Abstract
Serine hydrolases (SHs) are a diverse enzyme class representing > 1% of all human proteins. The biological functions for most SHs remain poorly characterized due to a lack of selective inhibitors to probe their activity in living systems. Here, we show that a substantial number of SHs can be irreversibly inactivated by 1,2,3-triazole ureas, which exhibit negligible cross-reactivity with other protein classes. Rapid lead optimization by click chemistry-enabled synthesis and competitive activity-based profiling identified 1,2,3-triazole ureas that selectively inhibit enzymes from diverse branches of the SH superfamily, including peptidases (acyl-peptide hydrolase or APEH), lipases (platelet-activating factor acetylhyrolase-2 or PAFAH2), and uncharacterized hydrolases (α, β-hydrolase 11 or ABHD11), with exceptional potency in cells (sub-nM) and mice (< 1 mg/kg). We show that APEH inhibition leads to accumulation of N-acetylated proteins and promotes proliferation in T-cells. These data designate 1,2,3-triazole ureas as a pharmacologically privileged chemotype for SH inhibition that shows broad activity across the SH class coupled with tunable selectivity for individual enzymes.
Serine hydrolases (SHs) are one of the largest and most diverse enzyme classes in the eukaryotic and prokaryotic proteomes with a membership that includes lipases, esterases, thioesterases, peptidases/proteases, and amidases1. Mammalian SHs are near-equally divided into two major sub-groups: ~100 serine proteases, mostly from the chymotrypsin/trypsin class; and another ~110 ‘metabolic’ enzymes, mostly from the alpha/beta-hydrolase class [Pfam class assignment – AB_hydrolase (CL0028)] that hydrolyze metabolites, peptides, or post-translational ester/thioester modifications on proteins. The important biological roles played by SHs have resulted in clinically approved drugs that target members of this enzyme class to treat diseases such as obesity2, diabetes3, microbial infections4, and Alzheimer’s disease5. Despite these advances, most of the 200+ mammalian SHs remain poorly understood in terms of their biochemical and cellular activities. Pursuit of this knowledge would benefit from the development of selective inhibitors to probe the function of individual SHs in living systems. This constitutes an exciting, but challenging task that has been successfully accomplished for only a handful of SHs to date1,6.
All SHs possess a serine nucleophile required for catalytic activity, opening up the opportunity to develop mechanism-based inhibitors that inactivate these enzymes by covalent modification. Among the inhibitors that have been shown to react with the serine nucleophile of SHs, fluorophosphonates (FPs)7,8 and carbamates9 are exceptional in that they show negligible cross-reactivity with other nucleophilic enzymes such as cysteine hydrolases. FPs are highly reactive and provide broad, near-complete coverage of mammalian SHs6. This feature has promoted the use of reporter-tagged FPs for activity-based protein profiling (ABPP) of SHs10,11, but limits the utility of FPs as pharmacological probes for specific members of this enzyme class. Carbamates, on the other hand, can show excellent selectivity for individual SHs. These inhibitors have proven to be valuable research tools12–14 and, in certain cases, advanced to the stage of approved drugs [e.g., rivastigmine, which targets acetylcholine esterase (ACHE) to treat Alzheimer’s disease5]. Despite considerable screening efforts, however, potent and selective carbamate inhibitors have been identified for only a fraction of mammalian SHs6, pointing to the need for alternative chemical classes of SH inhibitors.
Here, we report the discovery and characterization of the 1,2,3-triazole urea as a versatile chemotype for SH inhibition that couples the desired properties of broad coverage across the enzyme class with tunable selectivity for individual members. Starting from a scaffold that inhibits numerous SHs in living cells, we employed an efficient, two-step synthetic strategy based on click chemistry coupled with a quantitative platform for competitive ABPP to generate selective inhibitors for individual enzymes from diverse branches of the SH superfamily, including peptidases (APEH, or acyl-peptide hydrolase), lipases (PAFAH2, or platelet-activating factor acetylhydrolase-2), and uncharacterized enzymes (ABHD11 or α,β-hydrolase 11). These 1,2,3-triazole urea inhibitors exhibited remarkable, sub-nM activity in cells and exceptional potency (< 1 mg/kg) in mice, indicating that they are suitable probes for the functional characterization of SH targets in a diversity of biological systems. We used the APEH inhibitor AA74-1 to characterize the N-acetylated protein substrates of this enzyme in T-cells, where blockade of APEH activity stimulated cellular proliferation.
RESULTS
Class-specific inhibition of SHs by 1,2,3-triazole ureas
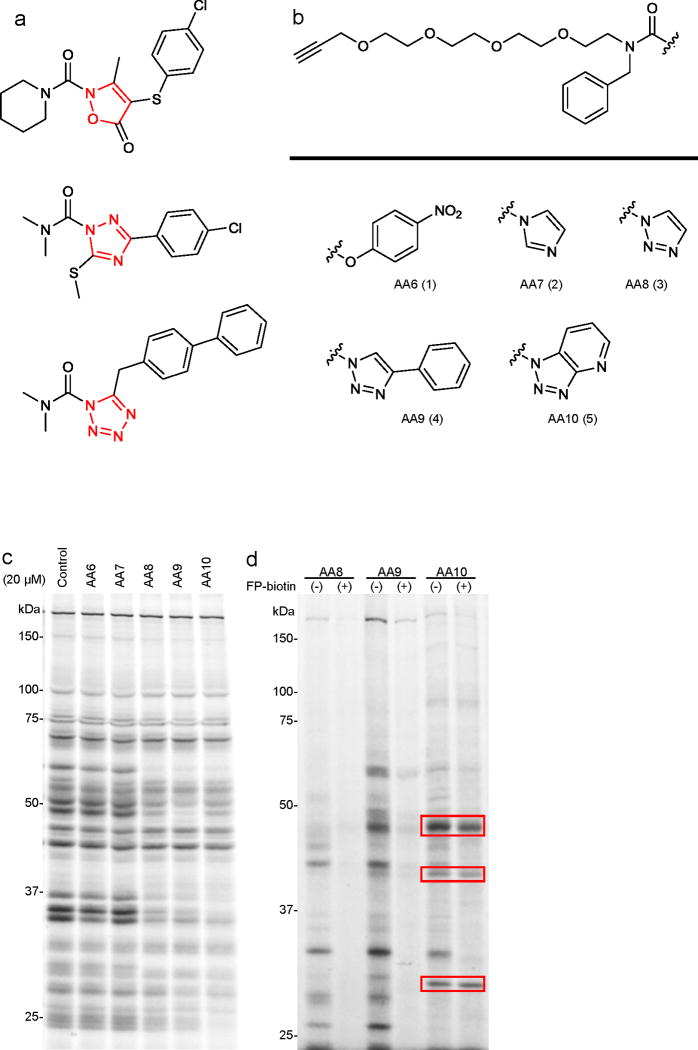
In the course of characterizing agents that perturb endocannabinoid uptake and metabolism, we discovered that the tetrazole urea LY218324015 (top, Fig. 1a) was a potent inhibitor of numerous SHs16. We confirmed that 1 inhibits the SH FAAH by covalent carbamoylation of the enzyme’s serine nucleophile16. There are a handful of other reports of N-heterocyclic ureas (NHUs) as SH inhibitors, including an isoxazolonyl urea17 (middle, Fig. 1a) and a 1,2,4-triazole urea18 (bottom, Fig. 1a), which are potent inhibitors of hormone-sensitive lipase (LIPE), but very limited selectivity data are available on these compounds. These initial findings suggested that the NHU might represent a useful scaffold for SH inhibitor design; however, several important questions remained unanswered. First, how does the electrophilicity of the NHU affect reactivity with SH enzymes, as well as potential cross-reactivity with other classes of proteins? Do NHUs inhibit SHs that are not susceptible to inactivation by other inhibitory scaffolds such as carbamates? Can one efficiently introduce structural diversity into the NHU scaffold by chemical synthesis? And, finally, can the properties of NHUs be optimized to achieve potent and selective inhibition of individual SHs in living systems?
Figure 1.
Competitive ABPP with clickable N-heterocyclic urea (NHU) activity-based probes AA6-AA10. (a) Structures of previously reported serine hydrolase inhibitors from the NHU class, including the endocannabinoid hydrolase inhibitor tetrazole urea LY218324015 (top) and the HSL inhibitors isoxazolonyl urea17 (middle) and 1,2,4-triazole urea18 (bottom). (b) Structures of carbamate- and NHU-alkyne probes with various leaving groups. (c) Competitive ABPP of AA6 - AA10 in the mouse brain membrane proteome. Brain membranes were incubated with 20 μM of AA6 - AA10 or DMSO for 30 min at 37 oC. Proteomes were then labeled with the SH-directed ABPP probe FP-Rh (2 μM, 30 min, 25 °C), separated by SDS-PAGE, and FP-Rh-labeled proteins detected by in-gel fluorescence scanning. This fluorescent gel and all gels in subsequent figures, are shown in grayscale. (d) Profiling the direct targets of AA6 - AA10 (20 μM, 30 min at 37 °C) in brain membranes in the presence or absence of the SH-directed probe FP-biotin (20 μM, 30 min at 37 °C). AA6 - AA10-labeled proteins were detected by reaction with an azide-Rh tag under click chemistry conditions following described protocols19,51. Targets of AA10 that are not competed by FP-biotin are highlighted with red boxes.
To answer the first question, we synthesized alkyne-modified agents AA6 - AA10 (1-5, Fig. 1b and Supplementary Fig. 1), which differ in electrophilicity due to variations in the leaving group. We then performed a competitive ABPP experiment by treating a mouse brain membrane proteome with AA6 - AA10 (20 μM, 30 min), followed by the SH-directed activity-based probe FP-rhodamine6,11 (FP-Rh, 2 μM, 30 min), separation by SDS-PAGE, and detection of FP-Rh-labeled proteins by in-gel fluorescence scanning (Fig. 1c). The carbamate AA6 and imidazole AA7 showed little to no detectable inhibition of SHs, whereas 1,2,3-triazoles AA8 - AA10 blocked the FP-Rh-labeling of several proteins. As expected, the reactivity of these compounds followed the trend of electrophilicity imparted by their leaving groups, with the pyridyl triazole AA10 being the most acidic and reactive NHU. To assess the cross-reactivity of 1,2,3-triazole ureas AA8 - AA10 with other protein classes, we performed a second, complementary competitive ABPP experiment. Here, we used the NHUs themselves as probes and asked whether their proteome reactivity profiles could be blocked by pre-incubation with FP-biotin10 (20 μM). Visualization of NHU-labeled proteins was achieved by click chemistry conjugation to an azide-Rh reporter tag19. FP-biotin was found to compete the labeling of all proteins modified by triazoles AA8 and AA9 (Fig. 1d), whereas several of the protein targets of the most electrophilic NHU AA10 were not sensitive to FP-biotin competition (Fig. 1d, red boxes), suggesting that they correspond to labeling of non-SH proteins. These data designated the monocyclic triazole ureas AA8 and AA9 as possessing the desired degree of electrophilicity to inhibit a number of SHs in proteomes, but, at the same time, avoid modification of proteins outside of the SH class.
1,2,3-triazole ureas inhibit many SHs in living cells
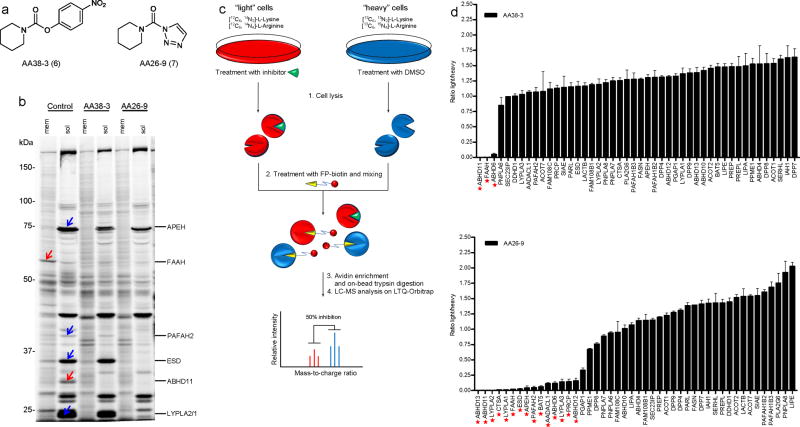
We next compared the proteome-wide SH reactivity of the 1,2,3-triazole urea with a more classical carbamate inhibitor. We synthesized carbamate AA38-3 (6) and triazole AA26-9 (7) (Fig. 2a), both based on a piperazine scaffold shown previously to inhibit SHs in the context of p-nitrophenoxy carbamate6,13,20, and treated mouse BW5147 T-cell hybridoma cells with each inhibitor at 20 μM for 4 h. Cells were then lysed and analyzed by competitive ABPP with the FP-Rh probe. Gel-based ABPP detected SHs that were inhibited by both AA38-3 and AA26-9 (Fig. 2b, red arrows), as well as a substantial number of additional SHs that were only inhibited by the triazole AA26-9 (Fig. 2b, blue arrows). To identify the inhibited enzymes, we utilized an advanced quantitative mass spectrometry (MS)-based platform that we term competitive ABPP-SILAC (Fig. 2c). Competitive ABPP-SILAC is essentially a merger of our described ABPP-MudPIT method7 with the stable isotope labeling of amino acids in culture (SILAC) technique21, which has been used to identify enzymes targets of activity-based probes22 and small-molecule-binding proteins in cell lysates23.
Figure 2.
Comparative ABPP of piperidine-based carbamate (AA38-3) and triazole urea (AA26-9) inhibitors. (a) Structures of AA38-3 and AA26-9. (b) Competitive ABPP of AA38-3 and AA26-9 in BW5147-derived murine T-cell hybridoma cells. Cells were cultured with 20 μM inhibitor or DMSO as a control for 4 h, lysed, separated into soluble and analyzed by competitive gel-based ABPP. Blue and red arrows mark SHs that were inhibited by AA26-9 and both AA26-9 and AA38-3, respectively. (c) Schematic representation of a competitive ABPP-SILAC experiment. Isotopically “light” (red) and “heavy” (blue) mouse T-cells are treated with inhibitor and DMSO, respectively, for 4 h. Cells are lysed, proteomes are treated with FP-biotin, and combined in a 1:1 ratio. Biotinylated proteins are enriched, trypsinized, and analyzed by LC-MS/MS. SH activities are quantified by comparing intensities of light and heavy peptide peaks. (d) Identification of SH targets for AA38-3 (top) and AA26-9 (bottom) in mouse T-cells by ABPP-SILAC. Cells were cultured with inhibitor (20 μM) or DMSO as a control for 4 h prior to analysis by competitive ABPP-SILAC. Asterisks mark SHs that were inhibited by > 75%. Bars represent the means ± s.e.m of light/heavy-ratios of identified tryptic peptides in both soluble and membrane proteomes for two independent biological replicates.
Briefly, mouse T-cells were cultured under ‘light’ (with 12C614N2-lysine and 12C614N4-arginine containing medium) and ‘heavy’ conditions (with 13C615N2-lysine and 13C615N4-arginine), treated with inhibitor and DMSO, respectively, and then harvested, lysed, separated into soluble and membrane fractions, and treated with FP-biotin (5 μM, 90 min). Light and heavy fractions were then mixed, enriched with avidin, digested on-bead with trypsin, and analyzed by LC-MS/MS using an LTQ-Orbitrap instrument. Light and heavy signals were quantified from parent ion peaks (MS1) and the corresponding proteins identified from product ion profiles (MS2) using the SEQUEST search algorithm. The depicted bar graphs represent the average ratios of light/heavy tryptic peptides for each of the 46 SHs identified in mouse T-cells (Fig. 2d and Supplementary Table 1). While in situ treatment with carbamate AA38-3 inhibited three SHs (ABHD6, ABHD11, and FAAH), 15 SHs were inhibited by triazole urea AA26-9 (including the aforementioned three targets of AA38-3) (Fig. 2d). Notably, AA26-9-inhibited enzymes originated from diverse functional subclasses of SHs, including lipases/phospholipases (AADACL1, ABHD6, ESD, FAAH, PAFAH2, LYPLA3), peptidases (APEH, PRCP, CTSA), thioesterases (LYPLA1, LYPLA2), and uncharacterized enzymes (ABHD11, ABHD13, BAT5) (Supplementary Table 2). Interestingly, 9 of these 15 triazole targets were not inhibited by any members of a 150+ carbamate library recently screened against mammalian SHs by our laboratory6. Taking into account the predicted molecular masses of AA26-9- and AA38-3-inhibited SHs, as well as their sensitivity to one or both inhibitors, allowed for confident assignment of many of the SH signals on ABPP gels (Fig. 2b). We confirmed by MS analysis that AA26-9 inhibited one of its enzyme targets LYPLA1 by covalent carbamoylation of the enzyme’s serine nucleophile (S114) (Supplementary Fig. 2). Finally, similar inhibitor sensitivity profiles were observed with live cells (Fig. 2b) and cell homogenates (Supplementary Fig. 3a), indicating that reductions in FP-Rh labeling of SHs reflected direct inhibition by AA26-9 and/or AA38-3 in situ, as opposed to indirect effects on the expression level of these enzymes.
These results, taken together, demonstrate that the 1,2,3-triazole urea is capable of inactivating numerous SHs, exhibiting a target profile that surpasses in breadth that of the structurally related carbamate.
Synthesis of 1,2,3-triazole ureas by click chemistry
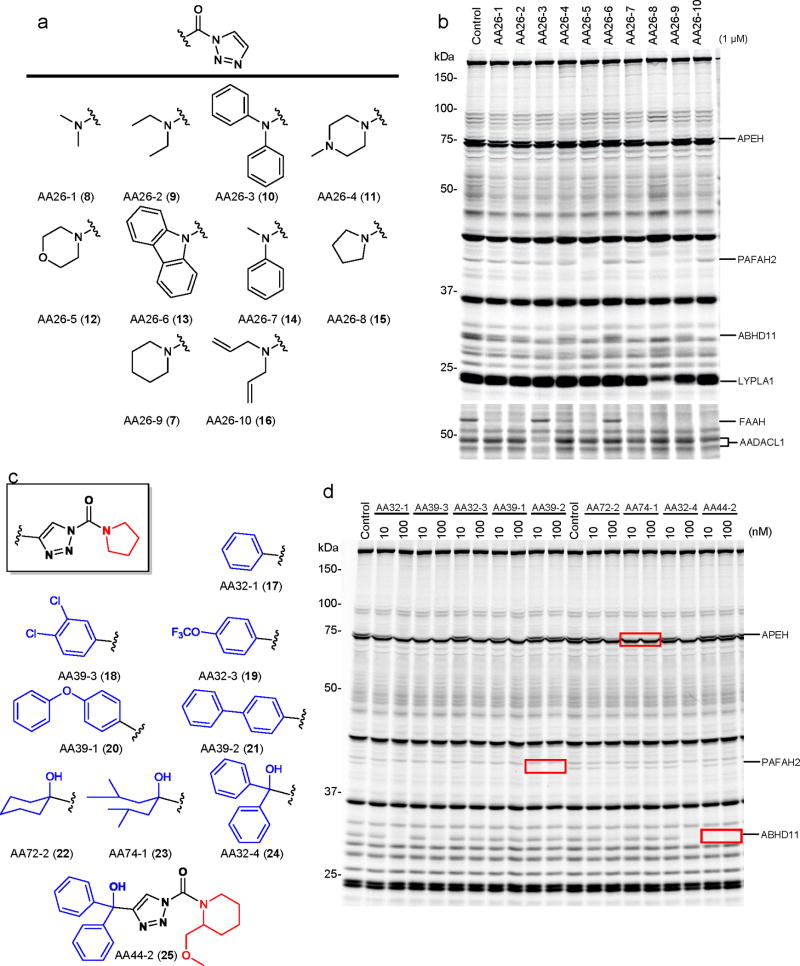
Having demonstrated that AA26-9 acts as a broad-spectrum inhibitor for SHs, the next major question was: can this simple 1,2,3-triazole urea scaffold be structurally modified to produce selective inhibitors for individual SH enzymes? There are two major sites for diversification on the 1,2,3-triazole urea scaffold: 1) the carbamoylating group, and 2) the 1,2,3-triazole leaving group. We first prepared 10 agents (AA26-1- AA26-10; 7-16) with distinct carbamoyl substituents combined with a uniform, unfunctionalized 1,2,3-triazole leaving group (Fig. 3a). Mouse T-cell proteomes were incubated in vitro with AA26-1 - AA26-10 (7-16, 1 μM, 30 min) and analyzed by competitive ABPP. The individual triazole ureas showed markedly different SH reactivity profiles, with one agent, in particular, the pyrrolidine urea AA26-8 (15) inhibiting several enzymes, including ABHD11, APEH, FAAH, PAFAH2, and LYPLA1 (Fig. 3b). Three of these enzymes, namely PAFAH2, ABHD11, and FAAH were also inhibited by morpholine urea AA26-5 (15). Notably, most of these enzymes were more potently inhibited by AA26-5 (12) and AA26-8 (15) compared to the piperidinyl analogue AA26-9 (7). We therefore selected the pyrrolidine- and morpholine-based scaffolds of AA26-8 (15) and AA26-5 (12) as starting points for constructing a focused library of 1,2,3-triazole ureas, where we hypothesized that introducing substituents onto the triazole group would enhance selectivity for individual SHs.
Figure 3.
Rapid optimization of triazole urea inhibitors by click chemistry-enabled synthesis and competitive ABPP. (a) Structures of ten 1,2,3-triazole ureas (AA26-1-AA26-10) with distinct carbamoyl substituents combined with a uniform, unsubstituted 1,2,3-triazole leaving group. (b) Reactivity profiles for AA26-1-AA26-10 in vitro. Soluble and membrane fractions of mouse T-cells were incubated with inhibitors (1 μM) for 30 min at 37 oC, after which the samples were analyzed by competitive gel-based ABPP. (c) Structures of representative pyrrolidine and piperidine compounds with functionalized 1,2,3-triazole leaving groups. (d) Competitive ABPP results for functionalized 1,2,3-triazole urea inhibitors in mouse T-cells in vitro (treated with inhibitors at the indicated concentrations for 30 min at 37 oC). Inhibitors AA39-2, AA74-1, and AA44-2 each inhibited only a single SH target in the T-cell proteome (highlighted with red boxes).
We developed a simple and efficient click chemistry approach to create substituted triazole ureas (Supplementary Fig. 3b). In this two-step procedure, substituted alkynes were reacted with in situ-formed azidomethanol to yield 4-substituted triazoles, which were then carbamoylated to give triazole urea products, typically as a 3:1 mixture of N1- and N2-carbamoylated regioisomers. The N1-carbamoyl triazole was separated by silica gel chromatography and used for subsequent experiments. We used this strategy to prepare a ~20 member library of 4-aryl- and 4-alkyl triazole derivatives of AA26-5 (12) and AA26-8 (15; representative structures 17-25 are shown in Fig. 3c; see Supplementary Fig. 3c for structures of the rest of the library), which were screened at concentrations of 10 and 100 nM in mouse T-cell proteomes (Fig. 3d, Supplementary Fig. 3d, and Supplementary Table 3). From this library, highly potent and selective inhibitors of APEH and PAFAH2 were identified. For instance, the biphenyl triazole AA39-2 (21) completely blocked PAFAH2 activity at 10 nM, while not inhibiting ABHD11, APEH, LYPLA1, or other SHs in T-cells at 100 nM. When substituted with bulky aliphatic groups, the triazole ureas generally showed higher preference for APEH, as exemplified by the 2,6-dimethylheptanol-substituted triazole in AA74-1 (23), which completely inhibited APEH at 10 nM, while not interacting with other SHs at 100 nM. We also identified 4-alkyl/aryl triazole ureas with good activity against ABHD11, including AA32-1 (17), AA39-3 (18), and AA32-4 (24), all of which completely blocked the activity of this enzyme at 100 nM, albeit non-selectively with respect to APEH and PAFAH2. Previous studies identified selective, moderate potency (IC50 values of ~100-200 nM) carbamate inhibitors of ABHD11 that possess a 2-substituted piperidine group6. Following this design, we replaced the pyrrolidine carbamoyl group of AA32-4 (24) with the larger and bulkier 2-(methoxymethyl)-piperidine to generate AA44-2 (25), which showed much improved potency for ABHD11 (complete inactivation at 10 nM) and no cross-reactivity with APEH, PAFAH2, or other SHs at 100 nM (Fig. 3d).
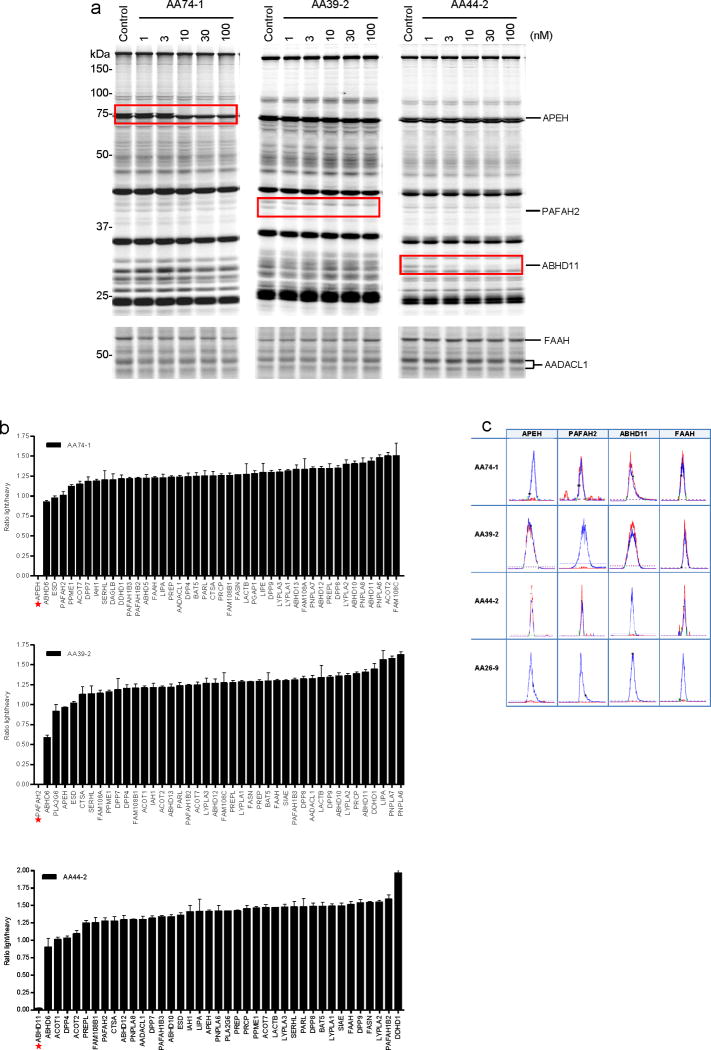
The triazole urea inhibitors, AA74-1 (23), AA39-2 (21), and AA44-2 (25), showed remarkable potency for their respective SH targets in mouse T-cell proteomes, exhibiting IC50 values of 5, 3, and 1 nM for APEH, PAFAH2, and ABHD11, respectively, in competitive gel-based ABPP assays (Fig. 4a and Supplementary Figs. 4a and 4b). No other SHs were visibly inactivated by the inhibitors in either soluble or membrane proteome of T-cells at concentrations up to 100 nM (Fig. 4a and Supplementary Fig. 4b). We confirmed that the 1,2,3-triazole ureas inhibited recombinant forms of their respective SH targets expressed by transient transfection in COS-7 or HEK-293 cells (Supplementary Fig. 4c). For AA74-1, we also measured a kobs/[I] value of 7300 +/−500 M−1s−1 (Supplementary Figs. 5a and 5b) and determined that it potently inhibited the human (IC50 = 11 nM) and rat (IC50 = 7 nM) orthologs of this enzyme (Supplementary Fig. 6).
Figure 4.
In vitro and in situ characterization of triazole urea inhibitors AA74-1, AA39-2, and AA44-2 in mouse T-cells. (a) Competitive ABPP results for the three inhibitors in soluble (top) and membrane (bottom) proteomes of T-cells after 30 min treatment at the indicated inhibitor concentrations. Inhibited SHs are highlighted with red boxes. The shown gels are representative of at least three independent biological replicate experiments. (b) ABPP-SILAC analysis of SH activities from inhibitor-treated mouse T-cells (in situ treatment with 3 nM AA74-1, AA39-2 or AA44-2 for 4 h). Asterisks mark the SH target of each compound, each of which was inhibited > 97%. Bars represent the means ± s.e.m of light/heavy-ratios for the multiple peptides observed for each enzyme; data are derived from both soluble and membrane proteomes for two independent biological replicates. (c) Orthogonal selectivity of inhibitors AA74-1, AA39-2, and AA44-2 illustrated by showing heavy and light MS1 peak pairs for representative tryptic peptides from APEH, PAFAH2, ABHD11, and FAAH. Note that unsubstituted inhibitor AA26-9 nonselectively inhibits all four SHs.
Taken together, these data demonstrate that potent and selective inhibitors can be identified for a diverse set of SHs from a modest-sized library of structurally varied 1,2,3-triazole ureas, which can be efficiently accessed by a click chemistry-based synthetic protocol.
Triazoles ureas are ultrapotent SH inhibitors in situ
We next asked whether the 1,2,3-triazole ureas maintain their potency and selectivity in living cells. Mouse T-cells were cultured with different concentrations of inhibitors for 4 h, lysed, and analyzed by competitive ABPP. All three inhibitors (AA74-1, AA39-2, AA44-2) inactivated their respective proteomic targets (APEH, PAFAH2, ABHD11) with exceptional subnanomolar potency (140-170 pM) in situ (Supplementary Fig. 5c). Although each inhibitor showed evidence of high target selectivity by gel-based competitive ABPP, we have previously found that LC-MS methods like ABPP-MudPIT provide a more comprehensive profile of SH activities in proteomes6,7,13. We therefore used ABPP-SILAC to assess the selectivity of AA74-1, AA39-2, and AA44-2 in living mouse T-cells (3 nM inhibitor, 4 h). All three inhibitors exhibited remarkable selectivity for their respective SH targets (Fig. 4b and Supplementary Table 1). AA74-1 and AA44-2 blocked > 95% of APEH and ABHD11 activity, respectively, while not affecting any of the other 40+ SHs detected in T-cells. AA39-2 was similarly effective at blocking its target PAFAH2 in T-cells, and only showed marginal cross-reactivity with a single SH ABHD6, which displayed ~40% reduction in activity. While it is possible that additional SH targets may be identified for these inhibitors when subjected to even broader profiling, we should note that AA74-1 and AA39-2 selectively inhibit PAFAH2 and APEH over their nearest sequence-neighbor enzymes PLA2G7 (AA39-2: IC50 for PAFAH2 = 3 nM; IC50 for PLA2G7= 100 nM; Supplementary Fig. 7) and the dipeptidylpeptidases (e.g., DPP4, DPP8, DPP9; Fig. 4b), respectively. As examples of the MS data used for quantitation, heavy and light MS1 peak pairs for representative peptides from APEH, PAFAH2, ABHD11, and the untargeted SH FAAH are shown in Fig. 4c to illustrate the orthogonal selectivity of inhibitors AA74-1, AA39-2, and AA44-2. For comparison, we also show the MS1 profiles for T-cells treated with the pan-SH inhibitor AA26-9, which inactivated all four of the displayed SHs (Fig. 4c).
Triazole AA74-1 is a selective APEH inhibitor in vivo
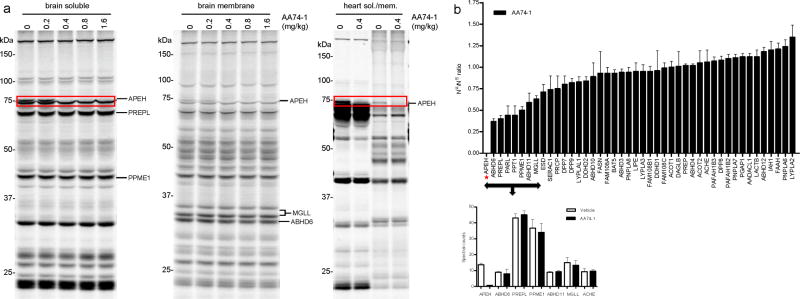
We next asked whether the potency and selectivity displayed by 1,2,3-triazole ureas in cell culture would also be observed in vivo. Mice were treated with the APEH inhibitor AA74-1 at various doses (0.2–1.6 mg/kg; PEG300 or 18:1:1 saline/ethanol/emulphor intraperitoneally) and sacrificed after 4 h. Brains and hearts were removed, homogenized, and analyzed by gel-based competitive ABPP. AA74-1 completely inhibited APEH in both brain and heart at doses as low as 0.4 mg/kg (equivalent to 10 μg AA74-1 per animal) (Fig. 5a and Supplementary Fig. 8). Complete inhibition of APEH at this dose was also confirmed by a substrate assay based on cleavage of the fluorogenic probe N-acetyl-L-alanine p-nitroanilide (Supplementary Fig. 9). No additional SH targets were observed for AA74-1 in either tissue by gel-based ABPP.
Figure 5.
Characterization of the activity and selectivity of APEH inhibitor AA74-1 in vivo. (a) Competitive ABPP results for soluble and membrane proteomes from brain and heart tissue of AA74-1-treated mice. Proteomes were prepared from mice injected with AA74-1 (0.2–1.6 mg/kg, i.p.) or vehicle (PEG300) for 4 h and analyzed by competitive gel-based ABPP, n = 3 mice per group. Inhibition of APEH is highlighted with red boxes. (b) ABPP-SILAM analysis of SH activities in brain tissue from mice treated with AA74-1 (0.8 mg/kg, i.p.) or vehicle (PEG300). Asterisk marks the ratio-of-ratio value for APEH, which was inhibited by greater than 97%. Bars represent the means ± s.e.m of ratios-of-ratios for the multiple peptides observed for each enzyme; data are derived from a single experiment.
We also explored the possibility of extending ABPP-SILAC for the characterization of inhibitor selectivity in vivo by taking advantage of recently described protocols for stable isotope labeling in mammals (SILAM)24,25. In this approach, brain tissues from vehicle- and AA74-1-treated mice were separately mixed with equivalent amounts of brain tissue from ‘heavy’ amino acid-fed mice, and the samples were then labeled with FP-biotin, enriched by avidin chromatography, analyzed by LC-MS, and SH activities quantified by measuring the ratio-of-ratios for observed peptide signals. Due to the limited availability and high cost of metabolically labeled mice, we conducted the ABPP-SILAM experiment only once. For comparison, we performed three biological replicates by the more classical competitive ABPP-MudPIT method with unlabeled mouse tissues using spectral counting as a measure of enzyme activity levels7. These analyses identified 44 SHs in brain and confirmed that, of these enzymes, only APEH was inhibited by AA74-1 (Fig. 5b and Supplementary Table 1). The ratio-of-ratio signals for APEH indicated greater than 90% inhibition of this enzyme in AA74-1-treated animals (Fig. 5b), consistent with average spectral count values (14 and 1 spectral counts for APEH in vehicle- versus AA74-1-treated mice, respectively; Fig. 5b and Supplementary Table 1). A handful of additional SHs (ABHD6, PREPL) showed ratio-of-ratio signals suggestive of partial inhibition (50–60%); however, we do not believe that these represent true changes, since the spectral count values (Fig. 5b) and gel-based ABPP signals (Fig. 5a) were not reduced for these enzymes in brain proteomes from AA74-1-treated animals nor were the corresponding enzymes affected in T-cells treated with AA74-1 (see Fig. 4b). Finally, we note that ACHE, an enzyme that is targeted by other reported NHU inhibitors18, was not inhibited by AA74-1 in either mouse brain (Fig. 5b) or transfected cell (Supplementary Fig. 10) proteomes.
These findings confirm that AA74-1 acts as an extremely potent and selective inhibitor of APEH in mice and furthermore provide the first example, to our knowledge, of using stable-isotope labeling methods to quantify inhibitor-enzyme interactions in living animals.
APEH inhibition alters protein N-acetylation in T-cells
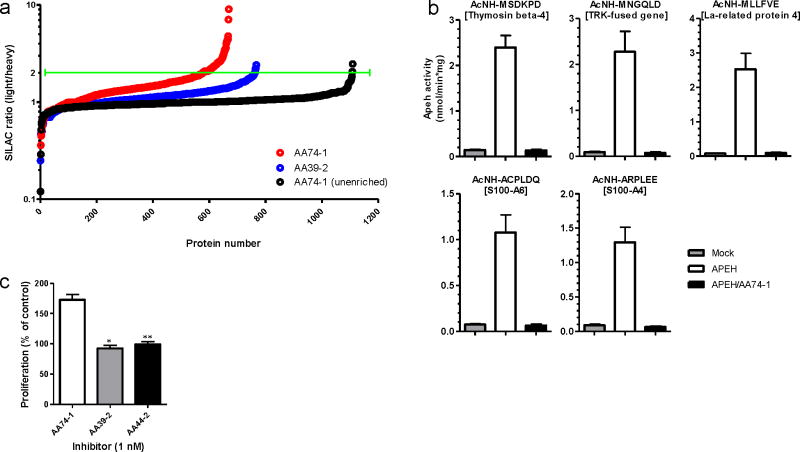
Despite having been postulated to serve as a key regulator of N-terminally acetylated proteins for many years25, very few endogenous substrates have been identified for APEH, nor have the biological effects of disrupting this enzyme been examined. The selective APEH inhibitor AA74-1 provided a pharmacological tool to investigate these questions. We measured changes in N-terminally modified proteins in AA74-1 (1 nM) versus DMSO-treated T-cells by SILAC combined with an established method for selective biotinylation of N-terminal amines26 (Supplementary Fig. 11a). Biotinylated proteins were enriched by avidin chromatography, digested on-bead with trypsin, and the resulting peptides analyzed by LC-MS on an LTQ-Orbitrap instrument. Data sets were filtered to identify proteins with multiple peptides that showed consistent two-fold or greater reductions in signals in AA74-1-treated cells. The resulting collection of ~25 proteins represented candidate APEH substrates (Fig. 6a, Table 1, and Supplementary Table 1). Importantly, none of these proteins showed altered N-terminal labeling profiles in T-cells treated with the PAFAH2 inhibitor AA39-2 (Table 1 and Supplementary Table 1), which produced a negligible number of total changes in the N-terminally modified protein profile (Fig. 6a). Furthermore, no changes were observed in total protein abundance for the subset of APEH substrates that could also be detected in unenriched proteomic profiles from AA74-1-treated cells (Supplementary Fig. 11b and Table 1), indicating that APEH inhibition affected the N-terminal modification state, but not overall stability of these proteins.
Figure 6.
Proteomic characterization of endogenous APEH substrates using N-terminal labeling and enrichment. (a) Measured SILAC ratios for N-terminally enriched and unenriched peptides from the soluble proteome of mouse T-cells treated in situ with AA74-1 or AA39-2 (1 nM, 6 h). Green line designates the two-fold signal change cut-off used to define candidate APEH substrates in AA74-1-treated cells. (b) In vitro APEH exopeptidase activity assay with synthetic N-acetylated hexapeptides. APEH was recombinantly expressed in HEK-293 cells. Whole cell lysates were pre-treated with DMSO or AA74-1 (3 nM, 30 min), incubated with peptides for 10 h, and release of the N-terminal N-acetylated amino acid was measured by LC-MS. Data are presented as means ± s.d. (n = 3). Mock corresponds to control cells transfected with an empty vector. (c) Stimulation of mouse T-cell proliferation by APEH inhibition. Mouse T-cells were treated in situ with the indicated inhibitors (1 nM) or DMSO for 12 h. Cell proliferation was measured using the colorimetric agent WST-1 (*p < 0.05 for AA74-1-versus AA39-2-treated cells; **p < 0.01 for AA74-1- versus AA44-2-treated cells). Data are presented as means ± s.d. (n = 4).
Table 1.
SILAC ratios for the top-25 most substantially altered proteins from N-terminal labeling proteomic analysis of AA74-1-treated T-cells. Shown are ratios for N-terminal-labeled and enriched peptides from AA74-1 or AA39-2-treated (1 nM, 6 h) versus DMSO-treated T-cells, as well as ratios for these peptides from unenriched proteomic analyses of AA74-1-treated T-cells. Asterisks mark proteins that have been previously determined to be N-terminal acetylated27,28. Bars represent the median values ± s.d. of light/heavy-ratios for the multiple peptides observed for each protein, n = 3.
| Name | Protein Description | Ratio AA74-1 (enriched) | Ratio AA74-1 (unenriched) | Ratio AA39-2 (enriched) |
|---|---|---|---|---|
| NDRG1 | N-myc downstream regulated gene 1 | 9.05 ± 1.5 | 0.98 ± 0.01 | 1.24 ± 0.28 |
| EG665937 | Similar to Ferritin light chain 1 | 7.07 ± 2.15 | 0.94 ± 0.08 | 1.05 ± 0.07 |
| CCDC97* | Coiled-coil domain containing protein 97 | 4.78 ± 0.22 | n.d. | 1.26 ± 0.16 |
| LARP4* | La-related protein 4 | 4.42 ± 0.23 | n.d. | 0.94 ± 0.05 |
| ARHGEF2 | Rho/Rac guanine nucleotide exchange factor 2 | 4.41 ± 0.55 | 1.08 ± 0.07 | 1.18 ± 0.13 |
| 5830457O10Rik | Chromosome transmission fidelity factor 8 homolog | 4.36 ± 1.17 | 1.25 ± 0.19 | 0.96 ± 0.02 |
| FBXO7 | F-box protein 7 | 4.27 ± 0.11 | 1.1 ± 0.04 | 0.88 ± 0.21 |
| NIBAN | Family with sequence similarity 129, member A | 4.22 ± 0.28 | 1.1 ± 0.04 | 1.30 ± 0.36 |
| S100A4* | S100 calcium binding protein A4 | 4.07 ± 0.3 | 1.01 ± 0.12 | 1.05 ± 0.08 |
| S100A6* | S100 calcium binding protein A6 | 3.93 ± 0.33 | 1.03 ± 0.15 | 1.07 ± 0.11 |
| PTPN18 | Protein tyrosine phosphatase, non-receptor type 18 | 3.58 ± 0.45 | 0.97 ± 0.09 | n.d. |
| NDUFV2 | NADH dehydrogenase (ubiquinone) flavoprotein 2 | 3.09 ± 0.52 | n.d. | 1.12 ± 0.19 |
| SERF2 | Small EDRK-rich factor 2 | 2.66 ± 0.15 | 1.05 ± 0.11 | 1.08 ± 0.08 |
| SKP1A | S-Phase kinase-associated protein 1 | 2.65 ± 0.24 | 1.04 ± 0.01 | 1.18 ± 0.26 |
| TMSB4X* | Thymosin beta 4, X-linked | 2.62 ± 0.12 | 1.19 ± 0.08 | 0.89 ± 0.17 |
| 1200013B08Rik | SAM and SH3 domain containing protein 3 | 2.61 ± 0.21 | 0.99 ± 0.16 | 1.15 ± 0.09 |
| PAIP2B | Poly(A) binding protein interacting protein 2B | 2.61 ± 0.63 | 0.92± 0.07 | 0.97 ± 0.01 |
| FDX1 | Adrenodoxin, mitochondrial | 2.6 ± 0.15 | 1.01 ± 0.11 | n.d. |
| PAIP2* | Poly(A) binding protein interacting protein 2 | 2.57 ± 0.29 | 0.9 ± 0.2 | 1.06 ± 0.14 |
| LARP1* | La ribonucleoprotein domain family, member 1 | 2.57 ± 0.44 | 1.08 ± 0.15 | 1.21 ± 0.26 |
| OXSR1 | Oxidative-stress responsive 1 | 2.52 ± 0.21 | 1.07 ± 0.04 | 1.09 ± 0.11 |
| TPI1 | triosephosphate isomerase 1 | 2.42 ± 0.35 | 1.12 ± 0.1 | 1.00 ± 0.07 |
| TFG* | TRK-fused gene | 2.36 ± 0.5 | 0.93 ± 0.15 | 1.13 ± 0.09 |
| UQCRH | Ubiquinol-cytochrome c reductase hinge protein | 2.27 ± 0.06 | 0.9 ± 0.26 | 1.22 ± 0.17 |
| EFHD2* | EF-hand domain family, member D2 | 2.18 ± 0.12 | 0.99 ± 0.14 | 0.97 ± 0.18 |
n.d., not detected.
Comparisons to previous proteomic27,28 studies revealed that about half of the APEH-regulated proteins have been confirmed to possess acetylated N-termini (asterisked proteins, Table 1). We selected five of these N-terminally acetylated proteins, which showed 2.5 to 4.6-fold changes in N-terminal labeling signals following AA74-1 treatment (Supplementary Fig. 11b), for further characterization. Hexameric peptides that match the N-terminally acetylated sequences for each protein were synthesized and tested as direct substrates with recombinantly expressed APEH. In each case, APEH-transfected cells were found to cleave the N-terminally acetylated residue to a much greater extent than mock-transfected cells and this cleavage was blocked by AA74-1 (Fig. 6b). These data thus provide the first global portrait of endogenous substrates for APEH and suggest that this enzyme plays a broad role in regulating the basal N-terminal acetylation state for many proteins in the proteome.
In the course of characterizing the effects of AA74-1 on APEH function in T-cells, we noted that this inhibitor also cause a significant increase in cellular proliferation (Fig. 6c). This stimulatory effect was not observed with triazole ureas targeting ABHD11 (AA44-2) or PAFAH2 (AA39-2) (Fig. 6c), supporting that it is a specific consequence of APEH blockade and likely related to changes in the enzyme’s N-acetylated substrates (see Discussion below).
DISCUSSION
ABPP has emerged as a versatile chemoproteomic strategy for enzyme characterization in complex biological systems29,30. ABPP probes have been developed for many enzyme classes and used to profile changes in the activity of enzymes in biological systems31–37. Another major utility of ABPP is for inhibitor discovery, where the technology enables compound profiling against many enzymes in parallel directly in complex proteomes38,39. We recently used ABPP to screen more than 70 mammalian SHs against a library of 150+ carbamates6. While this broad screening effort generated lead inhibitors for several SHs, it also underscored the need for new inhibitory chemotypes beyond the carbamate, considering that ~60% of the screened enzymes were unaffected by any of the tested compounds. Here, we show that the 1,2,3-triazole urea constitutes an attractive scaffold for SH inhibitor development that matches and, in some ways, surpasses the carbamate in terms of its breadth of interaction with mammalian SHs and potential for optimization into highly potent and selective inhibitors.
Our functional proteomic studies indicate that monocyclic 1,2,3-triazole ureas may be special among NHU scaffolds in possessing sufficient electrophilicity to react with a large number of SHs, but not excessive reactivity that would otherwise promote interactions with non-SH proteins. That a broad-spectrum 1,2,3-triazole urea AA26-9, which inhibited ~ 1/3 of the 40+ SHs found in T-cells, could be efficiently converted into compounds that potently and selectivity inactivate individual SHs can be attributed, at least in part, to the straightforward synthesis of structurally diverse 1,2,3-triazole ureas by click chemistry methods. We should further mention that most of the enzymes profiled in this study belong to the metabolic SHs, and it is therefore not yet clear whether 1,2,3-triazole ureas will also prove capable of inhibiting trypsin/chymotrypsin proteases. This question should be addressable in future investigations using competitive ABPP and proteomic sources that are more enriched in trypsin/chymotrypsin proteases7,8,31,33,40.
The remarkable ‘ultrapotency’ displayed by optimized 1,2,3-triazole ureas in living systems (sub-nM IC50 values in cells, sub-1 mg/kg potency in mice) was somewhat of a surprise and suggests that these compounds are readily taken up by cells and not susceptible to rapid metabolism. We used one of these agents, the APEH inhibitor AA74-1, to survey the endogenous substrate profile of this peptidase in T-cells and discovered dozens of proteins that show reduced N-terminal labeling signals following APEH inhibition. Considering that the majority of these proteins are known to exist as N-terminally acetylated species27,28, we interpret these results to reflect a constitutive role for APEH in regulating the stability of this modification. The proteomic changes caused by AA74-1 were accompanied by a significant increase in T-cell proliferation. Although we do not yet understand the mechanism underlying the pro-proliferative effect of APEH inhibition, some of the identified substrates for this enzyme, such as the S100A proteins, have been suggested to promote cellular proliferation41, and it is possible that changes in N-acetylation may impact their biological activity. The pro-proliferative effect of APEH inhibition is also noteworthy when considering that the APEH gene is deleted in certain cancers, where it has been proposed to serve as a potential tumor suppressor42,43.
The PAFAH2 and ABHD11 inhibitors developed in this study should also serve as valuable pharmacological tools. PAFAH2 has been shown through knockout mouse studies to play an important role in recovery from hepatic injury44, and it would be interesting to determine whether inhibition of this enzyme phenocopies this effect. ABHD11 is less well-characterized, but is located in a region of chromosome 7 (7q11.23) that is hemizygously deleted in Williams-Beuren syndrome, a rare genetic disease with symptoms that include vascular stenosis, mental retardation, and excessive sociability45. We recently discovered a highly selective carbamate inhibitor of ABHD11 termed WWL22246, and thus, AA44-2 represents a second chemically distinct scaffold suitable for pharmacological investigations of this enzyme. AA44-2 shows greatly improved potency for ABHD11 (IC50 in vitro: 1 nM versus 200 nM for WWL222) and therefore may prove of superior value for some biological studies where, for instance, complete and sustained enzyme blockade is required.
Like carbamates, 1,2,3-triazole ureas inhibit SHs through a covalent, irreversible mechanism involving carbamoylation of the catalytic serine nucleophile. While irreversible inhibitors have historically been viewed as having certain liabilities compared to reversibly acting compounds (e.g., cross-reactivity with other proteins, potential for promoting immune responses to adducted proteins), it is noteworthy that, among the enzymes that are targeted by marketed drugs today, ~30% are inhibited by at least one drug that acts by an irreversible mechanism47. As basic research probes, irreversible inhibitors also offer some tremendous advantages48,49, including a capacity to inactivate enzyme targets for extended periods of time in living systems (i.e. until the enzyme physically turns over). Additionally, assessment of target inhibition in living systems can be easily confirmed ex vivo for irreversible inhibitors by methods such as competitive ABPP. Finally, incorporation of latent affinity handles, such as alkynes or azides, into irreversible inhibitors permits a full assessment of their covalent protein interactions across the proteome9,50.
Projecting forward, it is worth pondering how many SHs might be susceptible to inhibition by 1,2,3-triazole ureas. While the SHs are certainly one of the most diverse enzyme classes in Nature, especially as pertains to substrate recognition, we found that the structurally simple 1,2,3-triazole urea AA26-9 was capable of inhibiting serine peptidases, lipases, amidases, and esterases/thioesterases. This result suggests that most, if not all of the major subgroups of SHs can react with 1,2,3-triazole ureas without requiring specific substrate-like recognition elements. Of course, progressing from reactivity with a broad-spectrum, lead inhibitor like AA26-9 to developing a selective chemical probe may prove challenging for some SH targets, even accepting our success in accomplishing this goal for APEH, ABHD11, and PAFAH2. Here, we expect that the synthetic tractability of 1,2,3-triazole ureas will prove beneficial, since large and structurally diverse libraries bearing this chemotype should be accessible by click chemistry with simple, commercially available starting materials. Finally, the exceptional activity displayed by 1,2,3-triazole ureas in cells and animals suggests that in vitro potency should translate into in vivo activity for most compounds of this class. These features, taken together, bolster our confidence that the 1,2,3-triazole urea will serve as a highly versatile scaffold for the future development of potent and selective inhibitors for the numerous SHs that populate eukaryotic and prokaryotic proteomes.
METHODS
Synthesis of 1,2,3-triazole urea inhibitors
See Supplementary Methods for details.
Gel-based ABPP of cell and tissue proteomes with FP probes
Gel-based ABPP experiments were performed using previously established methods6,7. See Supplementary Methods for details.
Competitive ABPP-SILAC
Isotopically “light” and “heavy” BW5147-derived murine T-cell hybridoma cells mouse T-cells were cultured with inhibitor and DMSO, respectively, for 4 h. Cells were lysed, proteomes were adjusted to a final protein concentration of 1.0 mg/mL and were labeled with 7 μM of FP-biotin (500 μL total reaction volume) for 1.5 hr at 25 °C. After incubation, heavy and light proteomes were mixed in 1:1 ratio. The proteomes were desalted with PD-10 desalting columns (Amersham Biosciences) and FP-labeled proteins were enriched with avidin beads. The beads were washed with 1% SDS in PBS, PBS, and H2O, then resuspended in 6M urea, reduced with DTT, and alkylated with iodoacetamide. On-bead digestions were performed for 12 hr at 37 °C with trypsin (Promega) in the presence of 2 mM CaCl2. Peptide samples were acidified to a final concentration of 5% formic acid, pressure-loaded on to a biphasic (strong cation exchange/reverse phase) capillary column and analyzed by two-dimensional liquid chromatography (2D-LC) separation in combination with tandem mass spectrometry using an Agilent 1100-series quaternary pump and Thermo Scientific LTQ Orbitrap ion trap mass spectrometer. See Supplementary Methods for more detailed description of the ABPP-SILAC experiment.
Fluorogenic APEH activity assay
APEH activity assay was performed using fluorogenic substrate N-Acetyl-L-alanine p-nitroanilide. See Supplementary Methods for details.
APEH activity assay with synthetic N-acetylated hexapeptides
Enzyme assays were performed with LC-MS by monitoring the release of the N-terminal N-acetyl amino acid. APEH was transiently overexpressed in HEK-293 cells and whole cell lysates were treated with DMSO or 3 nM AA74-1 for 30 min and adjusted to a final protein concentration of 0.3 mg/ml with PBS buffer. For each reaction 200 μM of hexapeptide were incubated with lysates for 10 h at 37°C before quenching with 0.8 mL MeOH. Samples were centrifuged for 10 min at 10,000 × g at 4 °C and the supernatant was dried down under vacuum. For LC-MS analysis samples were injected into an Agilent 1100 LC-MSD SL instrument applying the following gradient: Buffer A (MeCN) to Buffer B (95:5 H2O/MeCN + 50 mM NH4OAc + 0.2% NH4OH). Data represent the average±s.d. for three experiments. See Supplementary Methods for more detailed description of the substrate assay.
Supplementary Material
Acknowledgments
We would like to thank Jeffrey Savas and Daniel McClatchy for generously providing 15N-labeled mice and Tianyang Ji for technical assistance. This work was supported by the NIH (CA132630, DA028845, MH084512, CA151460 (K99 award to B.R.M.)), the Deutscher Akademischer Austausch Dienst (Postdoctoral Fellowship to A.A.), the National Science Foundation (Predoctoral Fellowship to D.A.B.), and the Skaggs Institute for Chemical Biology.
Footnotes
AUTHOR CONTRIBUTIONS
A.A. and B.F.C. designed the experiments; A.A., B.R.M., and K.L.H. performed the experiments; A.A., D.A.B., S.N., and H.H. contributed new reagents/analytic tools; A.A., C.W., and B.F.C. analyzed data; and A.A. and B.F.C. wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J Biol Chem. 2010;285:11051–5. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henness S, Perry CM. Orlistat: a review of its use in the management of obesity. Drugs. 2006;66:1625–56. doi: 10.2165/00003495-200666120-00012. [DOI] [PubMed] [Google Scholar]
- 3.Thornberry NA, Weber AE. Discovery of JANUVIA (Sitagliptin), a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. Curr Top Med Chem. 2007;7:557–68. doi: 10.2174/156802607780091028. [DOI] [PubMed] [Google Scholar]
- 4.Kluge AF, Petter RC. Acylating drugs: redesigning natural covalent inhibitors. Curr Opin Chem Biol. 2010;14:421–7. doi: 10.1016/j.cbpa.2010.03.035. [DOI] [PubMed] [Google Scholar]
- 5.Birks J, Grimley Evans J, Iakovidou V, Tsolaki M, Holt FE. Rivastigmine for Alzheimer’s disease. Cochrane Database Syst Rev. 2009:CD001191. doi: 10.1002/14651858.CD001191. [DOI] [PubMed] [Google Scholar]
- 6.Bachovchin DA, et al. A superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc Natl Acad Sci U S A. 2010;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jessani N, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–7. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 8.Okerberg ES, et al. High-resolution functional proteomics by active-site peptide profiling. Proc Natl Acad Sci U S A. 2005;102:4996–5001. doi: 10.1073/pnas.0501205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexander JP, Cravatt BF. Mechanism of carbamate inactivation of FAAH: implications for the design of covalent inhibitors and in vivo functional probes for enzymes. Chem Biol. 2005;12:1179–87. doi: 10.1016/j.chembiol.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci USA. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteome using fluorescent active site-directed probes. Proteomics. 2001;1:1067–1071. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 12.Kathuria S, et al. Modulation of anxiety through blockade of anandamide hydrolysis. Nat Med. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- 13.Long JZ, et al. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13:1041–50. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Moore SA, et al. Identification of a high-affinity binding site involved in the transport of endocannabinoids. Proc Natl Acad Sci U S A. 2005;102:17852–7. doi: 10.1073/pnas.0507470102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander JP, Cravatt BF. The putative endocannabinoid transport blocker LY2183240 is a potent inhibitor of FAAH and several other brain serine hydrolases. J Am Chem Soc. 2006;128:9699–704. doi: 10.1021/ja062999h. [DOI] [PubMed] [Google Scholar]
- 17.Lowe DB, et al. In vitro SAR of (5-(2H)-isoxazolonyl) ureas, potent inhibitors of hormone-sensitive lipase. Bioorg Med Chem Lett. 2004;14:3155–9. doi: 10.1016/j.bmcl.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Ebdrup S, Sorensen LG, Olsen OH, Jacobsen P. Synthesis and structure-activity relationship for a novel class of potent and selective carbamoyl-triazole based inhibitors of hormone sensitive lipase. J Med Chem. 2004;47:400–10. doi: 10.1021/jm031004s. [DOI] [PubMed] [Google Scholar]
- 19.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Long JZ, et al. Dual blockade of FAAH and MAGL identifies behavioral processes regulated by endocannabinoid crosstalk in vivo. Proc Natl Acad Sci U S A. 2009;106:20270–5. doi: 10.1073/pnas.0909411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–8. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 22.Everley PA, et al. Assessing enzyme activities using stable isotope labeling and mass spectrometry. Mol Cell Proteomics. 2007;6:1771–7. doi: 10.1074/mcp.M700057-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Ong SE, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci U S A. 2009;106:4617–22. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR., 3rd Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem. 2004;76:4951–9. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 25.McClatchy DB, Liao L, Park SK, Venable JD, Yates JR. Quantification of the synaptosomal proteome of the rat cerebellum during post-natal development. Genome Res. 2007;17:1378–88. doi: 10.1101/gr.6375007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timmer JC, et al. Profiling constitutive proteolytic events in vivo. Biochem J. 2007;407:41–8. doi: 10.1042/BJ20070775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orre LM, Pernemalm M, Lengqvist J, Lewensohn R, Lehtio J. Up-regulation, modification, and translocation of S100A6 induced by exposure to ionizing radiation revealed by proteomics profiling. Mol Cell Proteomics. 2007;6:2122–31. doi: 10.1074/mcp.M700202-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Helbig AO, et al. Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome. Mol Cell Proteomics. 9:928–39. doi: 10.1074/mcp.M900463-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cravatt BF, Wright AT, Kozarich JW. Activity-Based Protein Profiling: From Enzyme Chemistry to Proteomic Chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 30.Berger AB, Vitorino PM, Bogyo M. Activity-based protein profiling: applications to biomarker discovery, in vivo imaging and drug discovery. Am J Pharmacogenomics. 2004;4:371–81. doi: 10.2165/00129785-200404060-00004. [DOI] [PubMed] [Google Scholar]
- 31.Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer invasiveness. Proc Natl Acad Sci USA. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joyce JA, et al. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–53. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 33.Nomura DK, et al. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;14:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barglow KT, Cravatt BF. Discovering disease-associated enzymes by proteome reactivity profiling. Chem Biol. 2004;11:1523–31. doi: 10.1016/j.chembiol.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 35.Greenbaum DC, et al. A role for the protease falcipain 1 in host cell invasion by the human malaria parasite. Science. 2002;298:2002–6. doi: 10.1126/science.1077426. [DOI] [PubMed] [Google Scholar]
- 36.Blais DR, et al. Activity-based protein profiling identifies a host enzyme, carboxylesterase 1, which is differentially active during hepatitis C virus replication. J Biol Chem. 2010;285:25602–12. doi: 10.1074/jbc.M110.135483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaschani F, et al. Diversity of serine hydrolase activities of unchallenged and botrytis-infected Arabidopsis thaliana. Mol Cell Proteomics. 2009;8:1082–93. doi: 10.1074/mcp.M800494-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry. 2001;40:4005–4015. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- 39.Greenbaum D, et al. Chemical approaches for functionally probing the proteome. Mol Cell Proteomics. 2002;1:60–68. doi: 10.1074/mcp.t100003-mcp200. [DOI] [PubMed] [Google Scholar]
- 40.Jessani N, et al. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci U S A. 2004;101:13756–61. doi: 10.1073/pnas.0404727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazzucchelli L. Protein S100A4: too long overlooked by pathologists? Am J Pathol. 2002;160:7–13. doi: 10.1016/S0002-9440(10)64342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erlandsson R, et al. The gene from the short arm of chromosome 3, at D3F15S2, frequently deleted in renal cell carcinoma, encodes acylpeptide hydrolase. Oncogene. 1991;6:1293–5. [PubMed] [Google Scholar]
- 43.Scaloni A, et al. Deficiency of acylpeptide hydrolase in small-cell lung carcinoma cell lines. J Lab Clin Med. 1992;120:546–52. [PubMed] [Google Scholar]
- 44.Kono N, et al. Protection against oxidative stress-induced hepatic injury by intracellular type II platelet-activating factor acetylhydrolase by metabolism of oxidized phospholipids in vivo. J Biol Chem. 2008;283:1628–36. doi: 10.1074/jbc.M708622200. [DOI] [PubMed] [Google Scholar]
- 45.Schubert C. The genomic basis of the Williams-Beuren syndrome. Cell Mol Life Sci. 2009;66:1178–97. doi: 10.1007/s00018-008-8401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bachovchin DA, et al. A superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1011663107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robertson JG. Mechanistic basis of enzyme-targeted drugs. Biochemistry. 2005;44:8918. doi: 10.1021/bi050247e. [DOI] [PubMed] [Google Scholar]
- 48.Kodadek T. Rethinking screening. Nat Chem Biol. 2010;6:162–165. doi: 10.1038/nchembio.303. [DOI] [PubMed] [Google Scholar]
- 49.Johnson DS, Weerapana E, Cravatt BF. Strategies for discovering and derisking covalent, irreversible enzyme inhibitors. Future Med Chem. 2010;2:949–964. doi: 10.4155/fmc.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen MS, Hadjivassiliou H, Taunton J. A clickable inhibitor reveals context-dependent autoactivation of p90 RSK. Nat Chem Biol. 2007;3:156–160. doi: 10.1038/nchembio859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Amer Chem Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.