Abstract
Heat shock proteins (Hsp) are known to protect cells from several stressors. Nucleotide changes in the flanking regions [5′- and 3′-untranslated region (UTR)] of Hsp gene might affect inducibility, degree of expression, or stability of Hsp70 mRNA. The present study aimed to investigate the association between inducible Hsp70.1 single nucleotide polymorphisms (SNPs) and heat shock (HS) response of peripheral blood mononuclear cells (PBMC) in dairy cows. Four hundred forty-six Italian Holstein cows were genotyped for four Hsp70.1 SNPs: g895 C/- and g1128 G/T in 5′-UTR, and g2154 G/A and g64 G/T in 3′-UTR. Genetic polymorphisms in 3′-UTR of bovine Hsp70.1 gene resulted monomorphic. Distribution of alleles of the nucleotide sequence polymorphism within the 5′-UTR of the bovine Hsp70.1 gene were 81.2% and 18.8% for C and -, respectively, and 77.8% and 22.2% for G and T, respectively. Among the 446 genotyped animals, a group of cows balanced for days in milk and parity was selected to be representative of the following genotypes: CC (n = 8), C- (n = 7), and -- (n = 7) and GG (n = 8), GT (n = 11), and TT (n = 3) in 5′-UTR. PBMC were isolated from blood samples and heated at 43°C in thermal bath for 1 h and then incubated at 39°C in atmosphere of 5% CO2 for 1, 2, 4, 8, 16, and 24 h (recovery times). Cell viability was determined by XTT assay. Gene and protein expression of Hsp70.1 was determined by real-time reverse transcription-polymerase chain reaction and by ELISA assay, respectively. For the two SNPs detected, one allele was the most frequent (C, 66.8% and G, 56.8%). Genotypes -- and TG showed higher (P < 0.05) viability compared with CC and GG, respectively. Genotypes C- and TT had intermediate viability. Gene expression of Hsp70.1 showed higher (P < 0.001) levels in -- and TG genotype compared with their counterparts. Genotypes -- and TG showed the higher level of inducible Hsp70.1 protein in respect to C-, TT and CC, GG. In conclusion, exposure to HS differently affected cell viability and gene and protein expression of Hsp70.1 in the selected genotypes. These results indicate that the presence of SNPs (C/- and G/T) in the 5′-UTR region of inducible Hsp70.1 ameliorates HS response and tolerance to heat of bovine PBMC. These mutation sites may be useful as molecular genetic markers to assist selection for heat tolerance.
Keywords: Hsp70.1, Gene polymorphism, Peripheral blood mononuclear cells, Thermotolerance, Dairy cows
Introduction
The reduction of productivity with devastating economic consequences to the global dairy industry due to warm environment has been documented (St-Pierre et al. 2003; Bernabucci et al. 2010). One possible approach for reducing the impact of heat stress on cattle productivity is to improve genetic programs that select animals with thermotolerance. Genetic differences in thermotolerance at the physiological and cellular levels are documented by a series of studies on Bos indicus and Bos taurus (Paula-Lopes et al. 2003; Hansen 2004; Lacetera et al. 2006). Those studies confirm a genetic linkage between species, breed, and individual differences to heat tolerance at the cellular level.
Cellular tolerance to heat stress is mediated by a family of proteins named heat shock proteins (Hsp). Among members of the Hsp family, Hsp70 (namely, Hsp70.1 and Hsp70.2) is the most abundant and temperature sensitive (Beckham et al. 2004). Hsp70 transcription is increased by heat shock as well as other stress stimuli such as oxidative stress, ischemia, inflammation, or aging (Favatier et al. 1997) and can be an indicator of stress in cells (Sonna et al. 2002b). Hsp70 is a chaperone that promotes cell protection against heat damage, preventing protein denaturation and blocking apoptosis.
The regulation of Hsp production is critical to cell survival. Substantial evidence suggests that expression of the inducible Hsp70 gene is mainly under transcriptional control but also upon post-transcriptional mechanisms (Schwerin et al. 2002). While stress-induced synthesis of Hsp proteins represents a generalized molecular mechanism displayed by almost all cells, individual animals differ in their capacity to manage with stress. Moreover, nucleotide changes occurring naturally in the flanking regions [5′- and 3′-untranslated region (UTR)] of Hsp gene might affect inducibility, degree of expression, and/or stability of Hsp70 mRNA and contribute to the different individual stress tolerance at the cellular level.
Polymorphisms found in the functional promoter and 3′-UTR of the Hsp70.2 gene have been associated with mRNA stability and stress response in pigs (Schwerin et al. 2001, 2002). In human, Hsp70 gene polymorphism in the coding region, positively associated with human longevity and with survival advantage, was associated with an increased ability to respond to heat stress (Singh et al. 2006).
Association of polymorphisms of the Hsp70 gene with thermotolerance in dairy cattle is unknown. Therefore, studies to investigate genetic mechanisms associated with individual cellular response to heat shock (HS) may lead to genetic tools which will improve cattle tolerance to heat. The objective of the present study was to evaluate the association between inducible Hsp70.1 single nucleotide polymorphisms (SNPs) and HS response of peripheral blood mononuclear cells (PBMC) in dairy cows.
Materials and methods
Animals and sample collection
Four hundred forty-six Italian Holstein cows, originated from four commercial dairy units located in central Italy and under the same management regimen, were utilized as blood donors. Blood samples were obtained by jugular venipuncture using sodium heparin (10 IU/ml) as an anticoagulant. Immediately after collection, blood samples were stored in a portable refrigerator at 4°C, transported to the laboratory, and stored at −80°C until DNA extraction.
DNA isolation, amplification, and genotyping
Genomic DNA was extracted from blood samples using the Wizard DNA Genomic Kit (Promega Corporation, Madison, WI, USA). A Picogreen dsDNA Kit (Invitrogen, Carlsbad, CA, USA) was used to quantify DNA following purification.
Based on the identification of SNPs by Grosz et al. (1994) (g2154 G/A) and by Adamowicz et al. (2005) (g64 G/T) in the bovine Hsp70.1 3′-UTR of Holstein dairy breed, and on the identification of two SNPs (g895 C/- and g1128 G/T) by Starkey et al. (2007) in the bovine Hsp70 5′-UTR of Brahman meat breed, specific primers (Table 1) were used for SNPs characterization in the 446 cows.
Table 1.
Specific primers used for genotyping in 3′- and in 5′-untranslated regions (UTR)
| SNP | Primers | Region |
|---|---|---|
| Hsp70.1 (64 G/T and 2154 G/A) | Forward: 5′-GGATTGCTCATGTTTGTTATGG-3′ | 3′-UTR |
| Reverse: 5′-CTTGGAAGTAAACAGAAACGGG-3′ | ||
| Hsp70.1 (895 C/- and 1128 G/T) | Forward: 5′-GCCAGGAAACCAGAGACAGA-3′ | 5′-UTR |
| Reverse: 5′-CCTACGCAGG AGTAGGTGGT-3′ |
Large-scale genotyping of all animals was performed by outsourcing to a commercial genotyping company (http://www.Kbioscence.co.uk). Generally, accuracy greater than 99% was achieved. Quality control criteria were adopted (water as negative control, inter-plate duplicate testing of a known DNA, and intra-plate testing of a known DNA).
Among the 446 genotyped animals for SNPs in 5′-UTR, a group of cows balanced for days in milk and parity was selected to be representative of the following genotypes: CC (n = 8), C- (n = 7), and -- (n = 7) and GG (n = 8), GT (n = 11), and TT (n = 3). Therefore, venous blood samples were collected from these cows to perform the heat shock in vitro study described below.
Cell preparation
PBMC from the selected cows were isolated by density gradient centrifugation. Briefly, heparin-blood (10 ml) mixed with an equal volume of phosphate-buffered saline (PBS, Sigma, Milano, Italy) was gently layered over 16 ml of Ficoll-Paque Plus solution (APB, Milano, Italy). After centrifugation for 45 min at 1,720 rpm, the mononuclear cell band was recovered and washed twice in PBS. Residual red blood cells were eliminated by hypotonic shock treatment using redistilled water. PBMC recovery and viability were determined by a hemocytometer using the trypan blue exclusion method. Viability of PBMC typically exceeded 90%.
PBMC were resuspended at 1 × 106 viable cells/ml in RPMI 1640 medium containing 25 mM HEPES + 10% heat-inactivated FBS, 2 mM l-glutamine, 100 U of penicillin, 100 μg of streptomycin, and 0.25 μg of amphotericin B/ml (Sigma, Milano, Italy). The time between blood collection and establishment of cultures was always less than 6 h.
Heat shock treatment
An aliquot of cell suspension was utilized immediately to evaluate the cell viability and stored at −80°C until mRNA and protein analysis of Hsp70.1. This gave the measure of the basal (no-HS) level of viability and Hsp70.1 (time, −1 h). For measuring heat shock response, the cells were given HS for 1 h at 43°C in a water bath and then plated at a concentration of approximately 1 × 106 viable cells/ml in 24-well plates and incubated at 39°C in a 5% CO2 incubator for 24 h (recovery time).
Kinetics of Hsp70.1 gene expression and synthesis was studied after 0, 1, 2, 4, 8, 16, and 24 h of recovery time. At each recovery time, cells were utilized immediately to evaluate viability and stored at −80°C until mRNA and protein analysis of Hsp70.1.
Analysis of cell viability
Cell viability was determined using XTT assay with Cell Proliferation kit II (XTT, sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis (4-methoxy-6-nitro) benzene sulfonic acid hydrate; Roche Applied Science, Indianapolis, IL, USA) according to the manufacturers' instructions. Briefly, cells were seeded into 96-well microplates at an optimal density. At time −1 (no-HS) and after 0, 1, 2, 4, 8, 16, and 24 h of recovery time, 50 μl of XTT labeling mixture was added to each well. After 24 h incubation at 37°C, absorbance was measured using a microplate spectrophotometer at a wavelength of 450 nm. Background absorbance was subtracted from each row value.
RNA extraction and cDNA synthesis
Total RNA was isolated from PBMC using QIAzol Lysis reagent (Qiagen, Chatsworth, CA, USA) according to the manufactures' instructions. RNA was quantified using a fluorimetric method with Quant-iT kit (Invitrogen, Carlsbad, CA, USA). The integrity of the RNA was checked by visualization of 18s and 28s ribosomal bands on an agarose gel. One microgram of total RNA was reverse transcribed with ImProm-II reverse transcription system (Promega, Madison, WI, USA) in a total volume of 20 μl on a polymerase chain reaction (PCR)-Express thermal cycler (Hybaid, Ashford, UK). Controls without reverse transcription were performed to exclude the possibility of DNA contamination.
Real-time PCR
Real-time PCR was done for Hsp70.1 and a housekeeping gene (RPS9). Real-time PCR was performed in the LyghtCycler (Roche Applied Science, Indianapolis, IL, USA) using SYBR Green detection. Each reaction contained 10 μl of Quantitect SYBR Green PCR Master Mix (containing HotStart Taq DNA Polimerase, QuantiTect SYBR Green PCR Buffer, dNTP mix, SYBR Green I dye, ROX reference passive dye, and 5 mM MgCl2 (Qiagen, Chatsworth, CA, USA)), 1 μl of the sense and antisense specific primers, and 1 μl of cDNA in a final volume of 20 μl. A negative control without cDNA template was run in each assay.
The primers quantifying Hsp70.1 (GeneBank accession number U09861) were forward 5′-AACAAGATCACCATCACCAACG-3′ and reverse 5′-TCCTTCTCCGCCAAGGTGTTG-3′ with an annealing temperature of 59°C to amplify a 275-bp fragment. The RPS9 (GeneBank accession number DT860044) primers were forward 5′-CCTCGACCAAGAGCTGAAG-3′ and reverse 5′-CTTGTTTGCACTCCAGACCTCC-3′ with an annealing temperature of 59°C to amplify a 62-bp fragment. PCR products were subjected to a melting curve analysis on the LyghtCycler and subsequently 2% agarose/Tris-borate-EDTA gel electrophoresis to confirm amplification specificity and amplicon size.
To allow relative quantification after PCR, standard curves were constructed from the standard reactions for each target and housekeeping genes by plotting crossing point (Cp) values, i.e., the cycle number at which the fluorescence signal exceeds background vs. log cDNA dilution. The Cp readings for each for the unknown samples were then used to calculate the amount of either the target or housekeeping relative to the standard, using the Second Derivative Maximum Method with the LyghtCycler analysis software 3.5 (Roche Applied Science, Indianapolis, IL, USA). Bovine RPS9 was used to normalize gene expression.
Measurement of Hsp70.1 by ELISA
Frozen PBMC were rapidly thawed at 38°C, and concentrations of Hsp70.1 in PBMC were determined by an ELISA assay using a commercial kit (Stressgen, Bologna, Italy) at −1 (no-HS) and after 0, 1, 2, 4, 8, 16, and 24 h of recovery time. The intra-assay CV amounted to 1.3%.
Total protein level of PBMC lysate was measured with a BCA Protein Assay Kit from Pierce (Rockford, IL, USA). Hsp70.1 was adjusted to total protein levels included in the cell lysate.
Statistical analysis
Data for all variables measured were analyzed as repeated measures using the MIXED procedure of Statistica-7 software package (Stat Soft Inc., Tulsa, OK, USA). The model included fixed effects: Hsp70.1 gene polymorphisms, time (−1, 0, 1, 2, 4, 8, 16, and 24 h), interaction gene polymorphism × time, and the error term. Least square means were separated with the predicted difference (PDIFF) procedure. Data are reported as least-square means with standard errors. Significance was declared at P < 0.05.
Hardy–Weinberg equilibrium was calculated using Phase software, version 2.1, separately for each polymorphism (Stephens et al. 2001; Stephens and Scheet 2005).
Results
Identification of polymorphisms
Results from SNP genotyping in 3′-UTR of bovine Hsp70.1 gene showed that the population of 446 dairy cows under study resulted monomorphic (Table 2).
Table 2.
Distribution of genotypes and alleles of the two single nucleotide polymorphisms (SNPs) within the promoter region of the bovine Hsp70.1 gene in Holstein cows
| SNPs | Number of animals | Allele frequency (%) | Genotype frequency (%) | |||||
|---|---|---|---|---|---|---|---|---|
| 3′-UTR 64 G/T | GG | TG | TT | G | TT | GG | TG | TT |
| 446 | 0 | 0 | 100.0 | 0.0 | 100.0 | 0.0 | 0.0 | |
| 3′-UTR 2154 G/A | GG | GA | AA | G | A | GG | GA | AA |
| 0.0 | 2.0 | 444 | 0.2 | 99.8 | 0.0 | 0.4 | 99.6 | |
| 5′-UTR 895 C/- | CC | C- | -- | C | - | CC | C- | -- |
| 298 | 128 | 20 | 81.2 | 18.8 | 66.8 | 28.8 | 4.4 | |
| 5′-UTR 1128 G/T | GG | GT | TT | G | T | GG | GT | TT |
| 253 | 187 | 6 | 77.8 | 22.2 | 56.8 | 41.9 | 1.3 | |
Based on published SNPs information of bovine Hsp70.1 promoter region of Brahman (Starkey et al. 2007), a 539-bp segment of 5′-UTR Hsp70.1 was amplified and sequenced from genomic DNA samples. Two polymorphisms were detected in the population of 446 dairy cows under study, a deletion of cytosine at base 895 and a transversion from guanine to thymine at base 1128. Distribution of genotypes and alleles of the nucleotide sequence polymorphism within the 5′-UTR region of the bovine Hsp70.1 gene is reported in Table 2. For both SNPs, one allele was the most frequent, C for g895 (66.8%) and G for g1128 (56.8%) polymorphism. Each polymorphism in the 5′-UTR Hsp70.1 gene was found to be in Hardy–Weinberg equilibrium.
Cell viability
Viability changes in PBMC with different genotype were detected using XTT assay. XTT measures cell viability based on the activity of mitochondrial enzymes in live cells that reduce XTT and are inactivated shortly after cell death.
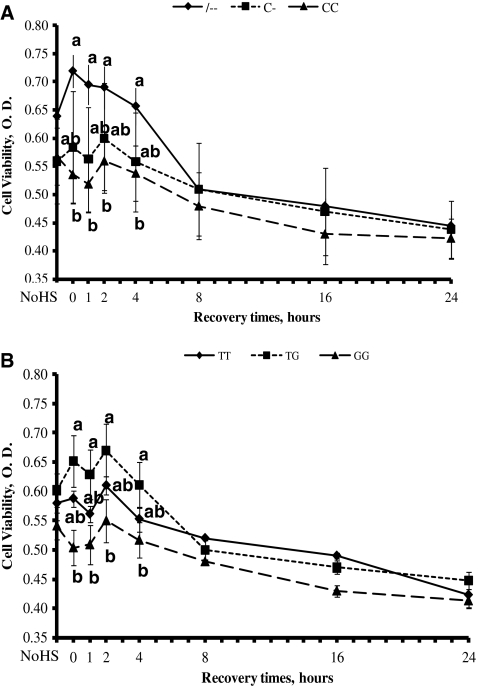
The genotypes -- and TG showed higher (P < 0.05) viability compared with CC and GG, respectively, during the first 4 h of the recovery time (Fig. 1a, b). PBMC with genotypes C- and TT had intermediate viability. No significant differences in the cell viability were seen in all genotypes after 4 h of recovery time when viability decreases in all cells.
Fig. 1.
Kinetics of viability in bovine peripheral blood mononuclear cells with two single nucleotide polymorphisms exposed to heat shock (HS). Cells were exposed to HS for 1 h at 43°C, then recovered at 39°C for 1, 2, 4, 8, 16, and 24 h and assayed using XTT absorbance. The values on the y-axis show the amount of optical density, which corresponds to the amount of cell viability from --, C-, and CC genotypes (a) and TT, GT, and GG genotypes (b). Results are means ± SE of triplicate determinations. Different letters indicate differences for P < 0.05 between genotypes within time. NoHS no heat shock
Hsp70.1 gene expression
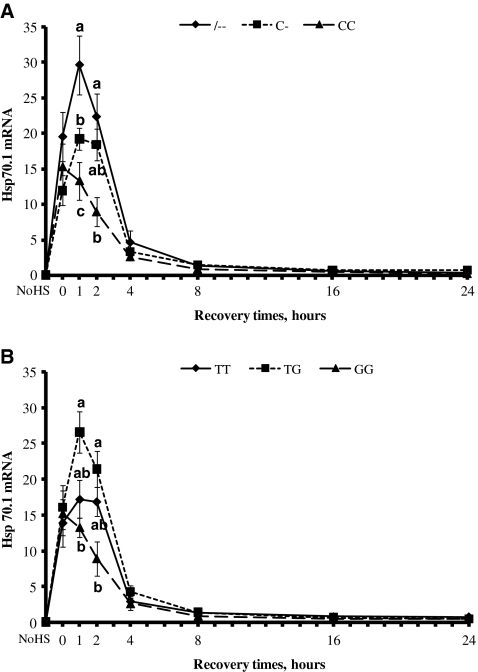
Kinetics of heat shock response in terms of induction of synthesis of Hsp70.1 mRNA was determined by real-time PCR in PBMC isolated from blood samples taken from cows representative of the all selected genotypes. Figure 2a and b show that Hsp70.1 mRNA levels increased significantly during the exposure to HS at 43°C, continued to increase until 1 h recovery time in --, C-, TG, and TT genotypes, and then declined toward the basal level after 8 h. In contrast, CC and GG genotypes Hsp70.1 mRNA decreased immediately after HS exposure. CC and GG genotypes showed lower (P < 0.05) Hsp70.1 mRNA levels at 1 and 2 h of recovery time.
Fig. 2.
Effects of heat stress on heat shock protein 70.1 (Hsp70.1) gene expression in bovine peripheral blood mononuclear cells. Cells with --, C-, and CC genotypes (a) and with TT, GT, and GG genotypes (b) were subject to heat shock (43°C × 1 h) and then recovered at 39°C for 1, 2, 4, 8, 16, and 24 h. Aliquots of cells were analyzed for relative mRNA level of Hsp70.1 and quantified by real-time RT-PCR at the indicated time points. Expression of mRNA was normalized using bovine RPS9 as housekeeping gene. Results are means ± SE of triplicate determinations. Different letters indicate differences for P < 0.05 between genotypes within time. NoHS no heat shock
Hsp70.1 concentration
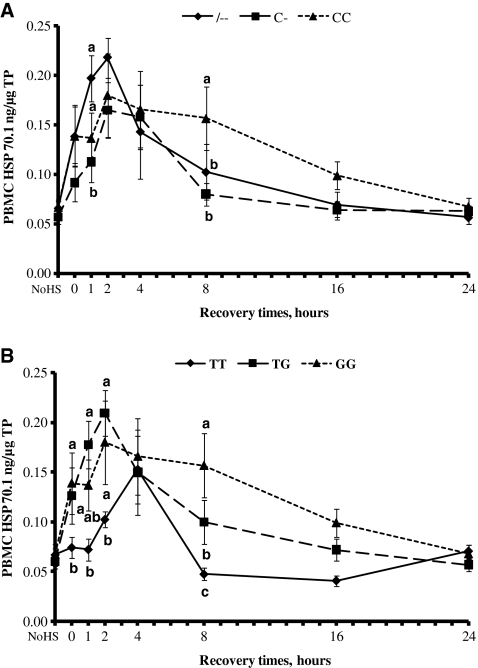
The ability of PBMC to synthesize Hsp70.1 was observed both before (basal) and after (induced) HS (Fig. 3a, b). The data showed no differences in the basal levels of Hsp70.1 in all genotypes.
Fig. 3.
Time course of heat shock protein 70.1 (Hsp70.1) protein expression in bovine peripheral blood mononuclear cells. Cells with --, C-, and CC genotypes (a) and with TT, GT, and GG genotypes (b) were subject to heat shock (43°C × 1 h) and then recovered at 39°C for 1, 2, 4, 8, 16, and 24 h. Aliquots of cells were analyzed for expression of Hsp70.1 proteins by ELISA at the indicated time points. Results are means ± SE of duplicate determinations. Different letters indicate differences for P < 0.05 between genotypes within time. NoHS no heat shock
Hsp70.1 levels increased significantly during the 1-h exposure to HS at 43°C, continued to increase until 2 h of the recovery time, and then declined toward the basal level after 8 or 16 h of recovery time. Genotype TT showed a different behavior: Hsp70.1 levels increased at 2 h and peaked at 4 h of recovery time and then started to decline. The higher Hsp70.1 protein concentration was reached by -- and TG genotypes at 2 h of recovery time.
Discussion
Our results on genetic polymorphisms in 3′-UTR of inducible bovine Hsp70.1 gene in Italian Holstein population showed monomorphic alleles for transversion from guanine to thymine at base 64 and for transition from guanine to adenosine at base 2154. These data do not agree with findings of Adamowicz et al. (2005), who reported polymorphic alleles for the same SNPs in Holstein Polish population. Results from our study and study from Adamowicz et al. (2005) support the concept that in the same breed but different population, the allele frequency of a specific SNP is not repeatable.
The significant novel aspect of our studies is that we have ascertained the presence of 5′-UTR variants in the inducible bovine Hsp70.1 gene in Holstein dairy cows.
The presence of deletion of cytosine at base position 895 and transversion from guanine to thymine at base 1128 of this gene were associated with upregulation of gene expression and synthesis of Hsp70.1 and increase of the ability of bovine mononuclear cells to respond to HS in terms of viability.
Cows homozygous with the cytosine deletion showed higher cell viability after HS and higher mRNA expression and protein concentration of Hsp70.1 when compared with heterozygous or homozygous cytosine cows. For SNPs found at base positions 1128, cows that were heterozygous with the minor allele had higher cell viability and an increase in Hsp70.1 levels of gene and protein expression after the HS, when compared with homozygous guanine or homozygous thymine cows. The SNP deletion of cytosine occurs within 350 bp of the putative transcription start site and is located in a region that has a critical role in heat shock gene induction (Burdon 1986; Lindquist and Craig 1988). The precise mechanism by which the polymorphisms described herein are associated with higher abundance of mRNA and synthesis of Hsp70.1 was not studied. Probably, the presence of promoter variants improved binding of corresponding transcription factors. In addition, the overproduction of Hsp70.1 indicates the activation of cellular protective mechanism that may be associated with increases in cell viability.
The evidence concerning genetic polymorphism in the genes coding the Hsp is rather sparse in dairy cows. Moreover, association of polymorphisms of the Hsp70 gene with tolerance to heat stress in bovine is unknown. The two SNPs within the 3′-UTR that were described by Grosz et al. (1994) and Adamowicz et al. (2005) in the bovine Hsp70.1 gene of Holstein breed were not related to any physiological or productive traits. In Chinese Holstein cows, a genetic polymorphism in the coding region of Hsp70.1 gene was associated with resistance to mastitis (Cheng et al. 2009). Starkey et al. (2007) observed that polymorphisms within the promoter region of Hsp70 gene were associated with crossbred Brahaman bull calf weaning weights. Banks et al. (2007) showed that the promoter region of bovine Hsp70 gene is polymorphic, and some SNPs indentified were associated with pregnancy rate in B. taurus/B. indicus crossbred cows. In addition, Rosenkrans et al. (2010) reported an association between SNPs in the promoter region of bovine Hsp70 gene and fertility in crossbred Brahaman cows. The only studies present in literature concerning the association between Hsp70 gene polymorphisms and tolerance to heat are in other species such as birds, pigs, and human. For example, Zhang et al. (2002) detected polymorphisms in regulatory and coding regions of the Hsp70 gene associated with different heat tolerance capability in broiler chickens. Huang et al. (2002) detected SNPs in the 5′ flanking region of the Hsp70.2 gene in boars. These SNPs have been associated with the quality of semen production during the summer. In pigs, a functional promoter and 3′-UTR variants of highly conserved inducible Hsp70.2 gene significantly affected mRNA stability and cell stress response. The authors suggested that both mutations could contribute to the different individual stress tolerance at the cellular level (Schwerin et al. 2001, 2002). In human peripheral mononuclear cells (Singh et al. 2006), polymorphisms in the coding region of Hsp70 gene, positively associated with human longevity and survival advantage, were associated with an increased ability to respond to heat stress. Those findings are consistent with our results in which variants in the Hsp70.1 gene induced difference in heat shock response between different genotypes as attested by differences in cell viability.
Studies have shown a specific association between the presence of Hsp70 and thermotolerance mechanisms in various types of cells. When vertebrate cells are exposed to high temperatures for a long period of time, the acclimatization properties were observed to increase due to synthesis of Hsp (Ulmasov et al. 1992; Hunter-Lavin et al. 2004). Hsp function as molecular chaperones in restoring cellular homeostasis and promoting cell survival (Horowitz 2001; Collier et al. 2008). Most often, the high temperature within the range of 42–45°C induces reversible damage, which involves a denaturation of labile proteins, acceleration of metabolism through the mitochondria, and activation of anti-apoptotic pathways (Pearce and Thomsen 1995; Stankiewicz et al. 2005). Several studies in mice, in bovine, and in human cells give evidence that constitutive elevation of the inducible Hsp level in gene and protein expression provides cytoprotection upon thermal stress (Horowitz 2001; Sonna et al. 2002a; Collier et al. 2006). In particular, Hsp70 represents the most highly inducible Hsp after thermal stress (Beckham et al. 2004). Those authors showed that Hsp70 is vitally important for thermotolerance in cell culture because in tandem to the role of Hsp70 in protein refolding, there are several mechanisms by which it can rescue cells from thermal insult. Hsp70 has been implicated in directing severely denatured proteins for degradation through the proteasomal pathway (Huang et al. 2001; Pirkkala et al. 2001), and Hsp70 intervenes in cell death by downregulating apoptotic signaling within the cell through its interaction with BAX (Bcl-2-associated X protein), a pro-apoptotic signaling protein (Stankiewicz et al. 2005; Steel et al. 2004). However, there are other pathways that contribute to thermotolerance such as Hsp90, Hsp110, Hsp25, and Hsp40 (Duncan 2005; Kampinga et al. 2003). Hsp90 and Hsp40 are known to work in conjunction with Hsp70 to refold proteins.
In conclusion, the genetic study with a series of functional analyses shows that genetic variants (base positions 895 and 1128) in the 5′-UTR of inducible Hsp70.1 are associated with an increased ability of PBMC to respond to HS in terms of gene expression and synthesis of Hsp70.1 and cell viability. However, many other mechanisms are likely to contribute to the above-mentioned difference in thermotolerance. Additional research will be necessary to establish the physiological mechanisms by which these polymorphisms alter gene and protein expression of Hsp70.1 and to determine the association between Hsp70.1 genotypes and cattle performances. However, the mutation sites ascertained in this study may be useful in the future as molecular genetic markers to assist selection for heat tolerance in cattle.
Acknowledgment
The research was financially supported by the SELMOL-Project (Italian Ministry of Agricultural, Food, and Forestry Policies).
References
- Adamowicz T, Pers E, Lechniak D. A new SNP in the 3′-UTR of the Hsp70-1 Gene in Bos taurus and Bos indicus. Biochem Genet. 2005;43:623–627. doi: 10.1007/s10528-005-9119-2. [DOI] [PubMed] [Google Scholar]
- Banks A, Looper ML, Reiter S, Starkey L, Flores R, Hallford D, Rosenkrans C., Jr Identification of single nucleotide polymorphisms within the promoter region of the bovine heat shock protein 70 gene and associations with pregnancy. Am Soc Anim Sci South Sect Meet. 2007;85(Suppl. 2):10. [Google Scholar]
- Beckham JT, Mackanos MA, Crooke C, Takahashi T, O'Connell-Rodwell C, Contag CH, Jansen ED. Assessment of cellular response to thermal laser injury through bioluminescence imaging of heat shock protein 70. Photochem Photobiol. 2004;79:76–85. [PubMed] [Google Scholar]
- Bernabucci U, Lacetera N, Baumgard LH, Rhoads RP, Ronchi B, Nardone A. Metabolic and hormonal adaptations to heat stress in domesticated ruminants. Animal. 2010;4:1167–1183. doi: 10.1017/S175173111000090X. [DOI] [PubMed] [Google Scholar]
- Burdon R. Heat shock and the heat shock proteins. Biochem J. 1986;240:313–324. doi: 10.1042/bj2400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WJ, Li QL, Wang CF, Wang HM, Li JB, Sun YM, Zhong JF. Genetic polymorphism of HSP70-1 gene and its correlation with resistance to mastitis in Chinese Holstein. Yi Chuan. 2009;31:169–174. doi: 10.3724/sp.j.1005.2009.00169. [DOI] [PubMed] [Google Scholar]
- Collier RJ, Stiening CM, Pollard BC, VanBaale MJ, Baumgard LH, Gentry PC, Coussens PM. Use of gene expression microarrays for evaluating environmental stress tolerance at the cellular level in cattle. J Anim Sci. 2006;84(E Suppl):E1–E13. doi: 10.2527/2006.8413_supple1x. [DOI] [PubMed] [Google Scholar]
- Collier RJ, Collier JL, Rhoads RP, Baumgard LH. Genes involved in the bovine heat stress response. J Dairy Sci. 2008;91:445–454. doi: 10.3168/jds.2007-0540. [DOI] [PubMed] [Google Scholar]
- Duncan R. Inhibition of Hsp90 function delays and impairs recovery from heat shock. FEBS J. 2005;272:5244–5256. doi: 10.1111/j.1742-4658.2005.04921.x. [DOI] [PubMed] [Google Scholar]
- Favatier F, Bornman L, Hightower LE, Eberhand G, Polla BS. Variation in Hsp gene expression and Hsp polymorphism: do they contribute to differential disease susceptibility and stress tolerance? Cell Stress Chaperones. 1997;2:141–155. doi: 10.1379/1466-1268(1997)002<0141:VIHGEA>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosz MD, Skow LC, Stone RT. An AluI polymorphism at the bovine 70 kD heat shock protein-1 (HSP70-1) locus. Anim Genet. 1994;25:196. doi: 10.1111/j.1365-2052.1994.tb00113.x. [DOI] [PubMed] [Google Scholar]
- Hansen PJ. Physiological and cellular adaptations of zebu cattle to thermal stress. Anim Reprod Sci. 2004;82–83:349–360. doi: 10.1016/j.anireprosci.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Horowitz M. Heat acclimation: phenotypic plasticity and cues to the underlying molecular mechanisms. J Therm Biol. 2001;26:357–363. doi: 10.1016/S0306-4565(01)00044-4. [DOI] [Google Scholar]
- Huang L, Mivechi NF, Moskophidis D. Insights into regulation and function of the major stress-induced hsp70 molecular chaperone in vivo: analysis of mice with targeted gene disruption of the hsp70.1 or hsp70.3 gene. Mol Cell Biol. 2001;21:8575–8591. doi: 10.1128/MCB.21.24.8575-8591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SY, Chen MY, Lin EC, Tsou HL, Kuo YH, Ju CC, Lee WC. Effects of single nucleotide polymorphisms in the 5′ flanking region of heat shock protein 70.2 gene on semen quality in boars. Anim Reprod Sci. 2002;70:99–109. doi: 10.1016/S0378-4320(01)00202-0. [DOI] [PubMed] [Google Scholar]
- Hunter-Lavin C, Davies EL, Bacelar MM, Marshall MJ, Andrew SM, Williams JH. Hsp 70 release from peripheral blood mononuclear cells. Biochem Biophys Res Commun. 2004;324:511–517. doi: 10.1016/j.bbrc.2004.09.075. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Kanon B, Salomons FA, Kabakov AE, Patterson C. Overexpression of the cochaperone CHIP enhances Hsp70-dependent folding activity in mammalian cells. Mol Cell Biol. 2003;23:4948–4958. doi: 10.1128/MCB.23.14.4948-4958.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacetera N, Bernabucci U, Scalia D, Basiricò L, Morera P, Nardone A. Heat stress elicits different responses in peripheral blood mononuclear cells from Brown Swiss and Holstein cows. J Dairy Sci. 2006;89:4606–4612. doi: 10.3168/jds.S0022-0302(06)72510-3. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Paula-Lopes FF, Chase CC, Jr, Al-Katanani YM, Krininger CE, Rivera RM, Tekin S, Majewski AC, Ocon OM, Olson TA, Hansen PJ. Genetic divergence in cellular resistance to heat shock in cattle: differences between breeds developed in temperate versus hot climates in responses of preimplantation embryos, reproductive tract tissues and lymphocytes to increased culture temperatures. Reproduction. 2003;125:285–294. doi: 10.1530/rep.0.1250285. [DOI] [PubMed] [Google Scholar]
- Pearce J, Thomsen S. Optical-thermal response of laser-irradiated tissue. Lasers, photonics, and electro-optics. New York: Plenum Press; 1995. pp. 561–603. [Google Scholar]
- Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Rosenkrans C, Jr, Banksa A, Reitera S, Looperb M. Calving traits of crossbred Brahman cows are associated with heat shock protein 70 genetic polymorphisms. Anim Reprod Sci. 2010;119:178–182. doi: 10.1016/j.anireprosci.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Schwerin M, Maak S, Kalbe C, Fuerbass R. Functional promoter variants of highly conserved inducible hsp70 genes significantly affect stress response. Biochim Biophys Acta. 2001;1522:108–111. doi: 10.1016/s0167-4781(01)00313-x. [DOI] [PubMed] [Google Scholar]
- Schwerin M, Maak S, Hagendorf A, Lengerken G, Seyfert HM. A 3′-UTR variant of the inducible porcine hsp70.2 gene affects mRNA stability. Biochim Biophys Acta. 2002;1578:90–94. doi: 10.1016/s0167-4781(02)00448-7. [DOI] [PubMed] [Google Scholar]
- Singh R, Kolvraa S, Bross P, Jensen UB, Gregersen N, Tan Q, Knudsen C, Rattan SIS. Reduced heat shock response in human mononuclear cells during aging and its association with polymorphisms in HSP70 genes. Cell Stress Chaperones. 2006;11:208–215. doi: 10.1379/CSC-184R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonna LA, Gaffin SL, Pratt RE, Cullivam ML, Angel KG, Lilly CM. Effects of acute heat shock on gene expression by human peripheral blood mononuclear cells. J Appl Physiol. 2002;92:2208–2220. doi: 10.1152/japplphysiol.01002.2001. [DOI] [PubMed] [Google Scholar]
- Sonna LA, Fujita J, Gaffin SL, Lilly CM. Invited review: effects of heat and cold stress on mammalian gene expression. J Appl Physiol. 2002;92:1725–1742. doi: 10.1152/japplphysiol.01143.2001. [DOI] [PubMed] [Google Scholar]
- Stankiewicz AR, Lachapelle G, Foo CP, Radicioni SM, Mosser DD. Hsp70 inhibits heat-induced apoptosis upstream of mitochondria by preventing Bax translocation. J Biol Chem. 2005;280:38729–38739. doi: 10.1074/jbc.M509497200. [DOI] [PubMed] [Google Scholar]
- Starkey L, Looper ML, Banks A, Reiter S, Rosenkrans C., Jr Identification of polymorphisms in the promoter region of the bovine heat shock protein gene and associations with bull calf weaning weight. Am Soc Anim Sci South Sect Meet. 2007;85(Suppl. 2):42. [Google Scholar]
- Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre NR, Cobanov B, Schnitkey G. Economic losses from heat stress by US livestock industries. J Dairy Sci. 2003;86(E-Suppl):E52–E77. doi: 10.3168/jds.S0022-0302(03)74040-5. [DOI] [Google Scholar]
- Ulmasov KA, Shammakov S, Karaev K, Evgen'ev MB. Heat shock proteins and thermoresistance in lizards. Proc Natl Acad Sci USA. 1992;89:1666–1670. doi: 10.1073/pnas.89.5.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Du H, Li J (2002) Single nucleotide polymorphism of chicken heat shock protein 70 gene. 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France