Abstract
The structure of ent-copalyl diphosphate synthase (CPS) reveals three α-helical domains (α, β, γ), as also observed in the related diterpene cyclase taxadiene synthase. However, active sites are located at the interface of the βγ domains in CPS but exclusively in the α domain of taxadiene synthase. Modular domain architecture in plant diterpene cyclases enables the evolution of alternative active sites and chemical strategies for catalyzing isoprenoid cyclization reactions.
Terpenoids (also known as terpenes or isoprenoids) comprise the largest family of natural products found on the earth, currently numbering more than 60,000 members according to the Dictionary of Natural Products (http://dnp.chemnetbase.com). These structurally and stereochemically diverse compounds are required for biological function, communication, and/or defense in all forms of life. Moreover, some terpenoids are commercially important as flavorings, fragrances, pesticides, fuels, and pharmaceuticals1–3. Terpenoid cyclases (also known as terpenoid synthases) are the enzymes responsible for catalyzing the cyclization of linear isoprenoid precursors to form the complex hydrocarbon skeletons of all cyclic terpenoids. The two different classes of terpenoid cyclases are distinguished by amino acid sequence motifs reflecting alternative chemical strategies for initiating catalysis4–7. Class I cyclases contain DDXXD and (N,D)DXX(S,T)XXXE motifs; boldface residues generally coordinate to three Mg2+ ions, which trigger the ionization of an isoprenoid diphosphate group to generate a reactive carbocation intermediate. Class II cyclases contain a DXDD motif, in which the “middle” aspartic acid protonates an isoprenoid C=C bond to generate a reactive carbocation intermediate. After initial carbocation formation in either case, the ensuing multi-step cyclization cascade is ultimately terminated by proton elimination from, or nucleophilic capture of, the final carbocation intermediate.
A number of X-ray crystal structures are available for class I terpenoid cyclases from bacteria, fungi, and plants. Bacterial and fungal sesquiterpene (C15) cyclases such as pentalenene synthase8 and trichodiene synthase9 are single-domain enzymes that adopt the α-helical protein fold first observed for farnesyl diphosphate synthase10. This fold is designated the “α” domain11. Plant monoterpene (C10) and sesquiterpene (C15) cyclases such as bornyl diphosphate synthase12 and 5-epi-aristolochene synthase13, respectively, contain a catalytically-active α domain and an unrelated, nonfunctional α-helical domain designated the “β” domain11, such that the overall domain architecture is αβ. In contrast, the class II triterpene (C30) cyclases squalene-hopene cyclase14 and oxidosqualene cyclase15 each contain two α-helical domains designated “β” and “γ” 11, which form an active site cavity at their interface. The DXDD motif is located in the β domain. Interestingly, the γ domain is an insertion between the first and second helices of the β domain4,14; 23% amino acid sequence identity and significant topological similarities between the β and γ domains of squalene-hopene cyclase suggest gene duplication and fusion early in the evolution of terpenoid biosynthesis4.
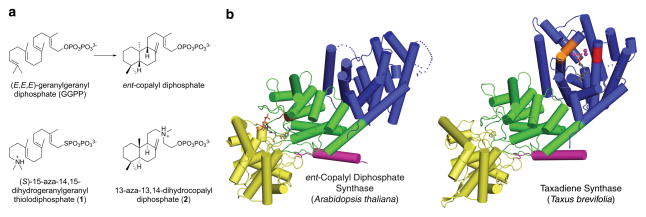
Strikingly, the recently determined structure of taxadiene synthase from Taxus brevifolia shows that a plant diterpene (C20) cyclase has domain composition αβγ and thereby contains the folds of both class I and class II terpenoid cyclases16. However, only the class I cyclase domain (α domain) is catalytically active; the class II cyclase (i.e., the βγ domains) is a nonfunctional vestige since it lacks the DXDD motif. In contrast, the diterpene cyclase ent-copalyl diphosphate synthase (CPS; also known as ent-kaurene synthetase A) from Arabidopsis thaliana has domain composition αβγ based on amino acid sequence analysis11, but catalyzes only the class II cyclization of geranylgeranyl diphosphate (GGPP) (Fig. 1a) initiated by general acid D379 in the D377IDD motif of the β domain17. CPS lacks metal binding motifs in the α domain, consistent with the lack of ionization-dependent class I cyclase activity17,18.
Figure 1. ent-Copalyl diphosphate synthase (CPS).
(a) The cyclization reaction catalyzed by CPS is the first committed step of gibberellin biosynthesis in Arabidopsis thaliana. Diterpene analogues (S)-15-aza-14,15-dihydrogeranylgeranyl thiolodiphosphate (1) and 13-aza-13,14-dihydrocopalyl diphosphate (2) partially mimic the substrate and product of this reaction. (b) Structure of CPS showing the vestigial class I cyclase active site in the α domain (blue) and functional class II cyclase active site at the interface of the β (green) and γ (yellow) domains. The N-terminal helix (magenta) and the D377IDD general acid motif (brown) in the β domain are indicated; substrate analogue 1 bound in the active site is shown as a stick figure. The domain structure of CPS is similar to that of taxadiene synthase, which is color-coded accordingly. However, taxadiene synthase lacks the general acid motif in the β domain and instead contains a fully active class I cyclase active site in the α domain, where D613DMAD and N757DTKTYQAE metal binding motifs are red and orange, respectively, three Mg2+ ions are magenta spheres, and a bound substrate analogue is shown as a stick figure.
We have determined the X-ray crystal structure of CPS complexed with (S)-15-aza-14,15-dihydrogeranylgeranyl thiolodiphosphate (1, Fig. 1a) at 2.25 Å resolution, and with 13-aza-13,14-dihydrocopalyl diphosphate (2, Fig 1a) at 2.75 Å resolution (Supplementary Results, Supplementary Table 1 and Supplementary Methods). While the modular αβγ architecture of CPS is structurally similar to that recently observed for taxadiene synthase (Fig 1b)16, the α domain of CPS (A534-V802) is a nonfunctional vestige since it cannot bind the metal ions required by a class I terpenoid cyclase. Additionally, helices D, F, G and H are shorter (helices G1 and α1 are nonexistent), the H–I loop is disordered, and the J–K loop is unusually short in comparison with the α domain of taxadiene synthase. Consequently, the nonfunctional class I active site of CPS is shallow and exposed (Supplementary Fig. 1).
The β domain of CPS (M84-I113 and P325-Q533) is structurally similar to the β domain of class I αβ terpenoid cyclases and the β domain of class II βγ triterpene cyclases. The γ domain of CPS (T114-F324) is structurally more similar to the catalytically nonfunctional γ domain of the class I diterpene cyclase taxadiene synthase than the catalytically functional γ domain of the class II triterpene cyclases squalene-hopene cyclase and oxidosqualene cyclase (Supplementary Fig. 2). This reflects the higher sequence identity between CPS and taxadiene synthase (Supplementary Table 2) as well as the lack of membrane association elements in CPS and taxadiene synthase that are present in the triterpene cyclases, which are monotopic membrane proteins14–16.
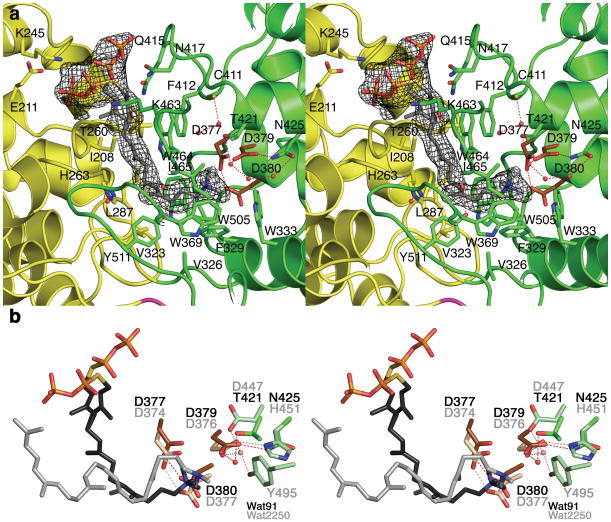
The active site of CPS resides in a deep cavity at the βγ interface and is more open and solvent-accessible than the active sites of squalene-hopene cyclase and oxidosqualene cyclase. The linear diterpene substrate analogue 1 binds to CPS such that the isoprenoid tail extends toward the base of the active site, and the thiolodiphosphate group binds in two alternate positions at the mouth of the active site (Fig. 2a). Parenthetically, we note that the bicyclic isoprenoid 2 does not bind in the active site, but instead binds at an interlattice site between two CPS molecules (Supplementary Fig. 3). Since the stereochemistry of 2 does not match that of the actual product of the cyclization reaction catalyzed by CPS, ent-copalyl diphosphate (Fig. 1a), this suggests that the active site contour exerts significant stereochemical control over isoprenoid ligand binding and product formation in catalysis. Comparison of the CPS-2 and CPS-1 complexes reveals that a conformational change of the loop containing K463 and W464 accompanies substrate analogue binding in the active site.
Figure 2. Binding of substrate analogue 1 in the active site of CPS.
(a) Simulated annealing omit map calculated with Fourier coefficients |Fo|−|Fc| in which 1 is omitted from the structure factor calculation, contoured at 4.0σ; selected residues are indicated. The thiolodiphosphate group adopts two alternate conformations. Residues from β and γ domains are green and yellow, respectively. Water molecules appear as red spheres. Conserved aspartic acid/aspartate residues of the D377IDD general acid motif are brown. (b) Least-squares superposition of the CPS and squalene-hopene cyclase (PDB 1UMP) active sites showing conserved features of aza-substrate analogue binding near the DXDD motifs. Atoms are color coded as follows: C = green (CPS) or light green (squalene-hopene cyclase), N = blue, O = red, P = orange; water molecules appear as red (CPS) or pink (squalene-hopene cyclase) spheres. Carbon atoms of the aspartic acids/aspartates in the DXDD motifs of CPS and squalene-hopene cyclase are dark and light brown, respectively. Carbon atoms of substrate analogues 1 and 2-azasqualene bound in the respective active sites are dark gray and light gray, respectively. Residue labels are black for CPS and gray for squalene-hopene cyclase. Hydrogen bond interactions are indicated by red and pink dashed lines, respectively.
The active site cavity of CPS is largely hydrophobic and is lined by numerous aliphatic and aromatic residues. In particular, the aromatic side chains of F329, W333, W369, and W505 are oriented such that they may stabilize carbocation intermediates in catalysis through cation-π interactions (Fig. 2a). The 15-aza group of 1 binds near D379 in the D377IDD general acid motif, in much the same fashion as the 2-aza group of 2-azasqualene binds near D376 in the D374VDD general acid motif of squalene-hopene cyclase19 (Fig. 2b). In the CPS-1 complex, given that the C14-N15 bond of 1 mimics the C14=C15 double bond of GGPP, the binding mode of 1 may mimic aspects of the initiation step of catalysis in a class II diterpene cyclase. However, given that 1 binds with a generally extended conformation, we conclude that it does not mimic the overall conformation of GGPP immediately prior to cyclization.
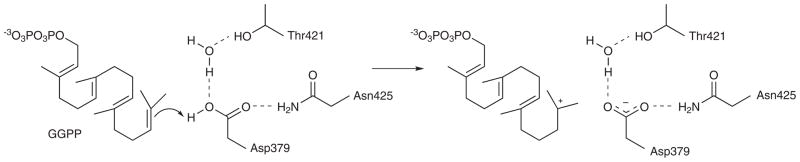
It is interesting that in both CPS and squalene-hopene cyclase, the conformation of the general acid residue is such that it would donate the superacidic anti-oriented proton to the isoprenoid substrate C=C bond; an anti-oriented carboxylic acid proton is ~104-fold more acidic than a syn-oriented proton20. This could enhance the general acid function of D379 in CPS and D376 in squalene-hopene cyclase (Fig. 3). General acid D376 in squalene-hopene cyclase is stabilized by a hydrogen bond with H45114,21; in CPS, general acid D379 is stabilized by a hydrogen bond with corresponding residue N425. These hydrogen bonding residues may facilitate general acid catalysis22. Sequence analyses of 18 class II diterpene cyclases indicate that N425 of CPS is conserved as asparagine, suggesting that an asparagine residue at this position signals diterpene cyclase function (Supplementary Fig. 4). Sequence analyses of 27 class II triterpene cyclases (16 squalene-hopene cyclases and 11 oxidosqualene cyclases) reveal greater variability at this position: residue 451 is conserved as histidine or arginine in squalene-hopene cyclases, but as alanine or serine in oxidosqualene cyclases (Supplementary Fig. 5).
Figure 3. Initiation of the class II diterpene synthase reaction by general acid catalysis.
Proposed first step of catalysis by CPS, highlighting substrate protonation by the anti-oriented carboxylic acid side chain of D379 to yield a carbocation intermediate.
A conserved water molecule is hydrogen bonded to the general acid in the active sites of class II cyclases: Wat91 of CPS is conserved as Wat2250 in squalene-hopene cyclase (Fig. 2b). In squalene-hopene cyclase, Wat2250 interacts with the general acid D376, Y495, and D447; Y495 is conserved in 26 of 27 triterpene cyclases, while D447 is conserved as D or E (Supplementary Fig. 5). This water molecule is proposed to assist Y495 in activating D376 for catalysis14,21. In CPS, Wat91 interacts with D379 and the hydroxyl group of T421; T421 is conserved in 17 of 18 class II diterpene cyclases (Supplementary Fig. 4) and replaces D447 of squalene-hopene cyclase. Y495 of the triterpene cyclases is not conserved in diterpene cyclases.
The thiolodiphosphate moiety and a portion of the isoprenoid chain of 1 are modeled in two alternate conformations due to apparent flexibility in the active site of CPS (Fig. 2a). The thiolodiphosphate group makes no well-defined hydrogen bond interactions in either conformation, although highly conserved residues K245 and K463 are partially disordered and about 3 Å away from thiolodiphosphate oxygen atoms (Fig. 2a). Slightly further away (~9 Å) are K241 (partially conserved) and R248 (highly conserved); accordingly, the diphosphate binding region is rather basic due in large part to the conserved “basic segment” K241-R248 (Supplementary Fig. 4). Possibly, these basic residues may interact with the substrate diphosphate group when bound in a fully productive conformation.
A single acidic residue, E211, is also found in this region, and is 5 Å away from the closest thiolodiphosphate oxygen atom of 1. This residue is conserved among 18 class II diterpene cyclases but is not conserved in class I diterpene cyclases (Supplementary Fig. 4). Since CPS requires divalent cations for optimal activity23, and since the chemistry of catalysis in a class II cyclase does not require metal ions (e.g., the protonation-dependent reactions of squalene-hopene cyclase and oxidosqualene cyclase do not require divalent cations24,25), a divalent cation important for enzyme activity may play a role in substrate binding, e.g., for binding the GGPP diphosphate group. Possibly, E211 could be part of the putative metal-binding site in CPS.
Alternatively, the metal binding site in plant class II diterpene cyclases is proposed to be the “EDXXD-like” motif found in the γ domain11. However, the corresponding E199DEND motif of CPS is ~18 Å away from the thiolodiphosphate group of 1, which is too far away to mediate an enzyme-metal-substrate interaction unless a significant protein structural change were to occur (Supplementary Fig. 6). No metal ions are observed in the CPS-1 complex even though crystals were prepared in presence of 2.5 mM Mn2+, although it is possible that metal binding is weakened by the relatively acidic conditions of the crystal structure determination (pH 5.4). Furthermore, Mn2+ does not functionally substitute for Mg2+ in catalysis by CPS23, so the origins of the 102–103-fold enhancement of CPS activity reported for Mg2+ and certain other divalent cations remain unclear. The structural basis for metal ion activation of catalysis by CPS will be explored in future studies.
Supplementary Material
Acknowledgments
We thank the NIH for grants GM56838 (D.W.C.), GM13956 (R.M.C.), and GM76324 (R.J.P.) in support of this research. Additionally, we thank the Advanced Photon Source at Argonne National Laboratory for beamline access.
Footnotes
Accession codes. Protein Data Bank: the atomic coordinates and the crystallographic structure factors of CPS complexed with 1 and 2 have been deposited with accession codes 3PYA and 3PYB, respectively.
Author contributions
M.K. and D.W.C. performed the X-ray crystallographic studies. H.H. and R.M.C. synthesized terpenoid diphosphate ligands. R.J.P. supplied the AtCPSd84 construct, and M.K. prepared the final CPS construct that yielded crystals. All authors contributed to the interpretation of the results and preparation of the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available online at http://www.nature.com/naturechemicalbiology/. Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Tholl D. Curr Opin Plant Biol. 2006;9:297–304. doi: 10.1016/j.pbi.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 2.Gershezon J, Dudareva N. Nat Chem Biol. 2007;3:408–414. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- 3.Bohlmann J, Keeling CI. The Plant J. 2008;54:56–669. doi: 10.1111/j.1365-313X.2008.03449.x. [DOI] [PubMed] [Google Scholar]
- 4.Wendt KU, Schulz GE. Structure. 1998;6:27–133. doi: 10.1016/s0969-2126(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 5.Wendt KU, Schulz GE, Corey EJ, Liu DR. Angew Chem Int Ed. 2000;39:812–2833. [PubMed] [Google Scholar]
- 6.Christianson DW. Chem Rev. 2006;106:412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 7.Christianson DW. Curr Opin Chem Biol. 2008;12:141–150. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesburg CA, Zhai G, Cane DE, Christianson DW. Science. 1997;277:820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 9.Rynkiewicz MJ, Cane DE, Christianson DW. Proc Natl Acad Sci USA. 2001;98:3543–13548. doi: 10.1073/pnas.231313098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarshis LC, Yan M, Poulter CD, Sacchettini JC. Biochemistry. 1994;33:0871–10877. doi: 10.1021/bi00202a004. [DOI] [PubMed] [Google Scholar]
- 11.Cao R, et al. Proteins: Struct Funct Bioinformatics. 2010;78:417–2432. [Google Scholar]
- 12.Whittington DA, et al. Proc Natl Acad Sci USA. 2002;99:5375–15380. [Google Scholar]
- 13.Starks CM, Back K, Chappell J, Noel JP. Science. 1997;277:815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 14.Wendt KU, Poralla K, Schulz GE. Science. 1997;277:811–1815. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]
- 15.Thoma R, et al. Nature. 2004;432:18–122. [Google Scholar]
- 16.Köksal M, Jin Y, Coates RM, Croteau R, Christianson DW. Nature. 2011;469:116–120. doi: 10.1038/nature09628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prisic S, Xu J, Coates RM, Peters RJ. ChemBioChem. 2007;8:869–874. doi: 10.1002/cbic.200700045. [DOI] [PubMed] [Google Scholar]
- 18.Sun TP, Kamiya Y. Plant Cell. 1994;6:509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinert DJ, Balliano G, Schulz GE. Chem Biol. 2004;11:121–126. doi: 10.1016/j.chembiol.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 20.Gandour RD. Bioorg Chem. 1981;10:169–176. [Google Scholar]
- 21.Wendt KU, Lenhart A, Schulz GE. J Mol Biol. 1999;286:175–187. doi: 10.1006/jmbi.1998.2470. [DOI] [PubMed] [Google Scholar]
- 22.Sato T, Hoshino T. Biosci Biotechnol Biochem. 1999;63:2189–2198. doi: 10.1271/bbb.63.2189. [DOI] [PubMed] [Google Scholar]
- 23.Prisic S, Peters RJ. Plant Physiol. 2007;144:445–454. doi: 10.1104/pp.106.095208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corey EJ, et al. J Am Chem Soc. 1997;119:1277–1288. [Google Scholar]
- 25.Ochs D, Tappe CH, Gärtner P, Kellner R, Poralla K. Eur J Biochem. 1990;194:75–80. doi: 10.1111/j.1432-1033.1990.tb19429.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.