Abstract
Objective
USA300 methicillin-resistant Staphylococcus aureus (MRSA) is increasing as a cause of severe community-associated bacteremic infections. We assessed severe sepsis in response to infection in patients with USA300 MRSA compared to non-USA300 MRSA bacteremia.
Methods
A cohort study was conducted from 1997–2008 comparing sepsis in response to infection in 271 patients with MRSA bacteremia from four VA hospitals.
Results
Sixty-seven (25%) patients with MRSA bacteremia were USA300 MRSA; 204 (75%) were non-USA300 MRSA. The proportion of MRSA bacteremia caused by USA300 MRSA increased over time (χ2 p<0.0001). Adjusting for age and nosocomial infection, patients with USA300 MRSA bacteremia were more likely to have severe sepsis or septic shock in response to infection than patients with non-USA300 MRSA bacteremia (adjusted Relative Risk=1.82; 95% CI: 1.16–2.87; p=0.01).
Conclusions
This suggests that patients with USA300 MRSA are more likely to develop severe sepsis in response to their infection, which could be due to host or bacterial differences.
Introduction
Patients with methicillin-resistant Staphylococcus aureus (MRSA) bacteremia generally have worse outcomes compared to patients with methicillin-susceptible S. aureus (MSSA) bacteremia.(Cosgrove et al. 2003; Cosgrove 2006) The predominant strain of MRSA causing community-associated MRSA (CA-MRSA) skin and soft tissue infections, USA300 MRSA, now causes approximately one-third of all MRSA bloodstream infections (BSI).(McDougal et al. 2003; Seybold et al. 2006) Few studies have examined infections complicated by bacteremia due specifically to the USA300 MRSA clone.(Seybold, Kourbatova et al. 2006)
The USA300 MRSA clone has two unique virulence factors in its genome: the genes for the Panton-Valentine leukocidin (PVL) and the arginine catabolic mobile element (ACME).(Diep et al. 2006) Although the role of the PVL gene in the virulence of CA-MRSA strains is still debated, it is a reliable biomarker, along with ACME, of USA300 MRSA isolates.(Voyich et al. 2006; Labandeira-Rey et al. 2007) It is biologically plausible that infections due to USA300 MRSA could lead to worse outcomes (such as severe sepsis or septic shock in response to infection) due to these or other virulence factors. The objective of this study was to examine the association of USA300 MRSA infections complicated by bacteremia (hereafter referred to as USA300 MRSA bacteremia) and severe sepsis or septic shock in response to infection.
Materials and Methods
Study Population
We conducted a multi-center retrospective cohort study to estimate the association between USA300 MRSA bacteremia and severe sepsis or septic shock in response to infection. The study subjects were veterans enrolled in care from January 1997 – June 2008 at four participating Veterans Affairs Medical Centers (VAMCs) including: 1) Baltimore, MD (January 1997–June 2008), 2) Washington, DC (March 2007–May 2008), 3) Buffalo, NY (January 2004–December 2007), and 4) Richmond, VA (August 2007– February 2008). The Institutional Review Boards at each participating site approved this protocol. Patients were included in the study if they: 1) were at least 18 years old, 2) were enrolled in patient care services at one of the participating sites, 3) had a positive blood culture for MRSA, 4) had no known previous invasive infection due to S. aureus, and 5) provided a bacterial isolate from the infection for testing. Patients were excluded if their infection was polymicrobial or if their bacteremia was found to be clinically insignificant. Staphylococcal bacteremia was determined to be clinically significant if the patient had at least one blood culture positive for S. aureus, with clinical signs and symptoms of infection.(Soriano et al. 2000)
Laboratory Evaluations
All MRSA isolates were sent to the Baltimore VAMC for testing. S. aureus was identified by traditional microbiologic techniques. Resistance to methicillin was confirmed via growth on oxacillin screen agar. All MRSA isolates were screened for the presence of the genes for PVL and ACME, as previously described.(Lina et al. 1999; Diep, Gill et al. 2006) Sequencing of the protein A (spa) gene hypervariable region was also performed.(Harmsen et al. 2003) Patient sequences were compared with sequences found in the Ridom spa Server (http://www.ridom.de/spaserver/).
USA300 MRSA isolates were accurately identified using an algorithm previously described by Diep, et al.(Diep, Gill et al. 2006) MRSA isolates that tested positive for the genes for PVL and ACME, and were spa type motif MBQBLO, were classified as USA300 MRSA. A 40% random sample of the suspected USA300 MRSA isolates from all participating sites throughout the study period was selected through the use of a random digit table and confirmed as USA300 MRSA by pulsed-field gel electrophoresis (PFGE).(McDougal, Steward et al. 2003) MRSA isolates that tested negative for all three genetic factors were classified as non-USA300 MRSA. A 30% random sample of the suspected non-USA300 MRSA isolates from all sites throughout the study period was also selected through the use of a random digit table and confirmed as non-USA300 MRSA by PFGE. All MRSA isolates that tested positive for at least one of the three (PVL, ACME, or spa type motif MBQBLO) were further characterized by PFGE to determine if any were the USA300 MRSA strain. If pulsed-field type (PFT) USA300 by PFGE, the isolates were classified as USA300 MRSA. Electronic images of the gels were analyzed via the Fingerprinting II software (BioRad Laboratories, Hercules, CA). The banding patterns of each isolate were compared to the USA PFGE types described by McDougal, et al.; the similarity between isolates was assessed using the criteria established by Tenover, et al.(Tenover et al. 1995; McDougal, Steward et al. 2003)
Study Design and Data Collection
A retrospective cohort study was conducted using all subjects with MRSA bacteremia during the study period from all four participating VAMCs. Eligible patients with USA300 MRSA bacteremia were compared to those with non-USA300 MRSA bacteremia with respect to severe sepsis or septic shock in response to infection. Chart review was performed via a standardized form to collect information regarding demographics, clinical data, and additional risk factors by an infection control nurse who was blinded to the outcome of each patient. Sepsis in response to infection was measured using the American College of Chest Physicians/Society of Critical Care Medicine (ACCP-SCCM) definitions.(Bone et al. 1992) Employing this classification scheme, patients were graded on a scale representing increasing severity of sepsis from no sepsis to sepsis, then severe sepsis, and finally septic shock. For this particular study, the outcome of interest was severe sepsis or septic shock (vs. sepsis or no sepsis).
An infection was classified as nosocomial if the patient’s culture was positive for MRSA more than 48 hours following hospital admission, or if the patient had been transferred from another healthcare facility, or if the infection was central-line associated. An infection was defined as central-line associated if it originated from a primary source (with no evidence of infection at a secondary site) in a patient with a central line in place within 48-hours prior to the onset of bacteremia.(Horan et al. 2008) The modified Duke criteria were used to define endocarditis.(Durack et al. 1994) Secondary sources of bacteremia were defined as pneumonia, skin and soft-tissue infection (SSTI), urinary tract infection (UTI), or other, and had to meet the Centers for Disease Control and Prevention (CDC) National Nosocomial Infections Surveillance System (NNIS) criteria for that site, despite the fact that the infection could have been community-associated.(Garner et al. 1988)
Also collected were the risk factors outlined by the CDC as part of their epidemiologic definition for CA-MRSA infections, including whether the patient had been hospitalized, had surgery, resided in a long-term care facility (LTCF), or had hemodialysis in the year prior to infection, as well as if there was any foreign medical device present at the time of infection or the patient had a previous episode of colonization or infection with MRSA.(Naimi et al. 2003; CDC 2004) The presence of comorbid conditions was also assessed to calculate each patient’s Charlson score.(Charlson et al. 1987)
Statistical Analysis
Data were analyzed using SAS statistical software, version 9.1. The Pearson’s χ2 or Fisher’s exact tests were used to compare categorical variables; the Student’s t-test or Wilcoxon signed rank test for continuous variables. A p-value ≤0.05 was considered statistically significant. An unadjusted relative risk (RR) was calculated to estimate the association between USA300 MRSA bacteremia and severe sepsis or septic shock in response to infection. Covariates associated with both USA300 MRSA bacteremia and severe sepsis or septic shock in response to infection were assessed for effect modification and confounding using stratified analysis. Any variable with a Breslow-Day p-value <0.05 was considered as an effect modifier. A 10% difference between the unadjusted and adjusted RR was used to identify confounding. A generalized linear model using a log link and assuming a Bernoulli distribution was developed to measure the association between USA300 MRSA bacteremia and severe sepsis or septic shock in response to infection, controlling for identified confounders and/or effect modifiers.
Results
We identified 271 eligible patients with MRSA bacteremia at the four facilities during the study period. Strains having all three determinants (PVL, ACME, and spa type motif MBQBLO) were designated USA300 MRSA, and a random sampling of these isolates by PFGE showed 100% sensitivity and specificity. Strains having none of the three determinants were designated non-USA300 MRSA, again with a sensitivity and specificity of 100%, as determined by PFGE of a random sample. Strains possessing one or two of the determinants were designated as non-USA300 MRSA based on our algorithm. Eighteen percent of these isolates were USA300 on PFGE, resulting in a sensitivity and specificity of the laboratory algorithm used for identifying USA300 MRSA of 100% and 82%, respectively.
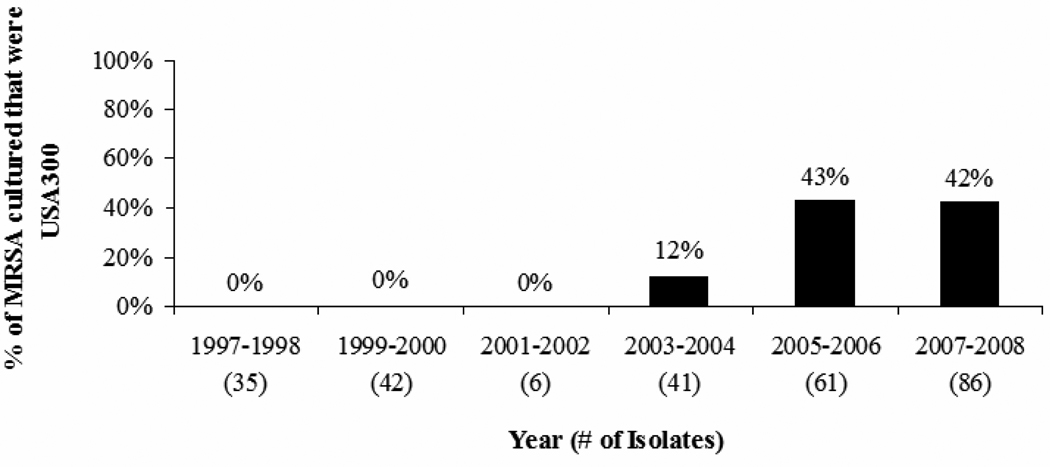
Sixty-seven (25%) of the infections were caused by USA300 MRSA; 204 (75%) by non-USA300 MRSA. Sixty-five (97%) of the 67 isolates classified as USA300 MRSA were positive for the PVL gene. In a sample of isolates confirmed as non-USA300 MRSA by PFGE, 56% were USA100 MRSA. The USA300 MRSA strain emerged in 2004, and remained the cause of a significant proportion of all MRSA infections complicated by bacteremia throughout the remainder of the study period (Figure 1; χ2 p<0.0001).
Figure 1.
Percent of MRSA infections complicated by bacteremia due to the USA300 MRSA strain in veterans at four VHA centers, 1997–2008.
Note: MRSA: methicillin-resistant Staphylococcus aureus; VHA: Veterans Health Administration.
a χ2 test for trend p<0.0001.
Patient, infection, and outcome characteristics of all subjects with MRSA bacteremia at the four participating sites are presented in Table 1. In the total study population, 63 (23%) subjects had severe sepsis or septic shock in response to their infection. The subjects had a mean age of 67 years and almost all were male. Infections were classified as nosocomial in 176 (65%) patients; 82 (47%) of those were central-line associated. Twenty-eight (10%) and 45 (17%) patients had infections in conjunction with endocarditis and pneumonia, respectively. Mortality within 90 days was identified in 121 (45%) patients; 83 (69%) of these deaths were attributed to the patient’s infection.
Table 1.
Patient, infection, and outcome characteristics of veterans with MRSA bacteremia at four VHA centers, 1997–2008, n=271.a
| Variable a | n (%) or mean ± SD |
|---|---|
| MRSA Isolate Characteristics | |
| USA300 MRSA | 67 (25) |
| Non-USA300 MRSA | 204 (75) |
| Patient Characteristics | |
| Age (in years) | 67 ± 13 |
| Male | 269 (>99) |
| Race | |
| White | 150 (55) |
| Black | 96 (36) |
| VHA Center isolated from | |
| Baltimore | 161 (59) |
| Washington DC | 23 (9) |
| Buffalo | 79 (29) |
| Richmond | 8 (3) |
| Year of presentation | |
| 1997–2003 | 87 (32) |
| 2004–2008 | 184 (68) |
| Charlson score | 5.0 ± 3.0 |
| Past history of colonization or infection with MRSA | 85 (31) |
| Hospitalized in the year prior to infection | 196 (72) |
| Residence in a LTCF in the year prior to infection | 46 (17) |
| Surgery in the year prior to infection | 117 (43) |
| Renal failure in the year prior to infection | 102 (38) |
| Infection Characteristics | |
| Nosocomial infection | 176 (65) |
| Presence of a central line at the time of infection | 82 (30) |
| Presence of permanent hardware at the time of infection | 98 (36) |
| Source of infection | |
| Primary | 101 (37) |
| Secondary | 167 (62) |
| Infection complicated by | |
| Endocarditis | 28 (10) |
| Pneumonia | 45 (17) |
| Outcome Characteristics | |
| Severe sepsis or septic shock in response to infection | 63 (23) |
| Mortality within 90 days of infection | 121 (45) |
| Infection caused or contributed to death | 83 (69) |
MRSA: methicillin-resistant Staphylococcus aureus; VHA: Veterans Health Administration; SD: standard deviation; LTCF: long-term care facility.
In bivariate analysis, patients with USA300 MRSA bacteremia were younger than patients with non-USA300 MRSA bacteremia (mean age: 63 ± 12 vs. 68 ± 13 years, p=0.003, Table 2) and were less likely to have been hospitalized (20% vs. 36%; p=0.008) or to have had surgery (15% vs. 32%; p=0.0007) in the year prior to infection. They were also less likely to have acquired their infection nosocomially (14% vs. 45%; p<0.0001) or to have died within 90 days of infection (17% vs. 31%; p=0.005). Older age (mean age: 70 ± 11 vs. 66 ± 14, p=0.02), a nosocomial acquisition of infection (RR=1.83; 95% CI: 1.08, 3.07; p=0.02), and death within 90 days of infection (RR=2.47; 95% CI: 1.56–3.92; p<0.0001) were all positively associated with severe sepsis or septic shock in response to their infection (Table 3). After controlling for age and nosocomial infection status, subjects with USA300 MRSA bacteremia were more likely to have severe sepsis or septic shock in response to infection than those with non-USA300 MRSA bacteremia (aRR: 1.82; 95% CI: 1.16–2.87; p=0.01; Table 4).
Table 2.
Patient and infection characteristics of veterans with MRSA bacteremia at four VHA centers associated with USA300 MRSA, 1997–2008, n=271. a
| Variable a | Type of MRSA | p-value c | |
|---|---|---|---|
| USA300 MRSA b n=67 |
Non-USA300 MRSA b n=204 |
||
| Patient Characteristics | |||
| Age in years, mean +/− SD | 63 ± 12 | 68 ± 13 | 0.003 |
| Race | |||
| Black | 34 (35) | 62 (65) | 0.003 |
| Other | 33 (19) | 142 (81) | |
| Charlson score, mean +/− SD | 5.2 ± 3.6 | 4.9 ± 2.8 | 0.60 |
| Hospitalized in the year prior to infection | |||
| Yes | 40 (20) | 156 (80) | 0.008 |
| No | 27 (36) | 48 (64) | |
| Residence in a LTCF in the year prior to infection | |||
| Yes | 10 (22) | 36 (78) | 0.61 |
| No | 57 (25) | 168 (75) | |
| Surgery in the year prior to infection | |||
| Yes | 17 (15) | 100 (85) | 0.0007 |
| No | 50 (32) | 104 (68) | |
| Renal failure in the year prior to infection | |||
| Yes | 42 (25) | 127 (75) | 0.95 |
| No | 25 (25) | 77 (75) | |
| Infection Characteristics | |||
| Nosocomial infection d | |||
| Yes | 25 (14) | 151 (86) | <0.0001 |
| No | 42 (45) | 52 (55) | |
| Presence of a central line at time of infection d | |||
| Yes | 11 (13) | 71 (87) | 0.004 |
| No | 56 (30) | 132 (70) | |
| Source of infection d | |||
| Primary | 19 (19) | 82 (81) | 0.07 |
| Secondary | 48 (29) | 119 (71) | |
| Infection complicated by endocarditis d | |||
| Yes | 9 (32) | 19 (68) | 0.34 |
| No | 57 (24) | 182 (76) | |
| Infection complicated by pneumonia d | |||
| Yes | 8 (18) | 37 (82) | 0.24 |
| No | 57 (26) | 161 (74) | |
| Outcome Characteristics | |||
| Severe sepsis or septic shock in response to infection d | |||
| Yes | 19 (30) | 44 (70) | 0.39 |
| No | 48 (25) | 146 (75) | |
| Mortality within 90 days of infection | |||
| Yes | 20 (17) | 101 (83) | 0.005 |
| No | 47 (31) | 103 (69) | |
MRSA: methicillin-resistant Staphylococcus aureus; VHA: Veterans Health Administration; SD: standard deviation; LTCF: long-term care facility.
Numbers represent number (row %), unless otherwise indicated.
Calculated via use of the Pearson’s χ2 or Fisher’s exact tests for categorical variables; the Student’s t-test or Wilcoxon signed rank test for continuous variables.
Any numbers not adding up to 271 are due to missing data.
Table 3.
Patient, infection, and treatment characteristics of veterans with MRSA bacteremia at four VHA centers associated with severe sepsis and/or septic shock, 1997–2008, n=257.a,b
| Variable a | Severe sepsis or septic shock c n=63 |
Sepsis or no sepsis c n=194 |
p-value d |
|---|---|---|---|
| MRSA Isolate Characteristics | |||
| USA300 MRSA | 19 (28) | 48 (72) | 0.39 |
| Non-USA300 MRSA | 44 (23) | 146 (77) | |
| Patient Characteristics | |||
| Age in years, mean +/− SD | 70 ± 11 | 66 ± 14 | 0.02 |
| Race | |||
| Black | 18 (20) | 74 (80) | 0.17 |
| Other | 45 (27) | 120 (73) | |
| Charlson score, mean +/− SD | 5.6 ± 3.2 | 4.8 ± 2.9 | 0.06 |
| Hospitalized in the year prior to infection | |||
| Yes | 46 (25) | 139 (75) | 0.83 |
| No | 17 (24) | 55 (76) | |
| Residence in a LTCF in the year prior to infection | |||
| Yes | 16 (37) | 27 (63) | 0.03 |
| No | 47 (22) | 167 (78) | |
| Surgery in the year prior to infection | |||
| Yes | 24 (22) | 84 (78) | 0.47 |
| No | 39 (26) | 110 (74) | |
| Renal failure in the year prior to infection e | |||
| Yes | 13 (23) | 43 (77) | 0.47 |
| No | 49 (25) | 146 (75) | |
| Infection Characteristics | |||
| Nosocomial infection e | |||
| Yes | 48 (29) | 115 (71) | 0.02 |
| No | 15 (16) | 78 (84) | |
| Presence of a central line at time of infection | |||
| Yes | 19 (26) | 54 (74) | 0.72 |
| No | 44 (24) | 140 (76) | |
| Source of infection e | |||
| Primary | 27 (28) | 69 (72) | 0.33 |
| Secondary | 36 (23) | 123 (77) | |
| Infection complicated by endocarditis e | |||
| Yes | 9 (33) | 18 (67) | 0.28 |
| No | 54 (24) | 173 (76) | |
| Infection complicated by pneumonia e | |||
| Yes | 23 (56) | 18 (44) | <0.0001 |
| No | 38 (18) | 171 (82) | |
| Outcome Characteristics | |||
| Mortality within 90 days of infection | |||
| Yes | 42 (37) | 73 (63) | <0.0001 |
| No | 21 (15) | 121 (85) |
MRSA: methicillin-resistant Staphylococcus aureus; VHA: Veterans Health Administration; SD: standard deviation; LTCF: long-term care facility.
There were 14 patients missing data regarding sepsis in response to infection;
Numbers represent number (row %), unless otherwise indicated.
Calculated via use of the Pearson’s χ2 or Fisher’s exact tests for categorical variables; the Student’s t-test or Wilcoxon signed rank test for continuous variables.
Any numbers not adding up to 257 are due to missing data.
Table 4.
Independent risk factors for severe sepsis or septic shock of veterans with MRSA bacteremia at four VHA centers, 1997–2008, n=257. a,b
| Variablea | Adjusted RR | 95% CI | p-value c |
|---|---|---|---|
| USA300 MRSA bacteremia | 1.82 | (1.16, 2.87) | 0.01 |
| Age (per 10 year increase) | 1.23 | (1.03, 1.47) | 0.02 |
| Nosocomial infection | 2.07 | (1.21, 3.54) | 0.01 |
MRSA: methicillin-resistant Staphylococcus aureus; VHA: Veterans Health Administration; RR: relative risk; CI: confidence interval.
There were 14 patients missing data regarding sepsis in response to infection;
Calculated via use of a generalized linear model using a log link and assuming a Bernoulli distribution.
Discussion
In this multi-center study, we demonstrated the emergence of USA300 MRSA in 2004 as a cause of MRSA bacteremia. We found that adults with USA300 MRSA bacteremia were nearly twice as likely to have severe sepsis or septic shock in response to their infection compared to adults with non-USA300 MRSA bacteremia when adjusted for age and nosocomial infection. This finding could be attributed to differences in host or bacterial characteristics.
The increase in the risk of sepsis could be a result of residual confounding after adjusting for age and nosocomial infection. For example, patients with USA300 MRSA bacteremia could be a generally sicker population, with a worse underlying severity of illness and more comorbid conditions. Alternatively, the currently circulating USA300 MRSA strain may be a more virulent clone compared to other strains of MRSA in patients with infections complicated by bacteremia.(Diep, Gill et al. 2006; Enright 2006) Nearly all of the USA300 MRSA strains in this evaluation were positive for the PVL gene. These data suggest that PVL-containing strains are associated with increased virulence in MRSA infections complicated by bacteremia.(Voyich, Otto et al. 2006; Labandeira-Rey, Couzon et al. 2007) While the role of PVL as a primary virulence factor is currently debated, our data support the hypothesis that it or other virulence factors should be considered as a possible explanation. Future studies are needed to address whether these findings are due to a combination of host and bacterial attributes.
To the authors’ knowledge, this study is the first to examine severe sepsis or septic shock as an outcome for USA300 MRSA bacteremic infections. We chose to use sepsis as the outcome because it was difficult to determine if patients who died within 90 days, died due to their infection. However, other studies using mortality as an outcome have had similar findings, with patients with USA300 MRSA bacteremia having worse outcomes compared to various control groups. For example, Kempker, et al. found that bacteremic patients infected with USA300 MRSA were at an increased risk of mortality compared to those infected with USA100 MRSA, with a HR of 1.79.(Kempker et al. 2010) One study by Popovich, et al. used a phenotypic rule based on antibiotic susceptibilities to predict the genotype of isolates as either community genotype (CG) or hospital genotype (HG). The results of this study showed that patients with BSIs due to CG strains had a higher risk of intensive care unit admission than those infected with HG strains.(Popovich et al. 2008)
This study has a number of limitations. First, the patient population included only veterans using VAMCs for healthcare, which could limit the generalizability of our findings. Veterans are a defined population, who are all adults, mostly male and African-American, older, and with a lower socio-economic status compared to similar adult populations in the U.S. However, it is unlikely that the relative severity of infections would be different in veterans compared to these similar adult populations, so the results should still be generalizable to the adult population. Our results are probably not generalizable to pediatric populations. Second, the control group of patients with non-USA300 MRSA bacteremia may have been a heterogeneous group, consisting of all other USA PFGE types. However, over half of the isolates in this group were found to be USA100 MRSA, which is the most common cause of MRSA infections acquired in the hospital setting.(McDougal, Steward et al. 2003)
Our study also had strengths. First, the use of more than one medical center improves generalizability. Second, we evaluated the robustness of the data by creating separate multivariate models excluding one VAMC at a time, and also restricting by year, to assess whether any of the associations observed may have been due to differences between the four VAMCs and time period. We found no difference in results between models, so chose to present the combined data. Third, the VHA uses an electronic medical record system across all of its sites, reducing the amount of missing data commonly encountered while performing chart reviews, and thus minimizing selection and information bias. Finally, the ability to provide molecular subtyping data for each MRSA isolate strengthens the findings of this study.
In conclusion, patients with USA300 MRSA bacteremia were more likely to have severe sepsis or septic shock in response to their infection than patients with non-USA300 MRSA bacteremia. These findings provide evidence that the USA300 MRSA strain may be more virulent than other strains of MRSA in patients with MRSA infections complicated by bacteremia. Our results underscore the need to prevent these infections.
Acknowledgements
This work was supported by a R36 grant award from the Centers for Disease Control and Prevention to K.M.K and by the University of Maryland General Clinical Research Center Grant M01 RR016500, General Clinical Research Centers Program, National Center for Research Resources (NCRR), NIH. It was also supported by a K12 RR023250-03 grant award from the National Institutes of Health to J.K.J. We thank Linda McGrail for her role in chart review of patient electronic medical records at the Baltimore VAMC. We also thank Jingkun Zhu for her efforts in mass electronic data extraction at the Baltimore VAMC and Alison Lydecker for the assistance she provided in statistical analysis. In addition, the support staff at the participating VAMC sites at Buffalo (Antonio Gonzalez), Washington, DC (Douglas Thomas and Sabiha Zubairi), and Richmond, VA (Rachel Bull) were helpful throughout the regulatory process and transfer of isolates to the Baltimore VAMC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Addresses Where Work Was Performed: VA Maryland Health Care System 10 North Greene Street Baltimore, MD 21201
VA Western New York Healthcare System 3495 Bailey Avenue Buffalo, NY 14215
Washington DC VA Medical Center 50 Irving Street NW Washington, DC 20422-0001
Hunter Holmes McGuire VA Medical Center 1201 Broad Rock Blvd Richmond, VA 23249
No conflicts of interest exist for any of the authors.
References
- Community-associated methicillin-resistant Staphylococcus aureus infections in Pacific Islanders--Hawaii, 2001–2003. MMWR Morb Mortal Wkly Rep. 2004;53:767–770. [PubMed] [Google Scholar]
- Bone RC, Sibbald WJ, Sprung CL. The ACCP-SCCM consensus conference on sepsis and organ failure. Chest. 1992;101:1481–1483. doi: 10.1378/chest.101.6.1481. [DOI] [PubMed] [Google Scholar]
- Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- Cosgrove SE. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis. 2006;42 Suppl 2:S82–S89. doi: 10.1086/499406. [DOI] [PubMed] [Google Scholar]
- Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 2003;36:53–59. doi: 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- Durack DT, Lukes AS, Bright DK. New criteria for diagnosis of infective endocarditis: utilization of specific echocardiographic findings. Duke Endocarditis Service. Am J Med. 1994;96:200–209. doi: 10.1016/0002-9343(94)90143-0. [DOI] [PubMed] [Google Scholar]
- Enright MC. Genome of an epidemic community-acquired MRSA. Lancet. 2006;367:705–706. doi: 10.1016/S0140-6736(06)68233-0. [DOI] [PubMed] [Google Scholar]
- Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections, 1988. Am J Infect Control. 1988;16:128–140. doi: 10.1016/0196-6553(88)90053-3. [DOI] [PubMed] [Google Scholar]
- Harmsen D, Claus H, Witte W, Rothganger J, Claus H, Turnwald D, Vogel U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36:309–332. doi: 10.1016/j.ajic.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Kempker RR, Farley MM, Ladson JL, Satola S, Ray SM. Association of methicillin-resistant Staphylococcus aureus (MRSA) USA300 genotype with mortality in MRSA Bacteremia. J Infect. 2010;61:372–381. doi: 10.1016/j.jinf.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labandeira-Rey M, Couzon F, Boisset S, Brown EL, Bes M, Benito Y, Barbu EM, Vazquez V, Hook M, Etienne J, Vandenesch F, Bowden MG. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315:1130–1133. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naimi TS, LeDell KH, Como-Sabetti K, Borchardt SM, Boxrud DJ, Etienne J, Johnson SK, Vandenesch F, Fridkin S, O'Boyle C, Danila RN, Lynfield R. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. Jama. 2003;290:2976–2984. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis. 2008;46:787–794. doi: 10.1086/528716. [DOI] [PubMed] [Google Scholar]
- Seybold U, Kourbatova EV, Johnson JG, Halvosa SJ, Wang YF, King MD, Ray SM, Blumberg HM. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis. 2006;42:647–656. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- Soriano A, Martinez JA, Mensa J, Marco F, Almela M, Moreno-Martinez A, Sanchez F, Munoz I, Jimenez de Anta MT, Soriano E. Pathogenic significance of methicillin resistance for patients with Staphylococcus aureus bacteremia. Clin Infect Dis. 2000;30:368–373. doi: 10.1086/313650. [DOI] [PubMed] [Google Scholar]
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyich JM, Otto M, Mathema B, Braughton KR, Whitney AR, Welty D, Long RD, Dorward DW, Gardner DJ, Lina G, Kreiswirth BN, DeLeo FR. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J Infect Dis. 2006;194:1761–1770. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]