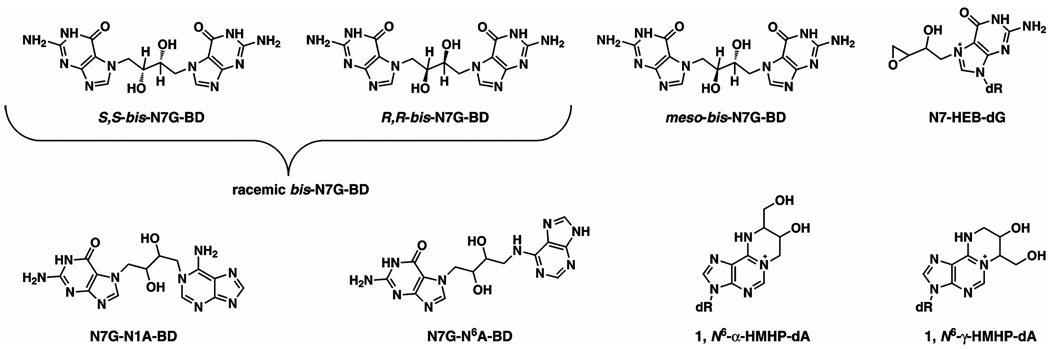
Abstract
1,3-butadiene (BD) is an important industrial and environmental chemical classified as a human carcinogen. The mechanism of BD-mediated cancer is of significant interest because of the widespread exposure of humans to BD from cigarette smoke and urban air. BD is metabolically activated to 1,2,3,4-diepoxybutane (DEB), which is a highly genotoxic and mutagenic bis-alkylating agent believed to be the ultimate carcinogenic species of BD. We have previously identified several types of DEB-specific DNA adducts, including bis-N7-guanine cross-links (bis-N7-BD), N6-adenine-N7-guanine cross-links (N6A-N7G-BD), and 1,N6-dA exocyclic adducts. These lesions were detected in tissues of laboratory rodents exposed to BD by inhalation (Goggin et al. Cancer Res. 2009;69:2479–2486). In the present work, persistence and repair of bifunctional DEB-DNA adducts in tissues of mice and rats exposed to BD by inhalation were investigated. The half-lives of the most abundant cross-links, bis-N7G-BD, in mouse liver, kidney, and lungs were 2.3–2.4 days, 4.6–5.7 days, and 4.9 days, respectively. The in vitro half-lives of bis-N7G-BD were 3.5 days (S,S isomer) and 4.0 days (meso isomer) due to their spontaneous depurination. In contrast, tissue concentrations of the minor DEB adducts, N7G-N1A-BD and 1,N6-HMHP-dA, remained essentially unchanged during the course of the experiment, with an estimated t 1/2 of 36–42 days. No differences were observed between DEB-DNA adduct levels in BD-treated wild type mice and the corresponding animals deficient in methyl purine glycosylase or the Xpa gene. Our results indicate that DEB-induced N7G-N1A-BD and 1,N6-HMHP-dA adducts persist in vivo, potentially contributing to mutations and cancer observed as a result of BD exposure.
Keywords: 1,3-butadiene; DNA adducts; DNA repair; mass spectrometry; transgenic mice
Introduction
1,3-Butadiene (BD)1 is a high volume industrial chemical used to synthesize plastics and rubber, resulting in occupational exposure of workers in the US and worldwide (1). It is also an environmental toxin present in automobile exhaust and in cigarette smoke, leading to a widespread exposure of human populations to BD (2,3). BD is classified as a known human carcinogen based on the results of laboratory animal exposures and human epidemiology studies (4,5). BD undergoes metabolic activation by cytochrome P450 monooxygenases to form 3,4-epoxybutene (EB), 3,4-epoxy-1,2-butanediol (EBD), and 1,2,3,4-diepoxybutane (DEB). Although it is a relatively minor metabolite of BD, DEB is considered the ultimate carcinogenic species of BD due to its potent genotoxic and mutagenic effects (6–8).
The adverse biological effects of DEB are attributed to its ability to form DNA-DNA cross-links at guanine and adenine nucleobases and exocyclic adenine adducts (Scheme 1) (9–12). We have previously quantified DNA-DNA cross-links, 1,4-bis-(guan-7-yl)-2,3-butanediol (bis-N7G-BD) and 1-(guan-7-yl)-4-(aden-1-yl)-2,3-butanediol (N7G-N1A-BD), and exocyclic adenine adducts, 1,N6-(1-hydroxymethyl-2-hydroxypropan-1,3-diyl)-2'-deoxyadenosine (1,N6-α-HMHP-dA) and 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2'-deoxyadenosine (1,N6-γ-HMHP-dA) (Scheme 1), in tissues of laboratory animals exposed to BD by inhalation (12–16). Immediately after exposure, bis-N7G-BD was the most abundant lesion, followed by N7G-N1A-BD and 1,N6-HMHP-dA adducts. Furthermore, mouse tissues contained 4–6-fold greater numbers of DEB-DNA adducts than tissues of rats exposed at the same conditions (15), consistent with a greater sensitivity of mice to BD-induced cancer (17,18). The stability of DNA adducts in cells greatly affects their biological impact since lesions that persist in DNA until the next round of replication have an opportunity to be converted to heritable mutations. Therefore, the goal of the present study was to investigate the persistence of DEB-induced DNA-DNA cross-links in vivo.
Scheme 1.
Structures of bifunctional DNA adducts induced by 1,2,3,4-diepoxybutane.
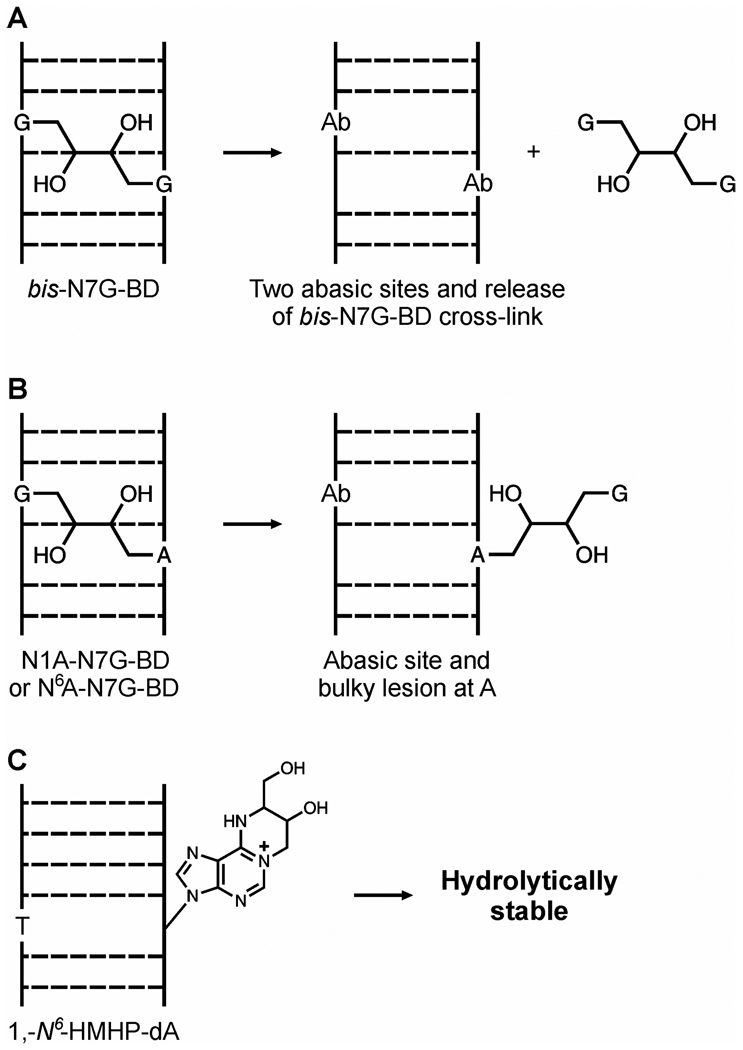
The ability of specific nucleabase adducts to persist in tissues is limited by their hydrolytic stability and their active repair by DNA repair machinery. The majority of N7-guanine adducts, including bis-N7G-BD, are hydrolytically unstable, as N-7 alkylation of guanine produces a positive charge on the modified nucleobase and destabilizes the β-glycosidic bond, leading to spontaneous depurination and the formation of abasic sites (9,13,19,20). Spontaneous hydrolysis of bis-N7G-BD releases free bis-N7-guanine conjugates and introduces two adjacent abasic sites (Scheme 2A). In vitro half-life of interstrand bis-N7G-BD adducts (which are the main type of cross-links induced by S,S DEB) in short synthetic DNA duplexes is 147 h, while the half-life of the corresponding intrastrand cross-links (originating from meso DEB) is 35 h (20). These differences in stability can be explained by a greater charge density associated with 1,2-intrastrand lesions as compared to 1,3-interstrand cross-links (20). In contrast to bis-N7G-BD, DEB-induced G-A cross-links, e.g. N7G-N1A-BD, are expected to be hydrolytically stable as they are only partially hydrolyzed (at the N7G), leaving an abasic site opposite a bulky lesion to the N1 of adenine (N7G-N1A-BDdep, Scheme 2B) (10). Exocyclic deoxyadenosine adducts of DEB, such as 1,N6-HMHP-dA, do not undergo spontaneous depurination (Scheme 2C) (12). Unless active repair mechanisms exist to remove N7G-N1A-BDdep and 1,N6-HMHP-dA in vivo, they are expected to persist in tissues, potentially contributing to mutagenesis.
Scheme 2.
Spontaneous hydrolysis products of bis-N7G-BD (A), N7G-N1A-BD (B) and 1,N6-α-HMHP-dA in DNA (C).
Multiple DNA repair pathways have evolved to help protect living cells against the nucleobase damage induced by alkylating agents. In base excision repair (BER), a glycosylase enzyme removes nucleobase adducts, and the resulting abasic sites are excised and replaced with normal nucleotides (21,22). An example of a BER protein is methyl purine glycosylase (MPG), which specifically excises alkylated purines, including 3-methyladenine, 3-methylguanine, 7-methylguanine (m7G), and etheno-dA (εA) (23–25). MPG protein participates in the repair of N7-guanine adducts induced by nitrogen mustards (26) and 1, N6-ethenoadenine (23,25), which are structurally similar to DEB-induced bis-N7G-BD and 1, N6-α-HMHP-dA adducts, respectively (Scheme 1).
The nucleotide excision repair (NER) pathway is a versatile DNA repair system that recognizes a wide variety of carcinogen-DNA adducts (27,28). The NER process in humans is complex, requiring the participation of over 30 proteins (28). A minimal set of human proteins involved in performing the NER repair reaction includes XPA, XPC-hHR23B, XPG, RPA, ERCC1-XPF, TFIIH, PCNA, DNA polymerase δ or ε, and DNA ligase I (29). Among these, the XPC-hHR23B and XPA proteins play a role in damage recognition/verification. Following damage recognition and separation of the DNA double helix at the lesion site, special excision endonucleases, XPG and ERCC1-XPF, hydrolyze two phosphodiester bonds on each side of the damaged base, releasing a short single stranded oligonucleotide fragment containing the lesion (27). DNA repair synthesis uses DNA polymerases δ or ε to fill in the resulting gap, replacing the excised strand, and DNA ligase I seals the nicks, restoring normal DNA.
Mammalian interstrand crosslink repair (ICL repair) has recently been reviewed by several authors (30–33). ICL repair is difficult because these lesions covalently link the complementary DNA strands, preventing duplex unwinding and strand separation (34). Furthermore, since both of the DNA strands are damaged, neither one is a good template for repair synthesis (34). While our understanding of mammalian ICL repair is somewhat limited, two predominant ICL repair pathways have been identified. In actively replicating cells, repair of ICLs is carried out by the Fanconi anemia (FA) protein complexes in conjunction with homologous recombination (31,33,35,36). In support of this mechanism, proliferating cells from individuals with FA mutations are highly sensitive to ICL-inducing agents (37). In addition, repair of some types of ICLs relies on proteins involved in NER (especially for repair of G0/G1 phase ICLs), as well as mismatch repair proteins (31,38,39) (32). In non-dividing cells, FA pathway and homologous recombination do not appear to be involved in ICL repair, and this pathway is termed recombination-independent ICL repair (36). Another mechanism for ICL repair in both recombination-dependent and recombination-independent pathways appears to rely primarily on translesion synthesis by specialized DNA polymerase enzymes (40).
In the present study, persistence and repair of three types of bifunctional DEB-DNA adducts, bis-N7G-BD, N7G-N1A-BD, and 1, N6-HMHP-dA (Scheme 1), was investigated in tissues of laboratory mice and rats exposed to BD by inhalation. Our results reveal significant differences between the stability of structurally distinct DEB-DNA adducts in vivo and suggest that N7G-N1A-BD and 1, N6-α-HMHP-dA lesions persist in liver, lung, and kidney tissues, potentially contributing to the adverse health effects of BD.
Experimental
Materials
Isotopically labeled internal standards, 15N10-bis-N7G-BD; 1, 2, 3-15N3, 2-13C1-N7G-N1A-BD, and 1,3,7,9-15N4-1,N6-HMHP-dA, were prepared as reported elsewhere (9,10,12). DEB isomers were synthesized in our laboratory using published methods (20). Phosphodiesterase I (PDE I), phosphodiesterase II (PDE II), and DNAase were purchased from Worthington Biochemical Corp. (Freehold, NJ). All other chemicals and solvents were obtained from Sigma (Milwaukee, WI) unless otherwise noted.
Animals and treatment
Persistence studies
BD inhalation exposures were performed at the Lovelace Respiratory Research Institute (Albuquerque, NM) as previously reported (15). B6C3F1 mice or F344 rats (5 per group) were randomly assigned to air-control and exposure groups by weight and were housed individually in hanging wire stainless steel cages according to the NIH guidelines (NIH Publication 86-23, 1985). All procedures involving the use of animals were approved by the LRRI Institutional Animal Care and Use Committee. Rodents (5 per group) were exposed to 625 or 1250 ppm BD, respectively, by inhalation for ten days (7 hours/day). Animals were euthanized by cardiac puncture either immediately at the end of the exposure period or following 3 or 10 day recovery period (mice) or 1, 3, or 6 days post exposure (rats). Liver, kidney, and lung tissues were collected, flash frozen, and shipped to the University of MN on dry ice, where they were stored at −80° C.
Repair studies
BD exposures of Mpg deficient mice and the corresponding wild-type controls were performed at the University of Texas-Medical Branch (Galveston, TX), as previously reported (41). The Mpg-deficient mice were a gift to Jeffrey K. Wickliffe from Leona D. Samson at the Massachusetts Institute of Technology. Animals were exposed to 62.5 ppm BD for 10 days. Female Xpa deficient mice and matched controls were exposed to 625 ppm BD for 10 days at the Lovelace Respiratory Research Institute (Albuquerque, NM) (15). Animals were euthanized immediately at the end of the exposure. Liver, lung, and kidney tissues were collected and stored at −80° C until DNA extraction.
DNA isolation
DNA was isolated using NucleoBond AXG500 anion exchange cartridges (Macherey-Nagel, Bethlehem, PA) according to the manufacturer’s instructions. In brief, tissues (0.1–0.4 g) were homogenized and incubated with RNAse and proteinase K at 50 °C for 2 – 4 hours. Samples were loaded on NucleoBond AXG cartridges, washed, and eluted according to kit instructions. DNA was precipitated by the addition of isopropanol. The DNA was spooled, washed with cold 70% ethanol (v/v), dried, and dissolved in Milli-Q water (500 µL). DNA amounts and purity were determined by UV spectrophotometry. Typical A260/A280 ratios were between 1.7 and 1.8, ensuring minimal protein contamination. Exact DNA amounts were established by HPLC-UV analysis of dG in DNA hydrolysates.
Neutral thermal hydrolysis and isolation of bis-N7G-BD adducts
Bis-N7G-BD were quantified by isotope dilution capillary HPLC-ESI+-MS/MS as described previously (15). In brief, DNA (100 µg) was spiked with S,S + R,R (racemic) and meso 15N10-labelled internal standards (300 fmol each) and incubated at 70 °C for 1 hour to release bis-N7G-BD as a free base conjugate. Partially depurinated DNA was removed by ultrafiltration with Nanosep 10K filters (Pall Life Sciences, Ann Arbor, MI) and saved for subsequent quantitation of N7G-N1A-BD and 1,N6-HMHP-dA as described below. The filtrates containing bis-N7G-BD and its internal standard were purified by offline HPLC on a Zorbax Eclipse XDB-C18 column (4.6 × 150 mm, 5 µm). The column was eluted with a gradient of 0.4% (v/v) formic acid in water (A) and acetonitrile (B). HPLC fractions containing bis-N7G-BD (12 – 13 min) were collected, dried under vacuum, and dissolved in 0.05% (v/v) acetic acid (25 µL) for HPLC-ESI+-MS/MS analysis (15).
Hydrolytic stability of bifunctional DEB-DNA adducts in vitro
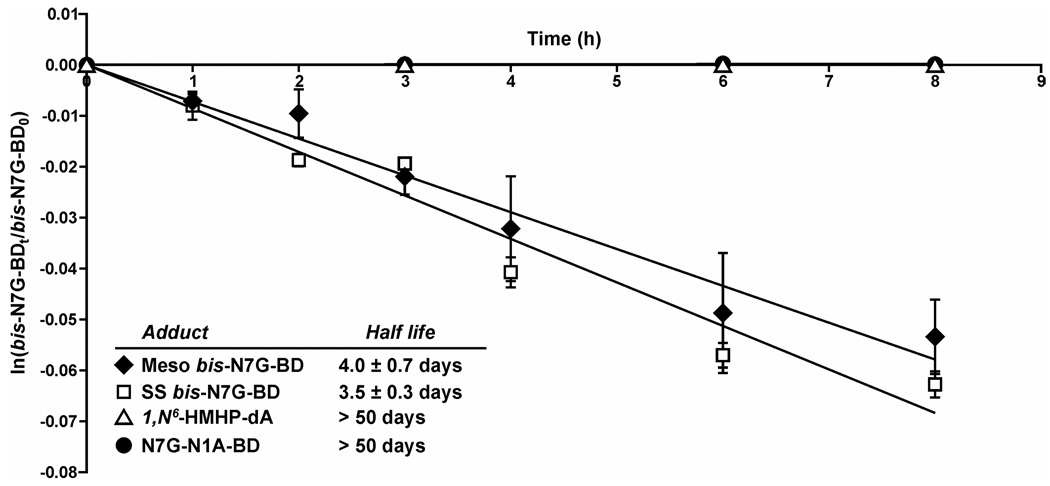
Calf thymus DNA (2 mg in 2 ml of Tris buffer, pH 7.2 containing 200 mM NaCl) was treated with S,S or meso DEB (1 mM) at 37° C for 24 h. The unreacted DEB was extracted with diethyl ether (3 × 400 uL), and DNA was precipitated with cold ethanol. DEB-treated DNA (600 µg) was dissolved in 600 µL of Tris buffer, pH 7.2, containing 200 mM NaCl and incubated in a water bath at 37° C. To determine starting amounts of bis-N7B-BD in treated DNA, a 50 µg aliquot of DNA was removed and spiked with 15N10- bis-N7G-BD internal standard (800 fmol). Following neutral thermal hydrolysis at 70° C for 1 h, bis-N7G-BD was quantified by HPLC-ESI+-MS/MS as described below. The remaining DNA was kept in a water bath at 37° C, and aliquots (50 µg) were removed following incubation for 0, 1, 2, 3, 4, 6, and 8 h. Following the addition of racemic and meso 15N10- bis-N7G-BD (800 fmol) (internal standards for mass spectrometry), samples were filtered through YM-10 Centricon filters or Nanosep 10K filters (Pall Life Sciences, Ann Arbor, MI), and nucleobase adducts released via spontaneous depurination were collected in filtrates. Samples were subjected to HPLC-ESI+-MS/MS analysis as described below to determine the amounts of bis-N7G-BD released via spontaneous hydrolysis. The number of bis-N7G-BD remaining in DNA at each time point (bis-N7G-BD t) was determined from the equation: bis-N7G-BD t = bis-N7G-BD total – bis-N7G-BD released at time t. The data was processed by plotting ln(bis-N7G-BD t/ bis-N7G-BD total) versus time to determine the adduct half life (t1/2 = ln(2)/-slope) (Figure 1).
Figure 1.
Kinetic analysis of spontaneous hydrolysis of R,S bis-N7G-BD, N7G-N1A-BD, and 1,N6-HMHP-dA adducts under physiological conditions (20 mM Tris buffer, pH 7.2 containing 200 mM NaCl).
Partially depurinated DNA was saved for capillary HPLC-ESI+-MS/MS analysis of N7G-N1A-BD and 1,N6-HMHP-dA lesions using previously published methodology (14,16). Data was processed using first order rate kinetics to determine adduct half-lives in vitro (Figure 1).
HPLC-ESI+-MS/MS analysis of bis-N7G-BD
Zorbax Extend C18 column (3.5 µm, 150 × 0.5 mm) was eluted with a gradient of 0.05% (v/v) acetic acid (A) and methanol (B). The solvent composition was linearly changed from 0 to 10 % B in 5 minutes, further to 60% in 10 minutes, and back to 0% B over 3 minutes. The column temperature was maintained at 40 °C, and the flow rate was 10 µL/min. A typical injection volume was 8 µL. With this solvent system, the retention time of S,S and R,R bis-N7G-BD was 9.9 min, while meso bis-N7G-BD eluted at 10.9 min.
Alternatively, a Synergi Hydro-RP HPLC column (4 µm, 0.5 × 250 mm) was eluted with a gradient of 0.05% acetic acid (v/v) (A) and methanol (B). The HPLC flow rate was 10 µL/min, the column temperature was maintained at 40 °C, and a typical injection volume was 8 µL. Solvent composition was linearly increased from 1% to 15% over 5 minutes and to 35% over 9 minutes and back to 1% in 4 minutes. With this solvent system, the retention time of S,S and R,R bis-N7G-BD was 15.51 min, while meso bis-N7G-BD eluted at 16.10 min.
The mass spectrometer was operated in the selected reaction monitoring mode by following the transitions corresponding to the loss of guanine from protonated molecules of bis-N7G-BD and the formation of protonated guanine: m/z 389.1 [M + H]+ → m/z 238.1 [M + H -Gua]+ and m/z 152.1 [Gua + H]+. The corresponding transitions for 15N10-bis-N7G-BD internal standard were m/z 399.1 [15N10-M + H]+ → m/z 243.1 [M + H - [15N5]Gua]+ and m/z 157.1 [15N5-Gua + H]+ (13).
nanoLC-nanoESI+-MS/MS analysis of bis-N7G-BD
Waters nanoAcquity UPLC system (Waters Corp., Millford, MA) interfaced to a Thermo-Finnigan TSQ Quantum Ultra mass spectrometer (Thermo Fisher Scientific Corp., Waltham, MA) was used in all nanospray MS experiments. HPLC solvents were 0.01% (v/v) acetic acid dissolved in CHROMASOLV® LC-MS water (Sigma-Aldrich, A) and HPLC-MS grade MeOH:Acetonitrile (1:1 volume ratio, from Sigma (Milwaukee, WI)) (B). Samples (4–8 µL) were loaded on a trapping column (Symmetry C18 nanoAcquity, 0.18 × 20 mm) for 1 min at 0% B at 15 µL/min. Chromatographic separation was achieved with a manually packed Zorbax SB-C18 column (75 µm × 180 mm) eluted at a flow rate of 0.35–0.4 µL/min and maintained at 35° C. Solvent composition was changed linearly from 0 to 10 % B in 2 min, further to 50% B in 8 min, and finally to 80% B over 5 min, before returning to 0% B for 14 min equilibration. Under these conditions, S,S + R,R and meso bis-N7G-BD eluted at 18 min, respectively.
Acid hydrolysis of DNA and isolation of N7G-N1A-BD
Partially depurinated DNA backbone recovered from the filters following neutral thermal hydrolysis of DNA (100 µg, see above) was spiked with 13C1,15N3-N7G-N1A-BD internal standard (300 fmol) and hydrolyzed in 0.1 M HCl (30 min at 70 °C) to release N7G-N1A-BD as a free base. The hydrolysates were filtered through YM-10 Centricon filters (Millipore, Billerica, MA) or Nanosep 10K filters (Pall Life Sciences, Ann Arbor, MI). Samples were heated in base (0.5 M NH4OH at 70 °C for 16 h) to induce Dimroth rearrangement of N7G-N1A-BD to N7G-N6A-BD and purified by solid phase extraction on C18 cartridges as reported previously (14). SPE fractions containing N7G-N6A-BD and its internal standard were concentrated under vacuum and re-dissolved in 0.05% (v/v) acetic acid (25 µL) for HPLC-MS/MS analysis. Please note that this methodology cannot distinguish between partially depurinated (N7G-N1A-BDdep) and intact G-A cross-links (N7G-N1A-BD, Scheme 2B), so the reported value is a sum of both adducts.
nanoLC-nanoESI+-MS/MS analysis of N7G-N1A-BD
N7G-N1A-BD adducts were quantified by isotope dilution nanoHPLC-ESI-MS/MS using previously published methods (15). HPLC solvents were 0.01% (v/v) acetic acid (Fluka, LC-MS grade) in water (A) and a 1:1 mixture of HPLC-MS grade methanol and acetonitrile (Sigma, Milwaukee, WI) (B). Samples (4–8 µL) were loaded onto a trapping column (Symmetry C18 nanoAcquity, 0.18 × 20 mm) for 1 min at 0% B. Chromatographic separation was achieved using an Atlantis dC18 column (75 µm × 100 mm) or a manually packed Zorbax SB-C18 column (75 µm × 180 mm). Both columns were eluted at a flow rate of 0.35 µL/min. The solvent composition was changed from 0 to 12 % B in 2 minutes, then to 25% B over 7 minutes, and further to 45% B over 15 minutes. The column was equilibrated at 0% B for at least 12 minutes prior each run. Under these conditions, N7G-N6A-BD adducts eluted as a single peak at 16.9 minutes (Atlantis dC18 column) or 14.7 minutes (Zorbax SB-C18 column). The mass spectrometer was operated in the selected reaction monitoring mode by following the transitions corresponding to loss of guanine from protonated molecules of N7G-N6A-BD and its internal standard: m/z 373.1 [M + H]+ → m/z 222.1 [M + H -Gua]+ and m/z 377.1 [13C1,15N3-M + H]+ → m/z 222.1 [M + H - [13C1,15N3]- Gua]+.
Enzymatic hydrolysis of DNA and isolation of 1,N6-α-HMHP-dA
Partially depurinated DNA recovered after neutral thermal hydrolysis (100 µg) was spiked with 15N4-1, N6-HMHP-dA internal standard (27.5 fmol) and enzymatically digested with DNase I (35 U/100 µg DNA), PDE I (70 mU/100 µg DNA), PDE II (80 mU/100 µg DNA), and alkaline phosphatase (14.6 U/100 µg DNA) in 10 mM Tris-HCl/15 mM MgCl2 at 37 °C for 18 h (16). 1, N6-HMHP-dA and its internal standard were isolated by solid phase extraction using Extract Clean Carbo cartridges (3 mL, from Grace Davidson, Deerfield, IL). SPE cartridges were washed with methanol (2 × 3 mL) and water (2 × 3 mL) prior to loading samples in water (1 mL). Samples were washed with water (3 mL) and 5 % methanol (3 mL) and eluted with 30 % methanol (3 mL). The 30 % methanol elution was dried under vacuum and re-dissolved in 25 µL 0.05% acetic acid. Typically, 8 µL of this solution was injected on column for HPLC-ESI+-MS/MS analysis.
Column Switching HPLC-ESI+-MS/MS analysis of 1,N6-HMHP-dA
DNA hydrolysates containing 1, N6-HMHP-dA were analyzed by column switching HPLC- ESI+-MS/MS methods as reported elsewhere (16). In brief, trapping was achieved with a 300 Å SCX column (Waters Corp., Milford, MA) using an isocratic flow of 0.5% methanol in 1mM ammonium hydroxide in water. Samples were then back-flushed with 0.05% acetic acid to transfer 1, N6-α-HMHP-dA and its internal standard onto a Synergi Hyrdo-RP (250 × 0.5 mm, Phenomenex) analytical column. The solvent system consisted of 0.05% (v/v) acetic acid (A) and methanol (B) delivered at a flow rate of 10 µL/min. Solvent composition was changed linearly from 0.5 % to 5% B in 5 min, and further to 20% B in 10 min. The mass spectrometer was operated in the selected reaction monitoring mode by following the neutral loss of deoxyribose from the [M+H]+ ions of 1, N6-HMHP-dA and the 15N4-internal standard (m/z 338.1 → 222.1 and m/z 342.1 → 226.1, respectively).
Results
In vitro stability of DEB-DNA adducts in the absence of DNA repair
To evaluate the hydrolytic stability of DEB-induced bis-N7G-BD, N7G-N1A-BD, and 1,N6-HMHP-dA adducts (Scheme 1) in the absence of active repair, aliquots of calf thymus DNA were treated with optically pure S,S or meso DEB. Following the removal of unreacted DEB, alkylated DNA was incubated under physiological conditions (200 mM NaCl, pH 7.2), and the spontaneous release of bifunctional DEB-DNA adducts was monitored by HPLC-ESI-MS/MS. We found that the half lives of S,S and meso bis-N7G-BD adducts in DNA in the absence of active repair were 3.5 and 4.0 days, respectively (Figure 1). In contrast, the concentrations of N7G-N1A-BDdep and 1,N6-HMHP-dA lesions were essentially unchanged under these conditions (estimated half life > 50 days, Figure 1). This difference in stability can be explained by spontaneous depurination of bis-N7G-BD from the DNA backbone, while N7G-N1A-BDdep and 1,N6-HMHP-dA are hydrolytically stable (Scheme 2).
Persistence of bifunctional DEB-DNA adducts in vivo
To evaluate the persistence and repair of DEB-induced DNA adducts in vivo, female B6C3F1 mice and F344 rats were exposed to 625 or 1250 ppm BD, respectively, by inhalation for ten days (7 hours/day). Animals were euthanized either immediately at the end of exposure or following a specified post exposure recovery period (1, 3, 6, or 10 days). DNA was extracted from liver, lung, and kidney tissues, and the concentrations of bis-N7G-BD, N7G-N1A-BD, and 1,N6-HMHP-dA were determined by isotope dilution HPLC-ESI-MS/MS methods reported previously (13–16). Immediately after exposure, bis-N7G-BD was the dominant adduct (3.95 per 107 nucleotides), followed by N7G-N1A-BD (0.27 per 107 nucleotides), and 1,N6-HMHP-dA (0.04 per 107 nucleotides) (Table 1).
Table 1.
Tissue concentrations of bifunctional DEB-DNA adducts in liver of B6C3F1 mice exposed to 625 ppm BD for 2 weeks. Animals were sacrificed 2 hours following exposure.
| Adduct | Adducts per 107 nucleotides | ||
|---|---|---|---|
| Liver | Kidney | Lung | |
| meso bis-N7G-BD | 0.62 ± 0.06 | 0.39 ± 0.06 | <0.29 |
| S,S+R,R bis-N7G-BD | 3.95 ± 0.89 | 1.36 ± 0.14 | 2.25 ± 0.33 |
| N7G-N1A-BD | 0.27± 0.01 | 0.08 ± 0.03 | 0.17± 0.03 |
| 1,N6-α,γ -HMHP-dA | 0.04± 0.002 | 0.01 ± 0.02 | <0.015 |
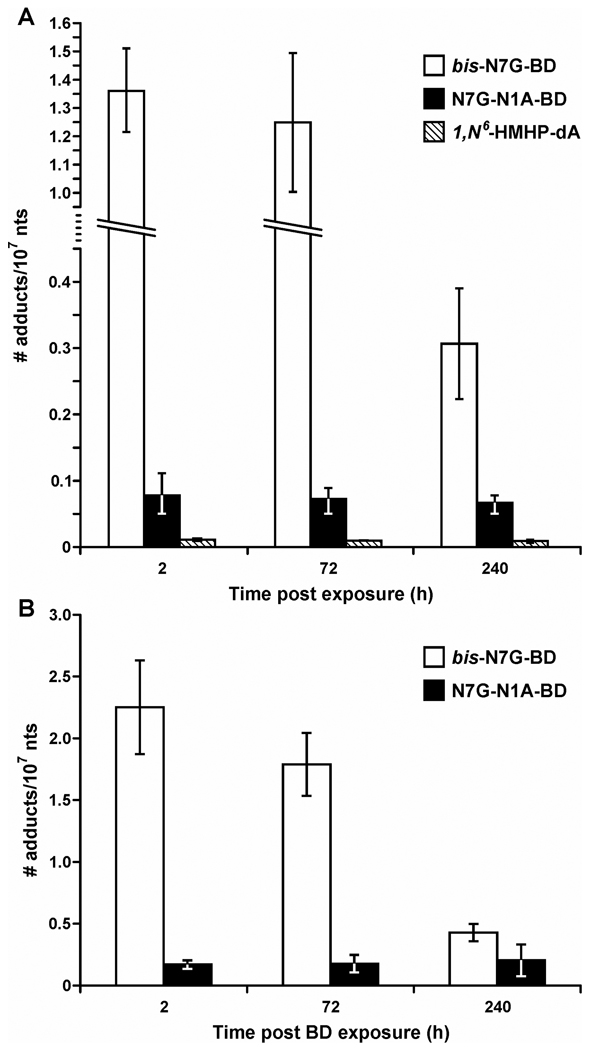
First order kinetic analysis was used to estimate the half life of each bifunctional DEB-DNA adduct in vivo (Supplementary Figures S1–S9). The stability of various DEB-DNA adducts in rodent tissues differed dramatically depending on their structure (Figures 2 and 3, Table 2). In particular, bis-N7G-BD concentrations decreased significantly with an increased post exposure recovery time. Specifically, bis-N7G-BD amounts in mouse liver DNA were approximately 20-fold lower 72 hours post exposure to BD than immediately after exposure (0.18 versus 3.9 adducts per 107 nucleotides, Figure 2). The half-lives of bis-N7G-BD adducts in mouse liver, kidney, and lungs were 2.3–2.4 days, 4.6–5.7 days, and 4.9 days, respectively (Table 2, Figure 1).
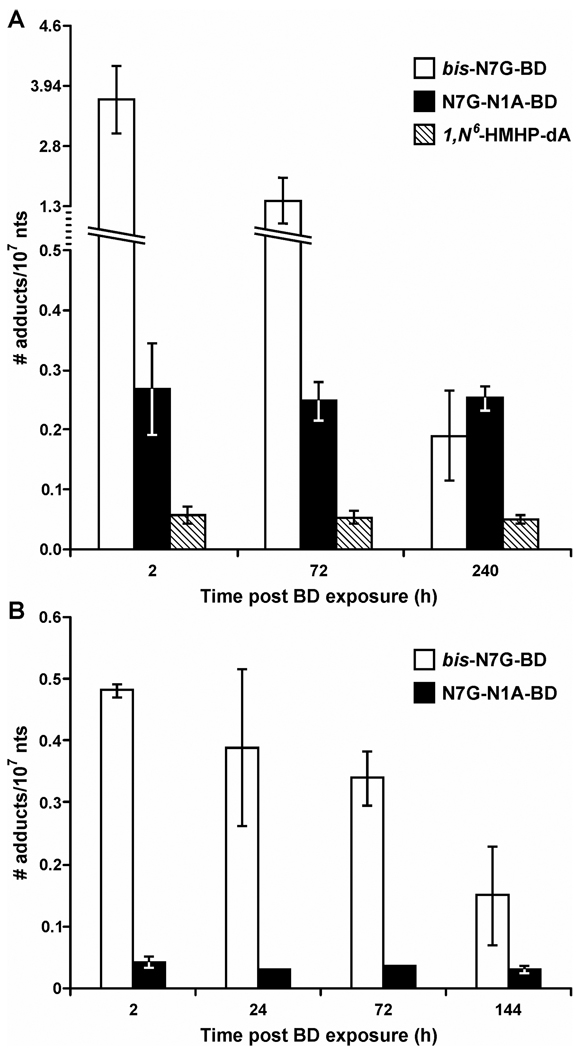
Figure 2.
Persistence of bifunctional DEB-DNA adducts in mouse and rat liver DNA. (A) Female B6C3F1 mice were exposed to 625 ppm BD for 2 weeks, and tissues were collected 2, 72, or 240 h post exposure. (B) Female F344 rats were exposed to 1250 ppm BD for 2 weeks, and tissues were collected 2, 24, 72, or 144 h post exposure. Bis-N7G-BD, N7G-N1A-BD, and 1,N6-HMHP-dA, were quantified by isotope dilution HPLC-ESI-MS/MS analysis of DNA hydrolysates.
Figure 3.
Persistence of bifunctional DEB-DNA adducts in mouse kidney (A) and lung DNA (B). Female B6C3F1 mice were exposed to 625 ppm BD for 2 weeks, and kidney tissues were collected 2, 72, or 240 h post exposure. bis-N7G-BD, N7G-N1A-BD, and 1,N6-HMHP-dA were quantified by isotope dilution HPLC-ESI-MS/MS analysis of DNA hydrolysates.
Table 2.
Estimated half lives of bifunctional DEB-DNA adducts in tissues of B6C3F1 mice exposed to 625 ppm BD for 2 weeks.
| Adduct | t½, days | ||
|---|---|---|---|
| Liver | Kidney | Lung | |
| meso bis-N7G-BD | 2.4 ± 0.8 | 5.7 ± 1.2 | - |
| S,S+R,R bis-N7G-BD | 2.3 ± 0.3 | 4.6 ± 0.12 | 4.9 ± 0.9 |
| N7G-N1A-BD | 36.7 ± 13.6 | 42.4 ± 23.6 | 41.03 ± 27. 8 |
| 1,N6-α,γ -HMHP-dA | 41.9 ± 5.7 | 37.5 ± 16.9 | - |
Unlike bis-N7G-BD, the concentrations of N7G-N1A-BD and 1, N6-HMHP-dA adducts in mouse liver, lung, and kidney tissues decreased very slowly post exposure, with an estimated t1/2 of 36.7–42.4 days (Figures 2, 3, and Table 2). As a result, while bis-N7G-BD adducts were the dominant lesions immediately following BD treatment (Table 1), N7G-N1A-BD became the main bifunctional DEB adducts in mouse liver 10 days post exposure (Figure 2). These results suggest that N7G-N1A-BD and 1, N6-HMHP-dA adducts are not actively repaired in tissues of female rats and mice. Furthermore, there appears to be no tissue-specific accumulation of DEB-DNA adducts, since similar half lives are observed for liver, lung, and kidney tissues (Table 2). Unfortunately, meso bis-N7G-BD and 1, N6-HMHP-dA could not be quantified in the lung since their concentrations were below the detection limits of our current methods.
Formation of bifunctional DEB-DNA adducts in tissues of Mpg and Xpa deficient mice
To investigate potential involvement of the BER and NER repair pathways in the removal of DEB-DNA adducts, the concentrations of bis-N7G-BD, N7G-N1A-BD, and 1, N6-HMHP-dA adducts were measured in liver DNA of female Mpg+/+ and Mpg−/− mice exposed to 62.5 ppm BD by inhalation for 10 days (Table 3) and in Xpa deficient and proficient animals exposed to 625 ppm BD (Table 4). HPLC-ESI-MS/MS analyses revealed little differences between adduct levels in the wild type and Mpg-deficient mice (p value > 0.18), suggesting that this repair pathway does not play a major role in the removal of DEB-induced lesions (Table 3). Similarly, no significant differences were observed between the concentrations of DEB-DNA adducts in liver DNA of Xpa deficient and proficient animals exposed to 625 ppm BD by inhalation (p > 0.2, see Table 4). Taken together, our results are not supportive of participation of the BER and NER pathways in repair of DEB-induced bifunctional DNA damage in tissues of female mice and rats.
Table 3.
Concentrations of BD-induced DNA adduct levels in liver DNA of mpg deficient and mpg proficient mice exposed to 62.5 ppm BD for 10 days.
| Racemic bis-N7G-BD /107 nts |
Meso bis-N7G-BD /107 nts |
N7G-N1A-BD /107 nts |
1,N6-HMHP-dA /107 nts |
|
|---|---|---|---|---|
| mpg+/+ (n = 4) | 1.13 ± 0.35 | 0.12 ± 0.033 | 0.086 ± 0.012 | 0.007 ± 0.009 |
| mpg−/− (n = 4) | 1.13 ± 0.23 | 0.096 ± 0.019 | 0.084 ± 0.011 | 0.011 ± 0.006 |
|
p-value mpg+/+vs. mpg−/− |
0.87 | 0.40 | 0.18 | 0.72 |
Table 4.
Comparison of bis-N7G-BD, N7G-N1A-BD and 1,N6-α-HMHP-dA adduct concentrations in liver DNA of XPA deficient and XPA proficient mice exposed to 625 ppm BD for 2 weeks.
| Racemic bis-N7G-BD /107 nts |
Meso bis-N7G-BD /107 nts |
N7G-N1A-BD /107 nts |
1, N6-HMHP-dA /107 nts |
|
|---|---|---|---|---|
| XPA +/+ (n = 4) | 1.19 ± 0.51 | 0.055 ± .022 | 0.14 ± 0.04 | 0.036 ± 0.017 |
| XPA −/− (n = 4) | 1.14 ± 0.53 | 0.094 ± 0.02 | 0.16 ± 0.02 | 0.041 ± 0.018 |
|
p-value XPA +/+ vs. XPA −/− |
0.63 | 0.20 | 0.44 | 0.67 |
Discussion
Although DEB is a relatively minor metabolite of BD, it is hypothesized to be the ultimate carcinogenic species of BD based on its proven genotoxic potency and the ability to induce bifunctional DNA adducts (7,42–44). In previous studies, we have identified three types of DEB-specific DNA adducts, bis-N7G-BD, N7G-N1A-BD, and 1,N6-HMHP-dA (9,10,12,20), and developed quantitative HPLC-ESI-MS/MS methods for their analyses in tissues of laboratory rodents treated with BD by inhalation (13,14,16). We found that immediately after BD exposure, bis-N7G-BD was the most abundant bifunctional adduct, followed by N7G-N1A-BD, and 1,N6-HMHP-dA, which were 15-fold and 100-fold less abundant, respectively (Table 1) (16).
In the present work, spontaneous hydrolysis and in vivo persistence of DEB-induced bifunctional DNA adducts were examined, and potential pathways responsible for their repair were investigated in a mouse model. Three types of bifunctional DNA adducts induced by butadiene treatment (bis-N7G cross-links, N1A-N7G cross-links, and exocyclic 1,N6-dA adducts) were found to have drastically different half lives in vivo (Figures 2 and 3, Table 2). This phenomenon is largely determined by differences in adducts' hydrolytic stability (Figure 1). While bifunctional alkylation of the N7 of guanine by DEB yields unstable bis-N7G-BD lesions that undergo spontaneous depurination, alkylation of the N1 and the N6 of adenine produces hydrolytically stable N7G-N1A-BDdep and 1,N6-HMHP-dA (Scheme 2) (9,19,19,20). We found that the half-lives of bis-N7G-BD adducts in rodent tissues (2.3–5.7 days) were similar to the rates of spontaneous depurination (3.5–4 days) (Table 2, Figure 1). In contrast, the concentrations of N7G-N1A-BD and 1,N6-HMHP-dA adducts in mouse liver, kidney, and lung tissues remained essentially unchanged following exposure to BD (estimated t1/2 = 36.7 – 42.4 days), indicating that these adducts persist in vivo and could contribute mutagenicity and cytotoxicity of BD.
In the second part of this study, we examined potential contributions of two DNA repair mechanisms, base excision repair (BER) and nucleotide excision repair (NER), to the in vivo removal of DEB-DNA adducts. Methylpurine glycosylase (MPG) is known to repair 7-methyl guanine adducts (23,25). We hypothesized that MPG may be also recognize N7-(2-hydroxy-3,4-epoxy-butyl)guanine monoadducts, which serve as precursors to bis-N7G-BD and N7G-N1A-BD cross-links (Scheme 1), potentially reducing the concentration of both adducts in vivo. Furthermore, MPG is also known to repair 1,N6-etheno-dA and 1,N6- ethano-dA adducts which are structurally similar to 1, N6-HMHP-dA adducts investigated here (45). However, similar concentrations of bis-N7G-BD, N7G-N1A-BD, and 1, N6-HMHP-dA adducts were observed in Mpg deficient and proficient mice exposed to BD by inhalation (Table 3). These results are consistent with in vitro data of Mattes et al. who reported that human and rat MPG proteins do not efficiently excise structurally analogous bis-N7-guanine cross-links induced by nitrogen-mustards (46).
The inability of the mouse MPG protein to remove 1, N6-HMHP-dA may be due to the presence of a six-membered exocyclic ring in its structure in contrast to the five-membered rings of known MPG substrates, EA and εA. In fact, a similar observation was made for exocyclic dG lesions. While 1,N2-ethene-G containing a five-membered exocyclic ring is repaired by Mpg, pyrimido[1,2-α]purin-10(3H)-one (M1G) containing a six-membered exocyclic ring is not a substrate (24). Another important difference between the structures of 1, N6-HMHP-dA and EA/εA is the presence of a positive charge on the N1 of adenine in 1, N6-HMHP-dA. A similar observation was made for 1-methyladenine lesions which also have a positive charge on the N1 of adenine and are not repaired by Mpg (25).
Previous studies involving structurally defined interstrand DNA-DNA cross-links suggested that repair of ICL lesions such bis-N7G-BD and N7G-N1A-BD can be accomplished via NER-mediated incision on each side of the adduct, followed by homologous recombination and ligation (47). In eukaryotic cells, the unhooking of the cross-linked DNA and the formation of repair-induced single strand breaks occurs at or near the site of a stalled or collapsed replication fork, leading to transient double strand breaks (48). Consistent with this mechanism, human fibroblasts deficient in XPA or XPF gene are more sensitive to UV-induced intrastrand cross-links, while XPF deficient cells are more sensitive to nitrogen mustards which are known to form 1,3-interstrand bis-N7G cross-links (49). In contrast, S. cerevisiae cells treated with nitrogen mustards give rise to cross-link induced DNA strand breaks in the absence of NER proteins (50,51). It appears that the preferred repair mechanism is dictated by the chemical structure of the cross-link, as differences have been observed between the enzymatic fate of ICLs induced by psoralen and nitrogen mustards, mitomycin C, and platinum compounds (50,51).
We found that the presence of a functional mouse Xpa gene did not affect the rates of disappearance of DEB-induced DNA-DNA cross-links and 1,N6-dA exocyclic adducts from mouse liver, kidney, and lung tissues (Table 4). These results are in contrast with an earlier observation of an increased mutation frequency in mice deficient in another NER-related gene, Xpc, following treatment with the metabolic precursor to DEB, 3,4-epoxy-1-butene (52). It is possible that NER pathway plays a role in the removal of EB-induced DNA adducts not considered here.
One important DNA repair system requiring further investigation in regard to its role in the removal of DEB-induced lesions is the recombinational repair pathway. Interstrand DNA-DNA cross-links induced by cisplatin are repaired by the replication-coupled Fanconi Anemia (FA) recombinational pathway (53). FA cells have been shown to be hyper-sensitive to DEB: in fact, DEB is used as a clinical tool to diagnose FA (37,54,55). Our preliminary cell culture studies with repair-proficient and deficient Chinese Hamster Lung cells suggest that the FA recombinational repair pathway contributes to repair of DEB-induced DNA-DNA cross-links (Malayappan et al., unpublished data). A possible involvement of this recombination repair pathway in the protection of cells against DEB-induced DNA damage remains to be investigated. However, our observation of a long-term persistence of N7G-N1A-BD and 1,N6-HMHP-dA in rodent tissues (at least 10 days) suggests that either recombination repair does not play a major role in repair of these lesions in liver, kidney, and lung tissues of female mice and rats, or that this repair pathway requires active DNA replication. Studies are now in progress in our laboratory to examine the effects of DEB-induced bifunctional DNA adducts on DNA structure and fidelity of DNA polymerases.
Supplementary Material
Acknowledgements
We thank Uthpala Seneviratne for synthesizing the 15N-labeled internal standard for 1,N6-γ-HMHP-dA, Bob Carlson (University of Minnesota Cancer Center) for preparing figures for this manuscript, Professor Roger Jones (Rutgers University) for providing isotopically labeled dG, Leona Sampson for providing the MPG deficient stock mice for this study, Brock Matter and Peter Vilalta (University of Minnesota Cancer Center) for helpful suggestions on nanoLC and nanoMS method development, and Cate Tsufis and Srikanth Kotapati (University of Minnesota Cancer Center) for help with DNA extractions.
Funding
This work is supported by grants from the National Cancer Institute (C.A. 100670), the National Institute of Environmental Health Sciences (ES 12689), the Health Effects Institute (Agreement 05-12), and the American Chemistry Council (OLF-163.0).
Footnotes
List of Abbreviations: BD, 1,3-butadiene; bis-N7G-BD, 1,4-bis-(guan-7-yl)-2,3-butanediol; DEB, 1,2,3,4-diepoxybutane; EA, 1,N6-ethano-dA; EB, 3,4-epoxy-1-butene; EBD, 3,4-epoxy-1,2-butanediol; EH, epoxide hydrolase; G-A, guanine-adenine; HPLC-ESI+-MS/MS, high performance liquid chromatography-electrospray ionization tandem mass spectrometry; ICL, interstrand cross-links; m7G, N7-methylguanine; N7G-N1A-BD, 1-(guan-7-yl)-4-(aden-1-yl)-2,3-butanediol; N7G-N6A-BD, 1-(guan-7-yl)-4-(aden-6-yl)-2,3-butanediol; 1,N6-α-HMHP-dA, 1,N6-(1-hydroxymethyl-2-hydroxypropan-1,3-diyl)-2'-deoxyadenosine; 1,N6-γ-HMHP-dA, 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2'-deoxyadenosine; MPG, methyl purine glycosylase; SD, standard deviation; NER, nucleotide excision repair; PDE, phosphodiesterase; SRM, selected reaction monitoring; THBG, N7-(2, 3, 4-trihydroxy-1-yl)guanine; XPA, xeroderma pigmentosum complementation protein A.
Supporting Information Available: kinetic analysis results for BD adduct removal in vitro and in vivo. This material is available free of charge via the Internet at http://pubs.acs.org.
Reference List
- 1.White WC. Butadiene production process overview. Chem. Biol. Interact. 2007;166:10–14. doi: 10.1016/j.cbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Hecht SS. Tobacco smoke carcinogens and lung cancer. J. Natl. Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 3.Pelz N, Dempster NM, Shore PR. Analysis of low molecular weight hydrocarbons including 1,3-butadiene in engine exhaust gases using an aluminum oxide porous-layer open-tubular fused-silica column. J. Chromatogr. Sci. 1990;28:230–235. doi: 10.1093/chromsci/28.5.230. [DOI] [PubMed] [Google Scholar]
- 4.Melnick RL, Huff JE. 1,3-Butadiene induces cancer in experimental animals at all concentrations from 6.25 to 8000 parts per million. IARC Sci. Publ. 1993:309–322. [PubMed] [Google Scholar]
- 5.Ward JB, Jr, Ammenheuser MM, Bechtold WE, Whorton EB, Jr, Legator MS. hprt mutant lymphocyte frequencies in workers at a 1,3-butadiene production plant. Environ. Health Perspect. 1994;102 Suppl 9:79–85. doi: 10.1289/ehp.94102s979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng Q, Redetzke DL, Hackfeld LC, Hodge RP, W DM, Walker VE. Mutagenicity of stereochemical configurations of 1,2-epoxybutene and 1,2:3,4-diepoxybutane in human lymphblastoid cells. Chem. Biol. Interact. 2007 doi: 10.1016/j.cbi.2006.06.001. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 7.Sasiadek M, Norppa H, Sorsa M. 1,3-Butadiene and its epoxides induce sister-chromatid exchanges in human lymphocytes in vitro. Mutat. Res. 1991;261:117–121. doi: 10.1016/0165-1218(91)90057-s. [DOI] [PubMed] [Google Scholar]
- 8.Recio L, Steen AM, Pluta LJ, Meyer KG, Saranko CJ. Mutational spectrum of 1,3-butadiene and metabolites 1,2-epoxybutene and 1,2,3,4-diepoxybutane to assess mutagenic mechanisms. Chem. Biol. Interact. 2001;135–136:325–341. doi: 10.1016/s0009-2797(01)00220-4. [DOI] [PubMed] [Google Scholar]
- 9.Park S, Tretyakova N. Structural characterization of the major DNA-DNA cross-link of 1,2,3,4-diepoxybutane. Chem. Res. Toxicol. 2004;17:129–136. doi: 10.1021/tx0342058. [DOI] [PubMed] [Google Scholar]
- 10.Park S, Hodge J, Anderson C, Tretyakova NY. Guanine-adenine cross-linking by 1,2,3,4-diepoxybutane: potential basis for biological activity. Chem. Res. Toxicol. 2004;17:1638–1651. doi: 10.1021/tx0498206. [DOI] [PubMed] [Google Scholar]
- 11.Zhang XY, Elfarra AA. Reaction of 1,2,3,4-diepoxybutane with 2'-deoxyguanosine: initial products and their stabilities and decomposition patterns under physiological conditions. Chem. Res. Toxicol. 2005;18:1316–1323. doi: 10.1021/tx0500979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seneviratne U, Antsypovich S, Goggin M, Quirk Dorr D, Guza R, Moser A, Thompson C, York DM, Tretyakova N. Exocyclic deoxyadenosine adducts of 1,2,3,4-diepoxybutane: synthesis, structural elucidation, and mechanistic studies. Chem. Res. Toxicol. 2010;23:118–133. doi: 10.1021/tx900312e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goggin M, Loeber R, Park S, Walker V, Wickliffe J, Tretyakova N. HPLC-ESI+-MS/MS analysis of N7-guanine-N7-guanine DNA cross-links in tissues of mice exposed to 1,3-butadiene. Chem. Res. Toxicol. 2007;20:839–847. doi: 10.1021/tx700020q. [DOI] [PubMed] [Google Scholar]
- 14.Goggin M, Anderson C, Park S, Swenberg J, Walker V, Tretyakova N. Quantitative high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry analysis of the adenine-guanine cross-links of 1,2,3,4-diepoxybutane in tissues of butadiene-exposed B6C3F1 mice. Chem. Res. Toxicol. 2008;21:1163–1170. doi: 10.1021/tx800051y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goggin M, Swenberg JA, Walker VE, Tretyakova N. Molecular dosimetry of 1,2,3,4-diepoxybutane-induced DNA-DNA cross-links in B6C3F1 mice and F344 rats exposed to 1,3-butadiene by inhalation. Cancer Res. 2009;69:2479–2486. doi: 10.1158/0008-5472.CAN-08-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goggin M, Seneviratne U, Swenberg JA, Walker VE, Tretyakova N. Column switching HPLC-ESI+-MS/MS methods for quantitative analysis of exocyclic dA adducts in the DNA of laboratory animals exposed to 1,3-butadiene. Chem. Res Toxicol. 2010 doi: 10.1021/tx900439w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson RF. Species differences in the metabolism of olefins: implications for risk assessment. Chem. Biol. Interact. 2001;135–136:53–64. doi: 10.1016/s0009-2797(01)00170-3. [DOI] [PubMed] [Google Scholar]
- 18.Thornton-Manning JR, Dahl AR, Bechtold WE, Henderson RF. Gender and species differences in the metabolism of 1,3-butadiene to butadiene monoepoxide and butadiene diepoxide in rodents following low-level inhalation exposures. Toxicology. 1996;113:322–325. doi: 10.1016/0300-483x(96)03466-x. [DOI] [PubMed] [Google Scholar]
- 19.Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens. New York and London: Plenum Press; 1983. [Google Scholar]
- 20.Park S, Anderson C, Loeber R, Seetharaman M, Jones R, Tretyakova N. Interstrand and intrastrand DNA-DNA cross-linking by 1,2,3,4-diepoxybutane: role of stereochemistry. J. Am. Chem. Soc. 2005;127:14355–14365. doi: 10.1021/ja051979x. [DOI] [PubMed] [Google Scholar]
- 21.Hollis T, Lau A, Ellenberger T. Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase. Mutat. Res. 2000;460:201–210. doi: 10.1016/s0921-8777(00)00027-6. [DOI] [PubMed] [Google Scholar]
- 22.Norbury CJ, Hickson ID. Cellular responses to DNA damage. Annu. Rev. Pharmacol. Toxicol. 2001;41:367–401. doi: 10.1146/annurev.pharmtox.41.1.367. [DOI] [PubMed] [Google Scholar]
- 23.Asaeda A, Ide H, Asagoshi K, Matsuyama S, Tano K, Murakami A, Takamori Y, Kubo K. Substrate specificity of human methylpurine DNA N-glycosylase. Biochemistry. 2000;39:1959–1965. doi: 10.1021/bi9917075. [DOI] [PubMed] [Google Scholar]
- 24.Hang B. Repair of exocyclic DNA adducts: rings of complexity. Bioessays. 2004;26:1195–1208. doi: 10.1002/bies.20130. [DOI] [PubMed] [Google Scholar]
- 25.Lee CY, Delaney JC, Kartalou M, Lingaraju GM, Maor-Shoshani A, Essigmann JM, Samson LD. Recognition and processing of a new repertoire of DNA substrates by human 3-methyladenine DNA glycosylase (AAG) Biochemistry. 2009;48:1850–1861. doi: 10.1021/bi8018898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McHugh PJ, Gill RD, Waters R, Hartley JA. Excision repair of nitrogen mustard-DNA adducts in Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:3259–3266. doi: 10.1093/nar/27.16.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sibghatullah, Husain I, Carlton W, Sancar A. Human nucleotide excision repair in vitro: repair of pyrimidine dimers, psoralen and cisplatin adducts by HeLa cell-free extract. Nucleic Acids Res. 1989;17:4471–4484. doi: 10.1093/nar/17.12.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reardon JT, Sancar A. Nucleotide excision repair. Prog. Nucleic Acid Res Mol. Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 29.Wang QE, Zhu QZ, Wani MA, Wani G, Chen JM, Wani AA. Tumor suppressor p53 dependent recruitment of nucleotide excision repair factors XPC and TFIIH to DNA damage. Dna Repair. 2003;2:483–499. doi: 10.1016/s1568-7864(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 30.Vasquez KM, Legerski RJ. DNA interstrand crosslinks: repair, cell signaling, and therapeutic implications. Environmental and Molecular Mutagenesis. 2010;51:491–492. doi: 10.1002/em.20598. [DOI] [PubMed] [Google Scholar]
- 31.Muniandy PA, Liu J, Majumdar A, Liu ST, Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Critical Reviews in Biochemistry and Molecular Biology. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood RD. Mammalian nucleotide excision repair proteins and interstrand crosslink repair. Environmental and Molecular Mutagenesis. 2010;51:520–526. doi: 10.1002/em.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mccabe KM, Olson SB, Moses RE. DNA interstrand crosslink repair in mammalian cells. Journal of Cellular Physiology. 2009;220:569–573. doi: 10.1002/jcp.21811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajski SR, Williams RM. DNA cross-linking agents as antitumor drugs. Chem Rev. 1998;98:2723–2796. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- 35.Hinz JM. Role of homologous recombination in DNA interstrand crosslink repair. Environmental and Molecular Mutagenesis. 2010;51:582–603. doi: 10.1002/em.20577. [DOI] [PubMed] [Google Scholar]
- 36.Legerski RJ. Repair of DNA interstrand cross-links during S phase of the mammalian cell cycle. Environmental and Molecular Mutagenesis. 2010;51:540–551. doi: 10.1002/em.20566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auerbach AD. Fanconi anemia and its diagnosis. Mutat. Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orelli B, McClendon TB, Tsodikov OV, Ellenberger T, Niedernhofer LJ, Scharer OD. The XPA-binding domain of ERCC1 is required for nucleotide excision repair but not other DNA repair pathways. J. Biol. Chem. 2010;285:3705–3712. doi: 10.1074/jbc.M109.067538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cantor SB, Xie J. Assessing the link between BACH1/FANCJ and MLH1 in DNA crosslink repair. Environmental and Molecular Mutagenesis. 2010;51:500–507. doi: 10.1002/em.20568. [DOI] [PubMed] [Google Scholar]
- 40.Ho V, Scharer OD. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environmental and Molecular Mutagenesis. 2010;51:552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- 41.Wickliffe JK, Ammenheuser MM, Salazar JJ, Abdel-Rahman SZ, Hastings-Smith DA, Postlethwait EM, Lloyd RS, Ward JB., Jr A model of sensitivity: 1,3-butadiene increases mutant frequencies and genomic damage in mice lacking a functional microsomal epoxide hydrolase gene. Environ. Mol. Mutagen. 2003;42:106–110. doi: 10.1002/em.10181. [DOI] [PubMed] [Google Scholar]
- 42.Melnick RL, Kohn MC. Mechanistic data indicate that 1,3-butadiene is a human carcinogen. Carcinogenesis. 1995;16:157–163. doi: 10.1093/carcin/16.2.157. [DOI] [PubMed] [Google Scholar]
- 43.Sasiadek M, Schlade K, Busza H, Czemarmazowicz H, Stembalska A. Classical and molecular cytogenetics in analysis of diepoxybutane- induced chromosome aberrations. Mutat. Res. 1998;419:155–161. doi: 10.1016/s1383-5718(98)00131-4. [DOI] [PubMed] [Google Scholar]
- 44.Stayner LT, Dankovic DA, Smith RJ, Gilbert SJ, Bailer AJ. Human cancer risk and exposure to 1,3-butadiene--a tale of mice and men. Scand. J Work Environ. Health. 2000;26:322–330. doi: 10.5271/sjweh.549. [DOI] [PubMed] [Google Scholar]
- 45.Ham AJ, Engelward BP, Koc H, Sangaiah R, Meira LB, Samson LD, Swenberg JA. New immunoaffinity-LC-MS/MS methodology reveals that Aag null mice are deficient in their ability to clear 1,N6-etheno-deoxyadenosine DNA lesions from lung and liver in vivo. DNA Repair (Amst) 2004;3:257–265. doi: 10.1016/j.dnarep.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Mattes WB, Lee CS, Laval J, O'Connor TR. Excision of DNA adducts of nitrogen mustards by bacterial and mammalian 3-methyladenine-DNA glycosylases. Carcinogenesis. 1996;17:643–648. doi: 10.1093/carcin/17.4.643. [DOI] [PubMed] [Google Scholar]
- 47.Van Houten B, Gamper H, Holbrook SR, Hearst JE, Sancar A. Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a defined position. Proc. Natl. Acad. Sci. U. S. A. 1986;83:8077–8081. doi: 10.1073/pnas.83.21.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem. Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clingen PH, Arlett CF, Hartley JA, Parris CN. Chemosensitivity of primary human fibroblasts with defective unhooking of DNA interstrand cross-links. Exp. Cell Res. 2007;313:753–760. doi: 10.1016/j.yexcr.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 50.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand crosslink-induced double-strand breaks. Mol. Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wickliffe JK, Galbert LA, Ammenheuser MM, Herring SM, Xie J, Masters OE, III, Friedberg EC, Lloyd RS, Ward JB., Jr 3,4-Epoxy-1-butene, a reactive metabolite of 1,3-butadiene, induces somatic mutations in Xpc-null mice. Environ. Mol. Mutagen. 2006;47:67–70. doi: 10.1002/em.20169. [DOI] [PubMed] [Google Scholar]
- 53.Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Auerbach AD. Diagnosis of Fanconi anemia by diepoxybutane analysis. Curr. Protoc. Hum. Genet. 2003 doi: 10.1002/0471142905.hg0807s37. Chapter. 8, Unit. [DOI] [PubMed] [Google Scholar]
- 55.Auerbach AD. Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp. Hematol. 1993;21:731–733. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.