Abstract
Autophagy is a lysosomal pathway by which intracellular organelles and proteins are degraded to supply the cell with energy and maintain cellular homeostasis. Recently lipid droplets have been identified as a substrate for macroautophagy. In addition to the classic pathway of lipid metabolism by cytosolic lipases, lipid droplets are sequestered in autophagosomes that fuse with lysosomes for the breakdown of lipid droplet components by lysosomal enzymes. The ability of autophagy to respond to changes in nutrient supply allows the cell to alter lipid droplet metabolism to meet cellular energy demands. Pathophysiological changes in autophagic function can alter cellular lipid metabolism and promote disease states. Autophagy therefore represents a new cellular target for abnormalities in lipid metabolism and accumulation.
Lipases and lysosomes
Recent investigations have demonstrated that lipid droplets (LDs) are not simple cytosolic structures that passively store triglycerides (TGs) and cholesterol, but rather complex intracellular organelles that perform a variety of biological functions [1]. These findings suggested that LDs may be regulated by standard pathways of lysosomal or proteasomal degradation. An important LD function is to store potentially toxic intracellular free fatty acids (FFAs) as TGs that can be broken down by lipolysis in times of limited nutrients. Lipolysis liberates FFAs for mitochondrial β-oxidation to supply the cell with ATP [2,3]. In times of adequate nutrient supply, lipolysis generates FFAs that are preferentially re-esterified back into TGs [4]. The metabolism of LD-stored TGs is under the control of cytosolic lipases [4], but the identity and sites of action of these lipases remain unclear, particularly in cells other than adipocytes. In addition to lipases that reside in the cytosol, lipases are present in lysosomes that are termed acid lipases. Acid lipases are known to hydrolyze the remnants of TG-rich lipoproteins taken up by receptor-mediated endocytosis [5]. Early attempts to study acid lipases through the use of nonspecific chemical lysosomal inhibitors such as ammonium chloride or chloroquine also suggested that lysosomal enzymes contribute to the lipolysis of intracellular lipids [6,7]. Recent investigations into the lysosomal pathway of autophagy have now delineated a mechanism for the lysosomal degradation of LD-stored TGs. This review will focus on new studies defining a critical function for autophagy in LD breakdown that markedly alters our understanding of LD metabolism in normal and pathological states.
Pathway of autophagy
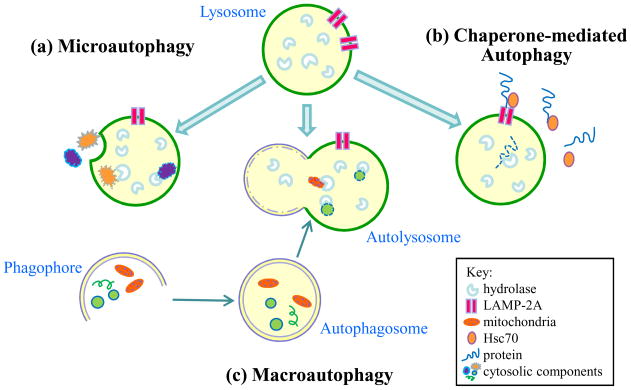
Autophagy, the literal meaning of which is self-eating, can be induced by starvation or other forms of nutrient deprivation to generate a variety of substrates for cellular energy generation [8]. Autophagy also serves as a catabolic pathway to recycle excessive or damaged intracellular organelles such as mitochondria. Largely through these two general functions, autophagy regulates a number of essential cellular processes including development and differentiation, immunity, apoptosis and aging [9–12]. ENREF_13 Three forms of autophagy have been defined: macroautophagy, chaperone-mediated autophagy (CMA) and microautophagy (Figure 1). In macroautophagy, the most physiologically important of the three in terms of the quantity of degradation, cytosolic organelles and protein complexes are sequestered in a double-membrane vesicle – the autophagosome [13]. Autophagosomes fuse with lysosomes to form an autolysosome in which the cargo of the autophagosome mixes with the hydrolytic enzymes of the lysosome for degradation and release into the cytoplasm for reuse [14]. In the absence of any definitive method to distinguish between an autophagosome and an autolysosome, the two structures are often grouped together and referred to as autophagic vacuoles. Although once considered non-selective, macroautophagy is now known to be specific in its targets [15–17], for example in cytosolic organelles such as mitochondria (mitophagy) [18] and the endoplasmic reticulum (reticulophagy) [19].
Figure 1.
The three types of autophagy. All three pathways result in the degradation of cytosolic components by lysosomal hydrolytic enzymes. In (a) microautophagy, an invagination of the lysosomal membrane allows for the internalization of cytosolic components such as organelles and proteins for their degradation by hydrolases. (b) Macroautophagy begins with the formation of a double membrane to form a structure termed a phagophore. The membrane elongates to sequester cellular elements including organelles and proteins within an autophagosome. The autophagosome traffics to a lysosome for fusion of the two into an autolysosome. The cargo of the autophagosome mixes with the hydrolytic enzymes of the lysosome for degradation. (c) Chaperone-mediated autophagy is specific for the removal of cytosolic proteins with a pentapeptide motif recognized by the chaperone Hsc70. Binding of this complex to the lysosomal LAMP-2A receptor leads to protein internalization and degradation.
In microautophagy organelles or proteins are taken up within an invagination of the lysosomal membrane for breakdown (Figure 1) [20]. Rather than targeting cellular organelles, CMA instead removes individual proteins with a specific peptide motif recognized by a chaperone protein Hsc70 (Figure 1). The chaperone-protein complex translocates to the lysosome where it binds to lysosome-associated membrane protein (LAMP) 2 for protein internalization and degradation [21]. Macroautophagy and CMA are induced by a number of stimuli in addition to limited nutrients [8], but all three forms of autophagy are constitutively active [22]. Studies to date have implicated only macroautophagy in the regulation of LDs, and macroautophagy (hereafter referred to as autophagy) will be the focus of this review.
Lipophagy degrades LDs
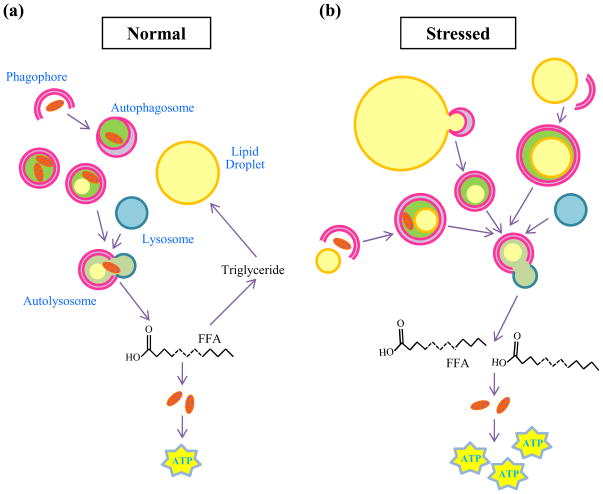
Lipolysis and autophagy share striking similarities. Both are essential catabolic pathways activated in response to nutrient deprivation. They are under identical hormonal control, i.e. inhibited by insulin or activated by glucagon [8]. Intracellular lipids were not previously considered autophagic substrates, but the similarities between lipolysis and autophagy, together with the existence of lysosomal lipases, suggested a possible link between the two pathways [23]. Indeed, an interrelationship between the two processes has recently been demonstrated by the finding that autophagy mobilizes lipids from LDs for metabolism through a process termed lipophagy (Figure 2) [24].
Figure 2.
Lipid droplet breakdown by lipophagy. (a) In the normal state, during times of sufficient nutrient supply, LDs are sequestered by autophagosomes and delivered to lysosomes for the degradation of TGs to FFAs. FFAs might contribute in a minor way to energy generation through mitochondrial β-oxidation to produce ATP or be re-esterified back into TGs for continued storage. Under these conditions the minority of autophagosomes contain lipid which is usually in combination with other cytosolic components. (b) When the cell is stressed either by nutrient deprivation or an excess of lipids, LD breakdown by this lysosomal pathway is increased. More autophagosomes contain lipid, frequently in the absence of other cargo. The increase in autophagy generates more FFA which are utilized preferentially to generate ATP.
Lipophagy was first described in hepatocytes which have the largest intracellular lipid stores of any cell except for adipocytes. Pharmacological inhibition of autophagy by 3-methyladenine or a genetic knockdown of the critical autophagy gene atg5 markedly increased TG and cholesterol content in hepatocytes challenged with an exogenous (oleate supplementation) or endogenous (culture in methionine/choline-deficient medium) lipid stimulus [24]. The accumulated lipid colocalized with LDs which increased in number and size with an inhibition of autophagy. LDs increased in the absence of a lipogenic stimulus as well, implicating autophagy in the basal control of cellular lipid content. LDs were visualized within autophagic vacuoles by electron microscopy (EM), and increased colocalization by fluorescence microscopy of neutral lipid and autophagosomal or lysosomal proteins in response to a lipid stimulus confirmed lipid movement into the autophagic pathway. Consistent with a function for autophagy in lipid degradation was that inhibition of autophagy did not alter TG synthesis but decreased rates of lipolysis and FFA β-oxidation. Administration of the chemical lipolysis inhibitor diethylumbelliferyl phosphate led to greater TG accumulation than that induced by 3-methyladenine, consistent with partial mediation of lipolysis by the lysosomal pathway. The precise extent of the contribution of autophagy to cellular lipid breakdown remains unclear. However, it appears that the involvement of autophagy is dictated by nutrient and lipid supplies, since a lipid stimulus led to an increase in the number of lipid-containing autophagosomes while limiting the sequestration of other substrates such as mitochondria. Thus, lipophagy is another form of selective organelle removal by autophagy.
The hypothesis that autophagy is responsible for hepatocyte LD degradation has also been confirmed in mice [24]. LDs alone, or mixed with other cytosolic cargo, were present in autophagic vacuoles in mouse livers by EM. LD size often exceeds that of an autophagosome suggesting that the lipid cargo represents portions of large LDs or small LDs engulfed whole. Starved mice were examined to determine the effect of the increased hepatic uptake of FFAs that results from elevated serum FFAs from starvation-induced adipose tissue lipolysis. The numbers of lipid-containing autophagic vacuoles were low in fed mice but increased with starvation as did the percentage of vacuoles containing lipid without any other substrate. These findings again indicate a selective movement of lipid into the autophagic pathway in response to a lipid challenge which was further demonstated by findings of increased amounts of TG and cholesterol in autophagic vacuole and lysosome cell fractions. These lipids were clearly derived from LDs as in addition to the increase in neutral lipids, LD-associated proteins adipose differentiation-related protein (ADRP) and tail-interacting protein of 47 kD (TIP-47) were detected by immunoblots of cell fractions of autophagic vacuoles and lysosomes. The presence of these proteins clearly indicates that LDs, or portions of LDs, are taken up into autophagosomes for lysosomal degradation.
Final evidence for the role of autophagy in hepatocyte lipid metabolism is that in vivo genetic ablation of hepatocyte autophagy led to lipid accumulation identical to findings in cultured hepatocytes [24]. Mice with a hepatocyte-specific knockout of the autophagy gene atg7 developed massively enlarged livers. The enlargement was due in part to a marked increase in TG and cholesterol content. These findings suggest that a general impairment in autophagy or a specific defect in lipophagy may in part underlie the development of steatotic liver diseases such as nonalcoholic fatty liver disease. Such an association remains to be demonstrated, but it is interesting that the incidences of nonalcoholic fatty liver disease and the metabolic syndrome increase with age [25], in parallel with a decrease in autophagic function [26]. In addition, a bidirectional interrelationship exists between autophagy and lipid stores as increased cellular lipid content decreases autophagic function [24,27]. Although the mechanisms of this effect remain unknown, the decrease could result from lipid alterations in membrane structures involved in autophagy. Supporting this possibility is that a defect in autophagosome/lysosome fusion has been described in high fat diet-fed mouse livers [28]. Alterations in membrane lipids resulting from the high fat diet presumably interfere with the normal fusion mechanism between the two membrane-bound organelles. Other accompanying factors such as hyperinsulinemia may act in concert with excessive lipids to decrease autophagy by downregulating the levels of autophagy genes [29]. These findings suggest that excessive lipid stores may lead to a further increase in LDs by inhibiting autophagic function which in turn leads to additional lipid retention in a self-perpetuating cycle.
The selective removal and degradation of lipids by lipophagy indicates that mechanisms exist to specifically target autophagosomes to LDs. Autophagy is known to increase in response to nutrient deprivation in part through mTOR signaling [30], so it is not surprising that LD breakdown occurs in response to limited nutrients. It remains to be established how increased cellular lipid content is sensed by the cell and converted into an increase in lipophagy. The selectivity of this process implies that LDs are recognized by components of the autophagic machinery that initiate formation of the autophagosome. Findings in other forms of selective autophagy suggest that organelle-specific proteins are required for the initiation of autophagosome membrane formation. Specific proteins mediating mitophagy are just being identified. Several forms of mitophagy are mediated by the Bcl-2 family member Nix [31–33]. Nix binds proteins required for autophagosome membrane formation [33], presumably recruiting the components necessary to assemble an autophagosome around mitochondria. It is likely that similar interactions between LD-associated proteins and components of the autophagic pathway exist, although these components remain to be identified. The autophagosome protein microtubule-associated protein light chain 3 (LC3) associates with LDs in the absence of a double-membrane structure in both fed and starved mice [24]. This finding suggests that LC3 conjugation may occur on the LD surface leading to formation of the limiting membrane in situ to sequester a portion of the LD. Other candidates for LD recognition are the LD-associated proteins. A number of LD-associated proteins have been identified by different techniques [34–37], and some of these proteins have been demonstrated to have roles in LD breakdown. For example, perilipin regulates standard lipolysis by altering LD exposure to cytosolic lipases [38] and mediates LD fragmentation [39]. An LD-associated protein likely serves as a recognition site for LD removal by autophagy. This situation would be analogous to that in mitophagy in which the mitochondrial protein Nix serves this function. Of the most prominent LD-associated proteins TIP-47 is a constitutively expressed protein whose levels do not change significantly with lipid accumulation, whereas ADRP levels increase markedly in proportion to elevations in lipid stores [40,41]. In contrast, in the livers of Atg7 null mice with excessive lipid accumulation ADRP levels rise only modestly but TIP-47 protein levels increase markedly [24]. This uncharacteristic increase in TIP47 in these mice indicates that autophagy normally functions to remove TIP-47 from LDs, suggesting that TIP-47 may be a LD protein recognized by the autophagic machinery. Whether TIP-47 or other LD-associated proteins mediate LD recognition and removal by autophagy is a critical area that requires further investigation in order to fully understand the process of lipophagy.
Autophagy regulates adipocyte LD formation through an effect on differentiation
Autophagy regulates many cellular functions suggesting that autophagy may affect LDs by mechanisms other than through LD breakdown. Studies of the effects of autophagy on adipocyte LDs have demonstrated a very different function for autophagy in these cells in contrast to that found in hepatocytes. Inhibition of autophagy by an Atg5 or Atg7 knockdown in 3T3-L1 pre-adipocytes blocked the LD formation and resultant TG accumulation that occurs with the chemically-induced differentiation of these cells into white adipocytes [42]. A similar effect occurred in Atg5 null murine embryonic fibroblasts induced to differentiate into adipocytes [43]. In 3T3-L1 cells the effect of a loss of autophagy on LDs was secondary to a failure of the cells to differentiate into white adipocytes [42]. Cells lacking autophagy failed to increase critical inducers of adipocyte differentiation, such as CCAAT/enhancer-binding protein-α and peroxisome proliferator-activated receptor-γ [42]. As a result, the cells lacked terminal markers of white adipocyte differentiation including fatty acid synthase, stearoyl-coenzyme A desaturase 1, aP2 and GLUT4. Thus, LDs were decreased from a lack of LD biogenesis secondary to a primary failure in adipocyte differentiation.
Genetic ablation of autophagy in vivo also profoundly affects adipocyte mass and differentiation [42,44]. In mice, an adipocyte-specific Atg7 knockout led to reduced body weight and increased insulin sensitivity because of a marked reduction in white adipose tissue mass. Fat depots typically composed of white adipocytes had increased morphological and molecular features of brown adipocytes. The cells had increased levels of uncoupling protein 1 and peroxisome proliferator-activated receptor-γ coactivator 1 which are markers of brown adipocytes. The cells also had multiple small LDs instead of the characteristic white adipocyte large unilocular LD. These findings together with an increase in the mass of brown adipocyte fat depots [42], indicated that the lack of autophagy favored formation of brown rather than white adipocytes. The absence of any effect of a loss of autophagy on white adipocyte stem cells suggests that the lack of autophagy may favor transdifferentiation of white adipocytes into brown [42], but the mechanism of this effect remains to be determined. The studies do demonstrate that autophagy is required for the formation of large LDs characteristic of white adipocytes. In contrast to the previously discussed decrease in autophagy that occurs in the liver with lipid accumulation [24,27], an increase in autophagy has been reported in human adipose tissue from obese individuals [45]. If this finding is confirmed, then augmented autophagy may be a mechanism of elevated LD fat storage and white adipocyte mass in obesity.
Studies to date have demonstrated two very different functions for autophagy in LD homeostasis. It is not surprising that adipocytes may differ from other cells in their mechanisms of LD formation and breakdown as these cells are uniquely constructed to store large amounts of lipid in a large single LD and have a number of lipases to supply FFAs for other tissues. The current findings have not ruled out a lipolytic function for autophagy in mature adipocytes as the loss of autophagy in differentiating adipocytes prevented LD formation. Investigations of autophagic inhibition in differentiated white adipocytes must be performed to address this possibility. Alternatively, the existence of a hepatocyte lipolytic pathway not operative in adipocytes may explain the ability of hepatocytes to handle large lipid stores similar to adipocytes despite the relative paucity of lipases in hepatocytes [46]. In the absence of robust lipolytic pathways such as that mediated by hormone-sensitive lipase, the hepatocyte can metabolize large LD stores through lipophagy. Autophagy does mediate LD breakdown in cells other than hepatocytes. Murine embryonic fibroblasts null for atg5 accumulate excessive TGs with lipid supplementation [24]. Autophagy-dependent LD regulation occurs in neurons as well. Increased neuronal lipid accumulation results from the Huntington’s disease mutation which causes a defect in the ability of autophagosomes to sequester cargo [47]. It is therefore likely that autophagy mediates LD breakdown in many if not all cell types, and the significance of this lysosomal degradative pathway in the metabolism of LDs will expand with studies of other cells.
Effects of autophagy on LDs in pathophysiological conditions
Autophagy regulates the development of fatty liver disease
The ability of autophagy to regulate LD accumulation in hepatocytes suggests that autophagic function in the liver may regulate the development of steatotic liver diseases such as nonalcoholic and alcoholic steatohepatitis [48]. These two highly prevalent liver diseases are characterized by increased storage of lipids in LDs and can progress to chronic liver injury and its complications such as fibrosis and hepatocellular cancer. Studies in an acute binge alcohol model have shown that alcohol increases autophagy to limit alcohol-induced accumulation of lipid in LDs and the development of a fatty liver [49]. Limiting LD stores may also prevent hepatocyte injury in this disease and in nonalcoholic steatohepatitis, as injury may result from steatosis by mechanisms such as the oxidation of excessive lipid stores. The increase in autophagy did not lead to a change in long-lived protein degradation suggesting that the hepatic autophagic response to alcohol was selective for lipophagy. Consistent with this conclusion is that increased numbers of autophagosomes in the livers of alcohol-fed animals contained LDs, demonstrating the selective movement of lipid into the autophagic compartment for degradation. Increased numbers of mitochondria were present in autophagosomes as well probably reflecting another selective autophagic response to remove mitochondria damaged by alcohol. Confirmatory of the function of autophagy in regulating LD accumulation from alcohol was that in vivo augmentation of autophagy by rapamycin decreased, and inhibition of autophagy increased, alcohol-induced steatosis [49]. These findings need to be confirmed in an animal model of chronic rather than acute alcohol ingestion that better reflects human alcoholic liver disease, but the results suggest that LD accumulation is regulated by autophagy in this liver disease. The findings also suggest that the highly variable extent of liver disease found among individuals with similar amounts of excessive alcohol intake may reflect differences in autophagic function. Future studies will examine whether genetic variation in autophagic function regulates the development of steatosis in human alcoholic and nonalcoholic steatohepatitis.
LD breakdown by autophagy is exploited for viral replication
The metabolic capabilities of autophagy may be usurped for purposes that are detrimental to the cell. The ability of certain viruses to alter cellular autophagy to promote their own replication may underlie a previously well known but poorly understood association between RNA flaviviruses and LD. The most well-established relationship is in chronic hepatitis C virus (HCV) infection in which the HCV core protein attaches to LD for unknown reasons but presumably to promote viral assembly [50]. Recent studies have demonstrated that up regulation of host cell autophagy by HCV is critical for the initiation of replication [51,52]. One possible mechanism for this effect may be that autophagy is essential for formation of the membranous web that is the site of viral replication. However, the discovery of lipophagy suggests that another possible mechanism for the regulation of HCV replication by autophagy is through a metabolic effect on LDs.
Recent investigations in another RNA virus, dengue virus, have demonstrated a metabolic effect of autophagy on viral replication. The site of dengue virus assembly is more controversial than for HCV and may occur in membrane structures adjacent to the ER [53], but evidence for LD-associated replication also exists [54]. Studies have demonstrated that dengue virus up regulates autophagy for the breakdown of LD-stored TGs that fuel dengue virus replication [55]. Dengue virus-infected hepatoma cells have increased numbers of autophagosomes that associate with LDs soon after infection and deliver lipids to lysosomes. As a result, autophagy mediated a 70% reduction in LD volume and a similar decrease in cellular TG content in infected cells. In contrast to reports of lipophagy regulating cholesterol [24], levels of cholesterol and cholesterol esters were unchanged, but their breakdown may have been masked by increased synthesis that occurs with viral infection [56]. Inhibition of autophagy blocked viral replication and a virus-induced increase in cellular FFA β-oxidation. β-oxidation was critical for viral replication and oleate supplementation overcame the block in replication induced by an inhibition of autophagy. The findings demonstrate that autophagy is critical for viral replication by degrading LDs to FFAs that can be utilized by β-oxidation to support viral replication. Presumably this process provides energy required for viral replication, but energy stores were not examined in these studies.
Changes in lipid metabolism in dengue infection may be more complex as these findings must be integrated with other studies demonstrating that dengue virus increases fatty acid synthase activity [57] and cholesterol synthesis required for viral replication [56]. A preliminary experiment with HCV indicated that this virus is also dependent on β-oxidation [55]. These findings suggest that an induction of autophagy is fundamental to flavivirus replication to mobilize LD stores. The ability of these viruses to associate with LD may serve to trigger lipophagy and the nature of this virus-LD interaction needs to be delineated. This finding may extend beyond this class of viruses as the DNA virus hepatitis B also exploits autophagy for its replication [58,59], ENREF_97 although this virus is not known to associate with LDs. Efforts to modify LDs or lipophagy may provide a novel avenue of anti-viral therapy.
Conclusions and future directions
Recent investigations have identified lipophagy as a new pathway for LD degradation. This finding necessitates a reevaluation of much of the knowledge and assumptions about LD metabolism in light of this new alternative pathway of lipolysis. Studies to date have only implicated macroautophagy in this process, but it is possible that the other forms of autophagy are involved as well. Microautophagy also removes organelles which potentially could include LDs. CMA could degrade LD-associated proteins that regulate cytosolic or lysosomal lipolysis. Suggestive of this possibility is that perilipin degradation in adipocytes is lysosome dependent [60] and therefore possibly CMA mediated. Critical areas of investigation for macroautophagy include how it is selective for LDs, what are the interacting proteins between LD and the autophagic pathway and how LD removal is increased in response to excess lipids. Although it seems that lipophagy is a pathway common to most cells, this remains to be proven. Lipophagy is likely to be an important metabolic pathway to supply energy for specific cellular functions. Other cell processes that rely on autophagy for LD breakdown for metabolic needs still must be determined. Little is known about genetic differences among individuals that lead to variations in autophagic function, but their existence may underlie the heterogeneity in manifestations of diseases marked by increased LD accumulation such as alcoholic and nonalcoholic steatohepatitis. The findings to date indicate that further investigations of lipophagy are likely to increase our understanding of the role of LD breakdown in cell physiology, and provide new avenues to treat diseases that result from defects in lipid metabolism or storage.
Acknowledgments
This work was supported by National Institutes of Health Grant DK61498 and AG031782.
Glossary
- Acute binge alcohol model
a rodent model of alcohol-induced liver injury in which the animal is given an excessive amount of alcohol over a one to two day period to induce LD accumulation and hepatocyte injury. These acute changes are reversible and may differ from the pathophysiology of chronic alcoholic liver disease that occurs in humans
- Alcoholic steatohepatitis
a liver disease in humans resulting from the excessive ingestion of alcohol that is characterized by the accumulation of LDs (steatosis) and the development of liver injury and inflammation (hepatitis)
- Autophagic vacuole
term referring to structures that are either autophagosomes or autolysosomes
- Autophagolysosome
cellular structure formed from the fusion of an autophagosome and a lysosome. The cargo from the autophagosome mixes with the hydrolytic enzymes of the lysosome and is degraded
- Autophagosome
double membrane vesicle that forms around cellular components for translocation to a lysosome as part of the process of macroautophagy
- Chaperone-mediated autophagy
one form of autophagy in which cytosolic proteins containing a pentapeptide motif are recognized by the chaperone Hsc70 for translocation to the lysosomal membrane and binding to the receptor LAMP-2A for protein internalization and degradation
- Lipophagy
specific type of macroautophagy in which portions of or whole LDs are sequestered alone or with other cytosolic constituents in autophagosomes for delivery to lysosomes for degradation
- Macroautophagy
type of autophagy in which cellular organelles and proteins are sequestered in autophagosomes which then fuse with lysosomes to form autolysosomes in which the cellular components are degraded and then released into the cytosol
- Microautophagy
a form of autophagy in which cellular components are taken up within an invagination of the lysosomal membrane for internalization and degradation
- Nonalcoholic fatty liver disease
a liver disease with histological similarity to alcoholic liver disease that occurs in the absence of excessive alcohol intake. The disease is a continuum from simple LD accumulation to steatosis together with hepatocyte injury and inflammation and eventual fibrosis. The mechanisms of this liver disease are unclear but it is strongly associated with insulin insensitivity and is a component of the metabolic syndrome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 2.Fujimoto T, et al. Lipid droplets: a classic organelle with new outfits. Histochem Cell Biol. 2008;130:263–279. doi: 10.1007/s00418-008-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiele C, Spandl J. Cell biology of lipid droplets. Curr Opin Cell Biol. 2008;20:378–385. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Gibbons GF, et al. Mobilisation of triacylglycerol stores. Biochim Biophys Acta. 2000;1483:37–57. doi: 10.1016/s1388-1981(99)00182-1. [DOI] [PubMed] [Google Scholar]
- 5.Wu X, et al. Exogenous VLDL stimulates apolipoprotein B secretion from HepG2 cells by both pre- and post-translational mechanisms. J Lipid Res. 1994;35:1200–1210. [PubMed] [Google Scholar]
- 6.Debeer LJ, et al. Lysosomal triacylglycerol lipase and lipolysis in isolated rat hepatocytes. J Biol Chem. 1979;254:8841–8846. [PubMed] [Google Scholar]
- 7.Duee PH, et al. Hepatic triglyceride hydrolysis and development of ketogenesis in rabbits. Am J Physiol. 1985;249:E478–E484. doi: 10.1152/ajpendo.1985.249.5.E478. [DOI] [PubMed] [Google Scholar]
- 8.Finn PF, Dice JF. Proteolytic and lipolytic responses to starvation. Nutrition. 2006;22:830–844. doi: 10.1016/j.nut.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 12.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 14.Tanida I. Autophagy basics. Microbiol Immunol. 2011;55:1–11. doi: 10.1111/j.1348-0421.2010.00271.x. [DOI] [PubMed] [Google Scholar]
- 15.Farre JC, et al. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraft C, et al. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 17.Kirkin V, et al. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 18.Tolkovsky AM. Mitophagy. Biochim Biophys Acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Bernales S, et al. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin Cell Dev Biol. 2010;21:719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 23.Czaja MJ, Cuervo AM. Lipases in lysosomes, what for? Autophagy. 2009;5:866–867. doi: 10.4161/auto.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ford ES, et al. Prevalence of the metabolic syndrome among US adults: findings from the third National Health and Nutrition Examination Survey. JAMA. 2002;287:356–359. doi: 10.1001/jama.287.3.356. [DOI] [PubMed] [Google Scholar]
- 26.Cuervo AM, et al. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- 27.Yang L, et al. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koga H, et al. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu HY, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–168. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandoval H, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schweers RL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartz R, et al. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- 35.Brasaemle DL, et al. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 36.Fujimoto Y, et al. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta. 2004;1644:47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 37.Sato S, et al. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J Biochem. 2006;139:921–930. doi: 10.1093/jb/mvj104. [DOI] [PubMed] [Google Scholar]
- 38.Brasaemle DL, et al. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J Biol Chem. 2000;275:38486–38493. doi: 10.1074/jbc.M007322200. [DOI] [PubMed] [Google Scholar]
- 39.Marcinkiewicz A, et al. The phosphorylation of serine 492 of perilipin A directs lipid droplet fragmentation and dispersion. J Biol Chem. 2006;281:11901–11909. doi: 10.1074/jbc.M600171200. [DOI] [PubMed] [Google Scholar]
- 40.Grasselli E, et al. PAT protein mRNA expression in primary rat hepatocytes: Effects of exposure to fatty acids. Int J Mol Med. 2010;25:505–512. doi: 10.3892/ijmm_00000370. [DOI] [PubMed] [Google Scholar]
- 41.Straub BK, et al. Lipid droplet-associated PAT-proteins show frequent and differential expression in neoplastic steatogenesis. Mod Pathol. 2010;23:480–492. doi: 10.1038/modpathol.2009.191. [DOI] [PubMed] [Google Scholar]
- 42.Singh R, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baerga R, et al. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5:1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovsan J, et al. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab. 2010 doi: 10.1210/jc.2010-1681. Epub. [DOI] [PubMed] [Google Scholar]
- 46.Zechner R, Madeo F. Cell biology: Another way to get rid of fat. Nature. 2009;458:1118–1119. doi: 10.1038/4581118a. [DOI] [PubMed] [Google Scholar]
- 47.Martinez-Vicente M, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Czaja MJ. Autophagy in health and disease. 2. Regulation of lipid metabolism and storage by autophagy: pathophysiological implications. Am J Physiol Cell Physiol. 2010;298:C973–C978. doi: 10.1152/ajpcell.00527.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding WX, et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLauchlan J. Lipid droplets and hepatitis C virus infection. Biochim Biophys Acta. 2009;1791:552–559. doi: 10.1016/j.bbalip.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 51.Dreux M, et al. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ait-Goughoulte M, et al. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welsch S, et al. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samsa MM, et al. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rothwell C, et al. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology. 2009;389:8–19. doi: 10.1016/j.virol.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 57.Heaton NS, et al. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sir D, et al. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci U S A. 2010;107:4383–4388. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang H, et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- 60.Kovsan J, et al. Regulation of adipocyte lipolysis by degradation of the perilipin protein: nelfinavir enhances lysosome-mediated perilipin proteolysis. J Biol Chem. 2007;282:21704–21711. doi: 10.1074/jbc.M702223200. [DOI] [PubMed] [Google Scholar]