Abstract
Lipin family proteins are emerging as critical regulators of lipid metabolism. In triglyceride synthesis, lipins act as lipid phosphatase enzymes at the endoplasmic reticular membrane, catalyzing the dephosphorylation of phosphatidic acid to form diacylglycerol, which is the penultimate step in this process. However, lipin proteins are not integral membrane proteins and can rapidly translocate within the cell. In fact, emerging evidence suggests that lipins also play critical roles in the nucleus as transcriptional regulatory proteins. Thus, lipins are poised to regulate cellular lipid metabolism at multiple regulatory nodal points. This review summarizes the history of lipin proteins and discusses the current state of our understanding of lipin biology.
Lipin proteins in the control of lipid homeostasis
The triglyceride core of lipid droplets is primarily synthesized through the sequential acylation of glycerol-3-phosphate in the pathway described by Kennedy in the 1950s [1, 2]. The last gene cloned in this pathway coded for the enzyme responsible for the Mg2+-dependent dephosphorylation of phosphatidic acid (PA) to form diacylglycerol (DAG) [3], which is the penultimate step in triacylglycerol (TAG) synthesis. In mammals, the enzymes that catalyze PA phosphatase (PAP) activity were encoded by a family of genes that had previously been cloned and named lipins (lipin 1, lipin 2, and lipin 3) [4]. In 2001, Karen Reue’s group used positional cloning to localize the cause of the disturbed metabolic phenotype of the fatty liver dystrophic (fld) mice to mutations in the gene encoding lipin 1 [4]. Fld mice are lipodystrophic and exhibit multiple defects in adipose tissue development and triglyceride content [5–7]. This phenotype fits well with the role of lipin 1 as a PAP enzyme and the subsequent identification of lipin 1 as a transcriptional regulator of fatty acid metabolism [8].
The relevance of lipin proteins to the lipid droplet biology theme of this review series is still emerging. Most obviously, the role that lipins play in synthesizing TAG, the principal constituent of lipid droplet cores, is critical. However, the synthesis of the phospholipids that are components of lipid droplets, including phosphatidylcholine (PC), requires DAG. Endoplasmic reticulum (ER) membrane DAG content, likely derived by dephosphorylation of PA by lipin, has also now been shown to enhance the recruitment of the perilipin family proteins to nascent lipid droplets [9]. Finally, many of the proteins that coat lipid droplets and serve as the interface with the cytosol are targets of the peroxisome proliferator-activated receptor (PPAR) family of transcription factors, which are known to be regulated by lipins [8, 10].
History of PAP enzyme activity and biochemistry
The first appearance of PAP as a potential enzymatic activity occurred in the 1950s, when Kennedy and colleagues linked the two halves of the phosphoglycerolipid synthesis pathway. It was known that enzyme systems in liver could generate phosphatidic acid from glycerol, and that DAG can be used to form PC (lecithin) and TAG. Kennedy’s insight led to the hypothesis that these enzymatic pathways could be completed by the dephosphorylation of PA to produce diacylglycerol. By demonstrating this enzymatic capacity in vitro, it was clear that phosphatidic acid could be used to generate lecithin [11]. This finding completed the enzymatic sequence of what became known as the Kennedy pathway of glyceride synthesis, and demonstrated that dephosphorylation of PA is essential for neutral and phospholipid synthesis in most mammalian cells.
The subsequent decades continued to reveal more about PAP enzyme activity, such as a requirement for Mg2+, and the surprising finding that the majority of PAP activity is in the soluble fraction of cell lysates [12]. In contrast, all of the other enzymatic reactions in the TAG synthesis pathway were catalyzed by integral membrane proteins. PAP activity in rat liver extracts was found to translocate from a cytosolic compartment to internal membranes in a dynamic fashion that was regulated by fatty acids and possibly phosphorylation [13]. Because of the lability of the enzyme and other technical problems, highly purified PAP was never obtained. In the late 1980s, however, yeast PAP enzymatic activity was purified to near homogeneity and biochemically characterized [14]. It was these same isolations of yeast PAP activity that were later resurrected from storage and, utilizing newly improved methods in protein mass spectrometry, allowed the identification of the gene encoding yeast PAP [3]. Almost exactly 50 years after the Kennedy paper proposing phosphatidic acid phosphatase as the name for the enzyme present in cell homogenates, the long-sought PAP was identified as Pah1p, the yeast ortholog of lipin 1 [3].
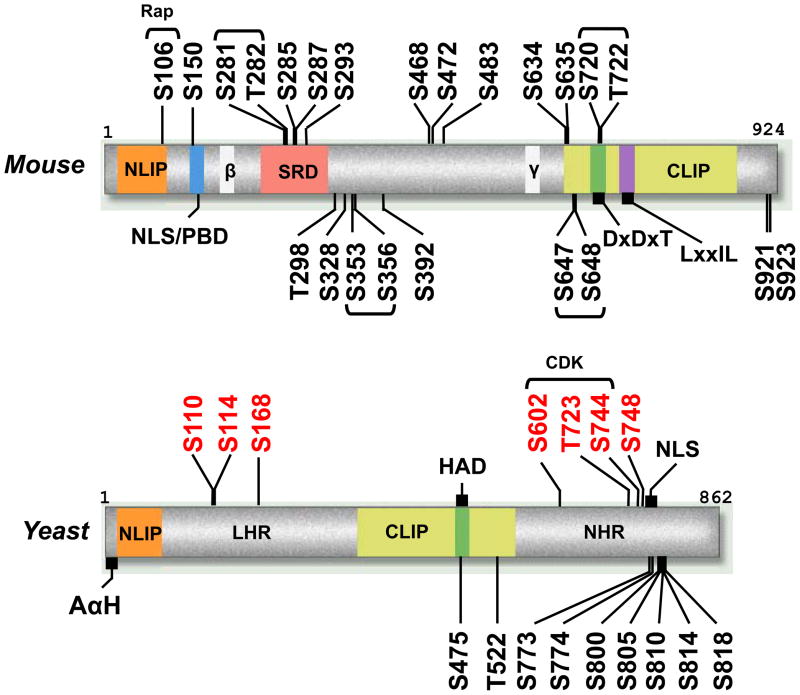
Most known vertebrate species have three lipin family genes (lipin 1, 2, and 3) that exhibit distinct patterns of tissue-specific expression. All three mammalian lipin proteins possess PAP activity, but the intrinsic enzymatic activity varies dramatically (Vmax lipin 1≫lipin 2>lipin 3) [15]. The relationship of the lipin proteins can be illustrated by a lipin family phylogenetic tree (Fig. 1) [16]. The principal feature that links all lipin proteins are highly-conserved NH2- and COOH-terminal lipin domains (NLIP and CLIP domains) (Fig. 2). The CLIP domain contains the haloacid dehalogenase (HAD) catalytic site required for PAP activity. Although the crystal structure of lipin proteins has not been determined, the amino acid sequence of the NLIP and CLIP domains is conserved across all lipin family proteins and throughout phylogeny suggesting a commonality of structure.
Fig. 1.
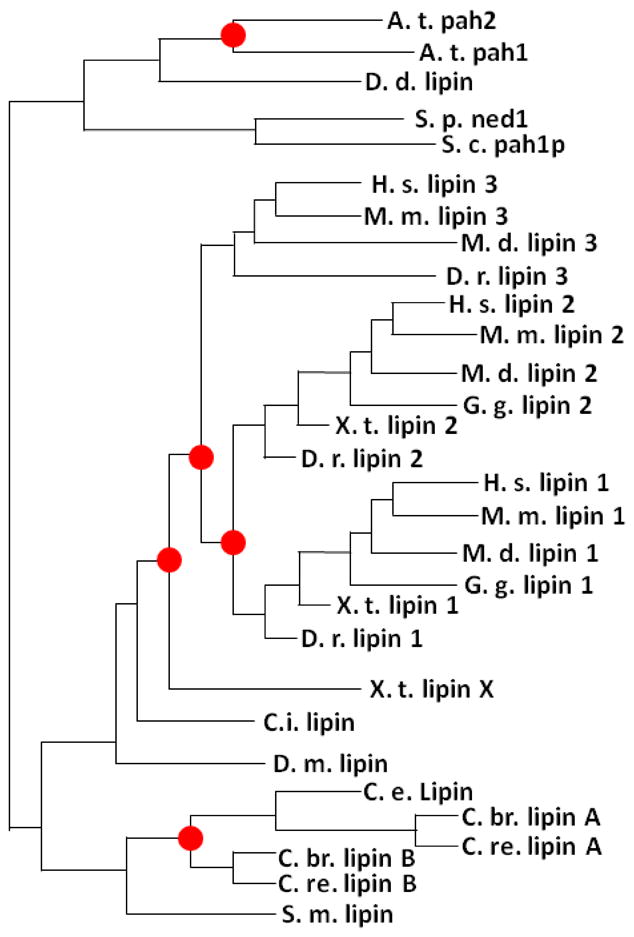
Phylogenetic tree of lipin family orthologs and paralogs (adapted from the Tree families database, http://www.treefam.org/). Red circles represent duplication events.
A.t. Arabidopsis thaliana
D. d. - Dictyostelium discoideum
S. p. - Schizosaccharomyces pombe
S. c. - Saccharomyces cerevisiae
H. s. - Homo sapiens
M. m. - Mus musculus
D. r. - Danio rerio
G. g. - Gallus gallus
X. t. - Xenopus tropicalis
C. i. - Ciona intestinalis
D. m. - Drosophila melanogaster
C. e. - Caenorhabditis elegans
C. br. - Caenorhabditis briggsae
C. re. - Caenorhabditis remanei
Fig. 2.
Cartoon of mouse lipin 1 and S. cerevisiae Pah1p illustrating phosphorylation sites and domain features. NLIP and CLIP are conserved NH2- and COOH-LIPin homology domains [4], LHR and NHR are Low and No homology domains within yeast lipin that have no homology with mammalian lipins [30], β and γ gamma represent alternatively spliced exons, DxDxT is haloacid dehalogenase domain [3], NLS is Nuclear Localization Sequence [4, 17, 40, 59]/polybasic domain (PBD) is the PA-binding motif in mammalian lipins [40], serine-rich domain (SRD) mediates the interaction with 14-3-3 proteins [59], AαH is the amphipathic α-Helix responsible for membrane association [19], residues in red are the amino acid residues mutated to alanine in PAH1-7A [19], CDK are the sites phosphorylated directly by CDK [20], Rap is the lipin 1 residue sensitive to rapamycin [30], bracketed phosphosites within lipin 1 indicate either site may be phosphorylated. Figure adapted from Harris et al, 2007.
Yeast (and other invertebrate) lipin
Yeast and most other invertebrates contain a single ortholog of lipin, while bony vertebrates contain multiple orthologs. Plants and some species of worms (but not C. elegans) are the exception, in that the two plant lipin paralogs, as well as the paralogs found in C. remanei and briggsae (termed Lipin A and Lipin B), are post-speciation duplication events and as such do not correspond to mammalian lipin 1 and 2. The existence of a single lipin ortholog in genetically tractable organisms has simplified the functional analysis of the role of lipin in these species.
As might be expected from the loss of a phosphatidic acid phosphatase, yeast Pah1p mutants display substantial increases in PA accumulation, accompanied by decreases in TAG and PC levels and elevated phosphatidylinositol and phosphatidylethanolamine [3]. Interpreting the direct effects of Pah1p enzymatic activity on neutral and phospholipid synthesis is complicated by the transcriptional consequences of PA accumulation in Pah1p mutants. Loss of Pah1p leads to defects in the regulation of genes involved in phospholipid synthesis through the Opi1p/Ino2p/4p feedback loop onto UASINO regulated genes [3, 17]. Opi1 is found only in yeast and is a transcriptional repressor of Ino2p/4p that is tethered to the ER by phosphatidic acid. Accumulation of PA in Pah1p mutants leads to derepression of Ino2p/4p and activation of UASINO genes. Therefore, the enzymatic activity of Pah1p can affect cellular phospholipid status directly, by eliminating the Kennedy pathway and forcing PC and PE synthesis through the CDP-DAG pathway; and indirectly, by derepressing the INO genes. In addition, Pah1p has been localized in the nucleus at the promoters of UASINO genes, suggesting a more direct action for Pah1p towards these genes [17]. Regardless of its potential role for directly modulating transcription, yeast Pah1p requires enzymatic activity to recover function in Pah1p mutant cells [18].
In yeast, Pah1p activity is regulated through membrane association with an amino-terminal amphipathic alpha helix. Phosphorylation of Pah1p prevents the amino-terminal amphipathic alpha helix from binding to membranes, while dephosphorylation by the Nem1/Spo7 phosphatase complex relieves this repression [19]. That Pah1p phosphorylation decreases its intrinsic functional enzymatic activity is demonstrated by alterations in phospholipids [20], the ability to repress the UASINO-containing INO1 gene, and nuclear membrane/ER morphology in phospho-deficient mutant strains [17]. Mutation of seven serine/threonine residues (PAH1-7A) has shown that these phosphorylation sites are required for proper control of Pah1p (Fig. 2) [21]. Three of these residues are phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase (CDK); however, even though substitution of the three CDK-phosphorylated residues with alanine decreases membrane association, this mutant does not fully recapitulate the phenotype of the PAH1-7A mutant [20]. Yeast accumulate phospholipids during exponential growth and TAG during the stationary phase. Decreasing Pah1p enzymatic activity by cell cycle kinases would decrease TAG synthesis and potentially explain how the shift in the lipid synthesis profiles occurs in growing cells.
First demonstrated in S. pombe, and then in S. cerevisiae, loss of Ned1/Pah1p causes dramatic abnormalities in the structure of the ER and the nuclear envelope as a result of increased membrane proliferation [17, 22]. Consistent with the disruption in ER structure, Pah1p mutant cells have defects in lipid droplet morphology with neutral lipid accumulating around the ER in stationary phase, similar to yeast lacking the ortholog of the human lipodystrophic gene seipin [23, 24]. Yeast do not have lipid droplet binding proteins analogous to the perilipin proteins found in mammalian cells, and whether the alterations in lipid droplet morphology arise from Pah1p-mediated alterations in phospholipid profiles or are an indirect consequence of PA-induced derepression of UASINO genes remains to be determined.
Loss of the single lipin homolog in C. elegans also disrupts the normal morphology of the ER and the nuclear structure [25, 26]. Unlike yeast, which undergo a closed mitosis, in metazoans, the nuclear envelope must degrade during division. In the absence of lipin, there is a defect in the ability of the nuclear envelope to breakdown, decreased formation of lipid droplets, as well as disorganization of the ER. The loss of both lipin homologs, named Pah1 and Pah2 after the yeast genes but not directly related to mammalian lipin 1 and 2, increases the level of phosphatidic acid, as well as most other phospholipids in Arabidopsis [27, 28]. Surprisingly, there were no alterations in TAG content in seeds [28]. The loss of both Pah1 and Pah2 also causes expansion of the ER and increased expression of phospholipid biosynthesis enzymes [28]. The work conducted in these tractable model systems has rapidly advanced our understanding of lipin 1 regulation and function and demonstrates that PAP in lower organisms regulates phospho- and neutral lipid levels, ER and nuclear membrane morphology, and transcriptional de-repression of phospholipid biosynthetic enzymes.
Lipins in higher organisms
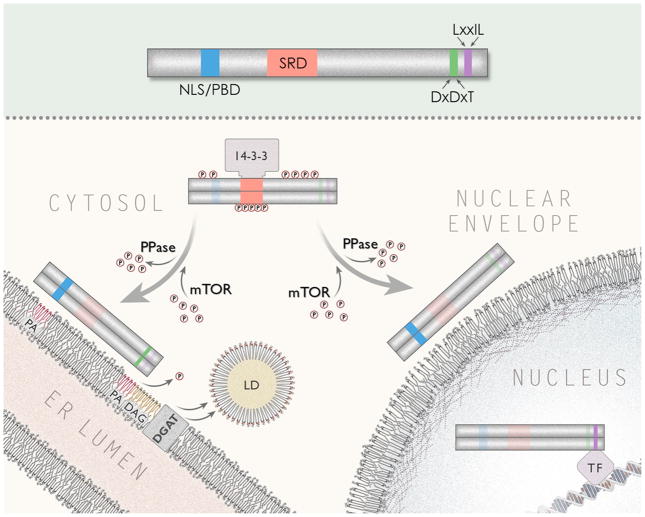
Several key differences in lipin protein biology between yeast and higher organisms have emerged. For example, phosphorylation alters intrinsic PAP activity in yeast but not in mammals [21, 29]. Instead, in vertebrates, phosphorylation seems to control vertebrate lipin activity by altering its cellular localization (Fig. 3) [30]. Also, many of the key phosphorylation sites that regulate activity in yeast lipins are not found in the mammalian orthologs (Fig. 2). Mammalian lipin proteins also form extremely stable oligomeric complexes, and lipin 1 exists primarily as a dimer or tetramer in its native state [31]. Lipin proteins interact through both N- and C-terminal domains in a head to head/tail to tail orientation [31], and lipin 1 can also form hetero-oligomers with lipin 2 and lipin 3. To our knowledge, it has not been determined whether yeast lipins also form oligomers. The full functional implications of oligomer formation on PAP activity and transcriptional regulation are still unclear. However, it is of interest that other enzymes involved in TAG synthesis also form tetramers [32], suggesting that these aggregates potentially confer an advantage in enzymatic efficiency.
Fig. 3.
This schematic depicts the intracellular trafficking of lipin 1 multimers. At top, lipin 1 protein with domains described in Fig. 2 is shown. At center, hyperphosphorylation of the SRD of lipin 1 leads to associations with 14-3-3 proteins and cytosolic retention. mTOR has been identified as a kinase that phosphorylates lipin 1 and regulates its localization. Dephosphorylation by unknown protein phosphatase(s) (PPase) direct lipin to the nucleus or ER compartments. The NLS/PBD targets lipin 1 to PA-rich membranes while the DxDxT catalytic motif is required for the dephosphorylation of PA to form DAG. DGAT then terminally acylates DAG to make TAG and form the core of nascent lipid droplets (LD). The NLS/PBD also mediates the nuclear localization of lipin 1. In the nucleus, lipin 1 interacts with DNA-bound transcription factors (TF) to regulate their activity.
Perhaps most importantly, evidence suggests that mammalian lipin proteins interact with DNA-bound transcription factors to regulate their transcriptional activity [8]. Lipin 1 was first shown to interact with PPARα and its coactivator protein, PPARγ coactivator-1α (PGC-1α), in hepatocytes [8]. Since that discovery, lipin 1 and lipin 2 have been shown to coactivate other nuclear receptor transcription factors, including PPARγ [10, 33], as well as non-nuclear receptor transcription factors including nuclear factor of activated T-cells c4 (NFATc4) [34] and myocyte enhancer factor 2 (MEF2) [35]. Lipin 1 likely enhances the activity of these transcription factor complexes by recruiting other coactivator proteins with the ability to enzymatically modify chromatin [8]. However, in other contexts, lipin 1 can also act as a transcriptional repressor by enhancing the recruitment of histone deacetylases. One example of this is lipin 1’s ability to represses the activity of NFATc4 to suppress proinflammatory cytokine signaling in adipocytes [34]. Thus, lipin 1 likely acts as a molecular scaffold that, depending upon the context, can lead to activation or repression of gene transcription.
The duality of function ascribed to mammalian lipin proteins is somewhat unique amongst lipid metabolic enzymes. Several enzymes involved in glucose metabolism, including glyceraldehyde-3-phosphate dehydrogenase, act in the nucleus to regulate gene expression by synthesizing acetyl-CoA for acetylation of histones [36–38]. However, less is known regarding how enzymes involved in lipid metabolism might affect gene expression, chromatin structure, or nuclear envelop morphology, though a role for sphingosine kinase 2-mediated production of sphingosine-1-phosphate in the regulation of histone acetylation has been demonstrated [39], Data provided thus far suggest that lipin-mediated PAP activity is not required for transcriptional effects in mammals. This differs from findings in yeast, where complementation studies have shown that this enzymatic activity is required for transcriptional effects [17, 18]. In mammalian cells, catalytic-dead lipin 1 mutants retain the ability to coactivate nuclear receptors and repress NFATc4 activity [8, 34], indicating that these two molecular effects are mechanistically separable and distinct in higher organisms. The recent finding that lipin family proteins form hetero-oligomers [31] suggests that an absolute requirement for catalytic activity cannot be completely excluded. Because of the relatively ubiquitous expression of the lipin proteins, the ability of overexpressed catalytic-dead lipin proteins to hetero-oligomerize with catalytically-active endogenous lipin proteins remains a possibility. It also cannot be excluded that the ability of lipin to enhance nuclear receptor transcriptional activity is somehow related to its ability to bind complex phospholipids [40] that might be ligands for its partner nuclear receptors. In these cases then, lipins could act as chaperone proteins, shuttling lipids to the cognate nuclear receptor – a function that would be independent of its catalytic activity. Thus, lipins could serve as sensors for the cellular content of these lipid ligands to control nuclear receptor activity. However, this is purely speculation at this point.
The known effects of lipin 1 on gene expression fit well with a coordinated physiologic response with regards to both the transcriptional and glycerolipid metabolic pathways. For example, lipin 1 and 2 are robustly induced by fasting in liver, which is associated with a marked increase in PPARα/PGC-1α activity to stimulate fatty acid oxidation gene expression [8, 41]. Fasting also dramatically increases the flux through hepatic TAG synthetic pathways to provide storage for fatty acids that can later be oxidized. It is likely that in this circumstance, the induction of lipin 1 serves to coordinately increase the capacity for both metabolic processes. In adipocytes, lipin 1 appears to enhance the activity of PPARγ to augment adipogenic gene expression profile [10], whilst providing PAP activity for the esterification of lipids into TAG, thus synchronizing the nucleo-cytoplasmic functions of lipin 1 to synergistically increase lipid storage. Other examples of lipin 1 bi-functional cooperativity may emerge as the full spectrum of nuclear targets and metabolic effects are identified.
Lipin 1
The first member of the lipin family is robustly expressed in tissues with high rates of lipid flux, including adipose tissue, skeletal muscle, and heart [4, 15]. Alternative splicing of the Lpin1 transcript gives rise to three distinct lipin 1 proteins (lipin 1α, lipin 1β, and lipin 1γ) [42, 43] that may each have unique cellular localization and intrinsic PAP activity. Lipin 1α appears to be predominantly nuclear, while lipin 1β resides mostly in the cytoplasm [42]. PAP activity of purified lipin 1α and lipin 1β is roughly similar, but the relative activity has varied between two published studies that used different methods to purify lipin 1 [15, 43]. PAP activity of lipin 1γ is almost negligible [43]. Consistent with its high enzymatic activity, mice lacking lipin 1 have extremely low PAP activity in fat, brain, kidney, lung, cardiac and skeletal muscle, and a 50% reduction in liver [15, 30].
Lipin 1 is highly regulated at the transcriptional level. Hepatic lipin 1 expression is induced by fasting [8], diabetes [8], high fat diet administration [44], cAMP signaling [45], statin administration [46], glucocorticoid injection [8, 45, 47], and chronic ethanol exposure [48]. Many of these stimuli were long known to increase hepatic PAP activity [49–53]. Activation of β-adrenergic receptor activation, likely through cAMP signaling, strongly activates lipin 1 expression [54, 55]. Finally, lipin 1 expression is activated by glucocorticoids as part of the adipogenic program [47, 56] and is down-regulated in response to pro-inflammatory cytokines [57] in adipocytes.
The transcriptional regulatory mechanisms that control lipin 1 expression continue to emerge. Several transcription factors interact with the Lpin1 promoter and regulate lipin 1 transcription directly, including the glucocorticoid receptor (GR) [47], NOR-1 [55], CCAAT enhancer binding protein (C/EBP) [10], sterol response element binding protein-1 (SREBP-1) [46], and cAMP response element binding protein (CREB) [44]. Important roles for two transcriptional coactivator proteins, PPARγ coactivator-1α (PGC-1α) [8] and CRTC2 [44] have also been identified. The general characteristics of the transcriptional regulators that activate lipin 1 expression fit well with the biological functions of this protein. Transcription factors that are activated by lipogenic stimuli (C/EBP, GR, SREBP-1) or signaling pathways related to increased need for oxidative metabolism (NOR-1, CREB, PGC-1α, or CRTC2) increase the expression of lipin 1 and increase the cellular capacity for lipogenesis or oxidative metabolism.
Lipin 1 is also highly modified at the post-translational level. Lipin 1 contains at least 19 and as many as 23 sites that are phosphorylated in response to insulin [30, 58]. Unlike yeast lipin, the intrinsic PAP activity of lipin 1 does not appear to be affected by phosphorylation [30], but instead, its activity is hypothesized to be modulated due to its location in the cell [13]. As an insoluble lipid, lipin 1’s substrate PA will be found within membranes. Serine/threonine phosphorylation of lipin 1 in response to insulin promotes its translocation away from the ER membrane and out of the nucleus into the soluble fraction (Fig. 3) [30]. The involvement of mTOR complex 1 (mTORC1) in lipin 1 phosphorylation is indicated by the at least partial sensitivity of insulin-stimulated lipin 1 phosphorylation to rapamycin [58], as well as rapamycin-induced nuclear localization [59]. Whereas phosphorylation of Pah1p by CDK to inhibit PAP activity is appropriate to control lipid synthesis in cycling cells, it is paradoxical that insulin, which increases TAG synthesis, leads to lipin 1 movement away from the site of TAG synthesis. It is possible that insulin-stimulated phosphorylation of lipin 1 merely weakens its association with membranes allowing lipin 1 to partition into the cytosolic fraction post-lysis [60]. The nuclear export of lipin 1 relies upon 14-3-3 proteins that associate with lipin 1 protein phosphorylated in a serine rich domain (SRD) in response to insulin (Fig. 3) [59]. Sumoylation also promotes lipin 1 nuclear retention [35]. Indeed, mutation of the consensus sumoylation sites of lipin 1 prevented its nuclear localization and abrogated its ability to coactivate MEF2. Cellular phosphatidic acid levels have been suggested to regulate nuclear versus cytosolic localization, since pharmacologic reductions in PA levels led to increased nuclear localization of lipin 1 [40]. This may somehow be explained by the fact that the polybasic string of amino acids that comprise the nuclear localization sequence also mediate an interaction with PA [40]. This domain in mammalian lipin 1 may therefore double as the interface for ER membrane-embedded PA and for nucleo-cytoplasmic transport (Fig. 3). The factors that determine whether lipin 1 proteins are directed to the ER or nucleus by this domain remain to be determined.
Loss of function mutations in lipin 1 produce the dramatic metabolic abnormalities observed in fld mice [4]. Two distinct mutations in lipin 1, a genomic rearrangement and a point mutation (glycine 84 mutated to arginine), were found to produce the same metabolic phenotype in two lines of fld mice. Fld mice are easily identifiable within a few days after birth due to growth retardation, hypertriglyceridemia, and the development of severe hepatic steatosis [5]. The loss of lipin 1 does not impair hepatic TAG synthesis, likely due to the high expression of lipin 2 in liver [41, 61]. Surprisingly, hepatic triglyceride levels return to normal by post-natal day 14, and the fld mice undergo a dramatic bout of compensatory growth that results in fld mice that are normal in outward appearance [62]. However, at necropsy, a near absence of abdominal and subcutaneous fat depots is obvious, and fld mice exhibit a rather severe lipodystrophic phenotype [6]. Conversely, transgenic mice overexpressing lipin 1 in adipose tissue exhibit increased adiposity [63]. This phenotype fits well with the roles of lipin 1 as an enzyme in the triglyceride synthesis pathway, and its role in the nucleus to regulate PPARγ-mediated increases in adipogenic gene expression [10]. In fact, both lipin 1-mediated PAP activity and its nuclear localization are required to complement adipogenic gene expression and the formation of lipid droplets in fld embryonic fibroblasts [40], highlighting the importance of both functions.
It should be noted that mutations in lipin 1 in humans are not associated with lipodystrophy. Instead, rare human LPIN1 mutations have been linked to acute, recurrent rhabdomyolysis (breakdown of skeletal muscle fibers) and susceptibility to statin-induced myopathy [64, 65]. These findings suggest either different metabolic effects of lipin 1 in humans or a higher expression level of lipin 1 in muscle in human subjects. On the other hand, a variety of studies have since linked variations in the LPIN1 gene to insulin resistance and diabetic phenotypes [66–68], and the expression of lipin 1 in adipose tissue and liver is strongly correlated with insulin sensitivity [67, 69–71], suggesting important roles for lipin 1 in maintaining systemic metabolic homeostasis in humans. Because most of this work is correlative, additional mechanistic work in model organisms in combination with translational studies conducted in human subjects may be required to fully understand the regulatory effects of lipin 1 on human intermediary metabolism.
Lipin 2
Lipin 2 is a liver-enriched lipin isoform, but also expressed at substantial levels in brain, lung, and kidney. In normal, ad libitum-fed mice, the expression of lipin 2 in liver is much higher than the expression of lipin 1 [33, 41]. Consistent with this, although the intrinsic PAP activity of lipin 2 is much lower than lipin 1 [15], siRNA-mediated knockdown of lipin 2 leads to a striking reduction in hepatocyte PAP activity [41]. Fld mice, which lack lipin 1, still have significant hepatic PAP activity [15, 30], and we have recently shown that hepatic lipin 2 protein is increased in fld mice as a compensatory mechanism [41]. Like lipin 1, lipin 2 also has the ability to regulate gene transcription and coactivates PPARγ in cultured cells [33].
Relatively less is known regarding the regulation of lipin 2. The hepatic expression of lipin 2 mRNA is up-regulated in response to fasting [41, 45]. Unlike lipin 1 however, neither PGC-1α, cAMP signaling, nor glucocorticoids influence lipin 2 expression [45]. Instead, lipin 2 seems to be a target of the peroxisome proliferator-activated receptor family [72]. Interestingly, hepatic lipin 2 protein levels seem to be under translational control in many physiological contexts [41]. For example, we have shown that the compensatory increase in lipin 2 protein that occurs in fld hepatocytes is mediated by an increased rate of lipin 2 translation and is independent of changes in steady state lipin 2 mRNA levels. Similarly, hepatic lipin 2 protein content is elevated in ob/ob and db/db mice independent of changes in mRNA expression [41]. The molecular mechanisms whereby this translational regulation occurs, however, remain to be determined.
Human mutations in the gene encoding lipin 2 (LPIN2) lead to an autoinflammatory bone disease known as Majeed Syndrome [73, 74]. Majeed syndrome is characterized by recurrent multifocal inflammation of bone and skin, fever, and dyserythropoietic anemia. The etiologic mechanisms whereby lipin 2 deficiency leads to Majeed syndrome are unclear since its expression is not particularly enriched in bone [74], but could be related to PA accumulation, which has been linked to activation of inflammatory signaling cascades [75].
Lipin 3
Lipin 3 is the least studied of the lipin family members. It seems to be expressed most highly in the stomach and intestine [15], perhaps suggesting a role in dietary fat absorption. However, intestinal absorption of dietary fat and esterification to TAG for packaging into chylomicrons is believed to be primarily mediated by the monoglyceride acyltransferase pathway, and this process would not require a PAP enzyme. Like the other mammalian lipin proteins, lipin 3 has PAP activity, but the specific activity and Vmax of lipin 3 are extremely low compared to lipin 1 [15]. A physical interaction between lipin 3 and PPARα has also been reported [8], but the effects of lipin 3 on transcription factor activity have not been determined. The physiologic relevance and regulated activity of lipin 3 has not been well-explored and this could potentially be an area of future interest.
Concluding remarks
The metabolic effects of lipin proteins play important roles in regulating lipid metabolism in multiple organ systems. However, much remains to be learned regarding how lipin activity is regulated and whether new and diverse biological pathways are also affected Box 1).
Box 1. Outstanding Questions.
How does phosphorylation regulate mammalian lipin translocation?
How are lipin-mediated transcriptional and enzymatic effects coordinated and regulated?
What is the molecular identity of the mammalian phosphatase that dephosphorylates lipin and do different phosphatases regulate nuclear versus ER localization?
Why are there three mammalian lipin genes?
Do the mammalian lipins regulate ER expansion, nuclear envelope morphology, and phospholipid biosynthesis genes?
How do lower organisms synthesize TAG in the absence of PAP activity?
Acknowledgments
The authors would like to thank Mark Pfister for the graphic design expertise and assistance with generating figures for this manuscript. This work was supported by NIH grant R01-DK078187 to BNF. TEH receives support from NIH grant R01-DK052753.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Kennedy EP. Biosynthesis of phospholipides. Fed Proc. 1957;16:847–853. [PubMed] [Google Scholar]
- 2.Kennedy EP. Synthesis of phosphatides in isolated mitochondria. J Biol Chem. 1953;201:399–412. [PubMed] [Google Scholar]
- 3.Han GS, et al. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterfy M, et al. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 5.Langner CA, et al. The fatty liver dystrophy (fld) mutation. A new mutant mouse with a developmental abnormality in triglyceride metabolism and associated tissue-specific defects in lipoprotein lipase and hepatic lipase activities. J Biol Chem. 1989;264:7994–8003. [PubMed] [Google Scholar]
- 6.Reue K, et al. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J Lipid Res. 2000;41:1067–1076. [PubMed] [Google Scholar]
- 7.Rehnmark S, et al. The fatty liver dystrophy mutant mouse: microvesicular steatosis associated with altered expression levels of peroxisome proliferator-regulated proteins. J Lipid Res. 1998;39:2209–2217. [PubMed] [Google Scholar]
- 8.Finck BN, et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Skinner JR, et al. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J Biol Chem. 2009;284:30941–30948. doi: 10.1074/jbc.M109.013995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koh YK, et al. Lipin1 is a key factor for the maturation and maintenance of adipocytes in the regulatory network with CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma 2. J Biol Chem. 2008;283:34896–34906. doi: 10.1074/jbc.M804007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy EP, et al. New synthesis of lecithin in an isolated enzyme system. Nature. 1956;178:594–595. doi: 10.1038/178594a0. [DOI] [PubMed] [Google Scholar]
- 12.Hubscher G, et al. Stimulation of biosynthesis of glyceride. Nature. 1967;216:449–453. doi: 10.1038/216449a0. [DOI] [PubMed] [Google Scholar]
- 13.Brindley DN. Intracellular translocation of phosphatidate phosphohydrolase and its possible role in the control of glycerolipid synthesis. Prog Lipid Res. 1984;23:115–133. doi: 10.1016/0163-7827(84)90001-8. [DOI] [PubMed] [Google Scholar]
- 14.Lin YP, Carman GM. Purification and characterization of phosphatidate phosphatase from Saccharomyces cerevisiae. J Biol Chem. 1989;264:8641–8645. [PubMed] [Google Scholar]
- 15.Donkor J, et al. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J Biol Chem. 2007;282:3450–3457. doi: 10.1074/jbc.M610745200. [DOI] [PubMed] [Google Scholar]
- 16.Li H, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santos-Rosa H, et al. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. Embo J. 2005;24:1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han GS, et al. The cellular functions of the yeast lipin homolog PAH1p are dependent on its phosphatidate phosphatase activity. J Biol Chem. 2007;282:37026–37035. doi: 10.1074/jbc.M705777200. [DOI] [PubMed] [Google Scholar]
- 19.Karanasios E, et al. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc Natl Acad Sci U S A. 107:17539–17544. doi: 10.1073/pnas.1007974107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi HS, et al. Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of SER(602), THR(723), AND SER(744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J Biol Chem. 286:1486–1498. doi: 10.1074/jbc.M110.155598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Hara L, et al. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J Biol Chem. 2006;281:34537–34548. doi: 10.1074/jbc.M606654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tange Y, et al. An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J Cell Sci. 2002;115:4375–4385. doi: 10.1242/jcs.00135. [DOI] [PubMed] [Google Scholar]
- 23.Fei W, et al. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol. 2008;180:473–482. doi: 10.1083/jcb.200711136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szymanski KM, et al. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci U S A. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golden A, et al. Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J Cell Sci. 2009;122:1970–1978. doi: 10.1242/jcs.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorjanacz M, Mattaj IW. Lipin is required for efficient breakdown of the nuclear envelope in Caenorhabditis elegans. J Cell Sci. 2009;122:1963–1969. doi: 10.1242/jcs.044750. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura Y, et al. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc Natl Acad Sci U S A. 2009;106:20978–20983. doi: 10.1073/pnas.0907173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eastmond PJ, et al. Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell. 22:2796–2811. doi: 10.1105/tpc.109.071423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimsey N, et al. Temporal and spatial regulation of the phosphatidate phosphatases lipin 1 and 2. J Biol Chem. 2008;283:29166–29174. doi: 10.1074/jbc.M804278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris TE, et al. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J Biol Chem. 2007;282:277–286. doi: 10.1074/jbc.M609537200. [DOI] [PubMed] [Google Scholar]
- 31.Liu GH, et al. Lipin proteins form homo- and hetero-oligomers. Biochem J. doi: 10.1042/BJ20100584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng D, et al. Human acyl-CoA:diacylglycerol acyltransferase is a tetrameric protein. Biochem J. 2001;359:707–714. doi: 10.1042/0264-6021:3590707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donkor J, et al. A conserved serine residue is required for the phosphatidate phosphatase activity but not the transcriptional coactivator functions of lipin-1 and lipin-2. J Biol Chem. 2009;284:29968–29978. doi: 10.1074/jbc.M109.023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim HB, et al. Lipin 1 represses NFATc4 transcriptional activity in adipocytes to inhibit secretion of inflammatory factors. Mol Cell Biol. 30:3126–3139. doi: 10.1128/MCB.01671-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu GH, Gerace L. Sumoylation regulates nuclear localization of lipin-1alpha in neuronal cells. PLoS One. 2009;4:e7031. doi: 10.1371/journal.pone.0007031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sirover MA. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- 37.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi H, et al. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 39.Hait NC, et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren H, et al. A phosphatidic acid binding/nuclear localization motif determines lipin1 function in lipid metabolism and adipogenesis. Mol Biol Cell. 21:3171–3181. doi: 10.1091/mbc.E10-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gropler MC, et al. Lipin 2 is a liver-enriched phosphatidate phosphohydrolase enzyme that is dynamically regulated by fasting and obesity in mice. J Biol Chem. 2009;284:6763–6772. doi: 10.1074/jbc.M807882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterfy M, et al. Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J Biol Chem. 2005;280:32883–32889. doi: 10.1074/jbc.M503885200. [DOI] [PubMed] [Google Scholar]
- 43.Han GS, Carman GM. Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J Biol Chem. 285:14628–14638. doi: 10.1074/jbc.M110.117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryu D, et al. TORC2 regulates hepatic insulin signaling via a mammalian phosphatidic acid phosphatase, LIPIN1. Cell Metab. 2009;9:240–251. doi: 10.1016/j.cmet.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Manmontri B, et al. Glucocorticoids and cyclic AMP selectively increase hepatic lipin-1 expression, and insulin acts antagonistically. J Lipid Res. 2008;49:1056–1067. doi: 10.1194/jlr.M800013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishimoto K, et al. Sterol-mediated regulation of human lipin 1 gene expression in hepatoblastoma cells. J Biol Chem. 2009;284:22195–22205. doi: 10.1074/jbc.M109.028753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang P, et al. Regulation of lipin-1 gene expression by glucocorticoids during adipogenesis. J Lipid Res. 2008 doi: 10.1194/jlr.M800061-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Z, et al. Involvement of adiponectin-SIRT1-AMPK signaling in the protective action of rosiglitazone against alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol. 298:G364–374. doi: 10.1152/ajpgi.00456.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vavrecka M, et al. The effect of starvation on the incorporation of palmitate into glycerides and phospholipids of rat liver homogenates. Biochem J. 1969;115:139–145. doi: 10.1042/bj1150139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lau DC, Roncari DA. Effects of glucocorticoid hormones on lipid-synthetic enzymes from different adipose tissue regions and from liver. Can J Biochem Cell Biol. 1983;61:1245–1250. doi: 10.1139/o83-160. [DOI] [PubMed] [Google Scholar]
- 51.Simpson KJ, et al. Hepatic phosphatidate phosphohydrolase activity in acute and chronic alcohol-fed rats. Biochem Soc Trans. 1989;17:1115–1116. doi: 10.1042/bst0171115. [DOI] [PubMed] [Google Scholar]
- 52.Brindley DN. Some aspects of the physiological and pharmacological control of the synthesis of triacylglycerols and phospholipids. Int J Obes. 1978;2:7–16. [PubMed] [Google Scholar]
- 53.Sturton RG, et al. Effects of starvation, corticotropin injection and ethanol feeding on the activity and amount of phosphatidate phosphohydrolase in rat liver. FEBS Lett. 1981;126:297–300. doi: 10.1016/0014-5793(81)80265-7. [DOI] [PubMed] [Google Scholar]
- 54.Higashida K, et al. Potential role of lipin-1 in exercise-induced mitochondrial biogenesis. Biochem Biophys Res Commun. 2008;374:587–591. doi: 10.1016/j.bbrc.2008.07.079. [DOI] [PubMed] [Google Scholar]
- 55.Pearen MA, et al. The orphan nuclear receptor, NOR-1, a target of beta-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinology. 2008;149:2853–2865. doi: 10.1210/en.2007-1202. [DOI] [PubMed] [Google Scholar]
- 56.Phan J, et al. Biphasic expression of lipin suggests dual roles in adipocyte development. Drug News Perspect. 2005;18:5–11. doi: 10.1358/dnp.2005.18.1.877165. [DOI] [PubMed] [Google Scholar]
- 57.Lu B, et al. LPS and proinflammatory cytokines decrease lipin-1 in mouse adipose tissue and 3T3-L1 adipocytes. Am J Physiol Endocrinol Metab. 2008;295:E1502–1509. doi: 10.1152/ajpendo.90323.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huffman TA, et al. Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proc Natl Acad Sci U S A. 2002;99:1047–1052. doi: 10.1073/pnas.022634399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterfy M, et al. Insulin-stimulated interaction with 14-3-3 promotes cytoplasmic localization of lipin-1 in adipocytes. J Biol Chem. 285:3857–3864. doi: 10.1074/jbc.M109.072488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moller F, Hough MR. Effect of salts on membrane binding and activity of adipocyte phosphatidate phosphohydrolase. Biochim Biophys Acta. 1982;711:521–531. doi: 10.1016/0005-2760(82)90068-6. [DOI] [PubMed] [Google Scholar]
- 61.Chen Z, et al. Alterations in hepatic metabolism in fld mice reveal a role for lipin 1 in regulating VLDL-triacylglyceride secretion. Arterioscler Thromb Vasc Biol. 2008;28:1738–1744. doi: 10.1161/ATVBAHA.108.171538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hall AM, et al. Dynamic and differential regulation of proteins that coat lipid droplets in fatty liver dystrophic mice. J Lipid Res. 51:554–563. doi: 10.1194/jlr.M000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Phan J, Reue K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005;1:73–83. doi: 10.1016/j.cmet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 64.Zeharia A, et al. Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am J Hum Genet. 2008;83:489–494. doi: 10.1016/j.ajhg.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Michot C, et al. LPIN1 gene mutations: a major cause of severe rhabdomyolysis in early childhood. Hum Mutat. 31:E1564–1573. doi: 10.1002/humu.21282. [DOI] [PubMed] [Google Scholar]
- 66.Wiedmann S, et al. Genetic variants within the LPIN1 gene, encoding lipin, are influencing phenotypes of the metabolic syndrome in humans. Diabetes. 2008;57:209–217. doi: 10.2337/db07-0083. [DOI] [PubMed] [Google Scholar]
- 67.Suviolahti E, et al. Cross-species analyses implicate Lipin 1 involvement in human glucose metabolism. Hum Mol Genet. 2006;15:377–386. doi: 10.1093/hmg/ddi448. [DOI] [PubMed] [Google Scholar]
- 68.Kang ES, et al. The 11482G >A polymorphism in the perilipin gene is associated with weight gain with rosiglitazone treatment in type 2 diabetes. Diabetes Care. 2006;29:1320–1324. doi: 10.2337/dc05-2466. [DOI] [PubMed] [Google Scholar]
- 69.Donkor J, et al. Adipose tissue lipin-1 expression is correlated with peroxisome proliferator-activated receptor alpha gene expression and insulin sensitivity in healthy young men. J Clin Endocrinol Metab. 2008;93:233–239. doi: 10.1210/jc.2007-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Croce MA, et al. Hepatic lipin 1beta expression is diminished in insulin-resistant obese subjects and is reactivated by marked weight loss. Diabetes. 2007;56:2395–2399. doi: 10.2337/db07-0480. [DOI] [PubMed] [Google Scholar]
- 71.Yao-Borengasser A, et al. Lipin expression is attenuated in adipose tissue of insulin-resistant human subjects and increases with peroxisome proliferator-activated receptor gamma activation. Diabetes. 2006;55:2811–2818. doi: 10.2337/db05-1688. [DOI] [PubMed] [Google Scholar]
- 72.Sanderson LM, et al. Peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) but not PPARalpha serves as a plasma free fatty acid sensor in liver. Mol Cell Biol. 2009;29:6257–6267. doi: 10.1128/MCB.00370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al-Mosawi ZS, et al. A splice site mutation confirms the role of LPIN2 in Majeed syndrome. Arthritis Rheum. 2007;56:960–964. doi: 10.1002/art.22431. [DOI] [PubMed] [Google Scholar]
- 74.Ferguson PJ, et al. Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome) J Med Genet. 2005;42:551–557. doi: 10.1136/jmg.2005.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nadra K, et al. Phosphatidic acid mediates demyelination in Lpin1 mutant mice. Genes Dev. 2008;22:1647–1661. doi: 10.1101/gad.1638008. [DOI] [PMC free article] [PubMed] [Google Scholar]