Abstract
Heat shock protein 70 (Hsp70) and heat shock protein 40 (Hsp40) function as molecular chaperones during the folding and trafficking of proteins within most cell types. However, the Hsp70–Hsp40 chaperone partnerships within the malaria parasite, Plasmodium falciparum, have not been elucidated. Only one of the 43 P. falciparum Hsp40s is predicted to be a cytosolic, canonical Hsp40 (termed PfHsp40) capable of interacting with the major cytosolic P. falciparum-encoded Hsp70, PfHsp70. Consistent with this hypothesis, we found that PfHsp40 is upregulated under heat shock conditions in a similar pattern to PfHsp70. In addition, PfHsp70 and PfHsp40 reside mainly in the parasite cytosol, as assessed using indirect immunofluorescence microscopy. Recombinant PfHsp40 stimulated the ATP hydrolytic rates of both PfHsp70 and human Hsp70 similar to other canonical Hsp40s of yeast (Ydj1) and human (Hdj2) origin. In contrast, the Hsp40-stimulated plasmodial and human Hsp70 ATPase activities were differentially inhibited in the presence of pyrimidinone-based small molecule modulators. To further probe the chaperone properties of PfHsp40, protein aggregation suppression assays were conducted. PfHsp40 alone suppressed protein aggregation, and cooperated with PfHsp70 to suppress aggregation. Together, these data represent the first cellular and biochemical evidence for a PfHsp70–PfHsp40 partnership in the malaria parasite, and furthermore that the plasmodial and human Hsp70–Hsp40 chaperones possess unique attributes that are differentially modulated by small molecules.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-010-0250-6) contains supplementary material, which is available to authorized users.
Keywords: Aggregation, ATPase, Codon harmonisation, Heat shock protein, Malaria, Molecular chaperone
Introduction
Plasmodium falciparum causes cerebral malaria in humans, and nearly all malaria deaths result from infection by this species (Snow et al. 2005). The parasite is adapted to survive in the stressful environment it encounters in the human host, and studies suggest that acute temperature increases, especially those associated with febrile episodes, augment parasite development and infectivity (Pavithra et al. 2004). These elevated temperatures invoke the increased expression of heat shock proteins. Through their action as molecular chaperones, it has been proposed that heat shock proteins enable the parasite to become resilient to subsequent physiological threats, thereby enhancing parasite pathogenicity (Pavithra et al. 2004). Consequently, molecular chaperones such as heat shock protein 70 (Hsp70) together with co-chaperone partners, such as Hsp40, are likely to provide a cytoprotective role in the adaptation of the parasite to its human host (Shonhai et al. 2007). It has also been reported that anti-malarial drugs invoke oxidative stress in the parasite, consequently inducing heat shock proteins, including a P. falciparum Hsp70 (Akide-Ndunge et al. 2009). Furthermore, P. falciparum Hsp70 and Hsp90 proteins associate with ferriprotoporphyrin IX (FPIX; a degradation product of the malaria parasite), in response to the effect of chloroquine (Famin and Ginsburg 2003). Overall, these observations suggest that heat shock proteins play a key role in the management of toxicity conferred by drugs to the parasite, and may augment drug resistance. In addition to their role within the parasite, growing evidence suggests that parasite heat shock proteins are involved in the trafficking of parasite-encoded proteins to the erythrocyte surface, thus playing a key role in the pathogenesis of P. falciparum malaria (de Koning-Ward et al. 2009; Külzer et al. 2010; Maier et al. 2008). Taken together, these observations suggest that heat shock proteins from P. falciparum are potential anti-malarial drug targets (Pesce et al. 2010; Shonhai 2010).
Hsp70 proteins are ubiquitous, highly conserved proteins found in all major organelles, and play well defined roles in nascent polypeptide folding, protein translocation, protein degradation and signal transduction (Kabani and Martineau 2008). Constitutive and inducible Hsp70s are vital for the maintenance of proteostasis under both normal and stressful conditions. A typical Hsp70 possesses a molecular mass of 70 kDa and consists of two distinct domains—the 45-kDa N-terminal ATP-binding domain and 25-kDa peptide-binding domain, which includes a C-terminal “lid” to entrap peptide substrates (Flaherty et al. 1990; Wang et al. 1993). When bound to ADP, Hsp70s display high affinity for substrate, but can release the substrate in their low-affinity ATP-bound form. In the absence of substrate and co-chaperone, ATP hydrolysis is a rate-limiting step because Hsp70s possess a low basal ATPase activity. To modulate peptide substrate binding and subsequent release, the ATPase domain of Hsp70 interacts with several co-chaperones that regulate its nucleotide-bound state, such as nucleotide exchange factors and Hsp40s (Brodsky and Bracher 2007; Hennessy et al. 2005). The binding of Hsp40 through a site in the underside of the ATPase domain of Hsp70 stimulates the ATP hydrolytic activity of Hsp70 to generate a stable Hsp70–ADP–substrate complex (Jiang et al. 2007; Landry 2003; Li et al. 2009; Wittung-Stafshede et al. 2003). Hsp40 is believed to bind to substrates, handing them over to Hsp70, thus regulating Hsp70 functional specificity. This model is corroborated by the fact that cells have fewer Hsp70s compared to Hsp40s.
The Hsp40 family of proteins is defined by the presence of a highly conserved, ~70 residue J domain (Hennessy et al. 2005). The J domain is the minimum region of an Hsp40 essential for Hsp70 interaction (Landry 2003). A highly conserved feature of the J domain, the HPD motif, is important for the interaction of Hsp40 with Hsp70. The Escherichia coli Hsp40 (also called DnaJ) is the prototypical Hsp40, and hence, other Hsp40s are classified based on their structural organisation relative to DnaJ (Cheetham and Caplan 1998). Type I Hsp40s possess the same structural components as DnaJ: they harbour the J domain, a glycine–phenylalanine (G/F)-rich region, a cysteine repeat domain and a C-terminal domain, all organised in a similar fashion as in DnaJ. Type II Hsp40s are similar to DnaJ over the J domain and the G/F-rich region. Type III proteins have only a J domain in common with DnaJ, which can be present anywhere in the protein. Type IV Hsp40s represent a newly described class of Hsp40s that possess a J-like domain in which the HPD motif is not conserved (Botha et al. 2007). Type I and type II Hsp40s are capable of targeting substrates to Hsp70s and stimulating ATP hydrolysis so as to ensure effective coupling of hydrolysis to substrate capture (Hennessy et al. 2005; Walsh et al. 2004).
The P. falciparum genome encodes six Hsp70 proteins, and 43 Hsp40 and Hsp40-like proteins (Botha et al. 2007; Shonhai et al. 2007). The major cytosolic Hsp70, which we will denote PfHsp70 (also called PfHsp70-1; PF08_0054), is a highly abundant protein expressed at all erythrocytic stages of the parasite lifecycle under normal conditions, with increased levels under heat shock conditions (Kumar et al. 1991). Biochemical evidence from in vitro and in vivo assay systems conducted in our laboratory has shown that PfHsp70 exhibits the properties of a bona fide Hsp70 molecular chaperone. In particular, it possesses ATPase activity and is capable of preventing protein aggregation (Matambo et al. 2004; Shonhai et al. 2005, 2008). Interestingly, the biochemical properties of PfHsp70 suggest that it is distinct from human Hsp70 (HsHsp70) and may be more responsive to fluctuations in ATP levels and co-chaperone modulation (Matambo et al. 2004; Shonhai et al. 2005).
Much of the pathology of P. falciparum infection is caused by increased “stickiness” of infected cells to endothelial cells (Miller et al. 2002). This so-called cytoadherence is mediated by proteins which are exported from the parasite to the host cell (Crabb et al. 1997). The mechanism by which the malaria parasite shuttles its proteins to the erythrocyte surface remains largely unclear, but seems to rely on export of a further number of parasite proteins, which appear to function as an “extracellular” secretory system (Maier et al. 2008). Amongst the approximately 250 proteins suggested to be exported to the host cell are 19 members of the Hsp40 family (Botha et al. 2007; Hiller et al. 2004; Marti et al. 2004). Specific members of this family are important for parasite proliferation, whilst others appear to be involved in pathogenesis (Maier et al. 2008; Silva et al. 2005). It has been suggested that certain of these exported Hsp40s, especially the type IIs, may work with HsHsp70 in the transport, folding and assembly of other exported malarial proteins (Külzer et al. 2010).
We and others have speculated on the existence of chaperone networks in P. falciparum-infected erythrocytes (Acharya et al. 2007; Pavithra et al. 2007; Shonhai et al. 2007). However, despite the potential central role of Hsp70–Hsp40 chaperone pathways in protein folding and trafficking in the malaria parasite, there is limited experimental evidence for any partnerships. Studies in our laboratory have provided the first evidence for an Hsp40 partner of PfHsp70. Pfj4, a specialised type II Hsp40, was found to have a functional J domain (Nicoll et al. 2007), to be heat-inducible, and to occur in a common complex with PfHsp70 in P. falciparum-infected erythrocytes (Pesce et al. 2008). P. falciparum encodes only two type I Hsp40s, neither of which are predicted to be exported (Botha et al. 2007). The type I Hsp40, Pfj1 (PFD0462w), was first described by Watanabe (1997), and represents a relatively large, atypical type I Hsp40 containing extensive N- and C-terminal extensions (Figure S1). Recently, Pfj1 was shown to be associated with the apicoplast genome (Kumar et al. 2010). Recombinant Pfj1 was found to have a functional J domain (Nicoll et al. 2007) and to stimulate the in vitro protein refolding activity of PfHsp70 (Misra and Ramachandran, 2009). However, since there is evidence that Pfj1 is localised to the apicoplast (Kumar et al. 2010), it is unlikely to associate with cytosolic PfHsp70 in vivo. The other type I Hsp40 (termed PfHsp40 here; PF14_0359) possesses all the canonical features of a typical cytosolic co-chaperone involved in assisting Hsp70s in maintaining proteostasis (Figure S1). Since PfHsp40 has not been characterised, we have investigated its potential to functionally interact with PfHsp70. Furthermore, we investigated the effect of small molecule modulators on the PfHsp70–PfHsp40 partnership in comparison to the human Hsp70–Hsp40 system. Our data provide the first evidence that PfHsp40 partners with PfHsp70, and that the plasmodial and human chaperone systems can be differentially inhibited.
Materials and methods
Antibody design
To raise antibodies specific to PfHsp40, a peptide immunogen was designed based on a region of high antigenicity and low conservation located in the C terminus of the protein (Figure S1). An epitopic region of PfHsp40 was selected using algorithms for determining peptide antigenicity, surface probability, hydropathy and chain flexibility as provided in the GeneRunner software (version 3.05; Hastings Software Inc.). The peptide [C]SPVDKEYIKVRVTK was synthesised, in which the [C] represents a cysteine residue preceding the target epitopic site. The peptide was subsequently conjugated to haemocyanin, and polyclonal antibodies were raised in rabbits in the laboratory of Professor Richard Zimmerman (Universität des Saarlandes, Germany).
Maintenance of parasite cultures
P. falciparum (strain 3D7) parasites were cultured in A+ human erythrocytes at a haematocrit of 4% in RPMI media supplemented with 10% A+ human serum, 200 μM hypoxanthine and 20 μg/ml gentamicin, at 37°C under N2 containing 5% (v/v) CO2 and 5% (v/v) O2. Cultures were synchronised during the ring stage of parasite development with sorbitol as previously described (Lambros and Vandeberg 1979). For western blot analyses, heat shock was performed by incubating subcultures at 37°C (control), 41°C (heat shock) and 43°C (heat shock) for 1 and 2 h. For immunofluorescence assays, heat shock was performed by incubating subcultures at 37°C (control) and 42°C (heat shock) for 2 h.
Preparation of P. falciparum lysates
The parasite-infected erythrocytes were treated with 0.1% (w/v) saponin in phosphate-buffered saline (PBS) to lyse the erythrocyte membranes. Subsequently, intact parasites were sedimented by centrifugation (2,800×g, for 5 min), the supernatant discarded, and the parasite pellet washed of excess saponin with PBS. All samples were prepared for sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and western blot analysis by reconstitution in PBS, before treatment by boiling in SDS–PAGE denaturing and reducing sample buffer. To ensure that lysate fractions were comparable, equivalent concentrations of proteins or cells were analysed. PfHsp40 was detected by western blot analysis using the rabbit polyclonal anti-PfHsp40 primary antibody (1:5,000 dilution), and horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (1:5,000 dilution; GE Healthcare). PfHsp70 was detected by western blot analysis as previously described (Pesce et al. 2008), and served as a positive control for the heat shock condition. Glycophorin (an erythrocyte membrane protein that associates with isolated parasites) was detected as a loading control using a mouse monoclonal anti-human glycophorin A/B antibody (1:500; Sigma), and an HRP-conjugated sheep anti-mouse secondary antibody (1:5,000; GE Healthcare).
Indirect immunofluorescence assays
Indirect immunofluorescence staining, microscopy and image manipulation were carried out as previously described (Külzer et al. 2010), using the purified IgG fraction of rabbit anti-PfHsp40 antibody (1:100) and cyanine 3 (Cy3)-conjugated goat anti-rabbit secondary antibody (1:2,000; Dianova). The samples were co-stained with Hoechst 33258 (50 ng/ml) to detect parasite nuclei. Background and nonspecific fluorescence signals were monitored in control experiments with rabbit pre-immunisation serum used in place of the primary antibody, and in the absence of the primary antibody.
Hsp40 and Hsp70 proteins
Recombinant hexahistidine-tagged (His6) PfHsp40 (PF14_0359) was purified as described below, and recombinant HsHsp70-His6 (HSPA1A) was purified as previously described (Chiang et al. 2009). Recombinant His6-PfHsp70 (PF08_0054) was purified under denaturing conditions as published (Matambo et al. 2004) with modifications (Chiang et al. 2009) for ATPase assays, and under nondenaturing conditions as previously described (Shonhai et al. 2008) for aggregation suppression assays. Yeast Ydj1 was kindly provided by Dr. D. Cyr (University of North Carolina Medical School, USA), whilst human Hdj2 was purchased (Assay Designs). The His6-Hsj1a-encoding expression plasmid (pQE30-Hsj1a) was prepared by polymerase chain reaction (PCR) amplification of the coding region for human Hsj1a using a previously described plasmid construct as a template (kindly provided by Dr M.E. Cheetham, University College London, UK; Cheetham et al. 1994), and insertion of the product between the Bam HI and Hind III sites of pQE30 (McNamara 2006). Recombinant His6-Hsj1a was purified using standard nickel affinity chromatography procedures under nondenaturing conditions (McNamara 2006).
Codon harmonisation and plasmid construction
The PfHsp40 coding region was codon-harmonised (Angov et al. 2008) for improved heterologous expression in E. coli by Dr. Evelina Angov (Walter Reed Army Institute of Research, MA, USA). The codon harmonisation was confirmed with use of the GeneSplitter software package, kindly provided by the developer, Professor Jaco de Ridder (University of Pretoria, South Africa). To facilitate insertion of the coding region into the pQE30 expression vector (Qiagen), Bam HI and Hind III restriction sites were engineered into the sequence. The codon-harmonised PfHsp40 coding sequence was synthesised by Inqaba Biotechnical Industries (South Africa) and inserted into the PCR-Smart cloning vector (Lucigen) to produce plasmid pPCR–Smart–PfHsp40. The insert was excised from the plasmid with Bam HI and Hind III restriction enzymes (Fermentas), and ligated into Bam HI/Hind III-digested pQE30 expression vector (Qiagen) to produce plasmid pQPfHsp40. DNA sequencing was employed to verify the integrity of the coding sequence.
Heterologous expression and purification
Recombinant His6-PfHsp40 protein for protein aggregation suppression assays was purified from E. coli XL1-Blue (pQPfHsp40) grown at 26°C in Luria–Bertani media (LB; 1% (w/v) tryptone, 0.5% (w/v) yeast extract, 1% (w/v) NaCl) supplemented with 200 μg/ml ampicillin. Protein expression was induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) in early log phase (A600 <0.6) in the presence of selection pressure, and the cells were harvested 16 h post-induction. Protein purification was performed in batch, under denaturing conditions (4 M urea) in the presence of 0.1% (v/v) PEI for protein solubilisation, 10 mM imidazole, 300 mM NaCl and 10 mM Tris–Cl (pH 8.0). Cell lysis was achieved by sonication and treatment with 1 mM lysozyme, which included a freeze–thaw step at −20°C, in the presence of proteinase inhibitors (1 mM phenyl methyl sulphonyl fluoride (PMSF), 0.5 μg/ml pepstatin A and 1 μg/ml leupeptin). The cell lysate was clarified by centrifugation (16,000×g, 20 min, 4°C), before being allowed to bind to nickel-nitrilotriacetic acid beads (Ni-NTA; Qiagen) at a ratio of 1:4,000 beads to culture volume for 4 h at 4°C with gentle agitation. The beads were subsequently washed consecutively with incremented concentrations of imidazole (50, 100 and 150 mM), 50 mM HEPES, 10 mM ATP and 1% (v/v) Triton X-100, respectively, in 300 mM NaCl and 10 mM Tris-Cl (pH 8.0). Protein elution, with 1 M imidazole, was subsequently performed in the absence of urea to permit protein refolding. The fractions containing PfHsp40 were dialysed overnight at 4°C against 50 mM Tris–HCl pH 7.4, 50 mM NaCl, 2 mM MgCl2, 0.8 mM DTT and 5% (v/v) glycerol, and the protein was concentrated with polyethylene glycol (PEG-3000; Sigma-Aldrich).
For ATPase assays, PfHsp40 was purified using a modified protocol. E. coli M15 (pRep4; Qiagen) cells transformed with pQPfHsp40 were grown at 37°C in 100 ml of LB supplemented with 50 μg/ml ampicillin and 25 μg/ml kanamycin to an A600 of ~0.6. Next, IPTG was added to a final concentration of 0.4 mM, and growth was continued at 30°C for 4 h. Cells were pelleted by centrifugation, quick-frozen, and stored at −80°C. To isolate the expressed PfHsp40, the cell pellets were thawed and resuspended in 10 ml of lysis buffer (10 mM Tris, pH 8.0, 300 mM NaCl, 10 mM imidizole, 8 M urea) and incubated for 1 h with periodic, gentle agitation to lyse the cells. The cell lysate was then cleared by centrifugation, applied to a 1-ml Ni-NTA (Qiagen) column equilibrated in lysis buffer at 4°C, and allowed to drain through the resin under gravity. The column was washed with 10 ml of the lysis buffer, and this step was followed with three 10-ml washes with decreasing concentrations of urea (4, 2 and 0 M) to allow the protein to refold on the column. The bound protein was eluted with the lysis buffer lacking urea but supplemented with 250 mM imidizole. The fractions containing PfHsp40 were dialysed overnight at 4°C against 50 mM Tris–HCl pH 7.4, 50 mM NaCl, 2 mM MgCl2, 0.8 mM DTT and 5% (v/v) glycerol, and aliquots were stored at −80°C.
In both protocols, the purity of the recombinant proteins was verified by SDS–PAGE, and protein integrity and identity were assessed by western blot analysis using a mouse monoclonal anti-His tag (1:5,000; GE Healthcare) or a mouse anti-penta-His (1:5,000; Qiagen) primary antibody and an HRP-conjugated sheep anti-mouse secondary antibody (1:5,000; GE Healthcare).
Single-turnover ATPase assays
Single-turnover and steady-state radioactive ATPase assays were employed to assess the basal and co-chaperone-stimulated ATPase activities of recombinant HsHsp70 and PfHsp70. The protocols for the ATPase assays were adapted from those previously described (Fewell et al. 2004; Wright et al. 2008). In brief, Hsp70–[α32P]ATP complex was prepared through the incubation of 25 μg of purified Hsp70 protein with 25 μM ATP (Sigma Scientific) and 100 μCi of [α32P]ATP (Perkin-Elmer Life Sciences) in single-turnover complex buffer (STCB; 110 mM MgOAc, 100 mM KCl, 25 mM HEPES, pH 7.5) at 4°C for 30 min. Unbound [α32P]ATP was eliminated by purification at 4°C on an illustra™ NICK Sephadex G-50 column (GE Healthcare) equilibrated in STCB. Eluted fractions of Hsp70–[α32P]ATP complex were detected with a Geiger counter, and a 5-μl aliquot was subsequently assayed in a scintillation counter to confirm peak activity. The fractions were pooled and supplemented with glycerol to a final concentration of 10%, and aliquots from the pooled fractions were quick-frozen. The complex was stored at −80°C for a period of no longer than 2 weeks prior to use. For each reaction in which the basal or co-chaperone-stimulated ATPase activity was determined, a 25-μl aliquot of Hsp70–[α32P]ATP complex was rapidly thawed at 30°C and added to 25 μL STCB in a 50–μl reaction. In the co-chaperone-stimulated reactions, purified Hsp40 (0.3 μM) was added 60 s after the start of the reaction. The reaction was allowed to proceed at 30°C, and aliquots of 3 μl were removed at selected time points throughout a 10-min time course. ATP hydrolysis was interrupted by treatment with 1 μl of stop solution (SS; 2 M LiCl, 4 M Formic Acid, 36 mM ATP) and a reduction of the reaction temperature to 4°C. Aliquots of the stopped reactions (1 μl) were spotted in duplicate on thin layer chromatography (TLC) plates and allowed to develop in TLC buffer (0.5 M LiCl, 1 M formic acid). The [α32P]ATP isotope was diluted 1:100 and similarly spotted and developed to determine spontaneous hydrolysis. Phosphorimager analysis (Fujifilm Phosphorimager BAS 2500 coupled to Image Gauge software, version 4.0) was employed to determine the percentage of ATP hydrolysed to ADP and Pi in each reaction. Curve fits and kinetic analyses were performed on the obtained data using KaleidaGraph (version 3.0.4, Synergy Software). Kinetic analyses for the co-chaperone stimulated reactions were performed such that the 60-s time point, which was when the co-chaperone was added (see above), was corrected to the zero time point.
Steady-state ATPase assays
Purified recombinant Hsp70 protein (2 μM) was pre-incubated in reaction buffer (50 mM NaCl, 2 mM MgCl2, 10 mM DTT, 50 mM HEPES, pH 7.4) for 15 min at 4°C. The reaction was started by the simultaneous addition of 50 μM ATP and 0.01 μCi [α32P]ATP and allowed to proceed at 30°C over a 50-min time course. At selected time points, 3-μl aliquots of each reaction were removed and added to 1 μl of SS at 4°C. The aliquots representing each time point were subjected to thin layer chromatography (in duplicate) and analysed as described above. The data from each time point were also corrected for spontaneous ATP hydrolysis. Co-chaperone stimulation of the basal Hsp70 activity was assessed by the addition of 1 μM of each Hsp40 to the pre-incubated reaction mixture at 4°C prior to the addition of ATP. Background ATP hydrolysis of the co-chaperone protein preparations was assessed and subtracted accordingly.
Assays of small molecule modulators of ATPase activity
To assess the effects of small molecule modulators that interfere with the functional interaction between Hsp70 and Hsp40 partners, MAL3-39 (benzyl 1-(4-((1-([1,1′-biphenyl]-4-yl)-2-(tert-butylamino)-2-oxoethyl)(2-morpholinoethyl)amino)-4-oxobutyl)-6-methyl-4-(4-nitrophenyl)-2-oxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate; C47H54N6O8; Mr 830.4; Fewell et al. 2004) and DMT002264 (benzyl 4-([1,1′-biphenyl]-4-yl)-1-(6-((2-(butylamino)-1-cyclohexyl-2-oxoethyl)(hexyl)amino)-6-oxohexyl)-6-methyl-2-oxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate; C49H66N4O5; Mr 790.50; Wright et al. 2008), provided by the Center for Chemical Methodologies and Library Development at the University of Pittsburgh, were added into the basal and co-chaperone-supplemented ATPase assays in the presence of HsHsp70 or PfHsp70. Single-turnover and steady-state ATPase assays were conducted as detailed above with the following exceptions. In single-turnover assays, the compounds MAL3-39 and DMT002264 were dissolved in dimethyl sulphoxide (DMSO) and added to a final concentration of 300 μM at the start of each reaction. In steady-state assays, pre-incubation of the assay components prior to the addition of ATP was achieved in the presence of 300 μM of each respective compound dissolved in DMSO. In all assays, data were compared to control reactions that contained an equal volume of DMSO.
Malate dehydrogenase aggregation suppression assays
This assay assessed the ability of individual chaperones and Hsp70–Hsp40 pairs to suppress the heat-induced aggregation of malate dehydrogenase (MDH) at 48°C over time. The MDH (0.72 μM; Roche Applied Sciences, Germany) and chaperone proteins of interest (0.3 μM respectively) were suspended in assay buffer (100 mM NaCl, 50 mM Tris–Cl, pH 7.4) pre-equilibrated to 48°C. The accumulation of protein aggregates was monitored by changes at A360 in a thermocontrolled spectrophotometer (Model 14NT-UV–VIS, Aviv Instruments Inc, Lakewood, NJ, USA). As a reference, MDH aggregation was monitored in the absence of chaperones at 48°C over time. Similarly, in the absence of MDH, the thermal stability of the chaperone proteins under investigation was assessed to ensure accurate monitoring of the MDH aggregation in the test experiments. Furthermore, BSA (0.3 μM) was used to ensure that the observed suppression of MDH aggregation was chaperone-mediated. In previous assays (Shonhai et al. 2008), ATP has been included to demonstrate the nucleotide dependence of the MDH aggregation suppression; this control was not repeated here. Protein concentrations were calculated assuming the monomeric forms of the relevant proteins.
Results
PfHsp40 protein levels are upregulated by heat shock
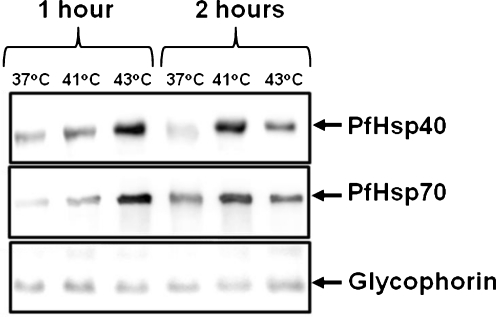
Antibodies generated to a C-terminal peptide region of PfHsp40 (Figure S1) were tested by western analyses and found to specifically detect PfHsp40 (data not shown). Using these anti-PfHsp40 antibodies, we conducted a western blot analysis on total protein extracts from parasites isolated from mixed-stage P. falciparum-infected erythrocytes grown in vitro under normal (37°C) and heat shock (41°C and 43°C) conditions. A single protein band of the expected molecular mass for PfHsp40 (49 kDa) was detected in each of the parasite extracts (Fig. 1, upper panel). As previously reported (Pesce et al. 2008), PfHsp70 was detected in parasite extracts after 37°C growth, and at increased levels after heat shock (Fig. 1, middle panel). PfHsp40 was detectable at 37°C, and similar to PfHsp70, increased protein levels were evident after heat shock at 41°C and 43°C. However, the expression of both proteins was slightly reduced at 43°C compared to 41°C after 2 h of heat shock compared to 1 h of heat shock. This was possibly due to parasite death after 2 h at 43°C. The differences in the levels of PfHsp40 and PfHsp70 for the various treatments were not due to differences in loading, as the levels of the loading control, glycophorin, remained relatively unchanged (Fig. 1, lower panel). Overall, these data indicate that the expression profiles of PfHsp40 and PfHsp70 are similar.
Fig. 1.
PfHsp40 is upregulated under heat shock conditions. P. falciparum-infected erythrocytes of mixed stages were grown at 37°C, 41°C and 43°C for 1 and 2 h, as indicated. Parasites were isolated from the infected erythrocytes by saponin lysis, and total protein from equal number of cells was subjected to SDS–PAGE, followed by western blot analysis using anti-PfHsp40 antibodies (upper panel) and anti-PfHsp70 antibodies (middle panel). The glycophorin signal was used as a loading control (lower panel). This experiment was repeated at least three times, and a representative series of western blots is shown
PfHsp40 is localised to the parasite cytosol in P. falciparum-infected erythrocytes
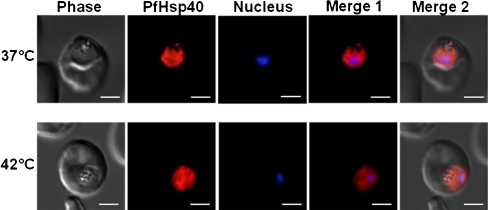
We next performed indirect immunofluorescence assays on P. falciparum-infected erythrocytes using purified IgG from the newly generated anti-PfHsp40 antiserum. As shown in Fig. 2, PfHsp40 localised mainly to the parasite cytosol at the trophozoite stage, and this localization was observed for parasites subjected to normal and heat shock conditions. A similar localization was detected for schizont-stage parasites (data not shown), and in all cases, the PfHsp40 localization profile was observed at an incidence of at least 90%. In comparison, PfHsp70 is primarily localised to the parasite cytosol and nucleus (Kappes et al. 1993; Pesce et al. 2008), and in the current study, we confirmed the localization of PfHsp70 to the parasite cytosol (data not shown). Interestingly, PfHsp40 was also occasionally detected externally to the parasite in the host cell, such as in punctate structures along the erythrocyte plasma membrane (data not shown). No fluorescence signal was detected when using the rabbit pre-immune serum, suggesting that the detection of PfHsp40 in the infected erythrocyte cytosol was specific. Overall, these data indicate that PfHsp40 resides primarily in the parasite cytosol.
Fig. 2.
PfHsp40 resides mainly in the parasite cytosol. Immunofluorescence staining to detect PfHsp40 was conducted on trophozoite stage P. falciparum-infected erythrocytes. Upper panels parasite-infected erythrocytes maintained at 37°C, lower panels parasite-infected erythrocytes incubated at 42°C for 2 h prior to fixation and staining. PfHsp40 was detected using the rabbit anti-PfHsp40 antibody and Cy3-conjugated goat anti-rabbit secondary antibody (indicated in red). Parasite nuclei were stained with Hoechst (indicated in blue). Columns: Phase phase-contrast image, Nucleus parasite nuclei, PfHsp40 PfHsp40 localization, Merge 1 merged image indicating PfHsp40 localization relative to the parasite nucleus, Merge 2 merged image indicating PfHsp40 localization relative to the parasite nucleus and phase-contrast image. A fluorescence signal was absent in negative controls that employed pre-immunisation serum as the primary antibody or that examined the signal when only a secondary antibody was used. The white size bars in each frame indicate 3 μm. This experiment was repeated at least three times, and a representative series of images is shown
Heterologous expression and purification of PfHsp40 and PfHsp70
To examine the biochemical properties of PfHsp40, the coding region for PfHsp40 was harmonised for heterologous expression in E. coli. Recombinant His6-PfHsp40 was expressed at moderate levels (Figure S2, lane 1), and its purification was successfully achieved by nickel affinity chromatography under denaturing conditions to yield approximately 1 mg of protein per litre of culture (Figure S2, lanes 2–5). This purification procedure yielded protein that was free of E. coli Hsp70 (DnaK). Although it was possible to partially purify His6-PfHsp40 using a nondenaturing purification procedure, the protein was typically contaminated with E. coli DnaK (data not shown). In parallel, recombinant His6-PfHsp70 protein was successfully overexpressed and purified using both denaturing and nondenaturing procedures. Purification of PfHsp70 using the denaturing procedure had the advantage of stripping the protein of any bound nucleotide, and therefore, this protein was ideal for use in the ATPase assays conducted in this study. Whilst PfHsp70 protein purified under both denaturing and nondenaturing conditions was successfully used in the protein aggregation suppression assays, to be consistent with our previous studies (Shonhai et al. 2008), the protein prepared under nondenaturing conditions was used in the aggregation assays in this study.
PfHsp40 stimulates the rate of ATP hydrolysis of PfHsp70 and HsHsp70
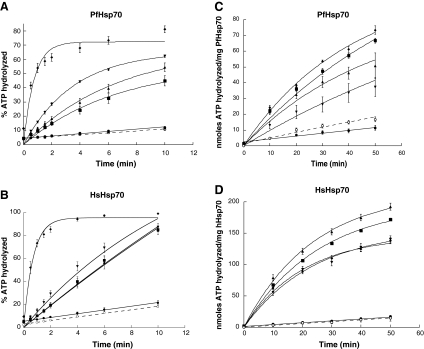
Hsp40 co-chaperones stimulate the rate-limiting ATP hydrolysis step in the Hsp70 catalytic cycle, which facilitates substrate capture. Using single-turnover assays in which this step is directly measured, we explored the effects of PfHsp40 and other Hsp40s on the ATPase activities of PfHsp70 and HsHsp70. Besides PfHsp40, the tested Hsp40s included both human (Hdj2 and Hsj1a) and yeast (Ydj1) Hsp70 co-chaperones. As anticipated, we first discovered that both PfHsp70 and HsHsp70 displayed relatively weak, basal rates of ATP hydrolysis (Fig. 3a, b). His6-Rtf1, a hexahistidine-tagged yeast protein involved in transcription (Stolinski et al. 1997) that lacks a J domain, served as a negative control and was unable to stimulate the ATPase activity of either PfHsp70 or HsHsp70. In contrast, PfHsp40, Hdj2, Ydj1 and Hsj1a stimulated the ATP hydrolytic rates of PfHsp40 and HsHsp70 when used at identical concentrations (Fig. 3a, b). The type I Hsp40s (PfHsp40, Hdj2 and Ydj1) stimulated each Hsp70 by comparable magnitudes, and with similar kinetics. The type II Hsp40, Hsj1a, simulated the ATP hydrolytic rates of both PfHsp70 and HsHsp70 to a greater magnitude and with very different kinetics to that observed for the type I Hsp40s (Fig. 3a, b).
Fig. 3.
PfHsp40 stimulates the rate of ATP hydrolysis of PfHsp70 and HsHsp70. The Hsp40-stimulated ATPase activities of PfHsp70 and HsHsp70 were monitored under single-turnover (a, b) and steady-state conditions (c, d). The ATPase activities of 2 μM PfHsp70 (a and c) and 2 μM HsHsp70 (b and d) were monitored for the Hsp70 alone (basal activity; filled circles) as well as in the presence of Hsp40 co-chaperones. The Hsp40 co-chaperone-stimulated ATP hydrolytic rates of PfHsp70 and HsHsp70 were monitored in the presence of 0.3 μM (single-turnover) or 1 μM (steady-state) of the co-chaperones PfHsp40 (filled downward triangles), Hdj2 (filled triangles), Ydj1 (filled squares), and Hsj1a (filled diamonds), as well as in the presence of a protein that lacks a J domain, Rtf1 protein (open circles), which was used as a negative control. For the single-turnover assays, the co-chaperones were added to the reaction mix after 60 s. Therefore, the results were plotted such that the 60-s time point was transposed as the zero time point, and all subsequent time points were considered relative to this time. Curve fits were applied using linear fits (PfHsp70 alone, HsHsp70 alone, and both Hsp70s with Rtf1), single-exponential fits (stimulation with PfHsp40, Ydj1, and Hdj2), or two-step exponential fits (stimulation with Hsj1a). For the steady-state assays, the Hsp70s alone or in the presence of the co-chaperones or Rtf1 were pre-incubated for 15 min at 4°C before the reaction was started upon the addition of ATP. The point at which ATP was added was set to zero, and curve fits were applied using linear (PfHsp70 alone, HsHsp70 alone, and both Hsp70s with Rtf1), or single-exponential (stimulation with PfHsp40, Ydj1, Hdj2, and Hsj1a) fits. Representative curve fits were generated in KaleidaGraph (version 3.0.4, Synergy Software). This experiment was repeated at least three times, and the standard deviations are shown as error bars for each point in all the curves
The ATP hydrolytic activities of PfHsp70 and HsHsp70 were also monitored in the presence of the Hsp40 proteins under steady-state conditions (Fig. 3c, d). Again, the control protein, His6-Rtf1, did not significantly stimulate the basal ATP hydrolytic activities of PfHsp70 or HsHsp70 (Fig. 3c, d). In contrast, PfHsp40 moderately stimulated that ATPase activity of PfHsp70, whilst Hdj2, Ydj1 and Hsj1a displayed a greater stimulatory effect (Fig. 3c). PfHsp40 and Hsj1a also moderately stimulated the ATPase activity of HsHsp70 to a similar extent, whilst Ydj1 and Hdj2 had a slightly greater stimulatory effect (Fig. 3d). Thus, although the relative stimulatory effects of each Hsp40 chaperone on PfHsp70 and HsHsp70 differ somewhat when the steady-state and single-turnover assays are compared, these data suggest that PfHsp40 interacts with PfHsp70 in the parasite. The data also indicate that PfHsp40, like some other Hsp40 co-chaperones (Hennessy et al. 2005), exhibits promiscuous functional interactions with evolutionarily diverse Hsp70s.
Pyrimidinone-peptoids differentially modulate Hsp40-stimulated ATPase activities of PfHsp70 and HsHsp70
Small molecule modulators have been identified that abrogate the ability of Hsp40s to activate the ATPase activity of Hsp70s, and some of these agents exhibit distinct effects on unique Hsp70–Hsp40 pairs (Fewell et al. 2004; Wright et al. 2008). Consistent with this mode of action, some members of this pyrimidinone class of Hsp70 modulators have been found to associate with Hsp70 at a region in the ATPase domain to which J domains bind (Wisén et al. 2010). Moreover, two structurally similar pyrimidinones, MAL3-39 and DMT002264, have been shown to modulate Hsp70 chaperone activity, and possess anti-malarial activity (Chiang et al. 2009). Because these molecules interfere with chaperone function by binding to a relatively large surface area on Hsp70 (Wisén et al. 2010), high concentrations of test compounds are required for these in vitro analyses. However, differences in their effects on unique chaperone classes can be used to infer distinct mechanisms of action.
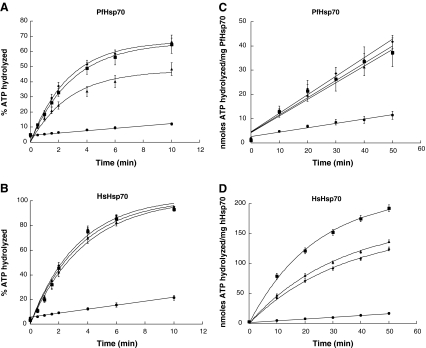
Based on these considerations, we examined the effects of MAL3-39 and DMT002264 on the Hsp40-stimulated ATPase activities of PfHsp70 and HsHsp70 (Fig. 4). In the presence of MAL3-39, PfHsp40-stimulated PfHsp70 ATPase activity was unaffected under both single-turnover (Fig. 4a) and steady-state conditions (Fig. 4c). MAL3-39 also had no effect on Hdj2-stimulated HsHsp70 activity under single-turnover conditions (Fig. 4b), but this agent had a strong inhibitory effect under steady-state conditions (compare filled diamonds to filled squares, Fig. 4d). The second compound, DMT002264, inhibited PfHsp40-stimulated PfHsp70 ATPase activity under single-turnover conditions (compare filled triangles to filled squares, Fig. 4a), but had no major effect under steady-state conditions (Fig. 4c). DMT002264 also had no effect on Hdj2-stimulated HsHsp70 ATPase activity under single-turnover conditions (Fig. 4b), but did have an inhibitory effect under steady-state conditions (compare filled triangles to filled squares, Fig. 4d). Overall, these data suggest that both MAL3-39 and DMT002264 are capable of inhibiting the Hdj2-stimulated ATPase activity of HsHsp70 under steady-state conditions, whilst the PfHsp40-stimulated ATPase activity of PfHsp70 is only inhibited by DMT002264 under single-turnover conditions. At present, the nature of the observed selectivity and the distinct effects on chaperone activity in steady-state versus single-turnover conditions is not completely clear (see “Discussion” section). However, we can conclude that the human and malarial chaperone systems are differentially inhibited by select members of this class of Hsp70 modulators. This result also suggests the possibility for enhanced, selective inhibition of PfHsp70–PfHsp40 over HsHsp70–Hsp40 function. Such selectivity represents the next step towards the development of chaperone-based, anti-malarial therapeutics.
Fig. 4.
The PfHsp70–PfHsp40 and HsHsp70–Hdj2 chaperone machineries can be differentially inhibited. The effects of the small organic molecules MAL3-39 and DMT002264 on the Hsp40-stimulated ATPase activities of PfHsp70 and HsHsp70 were monitored under single-turnover (a, b) and steady-state conditions (c, d). The assays were conducted in the presence of 2 μM of PfHsp70 alone (basal activity; filled circles), in the presence of PfHsp40 (1 μM; filled squares), and in the presence of PfHsp40 (1 μM) and MAL3-39 (300 μM; filled diamonds) or DMT002264 (300 μM; filled triangles; a and c). The assays were similarly conducted using 2 μM HsHsp70 alone (basal activity; filled circles), in the presence of Hdj2 (1 μM; filled squares), and in the presence of Hdj2 (1 μM) and MAL3-39 (300 μM; filled diamonds) or DMT002264 (300 μM; filled triangles; b and d). The curve fits were applied using linear (PfHsp70 and HsHsp70 alone), or single-exponential fits (stimulation with PfHsp40 and Hdj2 in the absence and presence of MAL3-39 and DMT002264). Representative curve fits were generated in KaleidaGraph (version 3.0.4, Synergy Software). This experiment was repeated at least three times, and the standard deviations are shown as error bars for each point in all the curves
PfHsp40 is able to suppress protein aggregation
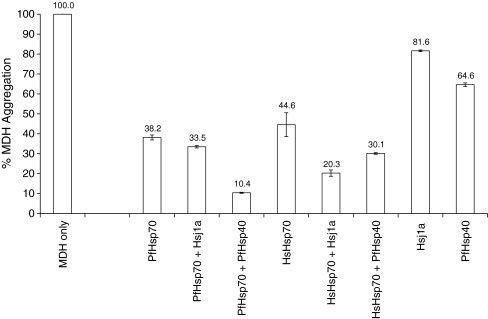
The ability of PfHsp70 to suppress the heat-induced aggregation of model proteins in vitro has been documented (Ramya et al. 2006; Shonhai et al. 2008). Consequently, we explored the ability of PfHsp40 to suppress protein aggregation alone (as reported for other type I Hsp40s; Lu and Cyr 1998; Walsh et al. 2004), as well as its ability to enhance the aggregation suppression activity of the Hsp70s. In these experiments, the aggregation of MDH in the absence of any added chaperone was set to 100% (Fig. 5). In the presence of a nonchaperone protein, BSA, we found MDH aggregation was unaffected (data not shown). Furthermore, none of the chaperones used in this study aggregated when tested alone under the assay conditions. In contrast, PfHsp40 suppressed MDH aggregation when used alone and was more effective than Hsj1a, which also suppressed MDH aggregation (Fig. 5; 64.6% versus 81.6% aggregation, respectively). As previously reported (Shonhai et al. 2008), PfHsp70 suppressed MDH aggregation, but when paired with PfHsp40, MDH aggregation was suppressed to a greater level (i.e. only 10% aggregation observed) than was due to the individual chaperones. Similarly, HsHsp70 suppressed MDH aggregation (44.6% aggregation), and in the presence of Hsj1a, there was greater suppression of MDH aggregation (Fig. 5; compare 44.6% versus 20.3% aggregation). Interestingly, the MDH aggregation suppression levels in the presence of each homologous Hsp70–Hsp40 pair were close to the sum of the individual aggregation suppression levels (Fig. 5). One interpretation is that the chaperone pair was acting independently in protein aggregation suppression. However, it cannot be excluded that there was synergistic cooperation between the chaperones. In comparison, the heterologous Hsp70–Hsp40 pairs exhibited MDH aggregation suppression activities that were somewhat less than the additive levels, especially in the case of the PfHsp70–Hsj1a pair where the aggregation suppression levels were similar to that of PfHsp70 alone (Fig. 5; compare 38.2% versus 33.5% aggregation). Overall, these data provide evidence for the first time that PfHsp40 alone can suppress protein aggregation, and that it may cooperate with PfHsp70 in the parasite.
Fig. 5.
PfHsp40 works more effectively with PfHsp70 than HsHsp70 in suppressing protein aggregation. The degree of heat-induced (48°C) aggregation of MDH was monitored in the presence or absence of the indicated chaperones. The aggregation of 0.72 μM MDH in the absence of chaperone proteins at saturation was assigned a value of 100%. The aggregation of MDH in the presence of the Hsp70s alone (0.3 μM), the Hsp40s alone (0.3 μM), and the Hsp70–Hsp40 combinations were assessed at saturation, and the percentage aggregation was determined relative to the MDH control. This experiment was repeated at least three times, and the average percent aggregation is indicated above each bar, with the standard deviations shown as error bars
Discussion
For the first time, we provide biochemical and cell biological evidence for a functional partnership between the canonical type I plasmodial Hsp40, PfHsp40, and the major plasmodial Hsp70, PfHsp70. Like PfHsp70, PfHsp40 localised mainly in the parasite, and its protein levels increased under heat shock conditions. Our findings are consistent with steady-state mRNA expression profiling data, which revealed that PfHsp40 is expressed during all stages of the intra-erythrocytic phase of parasite development, and that the expression pattern of PfHsp70 is similar (Le Roch et al. 2003). Importantly, our data also established that the protein expression profile of PfHsp40 mirrors the steady-state mRNA expression profile. Using purified recombinant protein, we found that PfHsp40 stimulated the basal ATP hydrolytic activity of PfHsp70 with similar magnitude and kinetics to that found for other type I Hsp40s with their Hsp70 partners. Furthermore, PfHsp40 suppressed protein aggregation and cooperated with PfHsp70 to inhibit the aggregation of a model protein. These results provide the strongest evidence to date for the existence of an Hsp70–Hsp40 pair in the malaria parasite, P. falciparum.
We also found that the type II Hsp40, Hsj1a, stimulated the ATP hydrolytic activities of both the human and plasmodial Hsp70s to relatively high levels, and with complex kinetics. These data are consistent with the fact that both Hsj1a and its splice variant, Hsj1b, have been shown to regulate the ATPase and substrate-binding activities of bovine Hsc70 (Cheetham et al. 1994). These chaperones also help mediate the degradation of select clients by the proteasome (Westhoff et al. 2005). In addition, whilst Hsj1a cooperated with HsHsp70 to suppress protein aggregation, the co-chaperone was unable to function with PfHsp70. Whilst Hsj1b has been shown to independently suppress protein aggregation (Schnaider et al. 2000), our experiments showed for the first time that Hsj1a similarly exhibited this chaperone-dependent property. Furthermore, Hsj1a was less effective than PfHsp40 in suppressing protein aggregation alone. Together, many of these results are consistent with published data that type I Hsp40s function better as chaperones on their own and are more effective at forming select chaperone partnerships with Hsp70 than type II Hsp40s (Lu and Cyr 1998; Rosser and Cyr 2007).
PfHsp40 is not the only Hsp40 co-chaperone that may partner with PfHsp70. The other type I Hsp40 in P. falciparum, Pfj1, has also been suggested to partner with PfHsp70 (Misra and Ramachandran 2009). However, as it is an atypical type I Hsp40 and resides in the apicoplast (Kumar et al. 2010), we do not favour the notion that Pfj1 is a PfHsp70 co-chaperone. However, we have previously shown that the type II Hsp40, Pfj4, is upregulated under heat shock conditions and is localised to the parasite cytosol and nucleus. Interestingly, in contrast to PfHsp40 (data not shown), we were able to detect Pfj4 in a complex with PfHsp70 after immunoprecipitation was performed from cell lysates (Pesce et al. 2008). This result suggests that the interaction between PfHsp70 and PfHsp40 is transient, as is commonly observed between chaperone and co-chaperone partners. In addition, and in accordance with our results, Hsp70s can be modulated by more than one Hsp40 partner (Hennessy et al. 2005; Walsh et al. 2004). This observation is important in light of the fact that Hsp40s serve as substrate scanners for Hsp70s (Rüdiger et al. 2001). Therefore, we suggest that the activity of PfHsp70 in the parasite is modulated by both PfHsp40 and Pfj4. There may even be additional P. falciparum Hsp40 partners for PfHsp70, which can now be tested using the tools developed in this report.
MAL3-39 and DMT002264 are known modulators of Hsp70 chaperone function and have been shown to exhibit anti-malarial activity (Chiang et al. 2009). The differential effects of these small molecules on the ATPase activities of PfHsp70–PfHsp40 compared to HsHsp70–Hdj2 strongly suggest that these chaperone partnerships are not identical, and provide the first side-by-side biochemical evidence for previous proposals that these plasmodial and human chaperone machineries are different (Matambo et al. 2004; Shonhai et al. 2008). The compounds exhibited unique effects in the steady-state and single-turnover ATPase assays—phenomena that have been observed for other chaperone pairs (Chiang et al. 2009). Possibly, the conformational change upon ligand binding in Hsp70 interferes differentially on nucleotide exchange versus ATP hydrolysis activity. Of note, the single-turnover assays only measure the kCAT (i.e. the ATP hydrolytic activity of Hsp70), whereas the steady-state assays quantify each step in the Hsp70 hydrolytic cycle, which includes nucleotide release.
Finally, whilst our assays were conducted in the presence of relatively high concentrations of compound, the data do provide a basis for the rational development of small molecule compounds that modulate Hsp70–Hsp40 interactions with greater selectivity and affinity (Wisén et al. 2010). Since molecular chaperones are emerging anti-malarial drug targets (Pesce et al. 2010; Shonhai 2010), compounds with greater selectivity and affinity will represent useful leads in novel drug discovery. However, more importantly, such compounds will also be useful as molecular probes for further dissecting the similarities and differences in the mechanism of action of plasmodial chaperones compared to their human homologues. The characterisation of chaperone partnerships, as performed in our study, provides a platform for the benchmarking of new “hit compounds” from recent large-scale screens (Guiguemde et al. 2010) for their potential effects on chaperone function.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Protein sequence alignment of PfHsp40, Pfj1 and type I Hsp40 proteins of eukaryotic and prokaryotic origin. Alignments were performed using type I Hsp40 protein sequences from Homo sapiens (Hdj2, NP_001530.1), Saccharomyces cerevisiae (Ydj1, CAA95937.1), E. coli (DnaJ, P08622.3) and P. falciparum (PfHsp40, PF14_0359/NP_702248.1; and Pfj1, PFD0462w/NP_702750.1). The N-terminal extensions of PfHsp40 and Pfj1, and the C-terminal extension of Pfj1 are shaded in red. The highly conserved J domain is shaded in blue with the conserved HPD, KFK and QKRAA motifs indicated in black boxes. The GF region is shaded in green with the conserved DIF motif highlighted in a black box (Cajo et al. [2006] J. Biol. Chem. 281: 12436–12444). The zinc-finger motifs of the zinc-binding domain (yellow shading) are similarly indicated in black boxes. Residues in bold and indicated by a downward arrow are proposed to be involved in substrate binding (Li and Sha [2005] Biochem. J. 386: 453–460). The CAAX-box motifs of PfHsp40, Ydj1 and Hdj2 are shaded in grey. The region of PfHsp40 used for the generation of peptide-based antibodies is shaded in purple. Conserved identical and similar residues are indicated in the consensus line by asterisks and dots, respectively. The alignment was performed using ClustalW (version 1.83; Chenna et al. [2003] Nucleic Acids Res. 31: 3497–3500) (DOC 46 kb)
Heterologous expression and purification of PfHsp40. Recombinant His6-PfHsp40 was expressed in E. coli XL1-Blue (pQPfHsp40), and purified by nickel affinity chromatography under denaturing conditions. The different stages of the purification process were examined by 10% SDS–PAGE (upper panels). Lane 1, E. coli XL1-Blue (pQPfHsp40) whole cell lysate fraction, 16 h post-induction; 2, E. coli XL1-Blue (pQPfHsp40) soluble fraction after sonication and treatment with urea and PEI; 3, unbound protein fraction (flow-through); 4, wash fraction using 150 mM imidazole; 5, elution fraction using 1 M imidazole, showing recovery of purified recombinant His6-PfHsp40 at 49 kDa. The presence of the His6-PfHsp40 protein in the various samples was verified by western blot analysis using anti-His tag antibodies (lower panels). This experiment was repeated at least three times (DOC 883 kb)
Acknowledgements
We thank the Arndt laboratory for the gift of purified Rtf1, and Dr. Douglas Cyr for supplying purified Ydj1. This research was funded in part by a NRF Focus Area grant (South Africa (GUN No: 2053542)), and a MRC grant awarded to GLB. The research was also partly funded by a DFG German-African Cooperation Projects in Infectology grant (DFG (Ref: LI 402/12-0)) awarded to GLB, AS, JMP and KL. MB was awarded NRF, Rhodes University and DAAD doctoral bursaries, and LLS a SAMI postdoctoral fellowship. SK is an associated member of the DFG Graduate school GK1216. JLB acknowledges support from the NIH (“The Pittsburgh Center for Kidney Research” (grant DK79307)). The Center for Chemical Methodologies and Library Development at the University of Pittsburgh provided DMT002264 and MAL3-39 and is supported by the NIH/NIGMS (P50 grant GM067082).
Abbreviations
- BSA
Bovine serum albumin
- DAPI
4′6-Diamidino-2-phenylindole
- DMSO
Dimethyl sulphoxide
- Hsp40
Heat shock protein 40
- Hsp70
Heat shock protein 70
- HRP
Horseradish peroxidase
- IPTG
Isopropyl-1-thio-β-d-galactopyranoside
- LB
Luria–Bertani media
- MDH
Malate dehydrogenase
- Ni-NTA
Nickel-nitrilotriacetic acid beads
- PBS
Phosphate-buffered saline
- PMSF
Phenyl methyl sulphonyl fluoride
- SDS–PAGE
Sodium dodecyl sulphate–polyacrylamide gel electrophoresis
- TRITC
Tetramethyl rhodamine isothiocyanate
References
- Acharya P, Kumar R, Tatu U. Chaperoning a cellular upheaval in malaria: heat shock proteins in Plasmodium falciparum. Mol Biochem Parasitol. 2007;153:85–94. doi: 10.1016/j.molbiopara.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Akide-Ndunge OB, Tambini E, Giribaldi G, McMillan PJ, Müller S, Arese P, Turrini F. Co-ordinated stage-dependent enhancement of Plasmodium falciparum antioxidant enzymes and heat shock protein expression in parasites growing in oxidatively stressed or G6PD-deficient red blood cells. Malar J. 2009;8:113. doi: 10.1186/1475-2875-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angov E, Hiller CJ, Kincad R, Lyon JA. Heterologous protein expression is enhanced by harmonizing the codon usage frequencies of the target gene with those of the expression host. PLoS ONE. 2008;3:e2189. doi: 10.1371/journal.pone.0002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botha M, Pesce E-R, Blatch GL. The Hsp40 proteins of Plasmodium falciparum: regulating chaperone power in the parasite and the host. Int J Biochem Cell Biol. 2007;39:1781–1803. doi: 10.1016/j.biocel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Bracher A (2007) Nucleotide exchange factors for Hsp70 molecular chaperones. In: Blatch GL (ed) Networking of chaperones by co-chaperones. Austin: Landes Bioscience; New York: Springer Science+Business Media; 26–37
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:SFAEOD>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham ME, Jackson AP, Anderton BH. Regulation of 70-kDa heat-shock-protein ATPase activity and substrate binding by human DnaJ-like proteins, HSJ1a and HSJ1b. Eur J Biochem. 1994;226:99–107. doi: 10.1111/j.1432-1033.1994.tb20030.x. [DOI] [PubMed] [Google Scholar]
- Chiang AN, Valderramos JC, Balachandran R, Chovatiya RJ, Mead BP, Schneider C, Bell SL, Klein MG, Huryn DM, Chen XS, Day BW, Fidock DA, Wipf P, Brodsky JL. Select pyrimidinones inhibit the propagation of the malarial parasite, Plasmodium falciparum. Bioorg Med Chem. 2009;17:1527–1533. doi: 10.1016/j.bmc.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb BS, Cooke BM, Reeder JC, Waller RF, Caruana SR, Davern KM, Wickham ME, Brown GV, Coppel RL, Cowman AF. Targeted gene disruption shows that knobs enable malaria-infected red cells to cytoadhere under physiological shear stress. Cell. 1997;89:287–296. doi: 10.1016/S0092-8674(00)80207-X. [DOI] [PubMed] [Google Scholar]
- Koning-Ward TF, Gilson PR, Boddey JA, Rug M, Smith BJ, Papenfuss AT, Sanders PR, Lundie RJ, Maier AG, Cowman AF, Crabb BS. A newly discovered protein export machine in malaria parasites. Nature. 2009;459:945–949. doi: 10.1038/nature08104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famin O, Ginsburg H. The treatment of Plasmodium falciparum-infected erythrocytes with chloroquine leads to accumulation of ferriprotoporphyrin IX bound to particular parasite proteins and to the inhibition of the parasite’s 6-phosphogluconate dehydrogenase. Parasite. 2003;10:39–50. doi: 10.1051/parasite/2003101p39. [DOI] [PubMed] [Google Scholar]
- Fewell SW, Smith CM, Lyon MA, Dumitrescu TP, Wipf P, Day B, Brodsky JL. Small molecule modulators of endogenous and co-chaperone-stimulated Hsp70 ATPase activity. J Biol Chem. 2004;279:51131–51140. doi: 10.1074/jbc.M404857200. [DOI] [PubMed] [Google Scholar]
- Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70-K heat shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Guiguemde WA, Anang A, Shelat AA, et al. Chemical genetics of Plasmodium facliparum. Nature. 2010;465:311–315. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005;14:1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller NL, Bhattacharjee S, Ooij C, Liolios K, Harrison T, Lopez-Estrano C, Haldar K. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Structural basis of the J cochaperone binding and regulation of Hsp70. Mol Cell. 2007;28:1–12. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, Martineau CN. Multiple Hsp70 isoforms in the eukaryotic cytosol: mere redundancy or functional specificity? Curr Genomics. 2008;9:338–348. doi: 10.2174/138920208785133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes B, Suetterlin BW, Hofer-Warbinek R, Humar R, Franklin RM. Two major phosphoproteins of Plasmodium falciparum are heat shock proteins. Mol Biochem Parasitol. 1993;59:83–94. doi: 10.1016/0166-6851(93)90009-M. [DOI] [PubMed] [Google Scholar]
- Külzer S, Rug M, Brinkman K, Cannon P, Cowman A, Lingelbach K, Blatch GL, Maier AG, Przyborski JM. Parasite encoded Hsp40 proteins define novel mobile structures in the cytosol of the P. falciparum infected erythrocyte. Cell Microbiol. 2010;12:1398–1420. doi: 10.1111/j.1462-5822.2010.01477.x. [DOI] [PubMed] [Google Scholar]
- Kumar N, Koski G, Harada M, Aikawa M, Zheng H. Induction and localization of Plasmodium falciparum stress proteins related to the heat shock protein 70 family. Mol Biochem Parasitol. 1991;48:47–58. doi: 10.1016/0166-6851(91)90163-Z. [DOI] [PubMed] [Google Scholar]
- Kumar A, Tanveer A, Biswas S, Ram EVSR, Gupta A, Kumar B, Habib S. Nuclear-encoded DnaJ homologue of Plasmodium falciparum interacts with replication ori of the apicoplast genome. Mol Microbiol. 2010;75:942–956. doi: 10.1111/j.1365-2958.2009.07033.x. [DOI] [PubMed] [Google Scholar]
- Lambros C, Vandeberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. doi: 10.2307/3280287. [DOI] [PubMed] [Google Scholar]
- Landry SJ. Structure and energetics of an allele-specific genetic interaction between dnaJ and dnaK: correlation of nuclear magnetic resonance chemical shift perturbations in the J-domain of Hsp40/DnaJ with binding affinity for the ATPase domain of Hsp70/DnaK. Biochemistry. 2003;42:4926–4936. doi: 10.1021/bi027070y. [DOI] [PubMed] [Google Scholar]
- Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA. Discovery of gene function by expression profiling of the malaria parasite lifecycle. Science. 2003;301:1487–1488. doi: 10.1126/science.1089799. [DOI] [PubMed] [Google Scholar]
- Li J, Qian X, Sha B. Heat shock protein 40: structural studies and their functional implications. Protein Pept Lett. 2009;16:606–612. doi: 10.2174/092986609788490159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the Hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- Maier AG, Rug M, O’Neill MT, Brown M, Chakravorty S, Szestak T, Chesson J, Wu Y, Hughes K, Coppel RL, Newbold C, Beeson JG, Craig A, Crabb BS, Cowman AF. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell. 2008;134:48–61. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- Matambo TS, Odunuga OO, Boshoff A, Blatch GL. Overproduction, purification, and characterization of the Plasmodium falciparum heat shock protein 70. Protein Expr Purif. 2004;33:214–222. doi: 10.1016/j.pep.2003.09.010. [DOI] [PubMed] [Google Scholar]
- McNamara C (2006) Rhodes University MSc thesis
- Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- Misra G, Ramachandran R. Hsp70-1 from Plasmodium falciparum: protein stability, domain analysis and chaperone activity. Biophys Chem. 2009;142:55–64. doi: 10.1016/j.bpc.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Nicoll WS, Botha M, Mcnamara C, Schlange M, Pesce ER, Boshoff A, Ludewig MH, Zimmermann R, Cheetham ME, Chapple JP, Blatch GL. Cytosolic and ER J-domains of mammalian and parasitic origin can functionally interact with DnaK. Int J Biochem Cell Biol. 2007;39:736–751. doi: 10.1016/j.biocel.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavithra SR, Banumathy G, Joy O, Singh V, Tatu U. Recurrent fever promotes Plasmodium falciparum development in human erythrocytes. J Biol Chem. 2004;279:46692–46699. doi: 10.1074/jbc.M409165200. [DOI] [PubMed] [Google Scholar]
- Pavithra SR, Kumar R, Tatu U. Systems analysis of chaperone networks in the malarial parasite Plasmodium falciparum. PLoS Comput Biol. 2007;3:1701–1715. doi: 10.1371/journal.pcbi.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce E-R, Acharya P, Tatu U, Nicoll WS, Shonhai A, Hoppe HC, Blatch GL. The Plasmodium falciparum heat shock protein 40, Pfj4, associates with heat shock protein 70 and shows similar heat induction and localisation patterns. Int J Biochem Cell Biol. 2008;40:2914–2926. doi: 10.1016/j.biocel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Pesce E-R, Cockburn IL, Goble JL, Stephens LL, Blatch GL. Malaria heat shock proteins: drug targets that chaperone other drug targets. Infect Disord Drug Targets. 2010;10:147–157. doi: 10.2174/187152610791163417. [DOI] [PubMed] [Google Scholar]
- Ramya TNC, Surolia NN, Surolia A. 15-Deoxyspergualin modulates Plasmodium falciparum heat shock protein function. Biochem Biophys Res Commun. 2006;348:585–592. doi: 10.1016/j.bbrc.2006.07.082. [DOI] [PubMed] [Google Scholar]
- Rosser MFN, Cyr DM (2007) Do Hsp40s act as chaperones or co-chaperones? In: Blatch GL (ed) Networking of Chaperones by Co-Chaperones. Austin: Landes Bioscience; New York: Springer Science+Business Media, 38–51
- Rüdiger S, Schneider-Mergener J, Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaider T, Soti C, Cheetham ME, Miyata Y, Yahara I, Csermely P. Interaction of the human DnaJ homologue, HSJ1b with the 90 kDa heat shock protein, Hsp90. Life Sci. 2000;67:1455–1465. doi: 10.1016/S0024-3205(00)00735-9. [DOI] [PubMed] [Google Scholar]
- Shonhai A. Plasmodial heat shock proteins: targets for chemotherapy. FEMS Immunol Med Microbiol. 2010;58:61–74. doi: 10.1111/j.1574-695X.2009.00639.x. [DOI] [PubMed] [Google Scholar]
- Shonhai A, Boshoff A, Blatch GL. Plasmodium falciparum heat shock protein 70 is able to suppress the thermosensitivity of an Escherichia coli DnaK mutant strain. Mol Genet Genomics. 2005;274:70–78. doi: 10.1007/s00438-005-1150-9. [DOI] [PubMed] [Google Scholar]
- Shonhai A, Boshoff A, Blatch GL. The structural and functional diversity of Hsp70 proteins from Plasmodium falciparum. Protein Sci. 2007;16:1803–1818. doi: 10.1110/ps.072918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonhai A, Botha M, Beer TAP, Boshoff A, Blatch GL. Structure-function study of Plasmodium falciparum heat shock protein 70 using three dimensional modeling and in vitro analysis. Protein Pept Lett. 2008;15:1117–1125. doi: 10.2174/092986608786071067. [DOI] [PubMed] [Google Scholar]
- Silva MD, Cooke BM, Guillotte M, Buckingham DW, Sauzet JP, Scanf C, Contamin H, David P, Mercereau-Puijalon O, Bonnefoy S. A role for the Plasmodium falciparum RESA protein in resistance against heat shock demonstrated using gene disruption. Mol Microbiol. 2005;56:990–1003. doi: 10.1111/j.1365-2958.2005.04603.x. [DOI] [PubMed] [Google Scholar]
- Snow RW, Guerra CA, Noor AM, Myint H, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolinski LA, Eisenmann DM, Arndt KM. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4490–4500. doi: 10.1128/mcb.17.8.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P, Bursac D, Law YC, Cry D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T-F, Chang J, Wang C. Identification of the peptide binding domain of hsc70. 18-kilodalton fragment located immediately after ATPase domain is sufficient for high affinity binding. J Biol Chem. 1993;268:26049–26051. [PubMed] [Google Scholar]
- Watanabe J. Cloning and characterization of heat shock protein DnaJ homologues from Plasmodium falciparum and comparison with ring infected erythrocyte surface antigen. Mol Biochem Parasitol. 1997;88:253–258. doi: 10.1016/S0166-6851(97)00073-X. [DOI] [PubMed] [Google Scholar]
- Westhoff B, Chapple JP, Spuy J, Höhfeld J, Cheetham ME. HSJI is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr Biol. 2005;15:1058–1064. doi: 10.1016/j.cub.2005.04.058. [DOI] [PubMed] [Google Scholar]
- Wisén S, Bertelsen EB, Thompson AD, Patury S, Ung PM-U, Chang L, Evans CG, Walter GM, Wipf P, Carlson HA, Brodsky JL, Zuiderweg ER, Gestwicki J. Binding of a small molecule at a protein-protein interface regulates the chaperone activity of Hsp70-Hsp40. ACS Chem Biol. 2010;5:611–622. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittung-Stafshede P, Guidry J, Horne BE, Landry SJ. The J domain of Hsp40 couples ATP hydrolysis to substrate capture in Hsp70. Biochemistry. 2003;42:4937–4944. doi: 10.1021/bi027333o. [DOI] [PubMed] [Google Scholar]
- Wright CM, Chovatiya RJ, Jameson NE, Turner DM, Zhu G, Werner S, Huryn DM, Pipas JM, Day BW, Wipf P, Brodsky JL. Pyrimidinone-peptoid hybrid molecules with distinct effects on molecular chaperone function and cell proliferation. Bioorg Med Chem. 2008;16:3291–3301. doi: 10.1016/j.bmc.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Protein sequence alignment of PfHsp40, Pfj1 and type I Hsp40 proteins of eukaryotic and prokaryotic origin. Alignments were performed using type I Hsp40 protein sequences from Homo sapiens (Hdj2, NP_001530.1), Saccharomyces cerevisiae (Ydj1, CAA95937.1), E. coli (DnaJ, P08622.3) and P. falciparum (PfHsp40, PF14_0359/NP_702248.1; and Pfj1, PFD0462w/NP_702750.1). The N-terminal extensions of PfHsp40 and Pfj1, and the C-terminal extension of Pfj1 are shaded in red. The highly conserved J domain is shaded in blue with the conserved HPD, KFK and QKRAA motifs indicated in black boxes. The GF region is shaded in green with the conserved DIF motif highlighted in a black box (Cajo et al. [2006] J. Biol. Chem. 281: 12436–12444). The zinc-finger motifs of the zinc-binding domain (yellow shading) are similarly indicated in black boxes. Residues in bold and indicated by a downward arrow are proposed to be involved in substrate binding (Li and Sha [2005] Biochem. J. 386: 453–460). The CAAX-box motifs of PfHsp40, Ydj1 and Hdj2 are shaded in grey. The region of PfHsp40 used for the generation of peptide-based antibodies is shaded in purple. Conserved identical and similar residues are indicated in the consensus line by asterisks and dots, respectively. The alignment was performed using ClustalW (version 1.83; Chenna et al. [2003] Nucleic Acids Res. 31: 3497–3500) (DOC 46 kb)
Heterologous expression and purification of PfHsp40. Recombinant His6-PfHsp40 was expressed in E. coli XL1-Blue (pQPfHsp40), and purified by nickel affinity chromatography under denaturing conditions. The different stages of the purification process were examined by 10% SDS–PAGE (upper panels). Lane 1, E. coli XL1-Blue (pQPfHsp40) whole cell lysate fraction, 16 h post-induction; 2, E. coli XL1-Blue (pQPfHsp40) soluble fraction after sonication and treatment with urea and PEI; 3, unbound protein fraction (flow-through); 4, wash fraction using 150 mM imidazole; 5, elution fraction using 1 M imidazole, showing recovery of purified recombinant His6-PfHsp40 at 49 kDa. The presence of the His6-PfHsp40 protein in the various samples was verified by western blot analysis using anti-His tag antibodies (lower panels). This experiment was repeated at least three times (DOC 883 kb)