Summary
The maturation of animal oocytes is highly sensitive to nutrient availability. During Drosophila oogenesis, a prominent metabolic checkpoint occurs at the onset of yolk uptake (vitellogenesis): under nutrient stress, egg chambers degenerate by apoptosis. To investigate additional responses to nutrient deprivation, we studied the intercellular transport of cytoplasmic components between nurse cells and the oocyte during previtellogenic stages. Using GFP protein-traps, we showed that Ypsilon Schachtel (Yps), a putative RNA binding protein, moved into the oocyte by both microtubule (MT)-dependent and -independent mechanisms, and was retained in the oocyte in a MT-dependent manner. These data suggest that oocyte enrichment is accomplished by a combination of MT-dependent polarized transport and MT-independent flow coupled with MT-dependent trapping within the oocyte. Under nutrient stress, Yps and other components of the oskar ribonucleoprotein complex accumulated in large processing bodies in nurse cells, accompanied by MT reorganization. This response was detected as early as 2 hrs after starvation, suggesting that young egg chambers rapidly respond to nutrient stress. Moreover, both Yps aggregation and MT reorganization were reversed with re-feeding of females or the addition of exogenous insulin to cultured egg chambers. Our results suggest that egg chambers rapidly mount a stress response by altering intercellular transport upon starvation. This response implies a mechanism for preserving young egg chambers so that egg production can rapidly resume when nutrient availability improves.
Keywords: Drosophila oogenesis, polarized transport, oskar RNP, starvation response, microtubule
Introduction
Female gamete development, or oogenesis, is closely linked to female physiology. Females devote significant energy and nutrition to egg production. To allocate nutrition resources between self-survival and production of progeny, the genetic program of oogenesis must be responsive to environmental stresses such as nutrient availability.
In Drosophila, a well-nourished female produces up to 60 eggs per day, which corresponds to about 40% of female body weight (King, 1970). To accomplish this remarkable level of fecundity, egg development is delegated to about 30 assembly lines called ovarioles contained within two ovaries. At the anterior tips of ovarioles, called germaria, a germ-line stem cell daughter called a cystoblast undergoes four rounds of mitosis, giving rise to one oocyte and 15 nurse cells. Cytokinesis is incomplete in all four mitoses, resulting in a cyst of 16 cells that are interconnected through cytoplasmic bridges called ring canals. Once packaged into the stage-1 egg chamber surrounded by somatic follicle cells, egg chambers exit the germarium and gradually move posteriorly within the ovariole as they grow (Fig. 1A; Ashburner, 1989; Spradling, 1993).
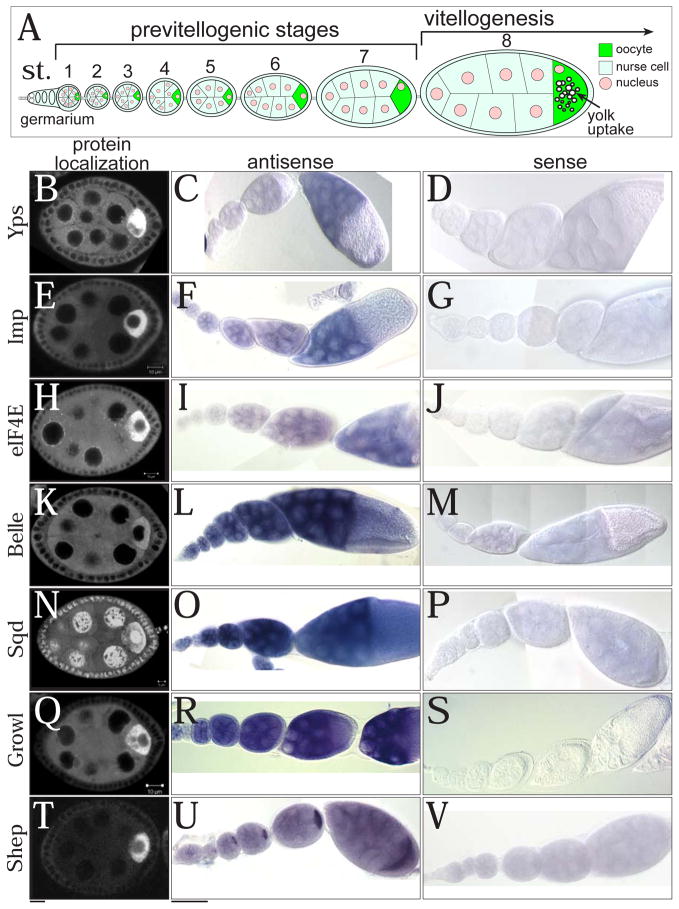
Fig. 1. The expression patterns of oocyte-enriched proteins.
(A) A schematic drawing of an ovariole from germarium to stage 8. Vitellogenesis (yolk uptake) starts at stage 8. GFP distributions from protein-trap lines (B, E, H, K, N, Q, and T) and mRNA localizations of GFP-trapped genes detected by RNA in situ hybridization (antisense probes: C, F, I, L, O, R, and U; sense probes: D, G, J, M, P, S, and V). (B–D) Ypsilon schachtel (Yps), (E–G) IGF-II mRNA binding protein (Imp), and (H–J) Eukaryotic initiation factor 4E (eIF4E), (K–M) Belle (Bel), (N–P) Squid (Sqd), (Q–S) Growl, and (T–V) Alan Shepard (Shep). Bars: (B, E, H, K, N, Q, and T) 10 μm; (C, D, F, G, I, J, L, M, O, P, R, S, U, and V) 50 μm.
Because the oocyte nucleus is transcriptionally inactive (King and Burnett, 1959), nurse cells supply the oocyte with maternal nutrients and cytoplasmic components by intercellular transport through ring canals. During stages 1–6, some of this nurse cell-to-oocyte transport depends on polarized microtubules (MTs) that have minus ends located in the oocyte and plus ends extending through ring canals into nurse cells (Koch and Spitzer, 1983; Theurkauf et al., 1993). For example, key maternal RNAs involved in embryonic axis patterning such as oskar (osk), bicoid (bcd), and gurken (grk) are synthesized in nurse cells and specifically transported to the oocyte. The minus-end directed motor Dynein and its adaptor proteins Egalitarian (Egl) and BicaudalD (BicD) mediate this transport (Bullock and Ish-Horowicz, 2001; Cha et al., 2001; Clark et al., 2007; Mach and Lehmann, 1997; Navarro et al., 2004; Pokrywka and Stephenson, 1991; Pokrywka and Stephenson, 1995; Theurkauf et al., 1993; Theurkauf et al., 1992). Curiously, though, the polarized MT cytoskeleton is disassembled in nurse cells after stage 6 and mostly reorganized to the oocyte during stages 7–9 (Steinhauer and Kalderon, 2006; Theurkauf et al., 1992). Previous live imaging studies reveal transport of particles in egg chambers during stages 7–9, but did not focus on specific intercellular transport during stages 1–6, when the polarized MT cytoskeleton is prominent between nurse cells and the oocyte. Moreover, less attention has been focused on the transport of proteins and organelles that support oocyte growth and metabolism during oogenesis.
Female reproduction is optimized for high fecundity under ideal conditions, but is also regulated in response to the nonoptimal conditions that flies can encounter in the wild. When females are deprived of yeast as the protein source, they protect themselves and their progeny from starvation by halting egg production. This is accomplished by apoptotic destruction of immature germline cells either before packaging into egg chambers in the germarium (region 2), or at the onset of vitellogenesis (stage 8) (Drummond-Barbosa and Spradling, 2001; Mazzalupo and Cooley, 2006; Pritchett et al., 2009). In addition, reduced stem cell proliferation slows the rate of egg chamber formation (Drummond-Barbosa and Spradling, 2001; Mazzalupo and Cooley, 2006; Pritchett et al., 2009). Importantly, previtellogenic egg chambers (stages 2–7) are preserved during starvation, although their growth rate is reduced (Drummond-Barbosa and Spradling, 2001). The cellular mechanisms induced in response to nutrient stress that slow, but do not destroy egg chambers are poorly understood.
In light of the importance of previtellogenic egg chamber development, we set out to investigate how intercellular protein movements are carried out and regulated between nurse cells and the oocyte. By examining a collection of GFP protein-trap lines (Kelso et al., 2004; Quiñones-Coello et al., 2007), we found evidence for the targeting of functionally related proteins to the oocyte. Surprisingly, we also found that in addition to MT-mediated polarized transport, the enrichment of RNA-binding proteins in the oocyte is mediated by extensive diffusion and MT-dependent trapping in the oocyte. Furthermore, we observed that previtellogenic egg chambers are exquisitely sensitive to nutrient stress, and respond by changing osk ribonucleoprotein (RNP) complex localization and reorganizing the MT cytoskeleton. These results indicate coordinated, and probably conserved, mechanisms to preserve egg chambers during starvation so that development can be rapidly resumed when nutrient conditions improve.
Material and methods
Fly strains
Drosophila cultures were maintained using standard procedures (Ashburner, 1989). w1118 was used as a wild-type control. The following protein-trap lines were used: ZCL1607/TM3 (GFP::Yps); ZCL1503 (GFP::Pdi); G00149 (GFP::Shep); G0080 (GFP::Imp); ZCL3169 (GFP::Growl); ZCL0734 (GFP::Sqd); YC0001 (GFP::eIF4E); YB0151 (GFP::RpL30). These lines are available at Flytrap (http://flytrap.med.yale.edu/). For germline-specific expression, matalpha4-GAL-VP16 (BL7063) or the “MTD” line expressing triple GAL4 drivers (pCOG-GAL4:VP16; NGT40; nos-GAL4:VP16) was used (Petrella et al., 2007). pUASp-GFP::tubulin (Grieder et al., 2000); pUASp-EB1::GFP (a gift from Dr. William Theurkauf); oskMS2, MS2-GFP/TM3 (Zimyanin et al., 2008); pUASp-GFP::LC3 (Rusten et al., 2004).
Construction of transgenes and generation of transgenic lines
A cDNA clone of yps (LD37574) was obtained from the Drosophila Genomics Resource Center (DGRC, Bloomington, IN). The full length or truncated forms of Yps were fused to mRFP and cloned into pUASp vector (see details in the legend of Fig. S1). A cDNA clone of Dendra2 was obtained from EUROGEN. Drosophila enbryo injection was performed by BestGene Inc. As YpsN::mRFP gave brighter fluorescence than YpsF::mRFP and localized to the oocyte similarly to YpsF::mRFP (Fig. S1), we used pUASp-YpsN::mRFP for live imaging.
RNA in situ hybridization
Whole mount in situ hybridization with ovaries was performed as followed by a standard protocol (Lehmann and Tautz, 1994). RNA probes were synthesized with a DIG-labeling kit (Roche). For the color reaction, we compared sample coloration with antisense probes and sense probes side by side.
Culture of previtellogenic egg chambers for live imaging
We followed a protocol from D. Montell’s lab with several modifications (Prasad et al., 2007). Young adult females (up to 3 days old) were fed on regular fly food with yeast paste for 2 days. For starvation experiments, females were transferred to a vial containing grape juice agar with or without yeast paste (the protein-rich/poor conditions). When dissected, ovarioles were carefully individually teased apart in Schneider’s Drosophila Medium (SDM) containing 10 % Fetal Bovine Serum plus 5 % Penicillin and Streptomycin and transferred to a glass-bottom culture dish (MatTek). For long-term imaging over 1 hr or recovery experiments, egg chambers were incubated in the medium with 0.2 mg/ml insulin. After dissection, FM4-64 (5 μg/ml, Molecular Probes) or colchicine (50 μg/ml, Sigma) was added. A cover glass was mounted to avoid desiccation of culture medium. Images were taken on a Zeiss LSM-510 META microscope or on a ZEISS Axiovert 200 equipped with a CARV II confocal imager (BD Bioscience) and CoolSnap HQ2 camera (Roper Scientific). Images were processed using ImageJ.
Fluorescence Loss In Photobleaching (FLIP)
FLIP was performed by photobleaching a region of interest (ROI; oocyte or nurse cells) and measuring the extent of fluorescence loss over time. The prebleach fluorescence intensity was designated as 100%, and the ratios of other intensities were plotted. See details in the legend of Fig. S2.
Immunohistochemistry
Ovaries from females deprived of yeast for 48 hrs were dissected and fixed with 4% paraformaldehyde for 20 min. Reagents used were anti-Dcp1 (1:20; Lin et al., 2006), anti-Pcm (1:500; Lin et al., 2008); anti-eIF4E (1:1000; Nakamura et al., 2004), anti-Stau (1:2000; St Johnston et al., 1991). For LysoTracker (Molecular Probes), ovaries were treated with 5 μM Lysotracker in SDM for 5 min.
Quantitative analysis
To measure the volume of GFP::Yps particles in 3-D, we used Imaris software (Bitplane, see Movie S3). To analyze MT organization, we examined the profiles of fluorescence intensity of GFP::tubulin with ImageJ (NIH). The position of membrane was normalized with FM4-64. To analyze the temporal profile of GFP::Yps aggregate formation, we extracted the pixel areas of GFP::Yps particles with brighter fluorescence than background. Pixel areas occupied in nurse cell cytoplasm areas were calculated. The area of nurse cell nuclei was subtracted from nurse cell area.
Results
The specificity of protein localization during previtellogenic stages
Efforts to produce fly lines that express GFP-fusion proteins provided new opportunities to examine the distribution of proteins in egg chambers (Kelso et al., 2004; Morin et al., 2001; Quiñones-Coello et al., 2007). Using a collection of GFP protein-trap lines, we classified proteins based on their localizations in egg chambers. Among 163 genes we examined, 48 produced proteins in germline cytoplasm. The remaining genes were not expressed in germline cytoplasm, although many produced germline nuclear proteins or follicle cell proteins. Of 48 germline proteins, 19 (40%) displayed oocyte enrichment during some stage of oogenesis. Strikingly, the oocyte-enriched proteins included nearly all of the RNA binding proteins and endoplasmic reticulum (ER) proteins in the collection (Table S1). In contrast, metabolic proteins, ribosomal proteins, and translation and protein folding factors were ubiquitously distributed in germline cytoplasm. These data provide evidence for active sorting of proteins between nurse cells and the oocyte based on their function. Cellular components needed for biosynthetic activity were ubiquitous in germline, and components needed for oocyte growth or patterning were enriched in the oocyte.
We focused on the RNA binding proteins to study how their oocyte enrichment is achieved. One mechanism is to localize and translate mRNAs encoding those proteins in the oocyte. Indeed, 10% of randomly chosen mRNAs were localized to the oocyte (Dubowy and Macdonald, 1998), and the oocyte enrichment of specific mRNAs has been documented including osk, bcd, grk, oo18 RNA-binding protein (orb), egl, and BicD (Costa et al., 2005; Mach and Lehmann, 1997; Suter and Steward, 1991). However, RNA in situ hybridization for ypsilon schachtel (yps), IGF-II mRNA-binding protein (Imp), eukaryotic initiation factor 4E (eIF4E), belle (bel), squid (sqd), growl, and alan shepard (shep) revealed that only shep mRNA was highly enriched in the oocyte, while the other mRNAs were distributed throughout germline cytoplasm (Fig. 1B–V). The yps expression pattern was consistent with a previous report (Mansfield et al., 2002), and similar patterns were reported for Dynein heavy chain (Dhc) and exuperantia (exu) mRNAs, whose proteins are abundant in the oocyte (Li et al., 1994; Macdonald et al., 1991). To identify a protein domain of Yps necessary for oocyte targeting, we created a series of transgenes expressing Yps fragments with the K10 3′UTR, which does not contain localization signals (Fig. S1). We found that full-length (YpsF) and the N-terminal half of Yps (YpsN) were enriched in the oocyte, and localized to the posterior pole of the oocyte at stage 10. This result strengthens the hypothesis that Yps protein, but not yps mRNA is transported to the oocyte.
Live imaging of young egg chambers detects few particle movements into the oocyte
Based on these results, we hypothesized that selected proteins are translated in nurse cells and specifically targeted to the oocyte during previtellogenic stages. To define the characteristics of intercellular transport between nurse cells and the oocyte, we devised a protocol of live imaging with previtellogenic egg chambers. While many live imaging studies have been carried out using vitellogenic egg chambers (stage 8 and older) (Bohrmann and Biber, 1994; Boylan et al., 2008; Cha et al., 2001; Clark et al., 2007; Forrest and Gavis, 2003; Mische et al., 2007; Nicolas et al., 2009; Prasad and Montell, 2007; Theurkauf and Hazelrigg, 1998; Wang and Hazelrigg, 1994; Weil et al., 2006; Zimyanin et al., 2008), fewer studies used previtellogenic egg chambers (stage 2–7) (Bohrmann and Biber, 1994; Clark et al., 2007). As previtellogenic stages are difficult to distinguish in living egg chambers, we measured the size of egg chambers to determine their stages (Fig. 1A and Table S2), similar to previous reports (Lin and Spradling, 1993; Spradling, 1993). To keep young egg chambers healthy in culture medium, we added Bovine insulin to Schneider’s Drosophila medium (Figs. 2A–C; Prasad et al., 2007). Without insulin, the shape of egg chambers became distorted and the oocyte area did not grow (Fig. 2C).
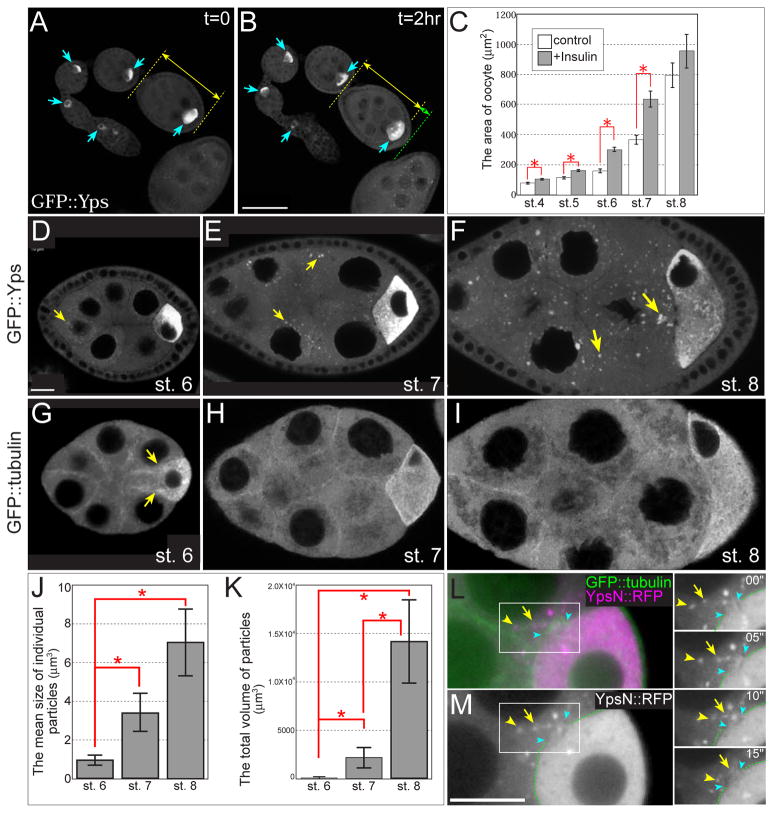
Fig. 2. Live imaging of young egg chambers in early oogenesis.
(A and B) An ovariole expressing GFP::Yps was cultured in Schneider’s Drosophila medium containing 0.2 mg/ml insulin. GFP::Yps was enriched in the oocyte during stages 1–6 (blue arrows). A double-headed yellow arrow indicates the length of a stage-5 egg chamber at t = 0 (A), and a yellow arrow plus a green arrow indicate its length after 2 hrs (B) (C) The areas of oocytes after 2 hrs of incubation with or without insulin. The oocyte area expanded more in egg chambers cultured in insulin-containing medium (gray bars) than in control medium (white bars). Asterisks show statistical significance (t-test, p<0.05). 10–20 egg chambers were analyzed in each stage from 3 independent experiments. (D–F) GFP::Yps particles form in nurse cells during stages 6–8. Few particles were detected at stage 6 (D, a yellow arrow), and then more were detected at stage 7 (E, yellow arrows) and stage 8 (F, yellow arrows). (G–I) The distribution of microtubules (MTs) was visualized with GFP::tubulin. MT bundles between nurse cells and the oocyte were most prominent at stage 6 (yellow arrows in G) and were less detected at stage 7 (H). At stage 8, most MTs were reorganized to the oocyte and long MT tracks were almost lost between nurse cells and the oocyte (I). (J and K) The mean size of individual particles and the total volume of particles increased during stages 6–8. Asterisks show statistical significance (t-test, p<0.05). 10 egg chambers were analyzed in each stage. (L and M) Overexpression of N-terminal half of Yps fused to mRFP (YpsN::mRFP, magenta) showed Yps-containing particles associated with MTs extending the ring canal during stage 6. Full-length Yps behaved similarly (data not shown). MTs were labeled with GFP::tubulin (green). Blue arrowheads indicate the rim of ring canal between a nurse cell and the oocyte. 4 time-lapse images taken at 5-second intervals are shown to the right. Arrowheads and arrows indicate individual particles that were moving along the MT toward the oocyte. See also Movie S1. Bars, 10 μm.
We used ovaries from females expressing GFP::Yps, in which the GFP-coding exon is inserted in the first intron of the yps gene. The distribution of GFP::Yps was almost identical to that of endogenous Yps (Wilhelm et al., 2000), indicating the presence of GFP did not affect the Yps localization pattern. Considering Yps is a component of osk RNP complex and Yps localization is closely related to its function, we assume that GFP::Yps mimics endogenous Yps not only in localization but also in function. During stage 6, GFP signals were highly enriched in the oocyte and evenly distributed throughout nurse cell cytoplasm with a few perinuclear particle-like signals (Fig. 2D) that exhibited very little movement toward the oocyte (data not shown). This observation was consistent with previous studies reporting that few particles were detected during stage 6 (Bohrmann and Biber, 1994; Wang and Hazelrigg, 1994). Particle-like signals gradually increased during stages 7 and 8 (Figs. 2E and F). The mean size of individual particles and the total volume of particles significantly increased over time (Figs. 2J and K). To compare the distribution of GFP::Yps with that of MTs, we used flies expressing GFP::tubulin. Arrays of MTs were detected during stages 1–6, with MTs abundant in the oocyte and in ring canals between nurse cells and the oocyte as previously described (Fig. 2G; Theurkauf et al., 1992). After stage 7, MT bundles were reorganized to the oocyte and became less obvious between nurse cells and the oocyte (Figs. 2H and I). Therefore, we confirmed that a polarized MT cytoskeleton is prominent at stage 6; however, since few GFP::Yps particles were present at this stage and the particle-to-dispersed signal ratio was low, it was difficult to track particle movement along these MTs. We could more easily follow overexpressed Yps fused to monomeric Red Fluorescent protein (mRFP) using the GAL4/UASp system. Several Yps particles moved in linear tracks toward the oocyte along MTs during stage 6 (Figs. 2L, M and Movie S1). This result demonstrates that Yps-containing particles are transported to the oocyte along polarized MTs during stage 6. However, it was not possible to follow the entire Yps population by particle tracking.
MT-dependent and -independent protein movements between nurse cells and the oocyte
As an alternative approach to monitor protein movements during stage 6, we developed a Fluorescence Loss In Photobleaching (FLIP) assay to evaluate overall cytoplasmic flow between nurse cells and the oocyte. GFP-fusion proteins were photobleached repeatedly in either the oocyte or a nurse cell adjacent to the oocyte, and images were recorded after each photobleaching (Figs. 3B and S2). We measured the level of fluorescence over time in the photobleached cell, neighboring cells connected by ring canals, distant cells at the anterior end of the egg chamber, and cells in an adjacent egg chamber to control for general photobleaching associated with imaging. FLIP experiments on egg chambers expressing Dendra2, which encodes an exogenous fluorescent protein (Gurskaya et al., 2006), revealed extensive bidirectional movement between nurse cells and the oocyte (Figs. 3Ba–c and S2 a–b). Photobleaching Dendra2 in the oocyte led to loss of fluorescence in the neighboring nurse cells (Figs. 3Bb and S2 a). The amount of fluorescence loss was greater in the nurse cells directly adjacent to the oocyte compared to a nurse cell at the anterior of the egg chamber. In contrast, when a nurse cell next to the oocyte was bleached, fluorescence was lost in both the oocyte and the neighboring nurse cell (Figs. 3Bc and S2 b). These data demonstrate that there is no physical barrier to diffusion through ring canals during stage 6.
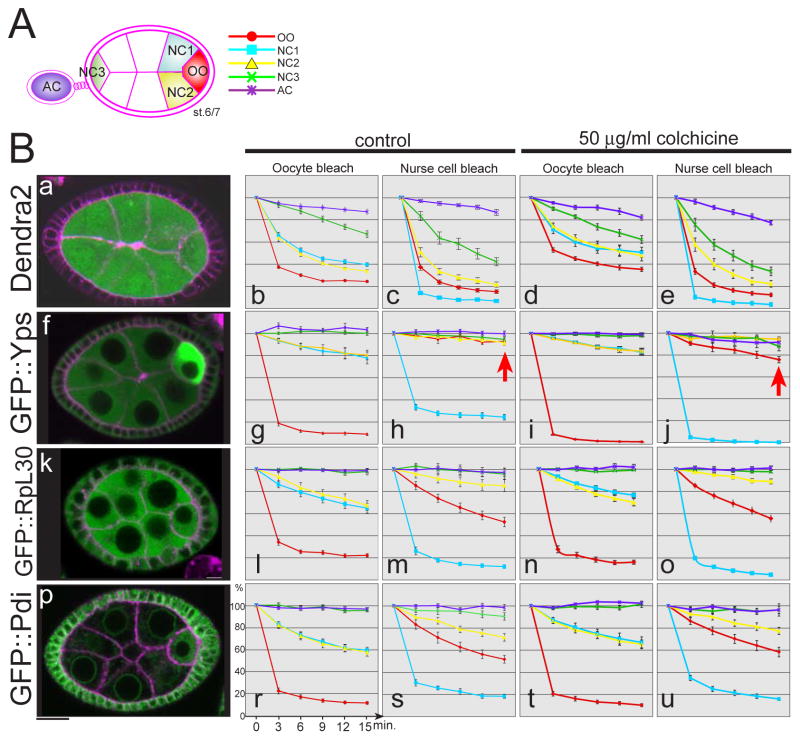
Fig. 3. FLIP analysis reveals intercellular cytoplasmic movements between nurse cells and the oocyte.
A schematic drawing of egg chambers with the regions of interest color coded: oocyte (red), neighboring nurse cells (blue and yellow), anterior nurse cell (green), and adjacent egg chamber (purple). Each color corresponds to lines of graphs in B. (B) Fluorescence loss in photobleaching (FLIP) analysis was performed with egg chambers expressing Dendra2 and GFP protein-trap lines. The level of fluorescence intensities (the Y-axis) after each photobleach was measured over time (the X-axis) with or without the MT depolymerizing drug, colchicine. The photobleach by laser scanning was performed every 3 minutes. (a–e) Dendra2, (f–j) GFP::Yps, (k–o) GFP::RpL30, (p–u) GFP::Pdi. Representative images before photobleach are shown in the left column. Membrane was labeled with FM4-64 (magenta). Each graph was obtained with 10 egg chambers at stage 6/7 from 3 independent experiments. Bar, 10 μm.
We performed the same analysis on several GFP-fusion proteins to compare the behaviors of representative proteins of the functional groups described above: GFP::Yps (RNA-binding protein), GFP::Protein disulfide isomerase (Pdi, ER protein), and GFP::Ribosomal protein L30 (RpL30, ribosomal protein). GFP::Yps and GFP::Pdi were enriched in the oocyte, while GFP::RpL30 was ubiquitously distributed in germline cytoplasm. Photobleaching of GFP::Yps, GFP::Pdi, or GFP::RpL30 in the oocyte resulted in a reproducible loss of fluorescence in adjacent nurse cells (Figs. 3Bg, l, and r), providing evidence that these proteins moved from nurse cells to the oocyte during stage 6. As the rate of fluorescence loss was more rapid for Dendra2 than other proteins, the movements of endogenous Drosophila proteins appeared restricted compared to Dendra2.
To assess movements out of the oocyte, we measured fluorescence loss in the oocyte after photobleaching an adjacent nurse cell. The fluorescence of GFP::Pdi or GFP::RpL30 in the oocyte decreased, suggesting that these proteins move from the oocyte to nurse cells (Figs. 3Bm and s). In sharp contrast, GFP::Yps fluorescence did not decrease in the oocyte (Fig. 3Bh, arrow). These results suggest the direction of Yps movement is specifically controlled; it can move into the oocyte, but not out. This is consistent with our live imaging analysis, showing polarized transport into the oocyte.
To examine whether these protein movements depend on MTs, we treated egg chambers with colchicine, an inhibitor of MT polymerization (Koch and Spitzer, 1983; Theurkauf et al., 1993; Wang and Hazelrigg, 1994) The effect of colchicine on MTs was confirmed by examining GFP::tubulin (Figs. 4E and H). With colchicine treatment, GFP::Yps was mislocalized to nurse cells and made large aggregates in cytoplasm (Figs. 4B and F, yellow arrows). This could be due to halting MT-dependent polarized transport and subsequent buildup of GFP::Yps in nurse cells. However, oocyte-bleaching experiments revealed only a small reduction in nurse cell-to-oocyte movement of GFP::Yps (Fig. 3Bi), suggesting that in addition to polarized transport along MTs, Yps can move to the oocyte in a MT-independent manner. This idea is consistent with previous studies reporting that transport through ring canals between nurse cells and the oocyte is MT-independent (Bohrmann and Biber, 1994; Theurkauf and Hazelrigg, 1998). Moreover, we found that the bidirectional movements of Dendra2, GFP::RpL30, and GFP::Pdi were hardly affected by colchicine treatment (Figs. 3Bd, e, n, o, t, and u). Together, these results reveal extensive diffusion of proteins through ring canals between the cells.
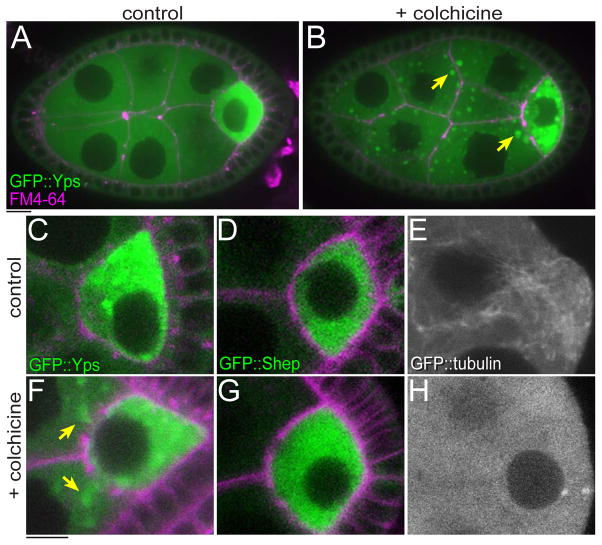
Fig. 4. The localization of GFP::Yps, but not GFP::Shep was affected by colchicine treatment.
Stage-6 egg chambers were incubated in culture medium without (control: A, C–E) or with 50 μg/ml colchicine (B, F–H) for 1 hr. GFP::Yps was enriched in the oocyte (A and C). When MTs were disrupted, GFP::Yps mislocalized in nurse cells and made large aggregates in cytoplasm (arrows in B and F). See also Movie S2. In contrast, the distribution of GFP::Shep was not detectably affected by colchicine (D and G). (E and H) Control and disrupted MTs were visualized by GFP::tubulin. Bars, 10 μm.
Interestingly, we found that GFP::Yps could move from the oocyte to nurse cells in the presence of colchicine (Fig. 3Bj, arrow). Time-lapse imaging confirmed that GFP::Yps flowed out of the oocyte into an adjacent nurse cell (Movie S2). Taken together, we conclude that MTs are required not only for polarized transport of a subset of Yps, but also to maintain Yps in the oocyte during stage 6. In contrast to Yps, the localization of GFP::Shep was not affected by colchicine (Figs. 4D and G). Since shep mRNA was enriched in the oocyte (Fig. 1U), GFP::Shep is likely translated mainly in the oocyte and then retained in the oocyte in a MT-independent manner. Thus, the enrichment of maternal factors in the oocyte is carried by both MT-dependent and -independent mechanisms.
Young egg chambers respond to protein deprivation by altering protein localization
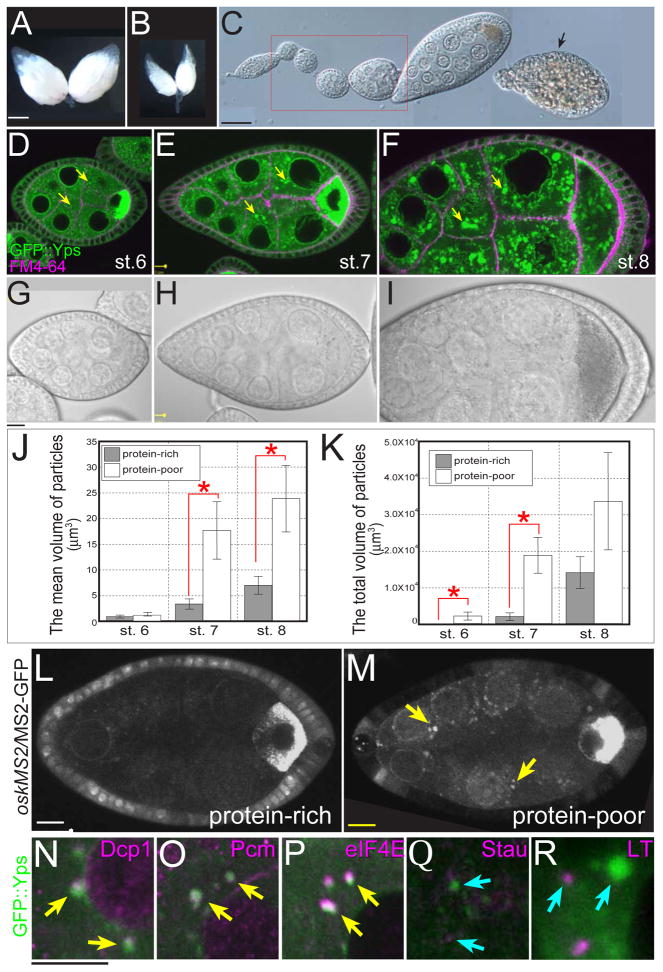
During the course of our live imaging experiments, we noticed that the distribution of GFP::Yps was sensitive to culture conditions and the age of females (Y.S. and L.C., unpublished data). These observations prompted us to ask if Yps localization was affected by nutrient stress. In the protein-rich environment of standard fly food, ovaries contained many eggs (Fig. 5A). However, when yeast was removed as the protein source, ovary size was dramatically decreased (Fig. 5B). In this protein-poor (“starved”) condition, egg chambers degenerated by apoptosis after stage 8 (Fig. 5C, arrow; Mazzalupo and Cooley, 2006; Pritchett et al., 2009), thus avoiding the metabolic cost of depositing yolk protein in eggs. In contrast, previtellogenic egg chambers (stages 2–7) remained intact with normal overall morphology (Fig. 5C, red box). However, we found that these egg chambers showed a striking redistribution of GFP::Yps into many large puncta in nurse cell cytoplasm with notable perinuclear accumulation (Figs. 5D–F). These puncta were also detected by anti-Yps antibody in the wild-type or GFP::Yps-expressing egg chambers, indicating that these puncta contain endogenous Yps as well as GFP::Yps (Figs. S3A–F, M–P). These dramatic changes in Yps distribution occurred in morphologically normal egg chambers, prior to apoptosis (Figs. 5G–I).
Fig. 5. Previtellogenic egg chambers respond to amino acid starvation.
(A) A pair of ovaries from a well-fed female. (B) Ovaries from a “starved” female deprived of protein-rich food for 5 days. (C) The DIC image of an ovariole from a starved female. Although apoptosis occurs after stage 8 (arrows), previtellogenic egg chambers are retained (red box). (D–I) The GFP::Yps fluorescence and DIC images of stage 6 (D and G), stage 7 (E and H), and stage 8 (F and I) egg chambers in the protein-poor condition for 24 hrs. GFP::Yps accumulated in the perinuclear regions and made large aggregates in nurse cells (yellow arrows). (J and K) The mean size of individual particles and the total volume of particles in the protein-rich (gray bars) and the protein-poor condition for 1 day (white bar). Asterisks indicate the statistical significance (t-test, p<0.05). 10 egg chambers were analyzed in each stage. (L and M) oskarMS2/MS2-GFP accumulated aberrantly in nurse cells of stage-6 egg chambers in the protein-poor condition for 24 hrs (yellow arrows). (N–R) GFP::Yps aggregates colocalized with P-body markers (yellow arrows in N–P) but not a stress granule marker or an autolysosome marker (blue arrows in Q and R). Green is GFP::Yps, while magenta is Dcp1 (N), Pcm (O), eIF4E (P), Stau (Q), and Lysotracker (R). Low-power images are placed in Fig. S5. Bars: (A and B) 0.5 mm; (C) 50 μm; (D–I, L–R) 10μm.
3-D analysis of GFP::Yps particles demonstrated that the mean size of individual particles and the total volume of particles increased over time (Figs. 5J and K; see also Movie S3). The total volume of particles was significantly increased in starved egg chambers from stage 6 to 7 (compare Figs. 2D and E with 5D and E), while the mean size of individual particles was significantly larger from stage 7 to 8 (compare Figs. 2E and F with 5E and F). Time-lapse imaging revealed that Yps particles continuously assembled and disassembled (Movie S4). Hereafter we describe large particles detected in protein-poor conditions as “GFP::Yps aggregates”.
Because Yps is a component of the osk RNP complex, we examined the localization of osk mRNA by using flies expressing oskMS2/MS2-GFP, in which osk mRNA is labeled with bound MS2-GFP to visualize osk mRNA in living egg chambers (Zimyanin et al., 2008). In protein-rich conditions, oskMS2/MS2-GFP was exclusively enriched in the oocyte (Fig. 5L). When females were deprived of protein for 24 hrs, some oskMS2/MS2-GFP accumulated in nurse cells (Figs. 5M and S4) where it colocalized with Yps aggregates (data not shown). Moreover, we detected aggregates similar to GFP::Yps particles with other components of the osk RNP complex including GFP::Exu, GFP::Imp, GFP::Me31B, and GFP::eIF4E (data not shown). These results indicate that the localization of the osk RNP complex in previtellogenic egg chambers was dramatically affected in response to amino acid starvation.
GFP::Yps aggregates resemble P-bodies assembled in response to nutrient stress
Yps has been shown to coimmunoprecipitate from ovary extracts with Decapping protein 1 (Dcp1) (Lin et al., 2006; Lin et al., 2008), which is a major component of processing bodies (P-bodies) involved in mRNA metabolism. P-bodies are cytoplasmic foci in which nontranslating mRNAs are degraded or stored for later translation (Parker and Sheth, 2007). Though present in healthy cells, they are enriched in cells exposed to a variety of stressors (Teixeira et al., 2005). Therefore, we hypothesized that nurse cell P-bodies became larger in response to starvation. To test this idea, we examined the localization of GFP::Yps and known P-body markers. The majority of GFP::Yps aggregates colocalized with dDcp1 (68.8 %, n = 378 particles; Figs. 5N and S5A), the 5′-3′ exonuclease Pacman (70.7 %, n = 232; Figs. 5O and S5B), and eIF4E (92.5 %, n = 212; Figs. 5P and S5C). These results suggest that GFP::Yps aggregates were enlarged P-bodies.
In contrast, we found no evidence that GFP::Yps aggregates were related to stress granules (SGs) or autophagosomes. SGs are transient, cytoplasmic sites containing untranslated mRNAs derived from disassembled polysomes (Anderson and Kedersha, 2008; Anderson and Kedersha, 2009). Although there is no reliable SG marker in Drosophila, Staufen is localized to SG in mammalian oligodendrocytes (Thomas et al., 2005). However, Staufen was not colocalized with GFP::Yps aggregates in nurse cells (0.3 %, n = 59; Figs. 5Q and S5D), although they were tightly colocalized in the oocyte (Fig. S5D). Phosphorylated eIF2α, present in the initial step of translation arrest induced by stress, was also not colocalized with GFP::Yps (Fig. S5E). Furthermore, neither a pH-sensitive dye LysoTracker nor GFP::LC3, a marker for autophagosome (Rusten et al., 2004), colocalized with Yps aggregates after 24 hrs of starvation (0.58 %, n = 1214; Figs. 5R, S5F, and not shown). From these results, we concluded that molecules in GFP::Yps aggregates most closely resemble P-bodies.
Microtubules are extensively reorganized in young egg chambers from starved females
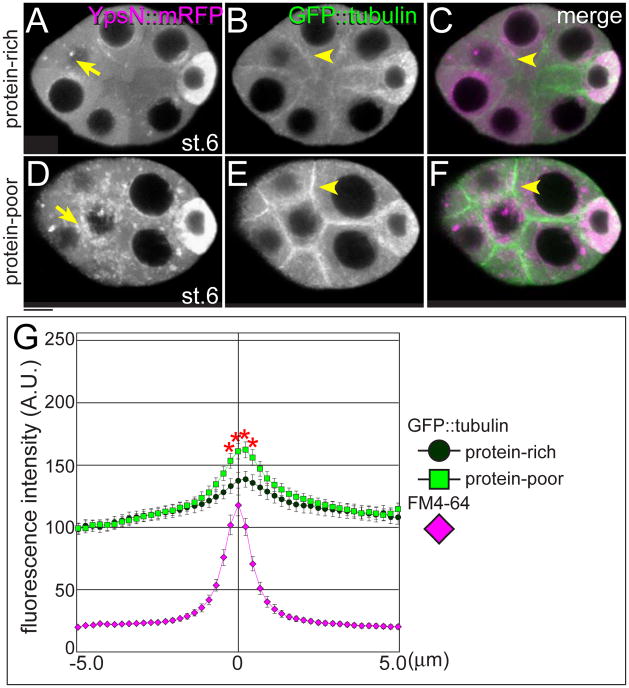
Strikingly, the response to poor nutrition included not only Yps aggregation but also a global rearrangement of the MT cytoskeleton. In the protein-rich condition, some YpsN::mRFP particles were detected in nurse cell cytoplasm (Fig. 6A). MTs were abundant in the oocytes and extended through ring canals into nurse cells where they were weakly associated with nurse cell boundaries (Fig. 6B). When female flies were deprived of protein for 24 hrs, YpsN::mRFP particles formed many aggregates in nurse cells (Fig. 6D). Strikingly, MTs shifted dramatically toward the cortical region of nurse cells (Figs. 6E–G). Both Yps aggregation and MT reorganization were detected during stage 6 in the protein-poor condition, prior to apoptosis. After stage 7, MTs were no longer obvious in nurse cell boundaries (data not shown). These results suggest a model where GFP::Yps localization is disrupted because MTs are reorganized.
Fig. 6. Microtubule organization is affected by poor nutrition.
Stage-6 egg chambers expressing GFP::tubulin (green) and YpsN::mRFP (magenta) dissected from females in the protein-rich (A–C) or the protein-poor condition for 24 hrs (D–F). In response to the protein-poor condition, YpsN::mRFP particles increased in nurse cells (arrows in D). Strikingly, GFP::tubulin showed a major shift in distribution from cytoplasmic MTs toward distinct cortical enrichment in nurse cells (arrowheads in E and F). (G) A quantitative analysis of MT distribution at nurse cell boundaries. The profiles of fluorescence intensities of GFP::tubulin were plotted within 5 μm distance from boundaries in the protein-rich (circle) and the protein-poor condition for 24 hrs (square). The position of membrane was normalized with FM4-64 (magenta). Nearly 40 boundaries from 30 egg chambers were analyzed in each condition. Asterisks show statistical significance (student t-test, p<0.05). Bar, 10 μm.
We asked if nutrient stress affects the plus-end dynamics of MTs by monitoring the behavior of EB1::GFP, which binds specifically to the growing ends of MTs and is involved in MT dynamics (Rogers et al., 2002). In stage-6 egg chambers from well-fed females, many distinct “comet”-like signals were moving in every direction in nurse cell cytoplasm, and some EB1::GFP comets were moving from the oocyte to nurse cells (Movie S5). When females were deprived of protein for 3 days, EB1::GFP comets were still present in cytoplasm and mostly moved along the cortical regions of nurse cells (Movie S6). This result indicates that at least some MTs remained dynamic, but MT elongation sites were reorganized in response to amino acid starvation.
The response to starvation is rapid, and can be reversed by providing food or exogenous insulin
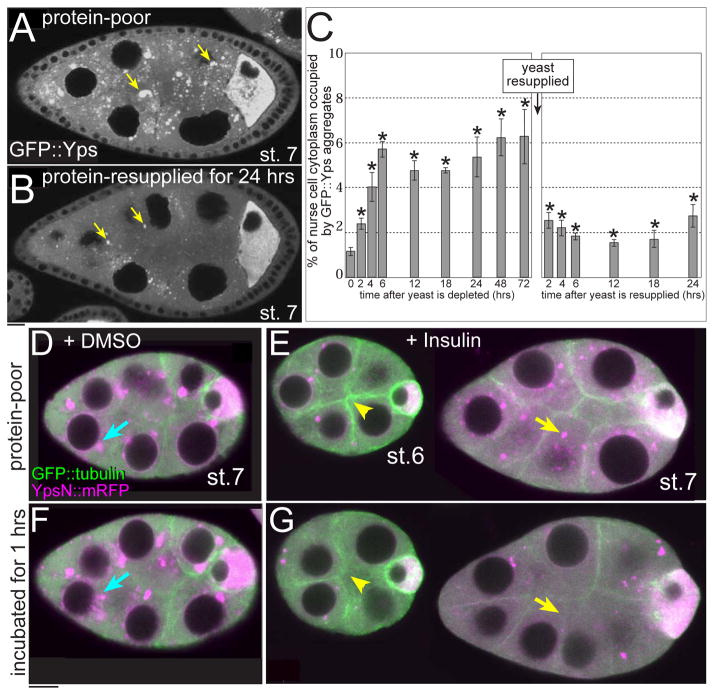
Unlike apoptotic degeneration of stage-8 egg chambers, the response of previtellogenic egg chambers to nutrient stress was reversible. When females reared on protein-poor food for 24 hrs were transferred back to protein-rich food for 24 hrs, the number of GFP::Yps aggregates decreased (Figs. 7A and B). To examine how quickly egg chambers respond to changes in nutrient conditions, we examined the temporal profile of GFP::Yps aggregate formation. Notably, GFP::Yps aggregates (Fig. 7C) and MT reorganization (not shown) were detected as early as 2 hrs after protein deprivation. Conversely, when flies were protein-deprived for 72 hrs and then resupplied protein, GFP::Yps aggregates decreased in 2 hrs. This result suggests that egg chambers respond just as quickly to the presence of protein-rich food as they do to protein deprivation.
Fig. 7. GFP::Yps aggregation and MT reorganization are reversible.
(A and B) GFP::Yps aggregates were detected in nurse cells of a stage-7 egg chamber in the protein-poor condition for 24 hrs. When females were starved for 24 hrs and then resupplied with protein-rich food for 24 hrs, GFP::Yps aggregates decreased (arrows). (C) The temporal profile of GFP::Yps aggregate formation in stage-6 egg chambers. The X-axis indicates time after starvation and after resupply of protein-rich food. The Y-axis represents the percentage of nurse cell cytoplasm area occupied by GFP::Yps aggregates. Asterisks indicate statistical significance when the area of GFP::Yps puncta at t = 0 was compared with those in other time points (2-sample Z-test for the proportion, p<0.05). 10–20 egg chambers were analyzed in each time point. (D–G) Egg chambers expressing YpsN::mRFP and GFP::tubulin were taken from females starved for 24 hrs and cultured in the medium with DMSO (vehicle: D and F) or 0.2 mg/ml insulin (E and G). Images were taken immediately after dissection (D and E) and after 1-hr incubation (F and G). Note that MTs are less prominent at the cortical regions (arrowheads) and the number of GFP::Yps aggregates was reduced (arrows) in culture with insulin. Bars, 10 μm.
The insulin/insulin-like growth factor signaling (IIS) pathway plays a key role in mediating responses to nutrition (Edgar, 2006; Toivonen and Partridge, 2009). To examine if the IIS pathway is involved in the response to nutrient stress, we tested whether Yps aggregation or MT reorganization can be reversed by addition of exogenous insulin. Egg chambers were dissected from females deprived of protein for 24 hrs and then cultured with or without insulin. Without insulin, the starvation phenotypes became more severe (Figs. 7D and F). In contrast, Yps aggregates dispersed and MT condensation at cell boundaries was reduced with insulin (Figs. 7E and G). Thus, insulin-like peptides may mediate the response to nutrient stresses in egg chambers.
Discussion
In this study, we focus on protein movements between nurse cells and the oocyte, showing that groups of RNA binding proteins and ER/Golgi proteins were selectively enriched in the oocyte prior to vitellogenesis. Furthermore, we found that osk RNP localization and MT organization were reversibly affected by protein deprivation. Our results suggest that previtellogenic egg chambers have active sorting and transport machineries adjustable to nutrient conditions, which ensure the quality of egg chambers entering vitellogenesis.
MT-dependent polarized transport and MT-independent protein movements in previtellogenic egg chambers
Several lines of evidence have indicated that intercellular cytoplasmic flow is a highly selective process (Bohrmann and Biber, 1994; Clark et al., 2007; Mahajan-Miklos and Cooley, 1994; Theurkauf et al., 1992). However, no direct observation of this cytoplasmic flow has been reported during stages 1–6. By using a collection of GFP protein-trap lines, we demonstrated that many proteins are specifically enriched in young oocytes in a mechanism that does not involve targeting of their encoding mRNAs to the oocyte. Oocyte-enriched proteins are likely required for oocyte growth and maturation. In contrast, metabolic enzymes and ribosomal proteins were ubiquitously distributed throughout germline cytoplasm, perhaps to ensure that the growth rates of nurse cells and the oocyte are coordinated during early oogenesis.
Live imaging techniques provided valuable insight into possible mechanisms for intercellular movement of proteins in young egg chambers. Although few particles were detected with GFP::Yps expressed at the endogenous level, overexpressed YpsN::mRFP particles allowed us to directly follow polarized transport from nurse cells to the oocyte (Fig. 2L and Movie S1). It is likely that these particles directionally move to the oocyte along MTs powered by Dynein motor complexes, as has been demonstrated in vitellogenic egg chambers (Clark et al., 2007; Mische et al., 2007; Theurkauf and Hazelrigg, 1998). In addition, our FLIP analysis revealed a surprisingly substantial level of MT-independent protein movements between nurse cells and the oocyte. Interestingly, Dendra2 was much more freely diffusible between nurse cells and the oocyte than Yps, RpL30, and Pdi (Fig. 3B). The rate of movements of the endogenous proteins may be restricted by the size of the complexes in which they reside (RNPs, ribosomes, ER vesicles) or their interactions with other molecules.
The FLIP assay also revealed very little retrograde movement of GFP::Yps from oocyte to nurse cells (Fig. 3Bh), strongly suggesting that Yps-containing particles are trapped within the oocyte. This maintenance in the oocyte is MT-dependent (Fig. 3Bj and Movie S2), and Dynein may act as the static anchor to MTs in the oocyte, as previously reported (Delanoue and Davis, 2005; Delanoue et al., 2007). Notably, the oocyte enrichment of Shep protein appears independent of MTs, implying that not all proteins are anchored in the oocyte in a MT-dependent manner. Taken together, these results indicate that polarized transport of specific RNP complexes takes place in the context of extensive bidirectional movements between nurse cells and the oocyte. Oocyte enrichment of the osk RNP, and likely many other maternal components, may be the combined result of trapping both complexes engaged with MT motors and complexes diffusing into the oocyte.
Young egg chambers rapidly respond to nutrient stress
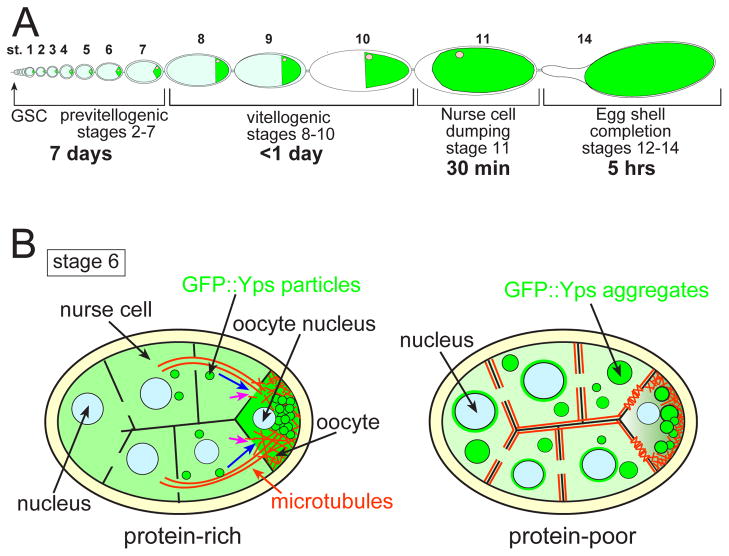
Previtellogenic egg chambers can mount a rapid and reversible response to amino acid starvation, which is in contrast to the irreversible process of apoptosis that eliminates germline cysts in the germarium and stage-8 egg chambers. We propose that the previtellogenic response to nutrient stress allows females to retain egg chambers that continue to grow at a minimal rate, and makes it possible to rapidly resume egg production when nutrient resources become available. In addition, development from germline stem cell through stage 7 takes about 7 days under normal conditions, while the remainder of oogenesis including the vitellogenic stages requires only about one additional day (Fig. 8A). Thus, slowing down rather than destroying previtellogenic egg chambers conserves the investment made in the initial assembly and patterning of egg chambers. Destruction of those egg chambers that reach stage 8 under nutrient stress saves the energy needed for yolk production, but only delays resumption of egg production one day. Our work reveals important mechanisms for the physiological response to environmental stress that function to coordinate organism survival and reproduction.
Fig. 8. A model of Drosophila egg chamber responding to nutrient stress.
(A) A schematic drawing of oogenesis. Green indicates oocytes. Development from germline stem cell (GSC) through stage 7 takes about 7 days, during which oocyte volume increases 55-fold. During the final day of oogenesis, oocyte volume increases more rapidly another 1,600 fold due to yolk uptake (stages 8–10, vitellogenesis) and the transfer of all remaining nurse cell cytoplasm to the oocyte (stage 11, nurse cell dumping). The eggshell forms in 5 hrs (stages 12–14). Thus, previtellogenic development requires the highest time investment, while the last day of oogenesis carries the higher metabolic cost. (B) Our working model. In the presence of protein-rich food, GFP::Yps-containing particles are directionally transported (blue arrows) from nurse cells to the oocyte along MT bundles (orange lines) during stage 6. A portion of GFP::Yps moves to the oocyte by diffusion (pink arrows). MTs are required for Yps to be retained in the oocyte. In the presence of protein-poor food, MTs shift toward the cortical region of nurse cells and polarized transport is disrupted. GFP::Yps assembles into aggregates and also accumulates in the perinuclear regions of nurse cells.
We propose that the primary response to nutrient stress is MT rearrangement, which disrupts polarized transport from nurse cells to the oocyte, leading to protein mislocalization in nurse cells (Fig. 8B). In our model, Yps-containing transport particles dissociate from MTs and assemble into aggregates upon starvation. Newly synthesized proteins are not loaded into transport particles, accumulating instead in the perinuclear regions of nurse cells. This model is based on the following observations: (1) the cortical redistribution of MTs and Yps aggregation occurred over the same time courses; (2) MT disruption by colchicine induced Yps aggregation; (3) MT-dependent localization of proteins was selectively affected by the protein-poor condition. In contrast to Yps, the localization of Shep or Pdi was hardly affected by the amino acid starvation or colchicine treatment. Although we cannot exclude the possibility that MT reorganization and GFP::Yps aggregation occurred independently in response to starvation, MT disruption and nutrient stress have a similar effect on promoting Yps aggregation.
Many types of cytoplasmic bodies have been reported in developing egg chambers: Balbiani bodies, nuage, sponge bodies, transport particles, processing bodies (P-bodies), Dynein aggregates, and uridine-rich bodies (U-bodies) (Bohrmann and Biber, 1994; Cox and Spradling, 2003; Lin et al., 2008; Liu and Gall, 2007; Nakamura et al., 2001; Navarro et al., 2009; Wilsch-Bräuninger et al., 1997). Not all of these structures are completely distinct, with some proteins associated with more than one type of cytoplasmic body.
Our immunostaining results indicated that Yps aggregates contained components of P-bodies, such as dDcp1, Pacman, and eIF4E (Fig. 5N–P). We hypothesize that Yps aggregates are enlarged P-bodies that function as storage sites for translationally repressed osk RNAs, similar to mRNA protection in yeast P-bodies (Brengues et al., 2005). Since MT disruption leads to aggregation of P-bodies in yeast (Sweet et al., 2007), it is feasible that starvation-induced MTs reorganization triggers Yps aggregation in egg chambers. Another possibility is that Yps aggregates associate with sponge bodies, as Yps coimmunoprecipitates with Exu (Wilhelm et al., 2000). Indeed, a recent study reporting that sponge body architecture changes in response to environmental conditions (Snee and Macdonald, 2009) is consistent with our observations of Yps aggregates. However, electron microscopy is required to determine whether Yps aggregates are associated with sponge bodies, in which ER-like cisternae are present.
The response of previtellogenic egg chambers to nutrient stress may represent a conserved mechanism for promoting cell survival in the germline. The primary response could be MT rearrangement since MT disruption by colchicine can induce Yps aggregation, similar to the aggregation of P-bodies upon MT destabilization in yeast (Sweet et al., 2007). In egg chambers, the net effect is disruption of oocyte enrichment of maternal components and slow growth. Interestingly, developing oocytes in C. elegans exhibit similar MT reorganization caused by depletion of sperm (Harris et al., 2006). MTs become concentrated at the cortex of arrested oocytes, and rapidly disperse in the cytoplasm in the presence of sperm or Major Sperm Protein (MSP). Furthermore, sperm depletion with aging also causes the formation of large cytoplasmic bodies containing components of both P-bodies and SGs, and similar large RNP foci form in response to heat shock, osmotic stress, or anoxia (Jud et al., 2008). These responses to sperm depletion are to those in egg chambers from starved females, suggesting that a related response would be elicited in C. elegans by nutrient stress. Thus, our work reveals a potentially conserved response to starvation that modulates the rate of oogenesis.
Information about environmental conditions has to be transmitted to the ovary to determine whether an egg chamber will progress into vitellogenesis. However, the underlying molecular mechanism is poorly understood. We found that exogenous insulin reversed the starvation phenotype of previtellogenic egg chambers, suggesting that the insulin/insulin-like growth factor signaling (IIS) pathway is involved in nutrient sensing in ovaries. Other evidence supporting a role for IIS signaling is that IIS pathway-defective mutant egg chambers fail to progress into vitellogenesis (LaFever and Drummond-Barbosa, 2005; Richard et al., 2005). The extensive reorganization of the MT cytoskeleton and P-bodies may explain the reduced growth rate reported for egg chambers mutant for IIS pathway. However, it is not known if IIS signaling directly regulates MT organization. Further studies are necessary to understand the molecular mechanism that trigger the response of previtellogenic egg chambers to nutrient stress conditions.
Supplementary Material
Acknowledgments
We are grateful to Drs. Wilhelm, Nakamura, Chou, Theurkauf, Stenmark, and St. Johnston for reagents; and members of Cooley lab for their helpful comments. We also thank the Bloomington Drosophila Stock Center and Drosophila Genome Resources Center for reagents, and Katsuo Furukubo-Tokunaga for use of his equipment. This work was supported by NIH grant (GM043301) to L.C. and by Grant-in-Aid for Scientific Research (KAKENHI) on Innovative Areas “Regulatory Mechanism of Gamete Stem Cells” (#23116701) to R.N. This work was also supported in part by Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science, and Technology of the Japanese Governmen. Y.S. is a recipient of fellowships from Human Frontier Science Program and the Japan Society for the Promotion of Science.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–50. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–6. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Drosophila. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1989. [Google Scholar]
- Bohrmann J, Biber K. Cytoskeleton-dependent transport of cytoplasmic particles in previtellogenic to mid-vitellogenic ovarian follicles of Drosophila: time-lapse analysis using video-enhanced contrast microscopy. J Cell Sci. 1994;107(Pt 4):849–58. doi: 10.1242/jcs.107.4.849. [DOI] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–9. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock SL, Ish-Horowicz D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 2001;414:611–6. doi: 10.1038/414611a. [DOI] [PubMed] [Google Scholar]
- Cha BJ, Koppetsch BS, Theurkauf WE. In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell. 2001;106:35–46. doi: 10.1016/s0092-8674(01)00419-6. [DOI] [PubMed] [Google Scholar]
- Clark A, Meignin C, Davis I. A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development. 2007;134:1955–65. doi: 10.1242/dev.02832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Wang Y, Dockendorff TC, Erdjument-Bromage H, Tempst P, Schedl P, Jongens TA. The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Dev Cell. 2005;8:331–42. doi: 10.1016/j.devcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Delanoue R, Davis I. Dynein anchors its mRNA cargo after apical transport in the Drosophila blastoderm embryo. Cell. 2005;122:97–106. doi: 10.1016/j.cell.2005.04.033. [DOI] [PubMed] [Google Scholar]
- Delanoue R, Herpers B, Soetaert J, Davis I, Rabouille C. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev Cell. 2007;13:523–38. doi: 10.1016/j.devcel.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Drummond-Barbosa D, Spradling AC. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev Biol. 2001;231:265–78. doi: 10.1006/dbio.2000.0135. [DOI] [PubMed] [Google Scholar]
- Dubowy J, Macdonald PM. Localization of mRNAs to the oocyte is common in Drosophila ovaries. Mech Dev. 1998;70:193–5. doi: 10.1016/s0925-4773(97)00185-8. [DOI] [PubMed] [Google Scholar]
- Edgar BA. How flies get their size: genetics meets physiology. Nat Rev Genet. 2006;7:907–16. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- Grieder NC, de Cuevas M, Spradling AC. The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development. 2000;127:4253–64. doi: 10.1242/dev.127.19.4253. [DOI] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–5. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- Harris JE, Govindan JA, Yamamoto I, Schwartz J, Kaverina I, Greenstein D. Major sperm protein signaling promotes oocyte microtubule reorganization prior to fertilization in Caenorhabditis elegans. Dev Biol. 2006;299:105–21. doi: 10.1016/j.ydbio.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Jud MC, Czerwinski MJ, Wood MP, Young RA, Gallo CM, Bickel JS, Petty EL, Mason JM, Little BA, Padilla PA, Schisa JA. Large P body-like RNPs form in C. elegans oocytes in response to arrested ovulation, heat shock, osmotic stress, and anoxia and are regulated by the major sperm protein pathway. Dev Biol. 2008;318:38–51. doi: 10.1016/j.ydbio.2008.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RC. Ovarian development in Drosophila melanogaster. Academic Press; New York: 1970. [Google Scholar]
- King RC, Burnett RG. Autoradiographic study of uptake of tritiated glycine, thymidine, and uridine by fruit fly ovaries. Science. 1959;129:1674–5. doi: 10.1126/science.129.3364.1674. [DOI] [PubMed] [Google Scholar]
- Koch EA, Spitzer RH. Multiple effects of colchicine on oogenesis in Drosophila: induced sterility and switch of potential oocyte to nurse-cell developmental pathway. Cell Tissue Res. 1983;228:21–32. doi: 10.1007/BF00206261. [DOI] [PubMed] [Google Scholar]
- LaFever L, Drummond-Barbosa D. Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science. 2005;309:1071–3. doi: 10.1126/science.1111410. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Tautz D. In situ hybridization to RNA. Methods Cell Biol. 1994;44:575–98. doi: 10.1016/s0091-679x(08)60933-4. [DOI] [PubMed] [Google Scholar]
- Li M, McGrail M, Serr M, Hays TS. Drosophila cytoplasmic dynein, a microtubule motor that is asymmetrically localized in the oocyte. J Cell Biol. 1994;126:1475–94. doi: 10.1083/jcb.126.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. Germline stem cell division and egg chamber development in transplanted Drosophila germaria. Dev Biol. 1993;159:140–52. doi: 10.1006/dbio.1993.1228. [DOI] [PubMed] [Google Scholar]
- Lin M-D, Fan S-J, Hsu W-S, Chou T-B. Drosophila decapping protein 1, dDcp1, is a component of the oskar mRNP complex and directs its posterior localization in the oocyte. Dev Cell. 2006;10:601–13. doi: 10.1016/j.devcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Lin M-D, Jiao X, Grima D, Newbury SF, Kiledjian M, Chou T-B. Drosophila processing bodies in oogenesis. Dev Biol. 2008;322:276–88. doi: 10.1016/j.ydbio.2008.07.033. [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Luk SK, Kilpatrick M. Protein encoded by the exuperantia gene is concentrated at sites of bicoid mRNA accumulation in Drosophila nurse cells but not in oocytes or embryos. Genes Dev. 1991;5:2455–66. doi: 10.1101/gad.5.12b.2455. [DOI] [PubMed] [Google Scholar]
- Mach JM, Lehmann R. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes Dev. 1997;11:423–35. doi: 10.1101/gad.11.4.423. [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Cooley L. Intercellular cytoplasm transport during Drosophila oogenesis. Dev Biol. 1994;165:336–51. doi: 10.1006/dbio.1994.1257. [DOI] [PubMed] [Google Scholar]
- Mansfield JH, Wilhelm JE, Hazelrigg T. Ypsilon Schachtel, a Drosophila Y-box protein, acts antagonistically to Orb in the oskar mRNA localization and translation pathway. Development. 2002;129:197–209. doi: 10.1242/dev.129.1.197. [DOI] [PubMed] [Google Scholar]
- Mazzalupo S, Cooley L. Illuminating the role of caspases during Drosophila oogenesis. Cell Death Differ. 2006;13:1950–9. doi: 10.1038/sj.cdd.4401892. [DOI] [PubMed] [Google Scholar]
- Mische S, Li M, Serr M, Hays TS. Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary. Mol Biol Cell. 2007;18:2254–63. doi: 10.1091/mbc.E06-10-0959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Navarro C, Puthalakath H, Adams JM, Strasser A, Lehmann R. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat Cell Biol. 2004;6:427–35. doi: 10.1038/ncb1122. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–46. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Petrella LN, Smith-Leiker T, Cooley L. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development. 2007;134:703–12. doi: 10.1242/dev.02766. [DOI] [PubMed] [Google Scholar]
- Pokrywka NJ, Stephenson EC. Microtubules mediate the localization of bicoid RNA during Drosophila oogenesis. Development. 1991;113:55–66. doi: 10.1242/dev.113.1.55. [DOI] [PubMed] [Google Scholar]
- Pokrywka NJ, Stephenson EC. Microtubules are a general component of mRNA localization systems in Drosophila oocytes. Dev Biol. 1995;167:363–70. doi: 10.1006/dbio.1995.1030. [DOI] [PubMed] [Google Scholar]
- Prasad M, Jang AC-C, Starz-Gaiano M, Melani M, Montell DJ. A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nature protocols. 2007;2:2467–73. doi: 10.1038/nprot.2007.363. [DOI] [PubMed] [Google Scholar]
- Pritchett TL, Tanner EA, McCall K. Cracking open cell death in the Drosophila ovary. Apoptosis. 2009;14:969–79. doi: 10.1007/s10495-009-0369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard DS, Rybczynski R, Wilson TG, Wang Y, Wayne ML, Zhou Y, Partridge L, Harshman LG. Insulin signaling is necessary for vitellogenesis in Drosophila melanogaster independent of the roles of juvenile hormone and ecdysteroids: female sterility of the chico1 insulin signaling mutation is autonomous to the ovary. J Insect Physiol. 2005;51:455–64. doi: 10.1016/j.jinsphys.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–84. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–92. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Snee MJ, Macdonald PM. Dynamic organization and plasticity of sponge bodies. Dev Dyn. 2009;238:918–30. doi: 10.1002/dvdy.21914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC. The developmental genetics of oogenesis. Cold Spring Harbor Laboratory Press; Plainview, N.Y: 1993. [Google Scholar]
- St Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- Steinhauer J, Kalderon D. Microtubule polarity and axis formation in the Drosophila oocyte. Dev Dyn. 2006;235:1455–68. doi: 10.1002/dvdy.20770. [DOI] [PubMed] [Google Scholar]
- Suter B, Steward R. Requirement for phosphorylation and localization of the Bicaudal-D protein in Drosophila oocyte differentiation. Cell. 1991;67:917–26. doi: 10.1016/0092-8674(91)90365-6. [DOI] [PubMed] [Google Scholar]
- Sweet TJ, Boyer B, Hu W, Baker KE, Coller J. Microtubule disruption stimulates P-body formation. RNA. 2007;13:493–502. doi: 10.1261/rna.355807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–82. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE, Alberts BM, Jan YN, Jongens TA. A central role for microtubules in the differentiation of Drosophila oocytes. Development. 1993;118:1169–80. doi: 10.1242/dev.118.4.1169. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE, Hazelrigg TI. In vivo analyses of cytoplasmic transport and cytoskeletal organization during Drosophila oogenesis: characterization of a multi-step anterior localization pathway. Development. 1998;125:3655–66. doi: 10.1242/dev.125.18.3655. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE, Smiley S, Wong ML, Alberts BM. Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Development. 1992;115:923–36. doi: 10.1242/dev.115.4.923. [DOI] [PubMed] [Google Scholar]
- Thomas MG, Martinez Tosar LJ, Loschi M, Pasquini JM, Correale J, Kindler S, Boccaccio GL. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol Biol Cell. 2005;16:405–20. doi: 10.1091/mbc.E04-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivonen JM, Partridge L. Endocrine regulation of aging and reproduction in Drosophila. Mol Cell Endocrinol. 2009;299:39–50. doi: 10.1016/j.mce.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Wang S, Hazelrigg T. Implications for bcd mRNA localization from spatial distribution of exu protein in Drosophila oogenesis. Nature. 1994;369:400–03. doi: 10.1038/369400a0. [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Mansfield J, Hom-Booher N, Wang S, Turck CW, Hazelrigg T, Vale RD. Isolation of a ribonucleoprotein complex involved in mRNA localization in Drosophila oocytes. J Cell Biol. 2000;148:427–40. doi: 10.1083/jcb.148.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–53. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.