Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is strongly linked to Kaposi's sarcoma, primary effusion lymphomas, and a subset of multicentric Castleman's disease. The mechanism by which this virus establishes latency and reactivation is unknown. KSHV Lyta (lytic transactivator, also named KSHV/Rta), mainly encoded by the ORF 50 gene, is a lytic switch gene for viral reactivation from latency, inasmuch as it is both essential and sufficient to drive the entire viral lytic cycle. Here we show that the Lyta promoter region was heavily methylated in latently infected cells. Treatment of primary effusion lymphoma-delivered cell lines with tetradecanoylphorbol acetate caused demethylation of the Lyta promoter and induced KSHV lytic phase in vitro. Methylation cassette assay shows demethylation of the Lyta promoter region was essential for the expression of Lyta. In vivo, biopsy samples obtained from patients with KSHV-related diseases show the most demethylation in the Lyta promoter region, whereas samples from a latently infected KSHV carrier remained in a methylated status. These results suggest a relationship among a demethylation status in the Lyta promoter, the reactivation of KSHV, and the development of KSHV-associated diseases.
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus (HHV) 8 , was first discovered in tissues obtained from Kaposi's sarcomas (KS; ref. 1) and later from multicentric Castleman's disease (MCD; ref. 2) and primary effusion lymphoma (PEL; ref. 3). KSHV is a newly recognized member of the γ-herpesvirus family and has a genetic similarity to herpesvirus saimiri and Epstein–Barr virus (EBV). In cells infected with herpesviruses, the regulation of gene expression generally follows an ordered cascade and can be classified as immediate-early (IE), early, or late, with the IE genes being transcribed first, after penetration of the virus. The early and late genes are transcribed thereafter (4). The expression of IE and early genes is independent of viral DNA replication, and some of these genes are involved in gene regulation and DNA replication. However, in vivo, viruses often have a more complicated pattern of expression that depends on the regulation of cellular as well as viral factors. KSHV transcripts in cell lines derived from primary effusion lymphomas, like BCBL-1, can also be divided into four classes of latent, IE, early, and late, based on their responsiveness to phorbol-ester treatment. KSHV-Lyta, a homologue of EBV-transactivator Rta, is mainly encoded by KSHV ORF 50 (5–7). Unlike EBV/Rta, KSHV-Lyta is essential and sufficient to drive the entire viral lytic cycle, and is the only lytic-switch gene for viral reactivation from latency (8). Consequently, the control of the expression of KSHV-Lyta should play an important role in the maintenance of viral latency. In this article, we report the role of promoter methylation in controlling Lyta expression. Methylation of DNA is involved in the regulation of gene expression (9). A methylation-free zone is required for initiation of transcription, and gene expression is often associated with demethylation in the vicinity of the promoter. Methylation is common among the γ-herpesvirus genomes, such as EBV and herpesvirus saimiri (10). KSHV belongs to γ2-herpesvirus (rhadinovirus) and is closely related to herpesvirus saimiri, the rhadinovirus most extensively studied before KSHV, and which is characterized by high 5′ cytosine-guanine (CpG) suppression and heavy methylation status in its genome. These facts strongly imply a similar methylation status in the KSHV genome. We report here our finding that the promoter region of KSHV-Lyta was heavily methylated in latency, and that demethylation in the Lyta promoter region was the essential event for the expression of Lyta, and subsequently for the whole lysis of the virus.
Materials and Methods
CpG Suppression Value.
In the case of double-stranded DNA, assessment of the CpG suppression value in a single sequence is measured by the formula P = 4 fCG/(fC + fG)2, where P denotes the CpG suppression value in a specific sequence, fCG represents the frequency of dinucleotide CG in the sequence, and fC or fG represents the frequency of mononucleotide C or G in the sequence (13). The P value of a random sequence should be 1. Any value less than 0.8 is regarded as CpG suppressed. The decreased frequency of CpG dinucleotides suggests that these viral genomes have been subjected to extensive methylation during coevolution with their host, resulting in the loss of CpG dinucleotides by mutation of methylated CpGs to 5′ thymine-guanine (TpGs). So, CpG suppression often reflects a methylation status in the genome.
Cell Lines and Drug Treatment.
BCBL-1 and BC-3, both KSHV-positive and PEL-derived cell lines, were cultivated with RPMI medium 1640 supplemented with 10% (vol/vol) FBS, 1 μM glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin. Ramos and Raji cells were cultured under the same conditions. 293T cells were cultivated with DMEM with 10% (vol/vol) FBS; 5-azacytidine (Sigma) was used to treat cells at a final concentration of 5 μM. Phorbol 12-tetradecanoate 13-acetate (TPA; Sigma) and phosphonoformic acid (Sigma) were used at a final concentration of 25 μg/ml and 400 μg/ml, respectively.
mAb and Indirect Immunofluorescence Antibody Assay (IFA).
mAb against gene products of ORF K9, ORF 59, and ORF K8.1 were developed by using TPA-treated BCBL-1 cells as described (12). The mAb to Lyta was developed by using an ORF 50 cDNA expression clone (6). IFAs were performed as described (6).
Southern Blotting.
Genomic DNA samples (5 μg) isolated from BCBL-1 (untreated or treated with TPA for 48 h) were digested in parallel with HpaII or MspI, electrophoresed on a 2% agarose gel, transferred to nylon membrane (Amersham Pharmacia), and probed with different digoxigenin (DIG)-labeled DNA fragments to assess the methylation status by using a DIG-DNA labeling and detection kit (Roche Molecular Biochemicals). DNA fragments of the Lyta and LANA promoter region were amplified by PCR. The primers for the Lyta promoter were 50.PF (5′-CTGCCCATGGGCGGGTGGGTGACAGTCCGC-3′) and 50.PR (5′-TGCGCCATGGTTGTGGCTGCCTGGACAGTA-3′). The primers for the LANA promoter were LANA.PF (5′-CGGCCCAGGTGACATTGGGGGGTGGG-3′) and LANA.PR (5′-TAAGTCAGCCGGACCAAGCTGCTGCTCTTG-3′). K9 and polymerase promoter fragments are described elsewhere (6).
Bisulfite Genomic Sequencing.
The bisulfite modification of the genomic DNA method first described by Frommer et al. (11) was used here with some modification. Briefly, 10 μg of DNA sample was digested with EcoRI, purified, and ethanol precipitated. Purified DNA was denatured in 0.3 M NaOH and treated with 5 M bisulfite/0.5 mM hydroquinone (pH 5.0) for 3 h at 55°C. DNA was purified, desulfonated in 0.3 M NaOH, neutralized with ammonium acetate, and precipitated. Purified DNA was dissolved in 100 μl of 10 mM Tris/1 mM EDTA (pH 8.0) buffer for PCR amplification. The conversion efficiency of nonCpG cytosines into thymines was over 99.5%. The PCR primers for promoter region of the Lyta were 50P.M1 (5′-TTAAAGCTTTTTAGGATATTGGTTATGTAT-3′) and 50P.M2 (5′-TACACCATGGTTATAACTACCTAAACAATA-3′). The primers for the LANA promoter were LAP.MF (5′-GGTTTAGGTGATATTGGGGGGTGGG-3′) and LAP.MR (5′-TAAATCAACCAAACCAAACTACTACTCTTA-3′). These primers were specific for the top strands of bisulfite-treated viral genomic DNA. For experiments on the time course of TPA treatment, PCR products were sequenced directly by an Applied Biosystems sequencer. For other experiments, PCR products were cloned into the pCR2.1 vector by using the TA cloning kit (Invitrogen) to separate single clones; then, plasmid DNA or colony-PCR products were sequenced by using a Li-Cor DNA sequencer (dye primer) or an Applied Biosystems sequencer (dye terminator).
Plasmids and Luciferase Assay.
The pGL3-Basic promoterless plasmid (Promega) containing the luciferase gene was used for construction of the Lyta promoter–reporter clone by using a PCR-based strategy. The PCR primers were the same as those used in Southern blotting. Deletion mutants of the Lyta promoter were generated by using a deletion kit (Takara Shuzo, Kyoto) and were described elsewhere (6). Other plasmids used in this report and luciferase assays performed are described elsewhere (6).
In Vitro DNA Methylation and Methylation Cassette Assay.
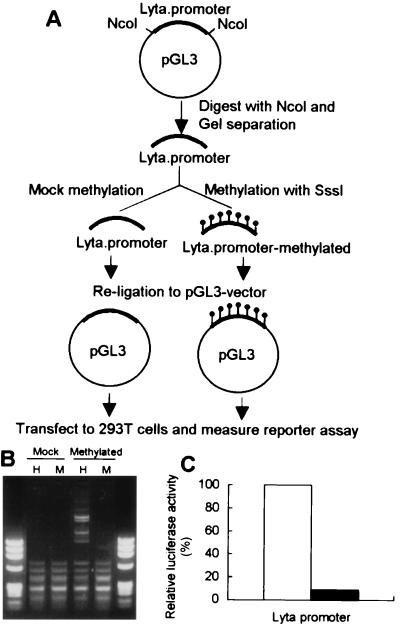
The methylation cassette assay was performed as described with some modification (ref. 14; outlined in Fig. 5a).
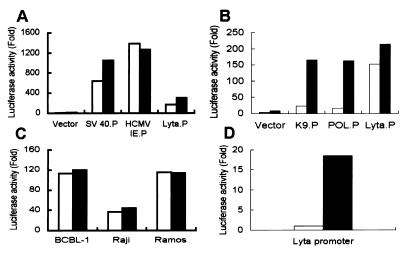
Figure 5.
Luciferase reporter assay of the Lyta promoter compared with other promoters and in different cell lines. Open bars in A, B, and C represent untreated TPA. Solid bars in A, B, and C represent TPA treated immediately after transfection. Cells were collected 48 h after transfection; luciferase activity values were normalized to a cotransfected internal control plasmid (pCMV-β-gal) and shown as folded, as compared with a promoterless vector (pGL3), which is defined as 1. (A) Compared with other virus IE promoters (human cytomegalovirus and simian virus 40) in 293T cells. (B) Compared with other gene promoters of KSHV [K9.P (ORF K9 promoter), POL.P (DNA polymerase promoter)] in 293T cells. (C) Lyta promoter activity in different cell lines with or without TPA treatment. (D) Lyta promoter activity in 293T cells cotransfected with pcDNA3.1 vector (open bar) or pcDNA3.1-ORF 50 cDNA clone (solid bar).
Clinical Biopsy Specimens.
Sixteen biopsy specimens (B1–B16) were collected from patients with KSHV-related diseases (Table 2). These biopsy specimens included four of MCD (B1–B4), four of PEL (B5–B8), and eight of KS (B9–B16). Samples were checked by IFA by using TPA-induced BCBL-1 cells (for KSHV) and specific antibodies (for LANA, vIL-6, vIRF, and ORF 65). As a control, samples of peripheral blood mononuclear cells (B17) and spleen (B18) were also collected from a latently infected KSHV carrier. Genomic DNA was extracted from these biopsy samples by conventional methods.
Table 2.
Biopsy samples from KSHV-related diseases—MCD, PEL, and KS
| Samples | KSHV | LANA | vIL-6 | vIRF | ORF65 |
|---|---|---|---|---|---|
| B1-MCD | + | + | + | + | ND |
| B2-MCD | + | + | + | + | + |
| B3-MCD | + | + | + | + | + |
| B4-MCD | + | + | + | + | ND |
| B5-PEL | + | + | + | − | − |
| B6-PEL | + | + | + | − | − |
| B7-PEL | ND | + | + | − | − |
| B8-PEL | + | + | + | ND | ND |
| B9-KS | + | ND | ND | ND | ND |
| B10-KS | + | ND | − | ND | ND |
| B11-KS | + | + | − | − | + |
| B12-KS | + | ND | − | ND | ND |
| B13-KS | + | + | − | − | + |
| B14-KS | + | + | − | ND | ND |
| B15-KS | + | ND | ND | ND | ND |
| B16-KS | + | ND | ND | ND | ND |
Results
Methylation Status in KSHV Genomes.
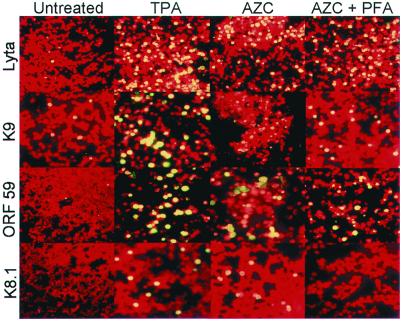
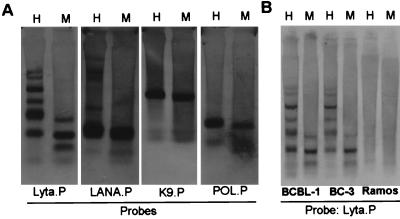
Table 1 shows the CpG suppression status in herpesvirus genomes. All γ-herpesviruses are CpG-suppressed, a characteristic that this herpesvirus group shares with human genome sequences, whereas CpG generally shows normal relative abundance in α- and β-herpesvirus genomes. The low CpG value of the HHV-7 genome may be caused by extensive CpG suppression in a 10-kb segment covering the immediate-early A (IE-A) region (PCG = 0.29). Like HHV-6, HHV-7 possesses some biological features of γ-herpesvirus such as lymphotropism. CpG suppression also existed in a 5-kb region covering the IE gene of HHV-6 (15). Compared with other γ-herpesviruses, the KSHV genome exhibits no obvious CpG suppression, although in the Lyta promoter region, the CpG suppression status is very low. One implication of this observation is that this region is methylated, and consequently, it raises the possibility that KSHV maintains its latent state by controlling the expression of Lyta. As shown in Fig. 1, treatment of BCBL-1 cells with 5-azacytidine, a DNA methyltransferase inhibitor (16), induced a KSHV lytic phase when tested by IFA by using mAb against Lyta (IE gene), ORF K9 (vIRF, early gene), ORF 59 (PF-8, early-late gene), and ORF K8.1 (late gene). The effect of 5-azacytidine was similar to TPA treatment. The activation of KSHV by 5-azacytidine in BCBL-1 was not caused by the activation of DNA synthesis, because treatment of 5-azacytidine plus the phosphonoformic acid (PFA) DNA-replication inhibitor still induced the expression of Lyta, whereas expression of the ORF K8.1 gene, a late glycoprotein gene, was completely blocked by PFA. Treatment of BC-3 cells, another PEL-derived cell line, with 5-azacytidine, showed results similar to BCBL-1 (data not shown). Southern blotting results showed that in latently infected cells, the restriction pattern of HapII and MspI was quite different (Fig. 2A), indicating that the Lyta promoter region was heavily methylated. The restriction pattern in the promoter regions of LANA, ORF K9, and DNA polymerase gene showed no difference. The Lyta promoter region in BC-3 cells showed the same characteristics (Fig. 2B). These results were compatible with the theoretical prediction of CpG-suppression status, indicating that there was no obvious methylation in the KSHV genome, but that the Lyta promoter region was heavily methylated.
Table 1.
CpG suppress status in herpesvirus genomes and KSHV gene promoters
| Sequence | PCG value |
|---|---|
| Herpesvirus genomes | |
| α-Herpesviruses | |
| HSV-1 | 1.01 |
| HSV-2 | 1.06 |
| VZV | 1.14 |
| β-Herpesviruses | |
| HCMV | 1.19 |
| HHV-6 | 1.13 |
| HHV-7 | 0.80 |
| γ-Herpesviruses | |
| EBV | 0.60 |
| Herpesvirus saimiri | 0.33 |
| KSHV | 0.81 |
| Murine herpesvirus 68 | 0.43 |
| Bovine herpesvirus 4 | 0.18 |
| KSHV individual gene promoters | |
| LANA promoter | 0.64 |
| Lyta promoter | 0.55 |
| K9 promoter | 0.73 |
| Polymerase promoter | 0.94 |
Figure 1.
Immunofluorescence assay (IFA) of BCBL-1 cells. Cells were collected untreated or treated (for 48 h) with TPA, 5-azacytidine (AZC), or AZC plus phosphonoformic acid (AZC + PFA), and checked by mAb against Lyta, ORF K9, ORF 59, and ORF K8.1 of KSHV. The negative cells were stained red (with Evans blue) and the positive cells, bright yellow.
Figure 2.
Southern blotting results. (A) Untreated BCBL-1 DNA were hybridized with different digoxigenin-labeled probes. The restriction enzyme HpaII (H) and MspI (M) are isoschizomers that cleave the same target sequence in DNA but have a different response to its state of methylation. MspI cleaves all CCGG sequences whether or not they are methylated at the second C, but HpaII cleaves only nonmethylated CCGG tetramers. (B) Lyta promoter probed with different cells shows the same pattern in BCBL-1 and BC-3 cells, but not in Ramos cells which are KSHV negative.
Bisulfite Genomic Sequencing Analysis.
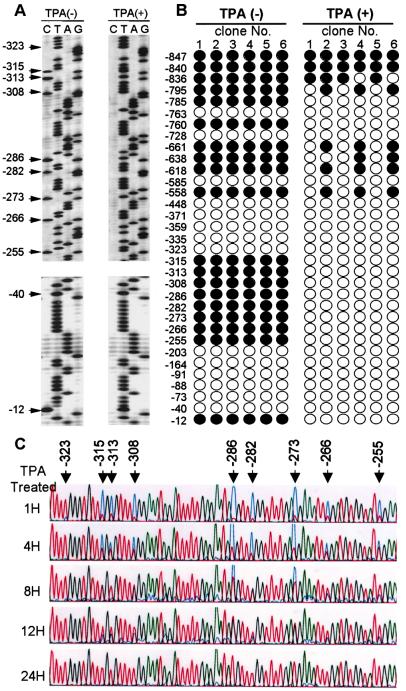
The bisulfite genomic sequence showed that in latently infected cells (DNA sample from untreated BCBL-1 cells), the Lyta promoter region was most methylated in CpG sites (Fig. 3A). Treatment of BCBL-1 cells by TPA resulted in demethylation in the Lyta promoter region (Fig. 3 A and B) and subsequently induced the activation of the virus. As shown in Fig. 3C, the time course of the methylation status in the Lyta promoter upon treatment with TPA showed that demethylation in the Lyta promoter began as soon as 1 h after TPA treatment, earlier than that of the transcription start of Lyta, which began 2 h after TPA treatment. The percentage of demethylated C (which converted into T by treatment with bisulfite) in the CpG sites was increased with TPA treatment, whereas the percentage of methylated C (which still remained as C after bisulfite treatment) was decreased. At 24 h after TPA treatment, all of the methylated C was demethylated and converted to T in the sequencing. By contrast, analysis of the promoter region of LANA, which is expressed constitutively in latently infected cells, using the same DNA samples from untreated BCBL-1 cells (latent), revealed the complete absence of methylation in the region extending from −361 bp through the transcription start site (Fig. 4). This region was sufficient for its promoter activity (17). Importantly, the coexistence of methylated and unmethylated sequences in the same DNA samples shows that the absence of methylation around the LANA promoter does not arise from virus replication, because no methylation was observed in the Lyta promoter region in the same sample. The unmethylated status in the LANA promoter suggests the presence of a cis element(s) that actively prevents methylation of this region and enables its constitutive transcription during latency. As with other changes in chromatin, it seems likely that the absence of methyl groups is associated with the ability to be transcribed rather than with the act of transcription itself, because only a portion of the cells are triggered into lytic phase. IFA assay shows that 20–30% of the BCBL-1 cells can be induced into lytic phase by TPA treatment (6).
Figure 3.
Methylation status in the Lyta promoter region. (A) Sequencing image shows the bisulfite modified sequence. All of the cytosines except for methyl-cytosine were converted to thymine when treated with bisulfite. Arrows indicate the CpG sites. Numbers indicate the position relative to the Lyta transcription start site. (B) Methylation status in the Lyta promoter region of BCBL-1 cells untreated or treated with TPA for 24 h. Each included six independent clones. Solid circles indicate methylated CpG, and open circles indicate demethylated CpG. (C) Time course of TPA treatment. DNA samples from BCBL-1 cells treated with TPA at time intervals of 1, 4, 8, 12, and 24 h were bisulfite-modified, PCR-amplified, and sequenced directly without the separation of a single clone. Arrows show the CpG sites, which were mixed with C (methylated C) and T (demethylated C). T, A, G, and C are shown as red, green, black, and blue, respectively, in the Applied Biosystems sequencing image.
Figure 4.
Methylation status in the LANA promoter region in latent (untreated) BCBL-1 cells. Solid circles indicate methylated CpG, and open circles indicate demethylated CpG. Numbers indicate the position relative to the transcription start site of LANA.
In Vitro Lyta Promoter Assays.
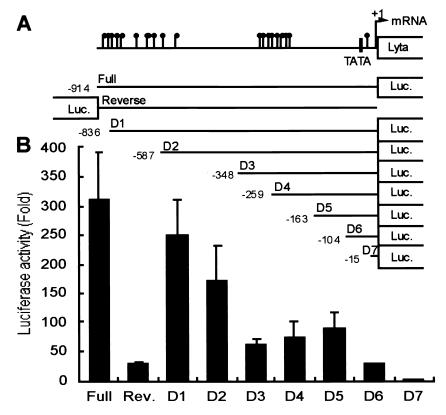
Fig. 5 shows the results of Lyta promoter luciferase reporter assay compared with different promoters and in different cell lines. Raw data showed that, although the Lyta promoter was responsive to TPA treatment, luciferase activity was increased only 3- to 4-fold (data not shown), much less than that of other KSHV gene promoters like ORF K9 or DNA polymerase (Fig. 5B). The Lyta promoter also has very high basal activity in vitro even in the absence of TPA (Fig. 5 A–C, open bar). Therefore, a mechanism of repression of the Lyta promoter may exist in latency in vivo. Its response to TPA treatment was similar to those of the human cytomegalovirus (HCMV) IE promoter and the simian virus 40 IE promoter, which are considered to be unresponsive to TPA treatment (Fig. 5A). We believe that the responsiveness of the Lyta promoter to TPA treatment was caused by general cell activation induced by TPA through the protein kinase C pathway. We used an HCMV IE promoter-driven reporter vector (pCMV-β-gal) as an internal control, and luciferase activity values were normalized by cotransfection of the internal control plasmid to eliminate the nonspecific activity. Interestingly, like many other herpesvirus IE proteins, Lyta promoter can be controlled by its own gene product (Lyta) through autoregulatory feedback (Fig. 5D). The basal activity of the Lyta promoter in vitro is caused by the nonmethylation of plasmid DNA when prepared from Escherichia coli, because digestion of the Lyta promoter–reporter construct DNA prepared from E. coli by Hap II and MspI showed the same pattern, indicating that the plasmid DNA was CpG-methylation free (Fig. 6B, lane Mock). Methylation cassette assay (ref. 14; Fig. 6A) showed that the Lyta promoter was sensitive to methylation. In vitro methylation of the Lyta promoter by a methylation enzyme (Sss I) showed that the Lyta promoter lost its promoter activity when methylated in the CpG sites of the promoter region (Fig. 6C). A series of deletion mutants of the Lyta promoter was generated to determine the promoter element (Fig. 7A). Luciferase reporter assay showed our cloned the Lyta promoter region indeed had an orientation-dependent promoter activity. A series of nested deletion mutants identified an element between D2 and D3 that may play an important role in promoter activity (Fig. 7B); in latently infected KSHV, a hypermethylated region was located downstream of this element.
Figure 6.
Methylation cassette assay of the Lyta promoter. (A) Outline of the methylation cassette assay. (B) Confirms the effect of methylation with SssI in vitro. Mock: plasmid DNA prepared from E. coli was mock methylated as a control. Methylated: the same equivalent DNA of the Lyta promoter fragment was methylated with a methylase-SssI. Both mock and methylated DNA were digested by HapII (H) and MspI (M) under the same conditions. (C) Lyta promoter ablates its luciferase activity when methylated by SssI in vitro. Open box: Mock-methylated Lyta promoter. Its reporter activity is shown as 100%. Solid box: the relative methylated Lyta promoter–reporter activity.
Figure 7.
Determination of elements in the Lyta promoter region. (A) Schematic diagram of the Lyta promoter and a series of nested deletion mutants. Numbers indicate the position relative to the transcription start site of Lyta (7). A hypermethylated region is shown in the promoter region between −315 to −255 base. Closed circles: methylated CpGs in latently infected KSHV. (B) Luciferase activation of the Lyta promoter and the corresponding deletion mutants. Rev.: Promoter orientation-reverse construct.
Methylation Status in the KSHV-Lyta Promoter in Vivo.
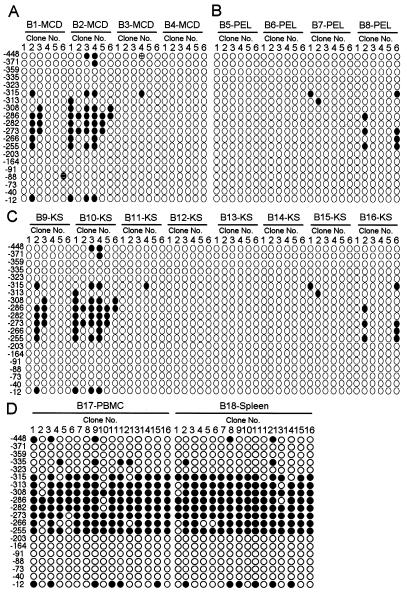
KSHV-infected cell lines (i.e., BCBL-1) provide an excellent model for the study of viral latency and reactivation, because they can be manipulated easily in vitro by treatment of cells with chemical reagents such as TPA and butyrate (18). But results from cultured cell lines must be extended to infected tissues. Biopsy specimens were collected from patients with MCD (lymphocyte), PEL (lymphoma), and KS (endothelial and spindle cells) (Table 2). Peripheral blood mononuclear cells and spleen cells were also collected from a latently infected KSHV carrier. The methylation status of the Lyta promoter in these biopsies was investigated by bisulfite genomic sequencing. All of the samples from tissues with MCD, PEL, and KS showed that most CpG sites were demethylated in the Lyta promoter (Fig. 8, A–C), similar to the reactivated status in BCBL-1 cells treated by TPA. As shown in Table 2, IFA results of these biopsy samples indicated that KSHV indeed was in lytic phase, although with different expression patterns, suggesting that the expression of KSHV protein in vivo is tightly restricted (19). By contrast, in KSHV latently infected samples (Fig. 8D), no complete demethylation was observed in the 16 clones we tested. These results suggest a relationship among demethylation status in the Lyta promoter, the reactivation of KSHV, and the development of KSHV-associated diseases. We believe that methylation in the the Lyta promoter region was critical for controlling the promoter activity, and that demethylation in the the Lyta promoter region was the essential event for the expression of Lyta and subsequently for the whole lysis of the virus.
Figure 8.
Methylation status of KSHV-lyta promoter in KSHV-related diseases. Solid circles indicate methylated CpG, and open circles indicate demethylated CpG. (A) MCD; (B) PEL; and (C) KS. (D) Peripheral blood mononuclear cells (PBMC) and spleen samples from a latent KSHV carrier.
Discussion
Methylation of CpG sites in the vicinity of the promoter usually prevents expression of the gene. Repression is caused by either of two methyl CpG binding proteins, MeCP1 (20) or MeCP2 (21). The protein MeCP1 requires the presence of several methyl groups to bind to DNA, whereas MeCP2 can bind to a single methylated-CpG base pair. The exact mechanism by which methylation of a promoter leads to repression of gene expression remains unclear, but current evidence suggests that chromatin remodeling plays an important role (22). MeCP2 can displace histone H1 from nucleosomes and then, (in part) by recruiting histone deacetylases, allows chromatin to rearrange into a tight structure that excludes transcription factors (23, 24). This observation provides a direct connection between two types of repressive modifications, methylation of DNA and acetylation of histones. Like Epstein–Barr virus, lytic replication of KSHV can be triggered by chemical agents including butyrate and TPA (14). Butyrate inhibits histone deacetylases, and TPA induces expression of histone acetyltransferases (25). We show here that a DNA methyltransferase inhibitor, 5-azacytidine, can also trigger KSHV into lytic phase, suggesting that derepression in gene transcription is a critical factor for initiation of lytic replication. A correlation between methylation of specific viral genes and transcriptional repression has been observed for other viruses, including Epstein–Barr virus (26), Marek's disease virus (27), and simian foamy virus type 3 (28). We believe that KSHV maintains latency through controlling the expression of some key gene, i.e., Lyta. In latency, methyl-CpG-binding protein may bind to methylation sites in the Lyta promoter region and associate with other corepressor complexes containing the transcriptional repressor mSin3A and histone deacetylase (to mediate transcriptional repression of Lyta and subsequently to maintain the latency of KSHV). Similar mechanisms may also exist in other latently infected herpesvirus genomes.
Acknowledgments
We thank Dr. Bernard Roizman for valuable comments on the preparation of this manuscript. This study was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan.
Abbreviations
- KSHV
Kaposi's sarcoma-associated herpesvirus
- HHV
human herpesvirus
- Lyta
KSHV lytic transactivator, also named KSHV Rta
- KS
Kaposi's sarcoma
- PEL
primary effusion lymphomas
- MCD
multicentric Castleman's disease
- TPA
phorbol 12-tetradecanoate 13-acetate
- IE
immediate-early
- IFA
immunofluorescence antibody assay
- CpG
5′ cytosine-guanine
References
- 1.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 3.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M N. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 4.Honess R W, Roizman B. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun R, Lin S F, Gradoville L, Yuan Y, Zhu F, Miller G. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Ueda K, Sakakibara S, Okano T, Yamanishi K. J Virol. 2000;74:8623–8634. doi: 10.1128/jvi.74.18.8623-8634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukac D M, Renne R, Kirshmer J R, Ganem D. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 8.Gradoville L, Gerlach J, Grogan E, Shedd D, Nikiforow S, Metroka C, Miller G. J Virol. 2000;74:6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewin B. In: Genes VII. Lewin B, editor. New York: Oxford Univ. Press; 2000. pp. 678–681. [Google Scholar]
- 10.Karlin S, Mocarski E, Schachtel G. J Virol. 1994;68:1886–1902. doi: 10.1128/jvi.68.3.1886-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frommer M, McDonald L E, Millar D S, Collis C M, Watt F, Grigg G W, Molloy P L, Paul C L. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haque M, Chen J, Ueda K, Mori Y, Nakano K, Hirata Y, Kanamori S, Uchiyama Y, Inagi R, Okuno T, Yamanishi K. J Virol. 2000;74:2867–2875. doi: 10.1128/jvi.74.6.2867-2875.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlin S, Doerfler W, Cardon L R. J Virol. 1994;68:2889–2897. doi: 10.1128/jvi.68.5.2889-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson K D, Ambinder R F. J Virol. 1997;71:6445–6454. doi: 10.1128/jvi.71.9.6445-6454.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin M E, Nicholas J, Thomson B J, Newman C, Honess R W. J Virol. 1991;65:5381–5390. doi: 10.1128/jvi.65.10.5381-5390.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson K D, Hayward S D, Ling P D, Samid D, Ambinder R F. Mol Cell Biol. 1995;15:6150–6159. doi: 10.1128/mcb.15.11.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarid R, Wiezorek J S, Moore P S, Chang Y. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parravicini C, Chandran B, Corbellino M, Berti E, Paulli M, Moore P S, Chang Y. Am J Pathol. 2000;156:743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cross S H, Meehan R R, Nan X, Bird A. Nat Genet. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 21.Nan X, Campoy F J, Bird A. Cell. 1997;21:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 22.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 23.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 24.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 25.Masumi A, Wang I M, Lefebvre B, Yang X J, Nakatani Y, Ozato K. Mol Cell Biol. 1999;19:1810–1820. doi: 10.1128/mcb.19.3.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson K D. Curr Top Microbiol Immunol. 2000;249:21–34. doi: 10.1007/978-3-642-59696-4_2. [DOI] [PubMed] [Google Scholar]
- 27.Hayashi M, Furuichi T, Ren S, Isogai E, Nonoyama M, Namioka S. J Vet Med Sci. 1994;56:287–291. doi: 10.1292/jvms.56.287. [DOI] [PubMed] [Google Scholar]
- 28.Schweizer M, Fleps U, Jackle A, Renne R, Turek R, Neumann-Haefelin D. Virology. 1993;192:663–666. doi: 10.1006/viro.1993.1084. [DOI] [PubMed] [Google Scholar]