Abstract
It has been frequently reported that seasonal changes in toxin production by cyanobacteria are due to changes in the proportion of toxic/nontoxic genotypes in parallel to increases or decreases in population density during the seasonal cycle of bloom formation. In order to find out whether there is a relationship between the proportion of genes encoding toxic peptide synthesis and population density of Planktothrix spp. we compared the proportion of three gene regions that are indicative of the synthesis of the toxic heptapeptide microcystin (mcyB), and the bioactive peptides aeruginoside (aerB) and anabaenopeptin (apnC) in samples from 23 lakes of five European countries (n=153). The mcyB, aerB, and apnC genes occurred in 99%, 99%, and 97% of the samples, respectively, and on average comprised 60 ± 3%, 22 ± 2%, and 54 ± 4% of the total population, respectively. Although the populations differed widely in abundance (10−3–103 mm3 L−1) no dependence of the proportion of the mcyB, aerB, and apnC genes on the density of the total population was found. In contrast populations differed significantly in their average mcyB, aerB, and apnC gene proportions, with no change between prebloom and bloom conditions. These results emphasize stable population-specific differences in mcyB, aerB, and apnC proportions that are independent from seasonal influences.
Keywords: cyanotoxins, eutrophication, Planktothrix, qPCR, spatial isolation, water monitoring
Introduction
Water-bloom-forming cyanobacteria threaten humans and livestock due to the production of toxic substances, most prominently the hepatotoxic heptapeptide microcystin. Microcystin and the closely related nodularin have been shown to inhibit eukaryotic protein phosphatases (PP) 1 and 2A. These toxins were responsible for tragedies, for example liver failure and the death of 52 patients in a hemodialysis center in Brazil (Carmichael et al., 2001). In addition to the microcystins, cyanobacteria have been shown to produce many other bioactive peptides (Welker & von Döhren, 2006). For example, aeruginosins are potent inhibitors of serine proteases, such as thrombin and trypsin (Ersmark et al., 2008), anabaenopeptins G, H, and T inhibit carboxypeptidase A (Itou et al., 1999), while the closely related oscillamide Y was shown to inhibit chymotrypsin (Sano & Kaya, 1995) and oscillamide C was shown to be a potent PP1 and 2A inhibitor (Sano et al., 2001). The synthesis of microcystin (MC), aeruginoside (AG), and anabaenopeptin (AP) is encoded by the nonribosomal peptide synthesis (NRPS) pathway (Tillett et al., 2000; Ishida et al., 2007; Rouhiainen et al., 2010;). The interspecific and intraspecific diversity in the production of bioactive nonribosomal peptides is known to be high, i.e. strains or colonies of cyanobacteria such as Microcystis sp. or Planktothrix spp. isolated from water samples show an impressive diversity in peptide production (Fastner et al., 2001; Welker et al., 2004;).
Quantitative real-time PCR (qPCR) has frequently been used to investigate environmental factors favoring harmful genotypes, i.e. genotypes indicative of the production of the toxic heptapeptide microcystin (Kurmayer & Kutzenberger, 2003). It was hypothesized that the seasonal succession of toxic (microcystin-producing) and nontoxic genotypes is a key mechanism determining microcystin concentrations in surface water (Kardinaal et al., 2007; Hotto et al., 2008;). Several studies reported the occurrence of major changes in the proportion of microcystin genotypes in relation to changes in environmental conditions, for example nitrate concentration (Yoshida et al., 2007), water temperature (Davis et al., 2009), and population density (Briand et al., 2008; Ye et al., 2009;). Some studies reported a decline in the ratio of toxic/nontoxic genotypes in the course of bloom development leading to a transition from an initial high toxicity to low toxicity at maximum cell numbers (Kardinaal et al., 2007; Briand et al., 2009; Bozarth et al., 2010;). Correspondingly, the analysis of clone libraries of variable ribosomal gene regions (Yoshida et al., 2005; Briand et al., 2009;) or the intergenic spacer of the phycocyanin operon (Bozarth et al., 2010) revealed significant changes in genotype composition that might explain the negative correlation between toxic genotype proportion and population density.
It is of interest to find out whether there is a general relationship between population density and the proportion of toxic genotypes or whether the toxic/nontoxic genotype ratio is influenced indirectly by seasonal shifts in genotype composition. In the latter case, it would be concluded that ecophysiological characters other than the ability to produce microcystin are selected for during bloom development. In order to quantify genes encoding microcystin, aeruginoside, and anabaenopeptin synthesis over a wide range in population abundance populations of the filamentous cyanobacterium Planktothrix were sampled from 23 lakes in Austria, Germany, the Netherlands, Denmark, and Norway. Planktothrix spp. reaches maximum cell numbers during summer, either in stratified layers of the water column (red-pigmented Planktothrix occurring in deep lakes of the Alps or in reservoirs) or in shallow turbid lakes (green-pigmented Planktothrix). Both ecotypes of Planktothrix are known to be highly adapted to low-light conditions and considered separate species (Suda et al., 2002).
Materials and methods
Sampling
During 2003 and 2004 four lakes, i.e. Lake Mondsee and Irrsee (Austria), Lake Wannsee (Germany), and Frederiksborg Slotssø (Denmark), were sampled monthly in the course of the EU project PEPCY (http://www.pepcy.de). Another 10 lakes were sampled at least three times a year during the growth season and another nine lakes were sampled once (Table 1). In addition, phytoplankton sampled monthly from Lake Wannsee during 1999 and 2000 was included (Kurmayer et al., 2003). Shallow polymictic lakes were sampled by collecting and combining 1 L each meter through the water column at maximum depth (e.g. Lake Wannsee and Frederiksborg Slotssø). Deep physically stratified lakes were sampled by combining 1 L from every 3 m to a depth of 20 m (e.g. Lake Mondsee, Wörthersee). Depending on the density of phytoplankton, 0.2–2 L were filtered onto glass fiber filters (Whatman GF/C) using low-vacuum filtration and filters were stored at −20 °C. DNA was extracted from the filters using the standard phenol–chloroform procedure (Kurmayer et al., 2003). Subsamples were Lugol fixed and analyzed for phytoplankton composition using the inverted microscope technique (Wetzel & Likens, 2000). The detection limit of Planktothrix spp. in the inverted microscope was one trichome per sedimentation chamber (50 mL volume). Planktothrix spp. was identified according to the morphological criteria described in Komárek (2003).
Table 1.
Origin of water samples
| Lake | Country | Latitude (°N) | Longitude (°E) | Surface area (km2) | Maximum depth (m) | Number of samples | Sampling period | Pigmentation of Planktothrix* | Abundance of Planktothrix† | Trophy |
|---|---|---|---|---|---|---|---|---|---|---|
| Intensively sampled lakes | ||||||||||
| Frederiksborg Slotssø | DK | 55°56′ | 12°18′ | 0.2 | 9 | 14 | Jul 03–Sep 04 | Green | A | Eutrophic |
| Irrsee | AT | 47°55′ | 13°18′ | 3.5 | 32 | 20 | Apr 03–Dec 04 | Red | A | Mesotrophic |
| Mondsee | AT | 47°49′ | 13°22′ | 13.8 | 68.3 | 27 | Apr 03 –Dec 04 | Red | A | Mesotrophic |
| Wannsee | DE | 52°25′ | 13°10′ | 2.7 | 10 | 27 | Jun 99–Oct 00 Aug 03–Dec 03 | Green | A | Eutrophic |
| Occasionally sampled lakes | ||||||||||
| Joppe | NL | 52°02′ | 04°50′ | 0.9 | 42 | 12 | Jul 03–Aug 05 | Green | R | Eutrophic |
| Klinkenberger Plas | NL | 52°04′ | 05°02′ | 0.3 | 35 | 4 | May 04–Aug 04 | Red/green | A | Eutrophic |
| Offensee | AT | 47°45′ | 13°50′ | 0.6 | 38 | 4 | Jul 03 Jul 04 | Red | A | Mesotrophic |
| Schwarzensee | AT | 47°45′ | 13°30′ | 0.5 | 54 | 6 | Apr 03–Oct 04 | Red | R | Oligotrophic |
| Slotermeer | NL | 52°09′ | 05°63′ | 12.4 | 6 | 3 | May 04–Oct 04 | Green | A | Eutrophic |
| Steinsfjorden | NO | 60°05′ | 10°19′ | 13.9 | 24 | 8 | Jun 03–Aug 04 | Red/green | A | Mesotrophic |
| Tjeukemeer | NL | 52°05′ | 05°50′ | 20 | 5 | 3 | May 04–Oct 04 | Green | A | Eutrophic |
| Wolfgangsee | AT | 47°45′ | 13°25′ | 12.8 | 113 | 5 | Jun 03–Nov 04 | Red | R | Oligotrophic |
| Wörthersee | AT | 46°37′ | 14°07′ | 19.4 | 85.2 | 4 | Aug 03–Nov 04 | Red | D | Mesotrophic |
| Zegerplas | NL | 52°13′ | 04°67′ | 0.7 | 34 | 7 | Jun 03–Aug 05 | Green | R | Eutrophic |
| Lakes sampled once | ||||||||||
| Albufera Lagoon | ES | 39°20′ | 0°21′W | 21 | 3 | 1 | Aug 04 | Green | A | Eutrophic |
| Hallwilersee | CH | 47°17′ | 08°12′ | 10 | 48 | 1 | Sep 04 | Red | D | Mesotrophic |
| Havel (Potsdam) | DE | 52°45′ | 12°10′ | - | 4 | 1 | Sep 04 | Green | A | Eutrophic |
| Lago Maggiore | IT | 45°28′ | 08°40′ | 212 | 372 | 1 | Sep 04 | Red | A | Mesotrophic |
| Miedwie | PL | 53°17′ | 14°54′ | 35.5 | 44 | 1 | Sep 04 | Red | A | Mesotrophic |
| Pozillo, Sicily | IT | 37°39′ | 14°35′ | 7.7 | 55 | 1 | Jan 06 | Red | D | Eutrophic |
| Prizzi, Sicily | IT | 37°43′ | 13°24′ | 1.3 | 44 | 1 | Feb 06 | Red | D | Eutrophic |
| Sapanca | TR | 40°69′ | 30°27′ | 46.8 | 55 | 1 | Sep 04 | Red | A | Mesotrophic |
| Talsperre Weida | DE | 50°42′ | 11°58′ | 0.9 | 22.5 | 1 | Sep 04 | Red | A | Mesotrophic |
Samples were grouped according to the visual inspection in the microscope: (1) red-pigmented; (2) green-pigmented; (3) red/green-pigmented.
A, abundant (<50%); R, rare (<10%); D, dominant (>50% in numbers according to the inspection in the microscope).
Design of the primers and probes
The TaqMan assay (TNA) was used to quantify (1) the intergenic spacer region of the phycocyanin operon (PC-IGS) to estimate the total population of Planktothrix, (2) the mcyBA1 gene encoding the first adenylation domain of the mcyB gene that is indicative of all Planktothrix cells containing the mcy gene cluster (Christiansen et al., 2003), (3) the aerB gene coding for an epimerase domain of the aeruginoside gene cluster in Planktothrix (Ishida et al., 2007), and (4) the apnC gene encoding the N-methyl transferase part of the apnCA2 adenylation domain that is involved in anabaenopeptin synthesis (Rouhiainen et al., 2010; R. Kurmayer and G. Christiansen, unpublished results).
Primers and the TaqMan probe that were specifically bound to PC-IGS or mcyBA1 of Planktothrix have been described previously (Schober & Kurmayer, 2006; Ostermaier & Kurmayer, 2009;). Although the mcyBA1 region was found to be, in part, affected by recombination events, a conserved region was identified based on 27 strains sequenced for the whole mcyBA1 domain previously (AJ890255–AJ890282; Kurmayer & Gumpenberger, 2006). For the design of the primers and probes to quantify the aerB gene coding for an epimerase domain of the aeruginoside gene cluster (Ishida et al., 2007) a 343-bp region was amplified and sequenced from 25 Planktothrix strains (GQ917083–GQ917107). From these sequences, the primers and probe were designed to amplify 89 bp of aerB (strain CCAP1459/36 had one substitution) using the primer express 2.0 software (Applied Biosystems, Vienna, Austria, Supporting Information, Tables S1, S2). In addition, the region encoding the N-methyl transferase part of the apnCA2 adenylation domain that is involved in anabaenopeptin synthesis (Rouhiainen et al., 2010) was amplified and sequenced from thirty strains (GQ917053–GQ917082). The primers and probe were designed from a region with a minimum sequence variation (142 bp): three strains (CYA126/8, CCAP1459/14, CCAP1459/31) showed a single-point mutation. The probes were labelled with a fluorescent reporter dye that was covalently attached to the 5′-end (FAM, 6-carboxyfluorescein) and a fluorescent quencher dye attached to the 3′-end (TAMRA, 6-carboxytetramethylrhodamine). Concentrations of the primers and probes were optimized according to the manufacturer's instructions (ABI TaqMan Universal PCR MasterMix, Table S1).
Quantitative real-time PCR (qPCR)
qPCR analysis was performed using the geneamp5700 as described (Kurmayer & Kutzenberger, 2003). Each sample was analyzed by four independent qPCRs set up from the same DNA extract. To establish the calibration curves, a dilution series of predetermined DNA concentrations from the extracts of Planktothrix strain PCC7821 (carrying the mcyB, aerB, and apnC gene) was prepared and the DNA content in the template (expressed in biovolume) was related to the threshold cycle (Ct) value (defined as the Ct to reach a manually set fluorescence) from 0.55 × 10−8 mm3 (4.1 cells)−0.55 × 10−5 mm3 (4100 cells) per template (Table 2). Proportions of mcyB, aerB, and apnC genes were calculated from the biovolume of the total population as determined via the PC-IGS region by qPCR from the same DNA extract. Calibration curves were not extrapolated beyond the highest dilution, which was defined arbitrarily as the limit of quantification corresponding to 0.55 × 10−8 mm3 in the template. The specificity and robustness of each TNA was tested by adding DNA originating from other organisms as a background. DNA extracted from the Microcystis strains HUB524, HUB53, and Synechococcus strain MW15#2SUB was added to the DNA of strain PCC7821 in two concentrations, 1.7 × 104 and 1.7 × 105 cells per template consisting of 13% of strain HUB53, 11% of strain HUB524, and 76% of strain MW15#2SUB, respectively. The addition of the background did not influence the Ct value for strain PCC7821 in the range of 0.55 × 10−8–0.55 × 10−5 mm3. All results on mcyB, aerB, and apnC abundance were compared using a nonparametric test (Kruskal—Wallis one way anova on Ranks). If an overall significant difference between lake groups was found (P<0.05), pair-wise post hoc comparisons were performed using the Dunn test (sigma plot for Windows Version 11.0).
Table 2.
Calibration curves used for qPCR in the present study
| qPCR | Gene region coding for | Calibration curve | E (%)* | R2 | n | P |
|---|---|---|---|---|---|---|
| PC-IGS | Intergenic spacer region of the phycocyanin operon | y=−3.895x+9.149 | 81 | 0.997 | 4 | < 0.001 |
| Pl mcyBA1 | First adenylation domain of the mcyB gene | y=−3.520x+11.372 | 92 | 0.995 | 4 | < 0.001 |
| Pl aerug | Epimerase of the aerB gene | y=−3.316x+11.601 | 100 | 0.982 | 4 | < 0.001 |
| Pl NMT | N-methyl transferase of the apnC gene | y=−4.032x+15.045 | 77 | 0.936 | 4 | < 0.001 |
The amplification efficiency (E) was calculated as follows: E=(10−1/slope−1) × 100.
y, the number of PCR cycles at the set fluorescence threshold level (Ct-value at 0.1); x, the amount of DNA in the template (as log 10 mm3 of biovolume).
Results
Biovolume of Planktothrix spp
The phytoplankton consisted of cyanobacteria, diatoms, chrysophytes, cryptophytes, dinophytes, and green algae. Average biovolume of the total phytoplankton differed according to the lakes' trophic state, i.e. the lakes located in the Alps and Lake Steinsfjorden in Norway were oligotrophic to mesotrophic, while lakes located in Denmark, Germany, and the Netherlands were classified as eutrophic (Table 1). In general, Planktothrix biovolume varied from 10−3 to 103 mm3 L−1 (n=146). In seven samples, no Planktothrix either by qPCR or by the microscope was detected. There was a clear linear correlation between microscopical detection and detection of Planktothrix by qPCR: y=0.909x−0.441 (R2=0.72; P<0.0001; n=101), where y was log10 biovolume as determined by the qPCR and x was log10 biovolume as determined by the microscope (Fig. S1). Surprisingly for samples from Lake Offensee (Austria) and Talsperre Weida (Germany), only irregular amplification curves by the PC-IGS assay were obtained. As these amplification curves made an estimate of the total population density impossible, these five samples were excluded from further analysis.
According to the pigmentation of Planktothrix, the samples were assigned to the following groups: (1) samples obtained from red-pigmented populations including lakes located within the Alps (n=65), for example Lake Mondsee, Lake Irrsee, and reservoirs (Pozzillo, Prizzi, Sapanca); (2) samples obtained from green-pigmented populations typically occurring in shallow eutrophic lakes (n=64), for example Lake Wannsee and Frederiksborg Slotssø; and (3) samples obtained from red/green-pigmented populations (n=12), for example Lake Steinsfjorden, Klinkenberger Plas (Table 1). Within the red or green pigmentation type, the samples were assigned to three categories according to population density: (1) dense populations: >1.0 mm3 L−1, minimum–median–maximum=1.1–2.7–903 mm3 L−1, n=6 (red), n=21 (green), n=4 (mixed); (2) moderately dense populations: 0.1–1.0 mm3 L−1, minimum–median–maximum=0.1–0.28–0.92, n=17 (red), n=16 (green), n=3 (mixed); and (3) sparse populations: <0.1 mm3 L−1, minimum–median–maximum=2.6 × 10−3–0.03–0.09, n=42 (red), n=27 (green), n=5 (mixed).
Abundances of the mcyB, aerB, and apnC genes
The mcyB, aerB, and apnC genes occurred in 99%, 99%, and 97% of all samples (n=119) that contained Planktothrix spp. above the quantification threshold (0.55 × 10−8 mm3 or 4.1 cells in the template). Below the quantification threshold of Planktothrix, the results were more variable and mcyB, aerB, and apnC genes occurred only in 96%, 19%, and 7% of the samples (n=27), respectively. It is likely that this lower frequency of aerB and apnC resulted from the lower sensitivity of the respective qPCR assays when compared with the qPCR assay for mcyB (Table 2).
Notably, the abundance of all three mcyB, aerB, and apnC genes was linearly correlated to the total Planktothrix biovolume as estimated by qPCR: for mcyB the linear regression curve was y=1.010x−0.441 (R2=0.78, n=133), for aerB the regression curve was y=1.253x−0.910 (R2=0.71, n=123), and for the apnC the regression curve was y=1.095x−0.545 (R2=0.76, n=122), where y was the log10 biovolume of the respective gene region and x was the log10 biovolume of the total population as estimated via the PC-IGS region by qPCR (Fig. 1).
Fig. 1.
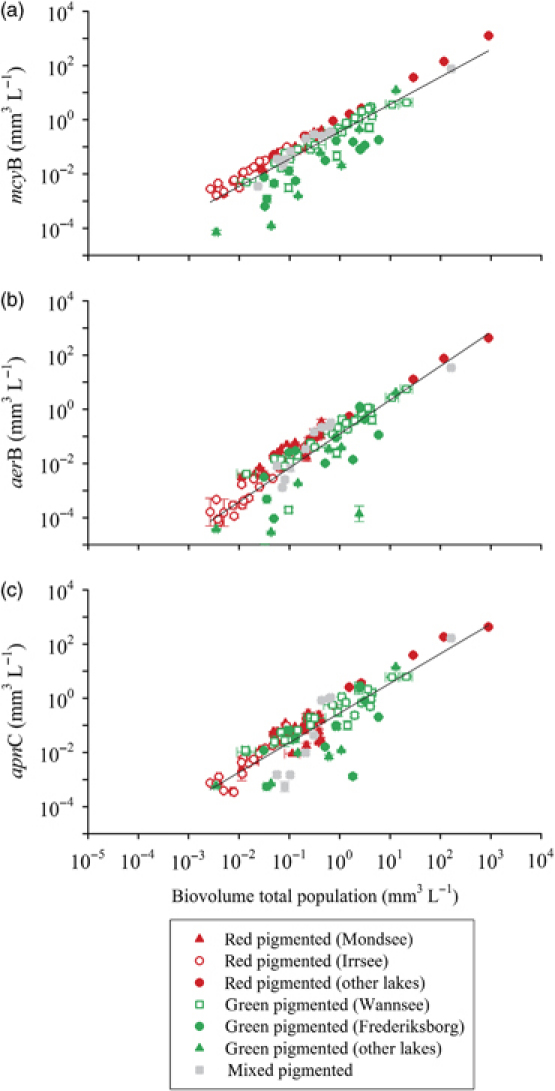
Relationship between Planktothrix spp. abundance and (a) the mcyB (microcystin) gene abundance, (b) the aerB (aeruginoside) gene abundance, and (c) the apnC (anabaenopeptin) gene abundance (mean ± 1 SE mm3 of biovolume L−1). For details on the regression curves see text.
Proportions of the mcyB, aerB, and apnC genes
The average proportions of the mcyB, aerB, and apnC genes were 60 ± 3%, 22 ± 2%, and 54 ± 4% respectively. On average the proportions of all three mcyB, aerB, and apnC genes were significantly higher among red-pigmented when compared with all the green-pigmented populations, i.e. red-pigmented: mcyB: 86 ± 3% (n=57); aerB: 26 ± 2% (n=55); apnC: 60 ± 6% (n=52) vs. green-pigmented: mcyB: 31 ± 4% (n=48); aerB: 16 ± 2% (n=46); apnC: 43 ± 5% (n=43), (Mann–Whitney rank-sum test, mcyB, aerB: P≤0.001, apnC: P=0.059, Figs 2 and 3). Within each pigmentation type, the mcyB proportion differed spatially between populations, for example, it was higher among samples from Lake Wannsee (43.0 ± 4%, n=26) than among samples from Lake Frederiksborg Slotssø (8 ± 2%, n=13, P<0.001, Fig. 2a). Analogously, the average aerB proportion differed significantly not only between red/green-pigmented populations, but also within a pigmentation type (P<0.001). When compared with Lake Mondsee (34 ± 3%, n=27) the aerB proportion in Lake Irrsee was significantly reduced (11 ± 2%, n=18), while the aerB proportion in Lake Wannsee (19 ± 2%, n=26) differed significantly from all other green-pigmented samples (6 ± 4%, n=7, Fig. 2b). Although less pronounced, a spatial difference was also found for the average apnC proportion within red/green-pigmented populations (P<0.001, Fig. 2c). It is concluded that Planktothrix populations differed in their average mcyB, aerB, and apnC proportion, which is not restricted to the difference between the red- or green-pigmented groups, but also occurs within the same pigmentation type.
Fig. 2.
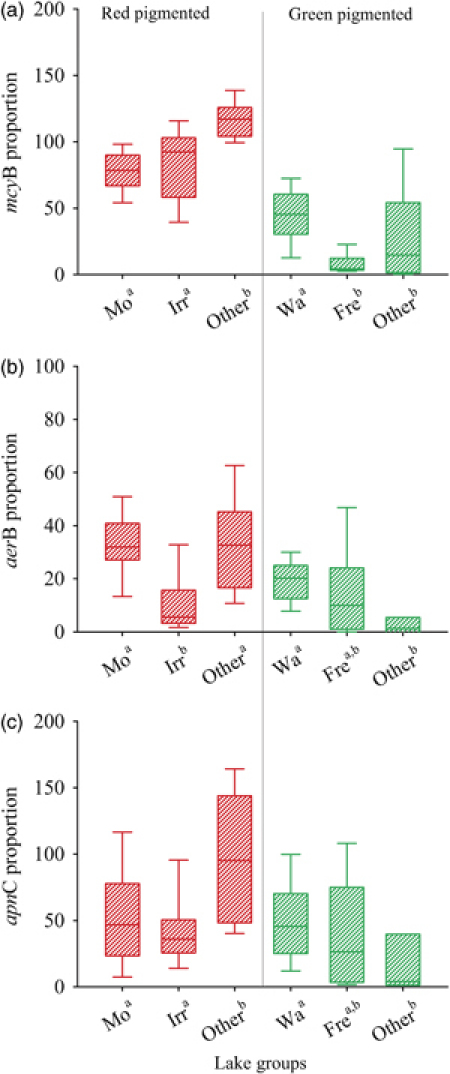
Proportions (%) of (a) mcyB, (b) aerB, and (c) apnC among red- or green-pigmented Planktothrix spp. Mo, Lake Mondsee (Austria); Irr, Lake Irrsee; Wa, Lake Wannsee; Fre, Frederiksborg Slotssø; other, occasionally sampled lakes and lakes sampled only once. The whiskers of each box indicate the 10th and 90th percentiles. If an overall difference between lake groups within a pigmentation type was found, superscripts (a, b) indicate homogeneous subgroups not significantly different at P<0.05.
Fig. 3.
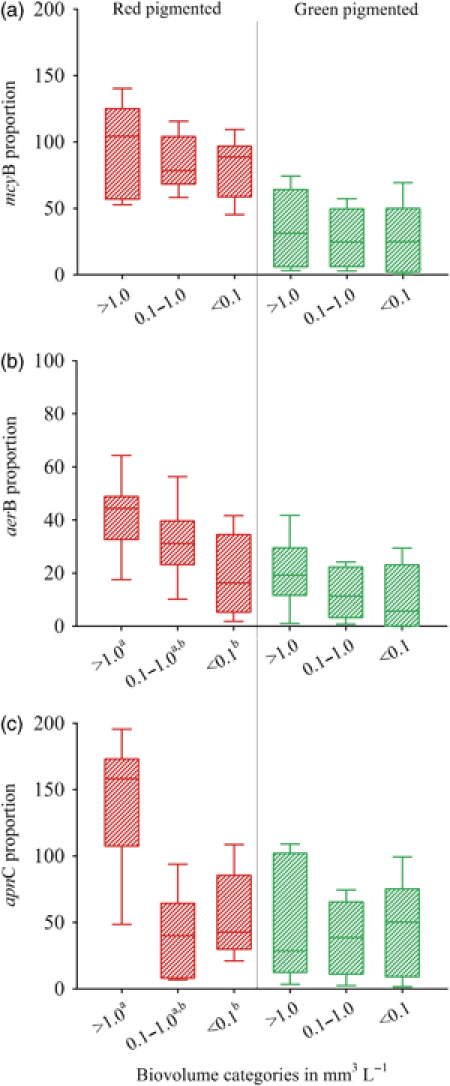
Proportions (%) of (a) mcyB, (b) aerB, and (c) apnC within three different population density categories of red- or green-pigmented Planktothrix spp.: dense,>1.0 mm3 L−1; moderately dense, 0.1–1.0 mm3 L−1; sparse,<0.1 mm3 L−1. The whiskers of each box indicate the 10th and 90th percentiles. If an overall difference between lake groups within a pigmentation type was found, superscripts (a, b) indicate homogeneous subgroups not significantly different at P<0.05.
In contrast to the spatial differences, the mcyB, aerB, and apnC proportions varied independently between dense (>1.0 mm3 L−1), moderately dense (0.1–1.0 mm3 L−1), and sparse (<0.1 mm3 L−1) population density categories among green-pigmented populations (P>0.05, Fig. 3). Among red-pigmented populations, the aerB and apnC proportion of the dense populations differed significantly from the moderately dense and sparse density categories (P<0.002, Fig. 3b and c). However, a Pearson product–moment correlation analysis revealed no relationship between mcyB, aerB, or apnC proportions and total population density within either green- or red-pigmented populations (R<0.5). Besides the variation in population abundance between populations, the total Planktothrix biovolume varied seasonally within lakes, i.e. 38-, 33-, 1650-, and 189-fold in Lake Mondsee, Lake Irrsee, Lake Wannsee, and Lake Frederiksborg Slotssø, respectively (Fig. 4). The Pearson product–moment correlation analysis did not reveal any significant relation between the mcyB proportion and total population density in all four lakes (R<0.5) and also no relationship between aerB and apnC proportion and population density in Lake Mondsee, Wannsee, and Frederiksborg Slotssø. Only among the samples obtained from Lake Irrsee a positive correlation between aerB and apnC proportion and the total population density was found (aerB: R=0.78; apnC: R=0.82). It is concluded that, within a pigmentation type, the mcyB, aerB, and apnC proportions were not related to population density between lakes and in the majority of samples also not within lakes during seasonal succession.
Fig. 4.
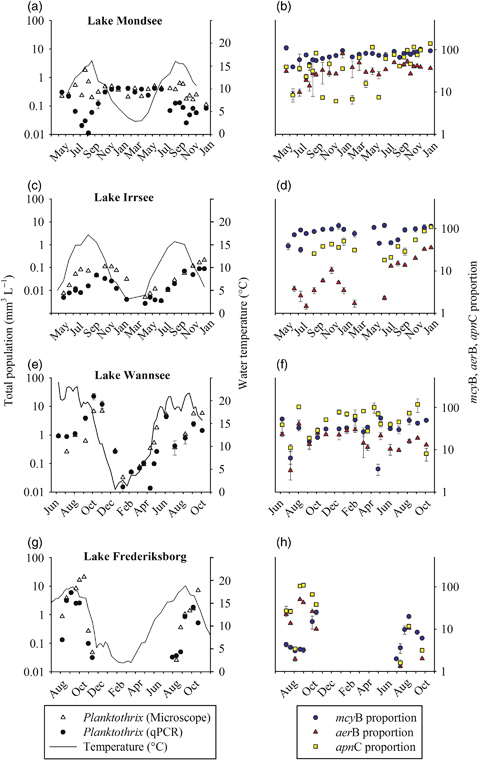
Total population density of Planktothrix spp. as counted in the microscope and estimated via qPCR and seasonal variability in the mcyB, aerB, and apnC proportion (mean ± SE in percent), in (a, b) Lake Mondsee, (c, d) Lake Irrsee, (e, f) Lake Wannsee, (g, h), and Lake Frederiksborg Slotssø. The straight line indicates water temperature integrated either from 0 to 20 m (Lake Mondsee, Lake Irrsee) or the total water column.
Discussion
Estimation of mcyB, aerB, and apnC gene abundance by qPCR
On a genetic level, recombination affecting particular gene regions can lead to an over- or underestimation of proportions of particular gene regions in a population. Both for PC-IGS and mcyBA1, recombination events have been described (e.g. Manen & Falquet, 2002; Kurmayer & Gumpenberger, 2006;). While for a particular gene region the influence of recombination can never be excluded, this potential bias has been minimized by sequencing the amplified gene regions in a larger number of strains isolated from nine European countries and 28 water bodies (Table S2). These strains did not show signs of past gene conversion (recombination) events. However, it cannot be excluded that the occurrence of single base pair substitutions within the amplified PC-IGS region resulted in the mcyB and apnC proportions over 100% as observed for the most dense populations of red-pigmented populations (>1.0 mm3 L−1, Fig. 3). It also seems likely that recombination events caused the irregular amplification curves of the PC-IGS assay as observed for samples from Lake Offensee (Austria) and Talsperre Weida (Germany). Two alternative qPCR reference gene assays to quantify the total population targeting either the 16S rRNA gene or the 16S rRNA gene internal transcribed spacer region were used to control the estimates by the PC-IGS region for the whole data set (data not shown). While both rRNA gene assays gave useful PCR amplification curves for the samples from Lake Offensee and Talsperre Weida, the application of these alternative qPCR assays did not eliminate the mcyB and apnC proportions over 100%. The three different reference gene assays were highly significantly related (R≥0.96, n=146); however, the PC-IGS assay was found to have the highest correlation coefficient in explaining the variation among Planktothrix biovolume as detected in the microscope (Fig. S1). It has been argued during the reviewing process of this paper that the mcyB, aerB, and apnC proportions should be related to one and the same reference gene assay. Nevertheless, it seems reasonable that combining several gene probes to amplify both the total population, as well as specific genes encoding toxin synthesis, will control the influence of unknown recombination events and therefore contribute to the development of a more robust qPCR protocol.
Abundances of the mcyB, aerB, and apnC genes
The results of this study imply that among European Planktothrix populations genes encoding toxin synthesis occurred frequently and varied independently in their proportion from the total population density over a wide range in population abundance as well as ecosystems that differ in trophy and morphometry, such as water depth (shallow lakes vs. deep reservoirs and deep lakes in the Alps). Indeed the abundance of the total population was a strong predictor of mcyB, aerB, and apnC abundance, implying that those populations containing such a gene are unlikely to lose it in the short term and become nontoxic in the course of seasonal development (Fig. 1). These results are relevant for our understanding of the likelihood of the occurrence of toxic algal blooms under specific environmental conditions.
Although genetic methods are only able to indicate the potential of toxic peptide synthesis, it has been shown previously that gene numbers estimated by qPCR can be used to infer concentrations of different peptide classes such as the nodularins (Koskenniemi et al., 2007) or the microcystins (Hotto et al., 2008; Okello et al., 2010; Ostermaier & Kurmayer, 2010;). On the other hand chemical methods such as MALDI-TOF MS or LC-MS typically have a much higher resolution and resolve a large number of individual peptide structural variants (Welker & Erhard, 2007; Rohrlack et al., 2008;). The drawback of the chemical methods is the requirement for purified standard substances in order to estimate individual peptide concentrations (e.g. Halstvedt et al., 2008). During this study's period, peptide concentrations have been determined by LC-MS for samples obtained from Lake Mondsee and Lake Irrsee on a relative scale (using MC-LR as an internal standard, R. Kurmayer, T. Rohrlack, NIVA, unpublished data). It is interesting to note that when related to Planktothrix biovolume, the peptide aeruginosin A (m/z 617) was found on average fourfold more abundant in samples from Lake Mondsee (n=40) when compared with samples obtained from Lake Irrsee (n=18). In contrast, the same ratio calculated for the sum of MC-RR (m/z 1024) and MC-LR (m/z 981) as well as the sum of AP-B (m/z 837) and AP-F (m/z 851) showed no difference between the two lakes. This difference in peptide concentrations corresponds with the mcyB, aerB, and apnC proportions reported in those two lakes (Figs 2 and 4). It is concluded that gene numbers and proportions estimated by qPCR can be used to estimate the population-specific production of bioactive and toxic peptides such as aeruginosides and anabaenopeptins in water bodies.
While Planktothrix populations showed relatively minor changes in their average toxic genotype proportion between prebloom and bloom conditions, the spatial divergence in the mcyB, aerB, and apnC proportion between populations was pronounced. This implies that although variation in the mcyB proportion within the same population during the season was recorded, the measurements on the mcyB ratio between populations were consistently different (Fig. 2). For example, Planktothrix in Lake Wannsee (Germany) not only never contained mcyB in all cells (as observed among red-pigmented populations) but also never contained such a low percentage of mcyB as observed in the samples of Lake Frederiksborg Slotssø (Denmark). We reported previously that the individual Planktothrix populations differed significantly in the occurrence of specific mcyB genotypes over several years (Kurmayer & Gumpenberger, 2006). Correspondingly, a pronounced spatial divergence in the proportion of the microcystin genotype has been observed for six spatially isolated populations of Microcystis sampled during 1 year in Uganda, East Africa (Okello et al., 2010). In a similar manner, Sabart et al. (2010) reported spatial differences in the microcystin genotype proportion of Microcystis sampled from several interconnected sites along the Loire River in France. The authors concluded that the relative selection of the microcystin-producing and non-microcystin-producing genotype occurs at the scale of each ecosystem and depends on many local environmental factors and processes. It is likely that as opposed to a single environmental factor, the proportion of mcyB, aerB, and apnC genes is influenced by multiple factors, both on a spatial scale and in the course of seasonal succession. Our conclusion is important in the light of experimental studies that attempt to find environmental factors that regulate the proportion of toxic genotypes in populations of toxic cyanobacteria such as Planktothrix or Microcystis. It has been shown earlier that nontoxic and toxic strains have diversified by factors not directly linked to microcystin production (Tanabe et al., 2007; Christiansen et al., 2008;). Probably, the benefits/costs solely attributable to microcystin, aeruginoside, and anabaenopeptin synthesis under varying population density conditions can only be estimated by comparing the growth between a generated mutant deficient in microcystin, aeruginoside, and anabaenopeptin production vs. the wildtype strain (Hesse et al., 2001). In the future, it would be required to perform competition experiments between Planktothrix strains and specific mutants under varying growth-limiting conditions in order to estimate the extent of benefits/costs of either of the three peptide classes on the cellular growth rate.
Acknowledgments
We are grateful to Gertraud Roidmayr, Michael Werndl, and Johanna Schmidt for microscopical counting, assisting in the laboratory, and taking samples in Lake Mondsee. Günther Bruschek, Karl Mayrhofer, and Reinhold Trutzka from the Federal Agency for Water Management in Scharfling (Upper Austria) sampled Lake Irrsee, Lake Offensee, Lake Schwarzensee, and Lake Wolfgangsee. Gabriele Wessel provided samples from Lake Wannsee during 2003. Trine Warming Perlt and Kirsten Christoffersen provided samples and phytoplankton data from Frederiksborg Slotssø. Thomas Rohrlack provided samples from Lake Steinsfjorden. We are also grateful for the comments from three anonymous reviewers to an earlier draft of this manuscript. This study was financially supported by the EU project PEPCY (QLK4-CT-2002-02634) and the Austrian Science Fund (FWF P18185, P20231).
Statement
Re-use of this article is permitted in accordance with the Terms and Conditions set out at: http://wileyonlinelibrary.com/onlineopen#OnlineOpen-Terms
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Comparison of the total Planktothrix spp. biovolume (mm3 L−1) estimated from lake water samples (Table 1) by counting under the microscope and by qPCR, mean ±1 SE.
Table S1. Oligonucleotides used for PCR and sequencing, and primers and probes used for qPCR to quantify six genotypes of Planktothrix spp.
Table S2. Planktothrix strains used in this study and sequences used to design TaqMan probes for quantitative real-time PCR.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Bozarth CS, Schwartz AD, Shepardson JW, Colwell FS, Dreher TW. Population turnover in a Microcystis bloom results in predominantly nontoxigenic variants late in the season. Appl Environ Microbiol. 2010;76:5207–5213. doi: 10.1128/AEM.00001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briand E, Gugger M, Francois J-C, Bernard C, Humbert J-F, Quiblier C. Temporal variations in the dynamics of potentially microcystin-producing strains in a bloom-forming Planktothrix agardhii (Cyanobacterium) population. Appl Environ Microbiol. 2008;74:3839–3848. doi: 10.1128/AEM.02343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briand E, Escoffier N, Straub C, Sabart M, Quiblier C, Humbert JF. Spatiotemporal changes in the genetic diversity of a bloom-forming Microcystis aeruginosa (Cyanobacteria) population. ISME J. 2009;3:419–429. doi: 10.1038/ismej.2008.121. [DOI] [PubMed] [Google Scholar]
- Carmichael W, Azevedo S, An J, Molica R, Jochimsen E, Lau S, Rinehart K, Shaw G, Eaglesham G. Human fatalities from cyanobacteria: chemical and biological evidence for cyanotoxins. Environ Health Perspect. 2001;109:663–668. doi: 10.1289/ehp.01109663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G, Fastner J, Erhard M, Börner T, Dittmann E. Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J Bacteriol. 2003;185:564–572. doi: 10.1128/JB.185.2.564-572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G, Molitor C, Philmus B, Kurmayer R. Nontoxic strains of cyanobacteria are the result of major gene deletion events induced by a transposable element. Mol Biol Evol. 2008;25:1695–1704. doi: 10.1093/molbev/msn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TW, Berry DL, Boyer GL, Gobler CJ. The effects of temperature and nutrients on the growth and dynamics of toxic and non-toxic strains of Microcystis during cyanobacteria blooms. Harmful Algae. 2009;8:715–725. [Google Scholar]
- Ersmark K, Del Valle JR, Hanessian S. Chemistry and biology of the aeruginosin family of serine protease inhibitors. Angew Chem Int Ed. 2008;47:1202–1223. doi: 10.1002/anie.200605219. [DOI] [PubMed] [Google Scholar]
- Fastner J, Erhard M, von Döhren H. Determination of oligopeptide diversity within a natural population of Microcystis (Cyanobacteria) by typing single colonies by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol. 2001;67:5069–5076. doi: 10.1128/AEM.67.11.5069-5076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstvedt CB, Rohrlack T, Ptacnik R, Edvardsen B. On the effect of abiotic environmental factors on production of bioactive oligopeptides in field populations of Planktothrix spp. (Cyanobacteria) J Plankton Res. 2008;30:607–617. [Google Scholar]
- Hesse K, Dittmann E, Börner T. Consequences of impaired microcystin production for light-dependent growth and pigmentation of Microcystis aeruginosa PCC 7806. FEMS Microb Ecol. 2001;37:39–43. [Google Scholar]
- Hotto A, Satchwell M, Berry D, Gobler C, Boyer G. Spatial and temporal diversity of microcystins and microcystin-producing genotypes in Oneida Lake, NY. Harmful Algae. 2008;7:671–681. [Google Scholar]
- Ishida K, Christiansen G, Yoshida WY, Kurmayer R, Welker M, Bonjoch J, Hertweck C, Börner T, Hemscheidt T, Dittmann E. Biosynthetic pathway and structure analysis of aeruginoside 126A and B, cyanobacterial peptide glycosides bearing an unusual 2-carboxy-6-hydroxyoctahydroindole moiety. Chem Biol. 2007;14:565–576. doi: 10.1016/j.chembiol.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itou Y, Suzuki S, Ishida K, Murakami M. Anabaenopeptins G and H, potent carboxypeptidase A inhibitors from the cyanobacterium Oscillatoria agardhii (NIES-595) Bioorg Med Chem Lett. 1999;9:1243–1246. doi: 10.1016/s0960-894x(99)00191-2. [DOI] [PubMed] [Google Scholar]
- Kardinaal WEA, Janse I, Kamst-Van Agterveld M, Meima M, Snoek J, Mur LR, Huisman J, Zwart G, Visser PM. Microcystis genotype succession in relation to microcystin concentrations in freshwater lakes. Aqu Microb Ecol. 2007;20:1–12. [Google Scholar]
- Komárek J. Planktic oscillatorialean cyanoprokaryotes (short review according to combined phenotype and molecular aspects) Hydrobiologia. 2003;502:367–382. [Google Scholar]
- Koskenniemi K, Lyra C, Rajaniemi-Wacklin P, Jokela J, Sivonen K. Quantitative real-time PCR detection of toxic Nodularia cyanobacteria in the Baltic Sea. Appl Environ Microbiol. 2007;73:2173–2179. doi: 10.1128/AEM.02746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmayer R, Gumpenberger M. Diversity of microcystin genotypes among populations of the filamentous cyanobacteria Planktothrix rubescens and Planktothrix agardhii. Mol Ecol. 2006;15:3849–3861. doi: 10.1111/j.1365-294X.2006.03044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmayer R, Kutzenberger T. Application of real-time PCR for quantification of microcystin genotypes in a population of the toxic cyanobacterium Microcystis sp. Appl Environ Microbiol. 2003;69:6723–6730. doi: 10.1128/AEM.69.11.6723-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmayer R, Christiansen G, Chorus I. The abundance of microcystin-producing genotypes correlates positively with colony size in Microcystis and determines its microcystin net production in Lake Wannsee. Appl Environ Microbiol. 2003;69:787–795. doi: 10.1128/AEM.69.2.787-795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manen J-F, Falquet J. The cpcB-cpcA locus as a tool for the genetic characterization of the genus Arthrospira (cyanobacteria): evidence for horizontal transfer. Int J Sys Evol Microbiol. 2002;52:861–867. doi: 10.1099/00207713-52-3-861. [DOI] [PubMed] [Google Scholar]
- Okello W, Ostermaier V, Portmann C, Gademann K, Kurmayer R. Spatial isolation favours the divergence in microcystin net production by Microcystis in Ugandan freshwater lakes. Water Res. 2010;44:2803–2814. doi: 10.1016/j.watres.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermaier V, Kurmayer R. Distribution and abundance of nontoxic mutants of cyanobacteria in lakes of the Alps. Microb Ecol. 2009;58:323–333. doi: 10.1007/s00248-009-9484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermaier V, Kurmayer R. Application of real-time PCR to estimate toxin production by the cyanobacterium Planktothrix sp. Appl Environ Microbiol. 2010;76:3495–3502. doi: 10.1128/AEM.02771-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrlack T, Edvardsen B, Skulberg R, Halstvedt CB, Utkilen HC, Ptacnik R, Skulberg OM. Oligopeptide chemotypes of the toxic freshwater cyanobacterium Planktothrix can form subpopulations with dissimilar ecological traits. Limnol Oceanogr. 2008;53:1279–1293. [Google Scholar]
- Rouhiainen L, Jokela J, Fewer DP, Urmann M, Sivonen K. Two alternative starter modules for the non-ribosomal biosynthesis of specific anabaenopeptin variants in Anabaena (Cyanobacteria) Chem Biol. 2010;17:265–273. doi: 10.1016/j.chembiol.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Sabart M, Pobel D, Briand E, Combourieu B, Salencon MJ, Humbert JF, Latour D. Spatiotemporal variations in microcystin concentrations and in the proportions of microcystin-producing cells in several Microcystis aeruginosa populations. Appl Environ Microbiol. 2010;76:4750–4759. doi: 10.1128/AEM.02531-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Kaya K. Oscillamide Y, a chymotrypsin inhibitor from toxic Oscillatoria agardhii. Tetrahedron Lett. 1995;36:5933–5936. doi: 10.1021/np9600210. [DOI] [PubMed] [Google Scholar]
- Sano T, Usui T, Ueda K, Osada H, Kaya K. Isolation of new protein phosphatase inhibitors from two cyanobacteria species, Planktothrix spp. J Nat Prod. 2001;64:1052–1055. doi: 10.1021/np0005356. [DOI] [PubMed] [Google Scholar]
- Schober E, Kurmayer R. Evaluation of different DNA sampling techniques for the application of the real-time PCR method for the quantification of cyanobacteria in water. Lett Appl Microbiol. 2006;42:412–417. doi: 10.1111/j.1472-765X.2006.01857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, Liu Y, Day JG. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Microbiol. 2002;52:1577–1595. doi: 10.1099/00207713-52-5-1577. [DOI] [PubMed] [Google Scholar]
- Tanabe Y, Kasai F, Watanabe MM. Multilocus sequence typing (MLST) reveals high genetic diversity and clonal population structure of the toxic cyanobacterium Microcystis aeruginosa. Microbiol. 2007;153:3695–3703. doi: 10.1099/mic.0.2007/010645-0. [DOI] [PubMed] [Google Scholar]
- Tillett D, Dittmann E, Erhard M, von Döhren H, Börner T, Neilan BA. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide–polyketide synthetase system. Chem Biol. 2000;7:753–764. doi: 10.1016/s1074-5521(00)00021-1. [DOI] [PubMed] [Google Scholar]
- Welker M, Erhard M. Consistency between chemotyping of single filaments of Planktothrix rubescens (Cyanobacteria) by MALDI-TOF and the peptide patterns of strains determined by HPLC-MS. J Mass Spec. 2007;42:1062–1068. doi: 10.1002/jms.1237. [DOI] [PubMed] [Google Scholar]
- Welker M, von Döhren H. Cyanobacterial peptides – Nature's own combinatorial biosynthesis. FEMS Microbiol Rev. 2006;30:530–563. doi: 10.1111/j.1574-6976.2006.00022.x. [DOI] [PubMed] [Google Scholar]
- Welker M, Christiansen G, von Döhren H. Diversity of coexisting Planktothrix (cyanobacteria) chemotypes deduced by mass spectral analysis of microystins and other oligopeptides. Arch Microbiol. 2004;182:288–298. doi: 10.1007/s00203-004-0711-3. [DOI] [PubMed] [Google Scholar]
- Wetzel RG, Likens GE. Limnological Analyses. 3rd edn. New York: Springer-Verlag; 2000. [Google Scholar]
- Ye WJ, Liu XL, Tan J, Li DT, Yang H. Diversity and dynamics of microcystin-producing cyanobacteria in China's third largest lake, Lake Taihu. Harmful Algae. 2009;8:637–644. [Google Scholar]
- Yoshida M, Yoshida T, Takashima Y, Kondo R, Hiroishi S. Genetic diversity of the toxic cyanobacterium Microcystis in Lake Mikata. Environ Toxicol. 2005;20:229–234. doi: 10.1002/tox.20102. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Yoshida T, Takashima Y, Hosoda N, Hiroishi S. Dynamics of microcystin-producing and non-microcystin-producing Microcystis populations is correlated with nitrate concentration in a Japanese lake. FEMS Microbiol Lett. 2007;266:49–53. doi: 10.1111/j.1574-6968.2006.00496.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


