repeat in human samples, though the increased genetic heterogeneity, and higher proportion of expanded CAG alleles in the adult onset rather than the juvenile onset range, will likely require a larger panel representing a denser series of CAG alleles across the continuum. Knowledge of the dominant molecular, biochemical and cellular responses that emanate from the quantitative impact of the polyglutamine repeat on full-length huntingtin, especially in cells of neuronal lineages, should provide a better understanding of the molecular mechanism that initiates the pathogenic process to which these cells are vulnerable.
In our study, the CAG knock-in and huntingtin-null panel members allowed an evaluation of alternate hypotheses for the impact of the polyglutamine repeat on full-length huntingtin: a simple gain of a novel function versus a dominant-negative or a mixed gain/loss of function. The CAG-correlated gene set was strikingly distinct from the huntingtin-null gene set, failing to support a model involving a loss of function. However, this outcome was completely consistent with a simple novel gain-of-function hypothesis, which posits different mechanisms for the two genetic paradigms and does not require any overlap in the two gene sets to be valid. This finding joins the previous genetic evidence supporting a mechanism of a simple gain of a novel function, wherein the structural alteration of the full-length huntingtin by the polyglutamine repeat produces a graded effect on some action of huntingtin, though it does not impair the protein's inherent function. Whether the gain of function involves the graded modulation of a property/task intrinsic to the full-length huntingtin or, alternatively, adds a novel non-intrinsic property/task into full-length huntingtin's biological repertoire remains to be determined. Simple enhancement of an intrinsic action of huntingtin by the polyglutamine repeat is supported by the interrelated nature of the pathways that emerged from the two genetic huntingtin paradigms. However, the polyglutamine repeat might also confer a new property, because, while the huntingtin-null changes do to a certain extent capture the CAG repeat-correlated changes, the converse is not the case. Thus, our results support the view that the polyglutamine repeat may confer enhanced intrinsic huntingtin activity and/or confer some additional non-intrinsic property on the full-length huntingtin.
Indeed, rather than revealing a common mechanism, our analyses indicate that the distinct CAG-correlated and huntingtin-null genes, defined by the structure–function and deficiency strategies, respectively, identify instead an interrelated hierarchy of biological pathways. The pathways that were specifically correlated with CAG size were not predictive of the pathways altered by huntingtin knock-out. However, together, the pathways clustered into about a dozen biological categories or networks in realms consistent with previous reports of huntingtin function (41), some influenced more prominently by the full-length huntingtin polyglutamine repeat (energy, cholesterol/sterol/lipid, nucleotide metabolism, regulation of cell cycle/death, RNA metabolism/ribosomal process and cell structure/adhesion) (18,32,42–45) and some by huntingtin deficiency (reproduction/development, protein metabolism, chromatin regulation, transcription and translation, immune process and signal transduction) (28,37,46–50).
The energy metabolism network revealed the effects of the polyglutamine-size in full-length huntingtin on glycolysis and of huntingtin deficiency on the tricarboxylic acid (Krebs) cycle, respectively. However, few processes were shared, implying that the elevated or decreased ATP/ADP ratios, respectively, that result from these insults may result from distinct rate-limiting steps in each case. The lipid/sterol/lipoprotein metabolism network, with unique impacts of polyglutamine size in full-length huntingtin on phospholipid/lipid biosynthesis and huntingtin deficiency on lipid catabolism (β-oxidation) and acetyl-CoA catabolism, respectively, featured a high proportion of shared processes, notably lipid, steroid and cholesterol biosynthesis. Thus, the two distinct full-length huntingtin mechanisms, polyglutamine expansion versus protein deficiency, which altered the expression of completely different gene sets, both converged at the pathway level on cholesterol biosynthesis, strongly implying that decreased levels of cholesterol biosynthetic intermediates in cells expressing either normal or mutant huntingtin likely result via different mechanisms due to the effects on distinct rate-limiting steps within the same network (34,51).
The CAG-correlated genes and pathways, which conform to genetically defined properties expected of the consequences of the HD disease-initiating mechanism, demonstrate the wide physiological impact of varying the polyglutamine repeat in full-length huntingtin. Though a common regulator is formally possible, it seems unlikely that targeting any single candidate regulator will effectively mitigate most of the many hundreds of interconnected consequences of the CAG repeat. Instead, our results strongly support a system-wide approach to the discovery and validation of factors that may moderate the dominant effects of extending the polyglutamine region in full-length huntingtin. Moreover, for therapeutic strategies aimed at lowering the expression of the expanded CAG allele, our results in this proof-of-concept experiment in murine ES cells strongly suggest that understanding the biological processes perturbed by the lack of full-length huntingtin may be important, as such treatments might instead exacerbate the physiologic effects of the expanded CAG repeat.
MATERIALS AND METHODS
ES cell culture
Wild-type parental, huntingtin null Hdhex4/5/ex4/5 and HdhneoQ20/7, HdhneoQ50/7 HdhneoQ91/7 HdhneoQ111/7 mouse ES cell lines have been reported previously (9,14,25,27). ES cells were maintained on feeder layers of irradiated mouse embryonic fibroblasts (Global Stem Sciences) at 37°C in 5% CO2 or on gelatin-coated plates (1% gelatin solution; Millipore). ES cell media (ESCM) typically contained KnockOut D-MEM (Invitrogen), 15% FBS (Hyclone), 50 I.U/ml of penicillin, 50 μg/ml of streptomycin (Cellgro), 0.2 mm GlutaMax (Invitrogen), 0.1 mm MEM non-essential amino acids (Invitrogen), 0.1 mm 2-mercaptoethanol (Sigma) and 1000 U/ml of leukemia inhibitory factor (LIF) (Millipore). For ES cell lines with targeted alleles, the media also contained G418 (150–450 μg/ml). Cells were passaged by trypsinization (0.05% trypsin, 0.53 mm EDTA) (Cellgro) every 3 days. For molecular analysis (protein, RNA, DNA), cells were grown on gelatin-coated plates in ESCM. Molecular analyses and analyses of nucleotide (ATP and ADP) levels were performed on independent cell cultures, grown under standard conditions.
To induce embryoid body (EB) formation, 5 × 106 ES cells were grown in the absence of feeder cells on 10 cm bacterial plates in ESCM without LIF. The day 4 EBs were characterized with germ-layer-specific markers using RT–PCR amplification assays as described below.
Adeno-Cre recombinase and identification of ES cells with neo-out CAG knock-in alleles
The Hdhneo20/7, Hdhneo50/7, Hdhneo91/7 and Hdhneo111/7 ES cells, in each case, were plated in 12-well plates at ∼2.5 × 105 cells/well, in G418 selection, and infected with 200 infectious units/cell Adeno-Cre (Ad5CMVCre). The following day, the selection was removed by changing to a medium lacking G418. Established colonies were subsequently plated into wells of duplicate 96-well plates, into media with G418 or media without G418. For each line, those G418-sensitive subclones that grew in the absence of G418, but not in the presence of G418, were propagated to establish PGKneo-minus lines for three to four subclones for each member of the allelic ES cell series. PCR amplification analysis confirmed the proper Cre-recombinase removal of the PGKneo cassette. DNA was purified using the Gentra Puregene Cell kit (Qiagen) and treated with RNase A, following the manufacturer's instructions. Primers to assess the presence of the PGKneo cassette were: Mouse mod-2 Neo-in F: 5′-GATCTCCTGTCATCTCACCTTG-3′ and 5′-UTR R: 5′-CAGGCAGGCAGCAGTAGTTA-3′. Cycling conditions were as follows: 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 90 s and a final extension at 72°C for 7 min. Primers to assess the absence of the PGKneo cassette were: Neo-out F: 5′-TCACTGCTTGGCTTTTTCCT-3′ and 5′-UTR R: 5′-CAGGCAGGCAGCAGTAGTTA-3′. Cycling conditions were as follows: 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 1 min and a final extension at 72°C for 7 min.
Quantitative RT–PCR amplification assays
For RNA extraction, washed ES cell pellets were homogenized in TRIzol (Invitrogen) and total RNA isolated using phenol/chloroform. Following digestion with DNaseI recombinant RNase-free (Roche), RNA was reverse-transcribed to produce cDNA using Superscript III according to the manufacturer's guidelines (Invitrogen). An oligo-dT primer was used to prime cDNA synthesis. Each quantitative real-time PCR amplification reaction was performed in a total volume of 40 μl using 2× iQ SYBR Green Supermix (Bio-Rad) and 0.2 pmol primer. Primers were designed to the 3′ end of each transcript analyzed and all products were 70–100 bp in length. Cycling parameters were: 95°C for 2 min to denature followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Melting temperature analysis was performed at the end of each run to validate the specificity of the PCR amplicon. Assays were conducted in a 96-well plate format with ‘no template’ and ‘no reverse transcriptase’ controls and run on the iQ cycler (Bio-Rad). Three technical replicates were performed for each sample and all genes were compared with the reference gene, mouse β-actin. Primers were designed using Primer3 software (http://frodo.wi.mit.edu/).
Primers to assess EB primary germ layers were: Afp F: 5′-AGCAAAGCTGCGCTCTCTAC-3′ and Afp R: 5′-CCGAGAAATCTGCAGTGACA-3′, Pdx1 F: 5′-GAAATCCACCAAAGCTCACG-3′ and Pdx1 R: 5′-TCTCCGGCTATACCCAACTG-3′, Bry F: 5′-CCGGTGCTGAAGGTAAATGT-3′ and Bry R: 5′-TGACCGGTGGTTCCTTAGAG 3′, Actc F: 5′ GCTTTGGTGTGTGACAATGG-3′ and Actc R: 5′-AGAGACAGCACTGCCTGGAT-3′, Pax6 F: 5′- AGGGGGAGAGAACACCAACT-3′ and Pax6 R: 5′ GGTTGCATAGGCAGGTTGTT-3′, Ncam1 F: 5′-AGCTGAAAACCAGCAAGGAA-3′ and Ncam1 R: 5′-TTTTGTTTGTGTGGCATCGT-3′. Cycling conditions for Afp, Actc, Pdx and Pax6 were 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 45 s and a final extension at 72°C for 7 min. Cycling conditions for Bry are: 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 50°C for 30 s, 72°C for 45 s and a final extension at 72°C for 7 min. Cycling conditions for Ncam1 are: 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 45 s and a final extension at 72°C for 7 min.
Primers to validate microarray results were: Mapt F: 5′-GCTCGTTAGGGAACATCCAT-3′ and Mapt R: 5′- TC GACTGGACTCTGTCCTTG-3′, Mll5 F: 5′-GAACAGTTT GAAGCAAATGGAT-3′ and Mll5 R: 5′-CCTCATTCCCA AAAGTCCTC-3′, Erdr1 F: 5′-AGTGATGTCACCCACG AAAG-3′ and Erdr1 R: 5′-GTGGGGATGGCAGAGAC AT-3′, Bhlhb2 F: 5′-CACCCCAAGATCCTTTCTGT-3′ and Bhlhb2 R: 5′-GGAGATGGAACCCTTTTTAGC-3′, Ccdc18 F: 5′-TGGTTTAAGTCAGGGAGGAAA-3′ and Ccdc18 R: 5′-AGCACAGTACCAACAATTCCA-3′, Abca1 F: 5′-TGT TGCATCCCTTTTTGTAGA-3′ and Abca1 R: 5′-AAAGCA CAAAACCAGCTTCA-3′, Mest F: 5′-AGTGAGGGAGG AGCTTGCTA-3′ and Mest R: 5′-GCAGCCGATGAAA CTAAATG-3′, Xlr F: 5′-GGAAGCCAGAAAGCTAATGG-3′ and Xlr R: 5′-GATTGAACACCATCCTTTGC-3′, Tgm2 F: 5′-AGAGCCTGGGAAGAATCAAA-3′ and Tgm2 R: 5′-AA AAGAAACAGAACCGTGTGG-3′, Lrp2 F: 5′-CTGAGC AAAAGGAAGCTGTG-3′ and Lrp2 R: 5′-CAGTGTA GCCTGGAGTCGAA-3′, Snapc1 F: 5′-TCCAACAGAA GAAAGGTGTCA-3′ and Snapc1 R: 5′-GCATGCTCTTCC ATCTCACA-3′, Utp14a F: 5′-AAGGCTGTGGATCTGA CCTT-3′ and Utp14a R: 5′-CCCTCAGGGGCTTTAAT AAG-3′, NfuI F: 5′-TCCATCATCACCCTGAAGAG-3′ and NfuI R: 5′-TTCGTCATCATCCATCACCT-3′, Hsd17b11 F: 5′-GAAAGGATCGTCCCTGAGAG-3′ and Hsd17b11 R: 5′- GCTGCGTCACTTGTCTTTGT-3′, Rpap2 F: 5′ GTTG CTGGAGGTGTCCACTA-3′ and Rpap2 R: 5′-AGGGGA GAGAGTGGGAAAGT-3′, Nicn1 F: 5′-AGTTAAGCCCCAC TGACAGG-3′ and Nicn R: 5′-GATCCTCTTTGCCCAAG TGT-3′, Ngdn F: 5′-CAGAGTCAGGAGGATCAGCA-3′ and Ngdn R: 5′-CCTTCGCAGTCCTTTCTCTC-3′, β-actin F: 5′-GACGGCCAGGTCATCACTAT-3′ and β-actin R: 5′-ATGCCACAGGATTCCATACC-3′. Cycling conditions are described previously.
Immunoblot analysis
Protein extracts were prepared from washed cell pellets by lysis on ice for 30 min in a buffer containing 20 mm HEPES (pH 7.6), 1 mm EDTA, 0.5% Triton X-100, protease inhibitor mixture (Roche) and 1 mm phenylmethyl sulfonyl fluoride. Lysates were mixed every 10 min during the incubation. The lysates were then cleared by centrifugation at 14 000g for 30 min and the supernatants collected. The protein concentration was determined using the Bio-Rad (detergent compatible) protein assay. Twenty-five micrograms of protein extract was mixed with 4× SDS sample buffer, boiled for 2 min and subjected to 6 or 10% SDS–PAGE. After electrophoresis, the proteins were transferred to nitrocellulose membranes (Schleicher & Schuell) and incubated for 30 min in a blocking solution containing 5% non-fat powdered milk in TBS-T (50 mm Tris–HCl, 150 mm NaCl, pH 7.4, 0.1% Tween-20). The membranes were probed overnight at 4°C with the primary antibody mAb2166 (Chemicon). After four 10 min TBS-T washes, the blots were incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-mouse antibodies. After an extensive 30 min wash, the membranes were processed using an ECL chemiluminiscence substrate kit (New England Biolabs) and exposed to autoradiographic film (Hyperfilm ECL; Amersham Bioscience). Notably, on SDS–PAGE immunoblots, anti-huntingtin antibodies are known to detect full-length huntingtin proteins differentially, based upon the length of the polyglutamine tract, though the reasons for this phenomenon are not yet known.
HPLC ATP/ADP measurement
Nucleotide extraction was as previously described (18,52). To normalize the amount of nucleotide, each pellet was solubilized with 1 N NaOH and the protein content was analyzed by Bio-Rad DC protein assay (Bio-Rad). HPLC-grade nucleotide standards (Fluka) were used to calibrate the signals. Internal standards were occasionally added to the samples to test recovery, which exceeded 90% for all nucleotides. Data were quantified by Breez software.
Microarray analysis
RNA extracted from ES cells using TRIzol reagent (Invitrogen) was further purified through RNeasy mini columns (Qiagen). All RNA samples passed quality control steps, including RNA purity check (OD 260/280 ratio) and Bioanalyzer analysis (Agilent). Total RNA (5 µg) was converted, using SuperScript II reverse transcriptase (Invitrogen), to cRNA, and labeled cRNA was prepared by in vitro transcription (Affymetrix). Twenty-five micrograms of labeled probe was hybridized to Affymetrix MG 430 2.0 arrays. Expression data were normalized using gcrma (R, 2.6.2; gcrma, 2.6.0), and significant probes were identified by nominal P-value (P < 0.001) and fold-change (absolute fold-change > 1.5) or Pearson's correlation coefficient (absolute correlation coefficient > 0.8). All microarray data have been deposited in NCBI's Gene Expression Omnibus (accession number GSE26001).
Gene set enrichment analysis
To test whether a set of significantly altered probes in knock-out ES cells was enriched in knock-in ES cells, GSEA (53) was performed using phenotype permutation procedures (1000 permutations). Similarly, we tested whether significantly correlated probes in knock-in ES cells were significantly enriched in knock-out ES cells using GSEA. For identification of significantly enriched pathways in knock-out and knock-in ES cells, pre-compiled gene sets from pathway databases (Biocarta and KEGG) and Gene Ontology were analyzed using the sigPathway program (54). Gene sets with sizes between 25 and 500 were selected, and gene sets showing undirectional enrichment were identified by gene set permutation (10 000) and phenotype permutation (1000) (false discovery rate, q < 0.01). To assess commonality between significant pathways in knock-out and CAG knock-in cells, the average of ranks of the top 20 knock-out significant pathways in the CAG knock-in data (i.e. enrichment score; 4.51) was compared with a distribution of averages of ranks obtained by randomly choosing the same number of pathways in knock-in data (10 000 permutations). Similarly, the average of ranks of the top 20 knock-in significant pathways in the knock-out data (i.e. enrichment score; 4.55) was compared with a distribution of averages of ranks of 20 randomly chosen pathways in knock-out data. Networks of significant pathways were hand-annotated, identifying shared pathway categories using the graphical view feature on the gene ontology website. Significant pathways from the KEGG database were annotated using the corresponding GO pathway. Networks were drawn using Cytoscape (version 2.7.0).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by National Institute of Neurological Disorders and Stroke (grant number NS32765 to M.E.M.), The Massachusetts HD Center Without Walls (grant number NS16367 to J.F.G. and M.E.M.), i2b2 (grant number LM008748-01 to I.S.K.), the Huntington's Disease Society of America Coalition for the Cure Normal Function Team (to M.E.M. and J.F.G.), and an Anonymous Donor. J.M.W. was the recipient of the Milton Wexler Postdoctoral Fellowship from the Hereditary Disease Foundation. G.G. was the recipient of the Sir Keith Murdoch Fellowship from the American Australian Association. J.J. is the recipient of the Philip Wrightson Fellowship from the Neurological Foundation of New Zealand.
Supplementary Material
REFERENCES
- 1.Vonsattel J.P., DiFiglia M. Huntington disease. J. Neuropathol. Exp. Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.McNeil S.M., Novelletto A., Srinidhi J., Barnes G., Kornbluth I., Altherr M.R., Wasmuth J.J., Gusella J.F., MacDonald M.E., Myers R.H. Reduced penetrance of the Huntington's disease mutation. Hum. Mol. Genet. 1997;6:775–779. doi: 10.1093/hmg/6.5.775. [DOI] [PubMed] [Google Scholar]
- 4.Quarrell O.W., Rigby A.S., Barron L., Crow Y., Dalton A., Dennis N., Fryer A.E., Heydon F., Kinning E., Lashwood A., et al. Reduced penetrance alleles for Huntington's disease: a multi-centre direct observational study. J. Med. Genet. 2007;44:e68. doi: 10.1136/jmg.2006.045120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenney C., Powell S., Jankovic J. Autopsy-proven Huntington's disease with 29 trinucleotide repeats. Mov. Disord. 2007;22:127–130. doi: 10.1002/mds.21195. [DOI] [PubMed] [Google Scholar]
- 6.Hendricks A.E., Latourelle J.C., Lunetta K.L., Cupples L.A., Wheeler V., MacDonald M.E., Gusella J.F., Myers R.H. Estimating the probability of de novo HD cases from transmissions of expanded penetrant CAG alleles in the Huntington disease gene from male carriers of high normal alleles (27–35 CAG) Am. J. Med. Genet. A. 2009;149A:1375–1381. doi: 10.1002/ajmg.a.32901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duyao M., Ambrose C., Myers R., Novelletto A., Persichetti F., Frontali M., Folstein S., Ross C., Franz M., Abbott M., et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat. Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- 8.Snell R.G., MacMillan J.C., Cheadle J.P., Fenton I., Lazarou L.P., Davies P., MacDonald M.E., Gusella J.F., Harper P.S., Shaw D.J. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat. Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 9.Auerbach W., Hurlbert M.S., Hilditch-Maguire P., Wadghiri Y.Z., Wheeler V.C., Cohen S.I., Joyner A.L., MacDonald M.E., Turnbull D.H. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum. Mol. Genet. 2001;10:2515–2523. doi: 10.1093/hmg/10.22.2515. [DOI] [PubMed] [Google Scholar]
- 10.Fossale E., Wheeler V.C., Vrbanac V., Lebel L.A., Teed A., Mysore J.S., Gusella J.F., MacDonald M.E., Persichetti F. Identification of a presymptomatic molecular phenotype in Hdh CAG knock-in mice. Hum. Mol. Genet. 2002;11:2233–2241. doi: 10.1093/hmg/11.19.2233. [DOI] [PubMed] [Google Scholar]
- 11.Heng M.Y., Detloff P.J., Paulson H.L., Albin R.L. Early alterations of autophagy in Huntington disease-like mice. Autophagy. 2010;6:1206–1208. doi: 10.4161/auto.6.8.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heng M.Y., Tallaksen-Greene S.J., Detloff P.J., Albin R.L. Longitudinal evaluation of the Hdh(CAG)150 knock-in murine model of Huntington's disease. J. Neurosci. 2007;27:8989–8998. doi: 10.1523/JNEUROSCI.1830-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menalled L.B. Knock-in mouse models of Huntington's disease. NeuroRx. 2005;2:465–470. doi: 10.1602/neurorx.2.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler V.C., Auerbach W., White J.K., Srinidhi J., Auerbach A., Ryan A., Duyao M.P., Vrbanac V., Weaver M., Gusella J.F., et al. Length-dependent gametic CAG repeat instability in the Huntington's disease knock-in mouse. Hum. Mol. Genet. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 15.Wheeler V.C., Gutekunst C.A., Vrbanac V., Lebel L.A., Schilling G., Hersch S., Friedlander R.M., Gusella J.F., Vonsattel J.P., Borchelt D.R., et al. Early phenotypes that presage late-onset neurodegenerative disease allow testing of modifiers in Hdh CAG knock-in mice. Hum. Mol. Genet. 2002;11:633–640. doi: 10.1093/hmg/11.6.633. [DOI] [PubMed] [Google Scholar]
- 16.Wheeler V.C., White J.K., Gutekunst C.A., Vrbanac V., Weaver M., Li X.J., Li S.H., Yi H., Vonsattel J.P., Gusella J.F., et al. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum. Mol. Genet. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- 17.Perlis R.H., Smoller J.W., Mysore J., Sun M., Gillis T., Purcell S., Rietschel M., Nothen M.M., Witt S., Maier W., et al. Prevalence of incompletely penetrant Huntington's disease alleles among individuals with major depressive disorder. Am. J. Psychiatry. 2010;167:574–579. doi: 10.1176/appi.ajp.2009.09070973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seong I.S., Ivanova E., Lee J.M., Choo Y.S., Fossale E., Anderson M., Gusella J.F., Laramie J.M., Myers R.H., Lesort M., et al. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum. Mol. Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 19.Andrade M.A., Bork P. HEAT repeats in the Huntington's disease protein. Nat. Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- 20.Li W., Serpell L.C., Carter W.J., Rubinsztein D.C., Huntington J.A. Expression and characterization of full-length human huntingtin, an elongated HEAT repeat protein. J. Biol. Chem. 2006;281:15916–15922. doi: 10.1074/jbc.M511007200. [DOI] [PubMed] [Google Scholar]
- 21.Seong I.S., Woda J.M., Song J.J., Lloret A., Abeyrathne P.D., Woo C.J., Gregory G., Lee J.M., Wheeler V.C., Walz T., et al. Huntingtin facilitates polycomb repressive complex 2. Hum. Mol. Genet. 2010;19:573–583. doi: 10.1093/hmg/ddp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takano H., Gusella J.F. The predominantly HEAT-like motif structure of huntingtin and its association and coincident nuclear entry with dorsal, an NF-kB/Rel/dorsal family transcription factor. BMC Neurosci. 2002;3:15. doi: 10.1186/1471-2202-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grinthal A., Adamovic I., Weiner B., Karplus M., Kleckner N. PR65, the HEAT-repeat scaffold of phosphatase PP2A, is an elastic connector that links force and catalysis. Proc. Natl Acad. Sci. USA. 2010;107:2467–2472. doi: 10.1073/pnas.0914073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ambrose C.M., Duyao M.P., Barnes G., Bates G.P., Lin C.S., Srinidhi J., Baxendale S., Hummerich H., Lehrach H., Altherr M., et al. Structure and expression of the Huntington's disease gene: evidence against simple inactivation due to an expanded CAG repeat. Som. Cell Mol. Genet. 1994;20:27–38. doi: 10.1007/BF02257483. [DOI] [PubMed] [Google Scholar]
- 25.Duyao M.P., Auerbach A.B., Ryan A., Persichetti F., Barnes G.T., McNeil S.M., Ge P., Vonsattel J.P., Gusella J.F., Joyner A.L., et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 26.Nasir J., Floresco S.B., O'Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., Hayden M.R. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 27.White J.K., Auerbach W., Duyao M.P., Vonsattel J.P., Gusella J.F., Joyner A.L., MacDonald M.E. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat. Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- 28.Zeitlin S., Liu J.P., Chapman D.L., Papaioannou V.E., Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat. Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 29.Myers R.H., Leavitt J., Farrer L.A., Jagadeesh J., McFarlane H., Mastromauro C.A., Mark R.J., Gusella J.F. Homozygote for Huntington disease. Am. J. Hum. Genet. 1989;45:615–618. [PMC free article] [PubMed] [Google Scholar]
- 30.Wexler N.S., Young A.B., Tanzi R.E., Travers H., Starosta-Rubinstein S., Penney J.B., Snodgrass S.R., Shoulson I., Gomez F., Ramos Arroyo M.A., et al. Homozygotes for Huntington's disease. Nature. 1987;326:194–197. doi: 10.1038/326194a0. [DOI] [PubMed] [Google Scholar]
- 31.Brooks S., Higgs G., Jones L., Dunnett S.B. Longitudinal analysis of the behavioural phenotype in Hdh((CAG)150) Huntington's disease knock-in mice. Brain Res. Bull. 2010 doi: 10.1016/j.brainresbull.2010.05.004. In press. [DOI] [PubMed] [Google Scholar]
- 32.Clabough E.B., Zeitlin S.O. Deletion of the triplet repeat encoding polyglutamine within the mouse Huntington's disease gene results in subtle behavioral/motor phenotypes in vivo and elevated levels of ATP with cellular senescence in vitro. Hum. Mol. Genet. 2006;15:607–623. doi: 10.1093/hmg/ddi477. [DOI] [PubMed] [Google Scholar]
- 33.Zuccato C., Belyaev N., Conforti P., Ooi L., Tartari M., Papadimou E., MacDonald M., Fossale E., Zeitlin S., Buckley N., et al. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington's disease. J. Neurosci. 2007;27:6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valenza M., Carroll J.B., Leoni V., Bertram L.N., Bjorkhem I., Singaraja R.R., Di Donato S., Lutjohann D., Hayden M.R., Cattaneo E. Cholesterol biosynthesis pathway is disturbed in YAC128 mice and is modulated by huntingtin mutation. Hum. Mol. Genet. 2007;16:2187–2198. doi: 10.1093/hmg/ddm170. [DOI] [PubMed] [Google Scholar]
- 35.Valenza M., Rigamonti D., Goffredo D., Zuccato C., Fenu S., Jamot L., Strand A., Tarditi A., Woodman B., Racchi M., et al. Dysfunction of the cholesterol biosynthetic pathway in Huntington's disease. J. Neurosci. 2005;25:9932–9939. doi: 10.1523/JNEUROSCI.3355-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuccato C., Tartari M., Crotti A., Goffredo D., Valenza M., Conti L., Cataudella T., Leavitt B.R., Hayden M.R., Timmusk T., et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 37.Futter M., Diekmann H., Schoenmakers E., Sadiq O., Chatterjee K., Rubinsztein D.C. Wild-type but not mutant huntingtin modulates the transcriptional activity of liver X receptors. J. Med. Genet. 2009;46:438–446. doi: 10.1136/jmg.2009.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borrell-Pages M., Zala D., Humbert S., Saudou F. Huntington's disease: from huntingtin function and dysfunction to therapeutic strategies. Cell. Mol. Life Sci. 2006;63:2642–2660. doi: 10.1007/s00018-006-6242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cattaneo E., Rigamonti D., Goffredo D., Zuccato C., Squitieri F., Sipione S. Loss of normal huntingtin function: new developments in Huntington's disease research. Trends Neurosci. 2001;24:182–188. doi: 10.1016/s0166-2236(00)01721-5. [DOI] [PubMed] [Google Scholar]
- 40.Zuccato C., Ciammola A., Rigamonti D., Leavitt B.R., Goffredo D., Conti L., MacDonald M.E., Friedlander R.M., Silani V., Hayden M.R., et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 41.MacDonald M.E. Huntingtin: alive and well and working in middle management. Sci. STKE. 2003;2003:pe48. doi: 10.1126/stke.2003.207.pe48. [DOI] [PubMed] [Google Scholar]
- 42.Caviston J.P., Holzbaur E.L. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell. Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faber P.W., Barnes G.T., Srinidhi J., Chen J., Gusella J.F., MacDonald M.E. Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 44.Godin J.D., Colombo K., Molina-Calavita M., Keryer G., Zala D., Charrin B.C., Dietrich P., Volvert M.L., Guillemot F., Dragatsis I., et al. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67:392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 45.Hilditch-Maguire P., Trettel F., Passani L.A., Auerbach A., Persichetti F., MacDonald M.E. Huntingtin: an iron-regulated protein essential for normal nuclear and perinuclear organelles. Hum. Mol. Genet. 2000;9:2789–2797. doi: 10.1093/hmg/9.19.2789. [DOI] [PubMed] [Google Scholar]
- 46.Kegel K.B., Sapp E., Alexander J., Valencia A., Reeves P., Li X., Masso N., Sobin L., Aronin N., DiFiglia M. Polyglutamine expansion in huntingtin alters its interaction with phospholipids. J. Neurochem. 2009;110:1585–1597. doi: 10.1111/j.1471-4159.2009.06255.x. [DOI] [PubMed] [Google Scholar]
- 47.Lumsden A.L., Henshall T.L., Dayan S., Lardelli M.T., Richards R.I. Huntingtin-deficient zebrafish exhibit defects in iron utilization and development. Hum. Mol. Genet. 2007;16:1905–1920. doi: 10.1093/hmg/ddm138. [DOI] [PubMed] [Google Scholar]
- 48.Truant R., Atwal R.S., Burtnik A. Nucleocytoplasmic trafficking and transcription effects of huntingtin in Huntington's disease. Prog. Neurobiol. 2007;83:211–227. doi: 10.1016/j.pneurobio.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 49.Valencia A., Reeves P.B., Sapp E., Li X., Alexander J., Kegel K.B., Chase K., Aronin N., DiFiglia M. Mutant huntingtin and glycogen synthase kinase 3-beta accumulate in neuronal lipid rafts of a presymptomatic knock-in mouse model of Huntington's disease. J. Neurosci. Res. 2010;88:179–190. doi: 10.1002/jnr.22184. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H., Das S., Li Q.Z., Dragatsis I., Repa J., Zeitlin S., Hajnoczky G., Bezprozvanny I. Elucidating a normal function of huntingtin by functional and microarray analysis of huntingtin-null mouse embryonic fibroblasts. BMC Neurosci. 2008;9:38. doi: 10.1186/1471-2202-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Battista N., Bari M., Tarditi A., Mariotti C., Bachoud-Levi A.C., Zuccato C., Finazzi-Agro A., Genitrini S., Peschanski M., Di Donato S., et al. Severe deficiency of the fatty acid amide hydrolase (FAAH) activity segregates with the Huntington's disease mutation in peripheral lymphocytes. Neurobiol. Dis. 2007;27:108–116. doi: 10.1016/j.nbd.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 52.Gines S., Seong I.S., Fossale E., Ivanova E., Trettel F., Gusella J.F., Wheeler V.C., Persichetti F., MacDonald M.E. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington's disease knock-in mice. Hum. Mol. Genet. 2003;12:497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- 53.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tian L., Greenberg S.A., Kong S.W., Altschuler J., Kohane I.S., Park P.J. Discovering statistically significant pathways in expression profiling studies. Proc. Natl. Acad. Sci. USA. 2005;102:13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.