Abstract
Multiple animal models have been developed to recapitulate phenotypes of the human disease, schizophrenia. A model that simulates many of the cognitive and sensory deficits of the disorder is the use of random variable prenatal stress (PS) in the rat. These deficits suggest a molecular origin in the hippocampus, a brain region that plays a role in the regulation of stress. To study both hippocampal gene expression changes in offspring of prenatally stressed dams and to address genetic variability, we used a random array of prenatal stressors in three different rat strains with diverse responses to stress: Fischer, Sprague-Dawley, and Lewis rats. Candidate genes involved in stress, schizophrenia, cognition, neurotrophic effects, and immunity were selected for assessment by real-time quantitative PCR under resting conditions and following a brief exposure to restraint stress. PS resulted in significant differences in gene expression in the offspring that were strain dependent. mRNA expression for the N-methyl D-aspartate receptor subtype 2B (Grin2b) was increased, and tumor necrosis factor-alpha (Tnfα) transcript was decreased in PS Sprague-Dawley and Lewis rats, but not in the Fischer rats. Expression of brain-derived neurotrophic factor (Bdnf) mRNA in the hippocampus was increased after an acute stress in all controls of each strain, yet a decrease was seen after acute stress in the PS Sprague-Dawley and Lewis rats. Expression of the glucocorticoid receptor (Nr3c1) was decreased in the Fischer strain when compared to Lewis or Sprague-Dawley rats, though the Fischer rats had markedly higher α7 nicotinic receptor (Chrna7) expression. The expression differences seen in these animals may be important elements of the phenotypic differences seen due to PS and genetic background.
Keywords: schizophrenia, rat model, prenatal stress, strain difference, Grin2b, Nr3c1, Chrna7, Tnfα, Bdnf
1. Introduction
Schizophrenia is a multifactorial illness affecting more than three million people, with contributions from both genetic and environmental factors. The disease is highly heritable [1], although thought to be multigenic [2,3]. Significant evidence of linkage to schizophrenia has been found at more than 15 chromosomal loci [4,5]. Implicated genes include brain-derived neurotrophic factor [6,7], glutamate receptors [8], and nicotinic receptors [9,10], all of which may play a role in the cognitive deficits seen in schizophrenia [11,12,13]. The environment also plays an important role in this developmental disorder [14]. Environmental factors such as stress, particularly during gestation, have been linked to schizophrenia [15,16,17,18].
Inherited endophenotypic traits, such as sensory processing of auditory stimuli [19] and eye-tracking deficits [20], are common in schizophrenic patients. Various models have been created to simulate these endophenotypes in animals [21]. One such animal model, the random variable PS paradigm, was developed to study the effects of prenatal stress (PS) on cognitive and sensory behavior and on gene expression in the offspring [22]. Administered in the last week of rat fetal development, it produces a phenotype with multiple features of schizophrenia including cognitive impairment, aberrant neuronal architecture, social withdrawal, altered stress responses, deficits in gating sensory information, disrupted pre-pulse inhibition, and increased response to amphetamines [23,24,25]. A previous microarray study of gene expression changes in this model was done in prefrontal cortex of Sprague-Dawley rats, demonstrating mRNA transcript alterations due to PS [26]. We have extended these studies to the hippocampus, following administration of the same random stress paradigm, because numerous studies have demonstrated pathologies in the hippocampus of patients with schizophrenia. Relevant candidate genes in five groups were evaluated: stress genes, candidate schizophrenia genes, genes involved in cognitive processes such as learning and memory, neurotrophins, and genes regulated in immune system signaling. In addition, three different rat strains with differing levels of stress responsivity were used to determine whether the genetic background of the animals would affect the outcome of expression changes. The strains were chosen based on their differing hormonal responses to stress: 1) Fischer rats exhibit hyperactivation of the hypothalamic-pituitary-adrenal (HPA) axis, 2) Lewis rats are known to be hypo-responsive to stress, and 3) Sprague-Dawley rats have intermediate corticosterone responses [27,28,29,30,31].
Six random stressors were administered during the last trimester to timed-pregnant rats. Male PS offspring at 56 days were either exposed to an acute stressor or not stressed, then sacrificed for study. The hippocampus was removed and utilized for gene expression assays. This region has been implicated in the pathology of schizophrenia [32,33], and in the cognitive effects of stress [34,35,36]. Of the genes assayed, significant effects of PS were found for Grin2b, Tnfα, and Bdnf, though the changes were strain dependent. Expression of Chrna7 and Nr3c1 was significantly different across rat strains, with no effects seen from PS.
2. Materials
Timed-pregnant rats were purchased from a breeding facility (Charles River, Kingston, NY) and arrived on the second gestational day (E2). Animals were given ad libitum access to food and water while on a 12 hour light/dark cycle. After parturition, dams and offspring were left undisturbed in large static cages until weaning on postnatal day (P) 23–25. There are no effects on litter size or birth weight with this random stress gestational model [25]. Males were then removed and housed with a single male littermate until the acute stress procedure and sacrifice on P56. All animal procedures were conducted in accordance with NIH policies and were approved by the University of Maryland, Baltimore Animal Care and Use Committee.
3. Methods
3.1. Prenatal stress procedure
Dams were randomly chosen to be placed in either a control or PS group. All dams in the PS group were exposed to a random variable stress paradigm for one week, from E14 to E21. Mild psychological and physiological stressors were given two or three times a day. All stressed dams received the same stressors at the same time of day. The stressors utilized were: 1) home cage placement in a cold room (4°C) for 6 hours, 2) 15 minutes of non-escapable swim, 3) one hour restraint in a Plexiglas restrainer (Harvard Bioscience, Boston, MA), 4) twelve hours of overnight food deprivation, 5) light cycle reversal, or 6) twelve hours of cage overcrowding.
3.2. Postnatal acute stress
Male pups were sacrificed for the collection of hippocampal tissue and trunk blood by decapitation on P56. Sacrifice of both control and PS groups was performed at one of two time points: 1) at baseline without acute stress (baseline), or 2) immediately after a 30 min restraint stress (acute). At sacrifice, blood was collected in tubes containing the anticoagulant EDTA and plasma was harvested following centrifugation. The plasma was stored frozen at −80°C until the corticosterone assay was performed.
3.3. Corticosterone measurement
Plasma corticosterone (CORT) concentrations were determined by radioimmunoassay according to manufacturer’s instructions (MP Biomedicals, Orangeburg, NY). The assay has a sensitivity of 3 ng/ml, and an intra- and inter-assay coefficient of variation of less than 10%.
3.4. Tissue Preparation and RNA Isolation
After rapid decapitation, hippocampi composed of both dorsal and ventral structures were extracted from the brains by freehand dissection and placed in 2 mls of RNAlater solution (Ambion, Carlsbad, CA) to stabilize RNA for gene expression quantification. Hippocampi were stored at −80°C prior to their transfer into tubes containing homogenization beads and TRIzol® lysis buffer (Invitrogen, Carlsbad, CA). Homogenization was accomplished using a Mini Beadbeater-8® (Biospec, Bartlesville, OK). Following tissue lysis, RNA was purified using an RNeasy mini kit (Qiagen, Valencia, CA). RNA quality was verified using an Agilent bioanalyzer 2100 (Agilent, Santa Clara, CA).
3.5. Quantification of mRNA levels by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
Gene specific primers were designed to cross intron-exon boundaries and obtained from Eurofins Operon (Huntsville, AL). Primers used for the study are listed in Supplemental Table 1.
First strand complementary DNA (cDNA) was generated by reverse transcription using random hexamers and Superscript III reverse transcriptase (Invitrogen). qRT-PCR was performed on a Bio-rad iCycler IQ® (Bio-rad, Hercules, CA) using SYBRgreen supermix(Bio-rad). Each sample was run in triplicate, followed by a heat dissociation step to identify nonspecific products. Triplicates of each sample were averaged for each gene of interest (GOI) and then normalized to the housekeeping gene Polr2a (Pol) [37] using mean normalized expression (MNE) and the actual efficiencies (eff) of each run: (effpol)CT/(effGOI)CT=MNE, as described [38].
3.6. Statistical Analysis
For all analyses, measures were expressed as means of each group ±SEM and compared using a mixed model analysis of variance (ANOVA) with fixed effects of strain, condition, and time group, plus a random effect of the offspring’s dam (SAS foundation software V9, Cary, NC). Each group consisted of five to six male rats with each rat being taken from a different litter. Statistical differences between experimental groups were considered significant when p<0.05.
4. Results
4.1 Corticosterone response
At baseline there were no significant differences in CORT, either between strains or in the PS animals (Fig. 1). Each strain displayed the typical response to an acute stress with elevated CORT following a 30 minute restraint stress. PS did not affect the CORT response in either the Fischer or Sprague-Dawley rats. However, in Lewis control and PS animals CORT levels were significantly lower than in the other two strains after the acute stress. Further, after the acute stress the Lewis PS animals had CORT levels that were significantly lower than the Lewis rat non-PS controls (p<0.05).
Figure 1. Corticosterone levels before and immediately after an acute stress.
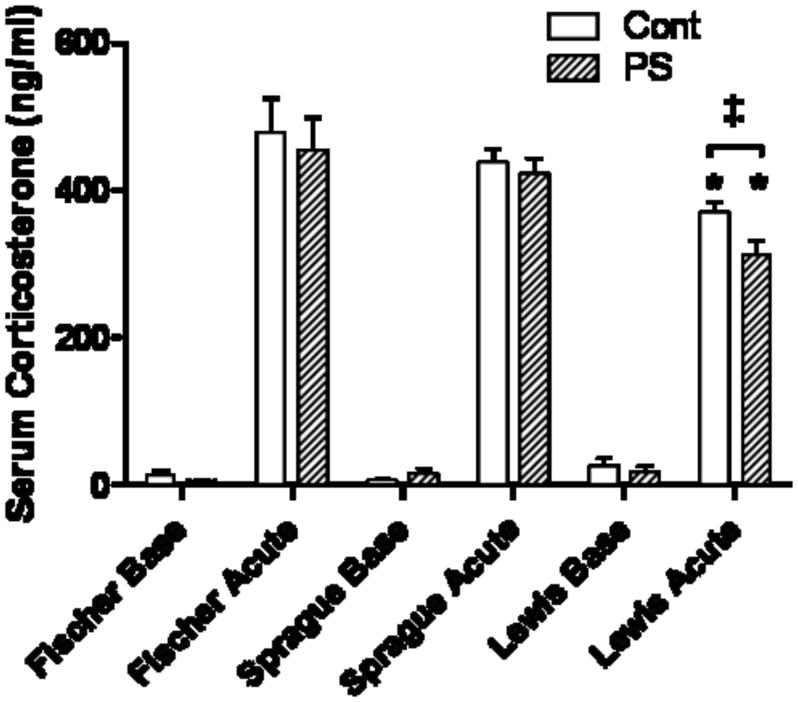
Lewis control and PS animals have significantly lower serum CORT after an acute stress compared to Fischer or Sprague-Dawley rats (*, p<0.05). CORT levels were further reduced in Lewis PS rats, compared to the non-PS control (‡, p< 0.05).
4.2 Gene expression in the hippocampus
Candidate genes were selected to survey changes generated by the PS and/or the acute stressor, and to evaluate any rat strain differences. The gene groups, shown in Table 1, include genes known to play a role in stress or cognition, schizophrenia candidate genes, neurotrophins involved in development, and genes in the immune system implicated in stress and infection. Two genes were expressed at different levels across the strains and three were affected by PS in a strain-specific manner. Expression for all other genes listed was not significantly different in the PS animals nor between the rat strains.
Table 1.
Genes analyzed by qRT-PCR in the hippocampus of each animal group.
| Stress | Schizophrenia | Cognition | Neurotrophins Immunity | |
|---|---|---|---|---|
| NR3C1* | CHRNA7* | GRIN1 | BDNF** | TNFA** |
| NR3C2 | RELN | GRIN2A | NGF | MR1 |
| CRH | DTNBP1 | GRIN2B** | NTF3 | B2M |
| CRHR1 | NRG1 | GRIN2C | NTF5 | IL1b |
| CRHBP | PPP3CA | GRIN2D | NTRK1 | IL6 |
| POMC | SYN2 | GRINA | NTRK2 | IL10 |
| HSPB1 | SYN3 | GRIA1 | NTRK3 | |
| COMT | GRIA2 | MECP2 | ||
| RGS4 | GRIA3 | |||
| DISC1 | GRIA4 | |||
| CAMK2A | ||||
| CAMK2B | ||||
| PRKACA | ||||
| PRKACB | ||||
| DLG4 | ||||
| LRRC7 | ||||
| NCAM1 | ||||
Significant differences were found for those in bold.
different in rat strains;
affected by PS
4.2. Grin2b, Tnfα, and Bdnf hippocampal expression is altered by prenatal stress in a strain specific pattern
4.2.1. Hippocampal Grin2b expression levels are increased in prenatally stressed Sprague-Dawley and Lewis Rats
Expression of Grin2b mRNA was increased in offspring of PS Sprague-Dawley rat dams (Fig. 2; p<0.05). The increase was found at both baseline and after acute stress, suggesting there was not a further increase due to the acute stress. In the Lewis rats, there was an increase in mRNA expression in the PS animals at baseline that did not reach significance. However, following exposure to an episode of restraint stress, an increase in hippocampal Grin2b mRNA levels was detected. Expression of Grin2b in the Fischer strain was not changed by PS or acute stress.
Figure 2. Grin2b mRNA levels in the hippocampus.
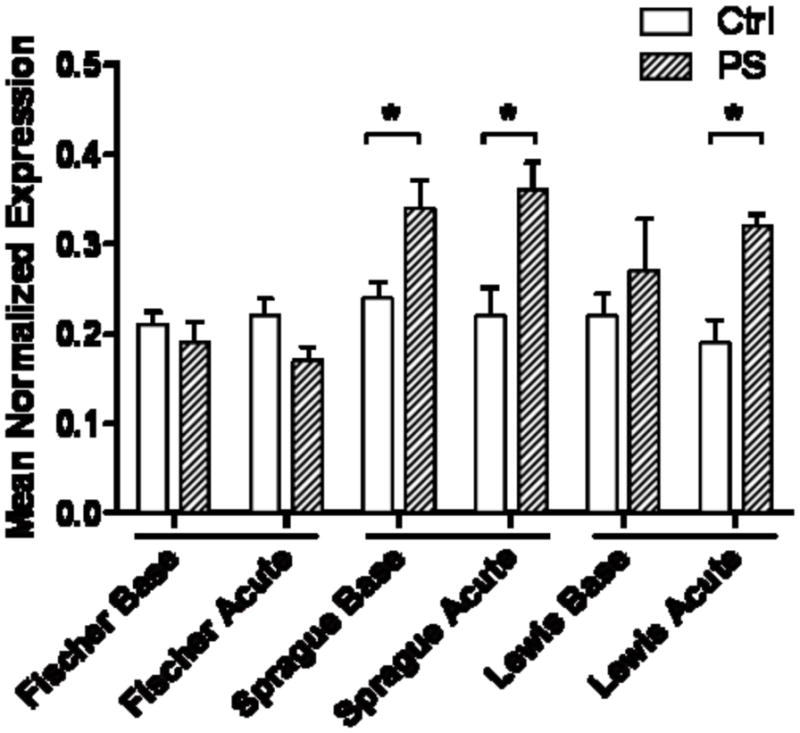
Sprague-Dawley baseline and acute, plus Lewis acutely stressed, have up-regulated expression when compared to their respective controls (*, p<0.05). Fischer rats were not changed.
4.2.2. Expression of Tnfα is decreased by acute stress in prenatally stressed offspring
Of the genes in the immunity group, only the expression of hippocampal Tnfα mRNA was decreased following acute stress in PS Sprague-Dawley and Lewis rat strains (Fig. 3; p<0.05). The decrease suggests that immune system responses in the hippocampus of PS animals are altered in these strains. There was no such change in expression of Tnfα mRNA in the PS Fischer rats.
Figure 3. Tnfα mRNA levels in the hippocampus.
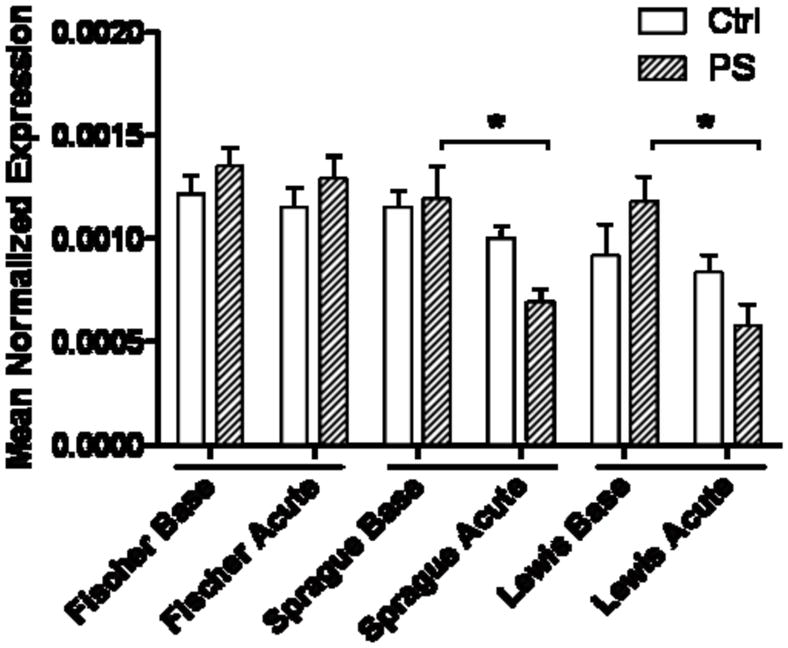
Sprague-Dawley and Lewis rats exposed to PS show a significant decrease in expression of Tnfα after an acute stress that is not seen in the Fischer animals (*, p<0.05).
4.2.3. Expression of Bdnf in the hippocampus is altered in the prenatally stressed Sprague-Dawley and Lewis strains, but not in the Fischer strain
Although the Bdnf gene is currently known to express multiple isoforms from its nine exons, there is one coding region identical to all transcripts which all translate to the same protein. We targeted our analysis to this coding region, and found that the expression of total Bdnf mRNA was significantly increased by an acute stress in all three rat strain controls that were not exposed to PS (Fig. 4; p<0.05), suggesting a hippocampal response to stressful events. However, the responses in the PS offspring were quite different and were strain dependent. Bdnf mRNA expression was significantly elevated at baseline in the PS Sprague-Dawley and Lewis rat strains (Fig. 4; p<0.05), but was unchanged in the Fischer rats. There was no further change in Bdnf expression in the PS Sprague-Dawley or Fischer rats following acute restraint stress. However, Bdnf expression markedly declined in the PS Lewis rats (Fig. 4; p<0.05) after acute stress, compared to respective baseline expression. The results suggest that Bdnf is differentially regulated in the three rat strains.
Figure 4. Bdnf mRNA expression in the hippocampus.
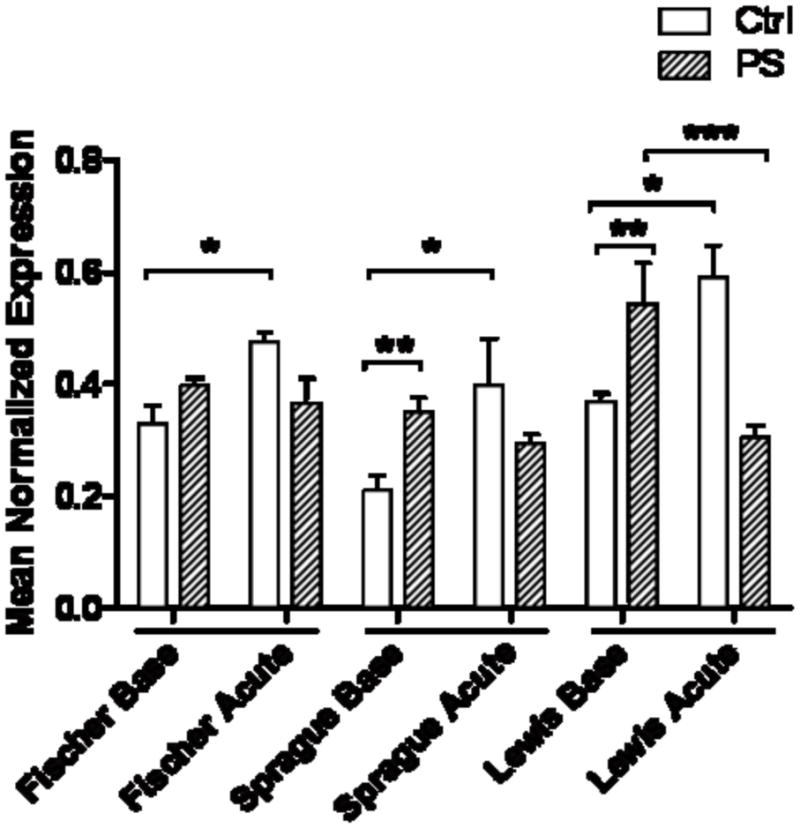
Non-PS control animals of each strain significantly up-regulate Bdnf mRNA after an acute stress (*, p<0.05). PS Sprague-Dawley and Lewis rats had higher baseline Bdnf expression (**, p<0.05), which was unchanged after acute stress in the Sprague-Dawleys, but significantly decreased in the Lewis rats (***, p<0.05). PS did not change expression in the Fischer strain.
4.3. Nr3c1 and Chrna7 mRNA expression is significantly different across rat strains
Both Nr3c1 and Chrna7 mRNA transcripts are differentially expressed in the hippocampus of the three rat strains. Nr3c1 mRNA was expressed at significantly lower levels in the Fischer rats at baseline, compared to both the Sprague-Dawley and Lewis rats (Fig. 5; p<0.05). There was no effect of PS on gene expression in any of the rat strains. Chrna7 was expressed at an approximately 50% higher level in the Fischer strain than in Sprague-Dawley or Lewis rats (Fig. 6; p<0.01). Again, there was no effect of PS or the acute stress on mRNA expression levels.
Figure 5. Nr3c1 mRNA expression in the hippocampus.
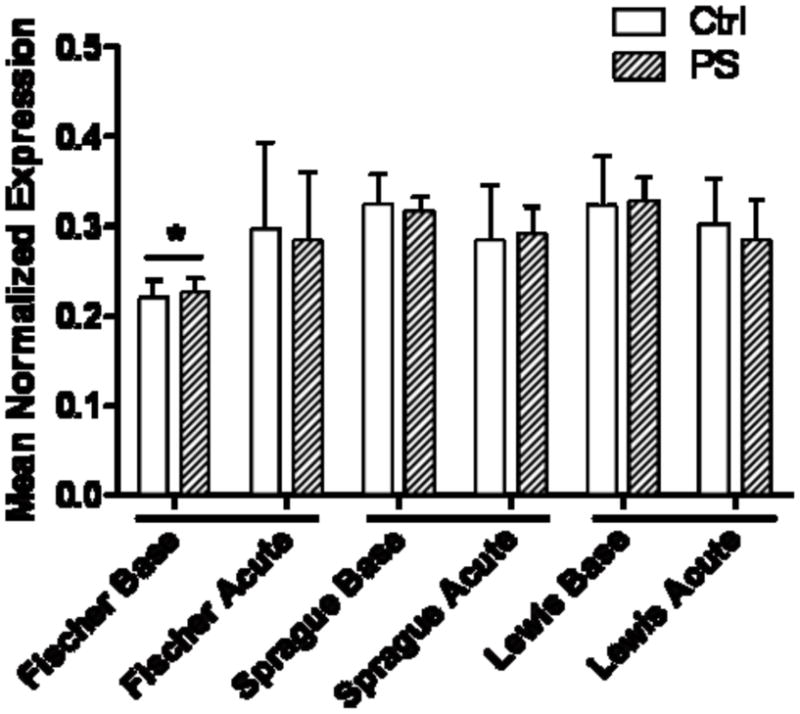
Fischer rats, regardless of PS or Ctrl group, have less Nr3c1 transcript than either Sprague-Dawley or Lewis strains (*, p<0.05).
Figure 6. Chrna7 mRNA expression in the hippocampus.
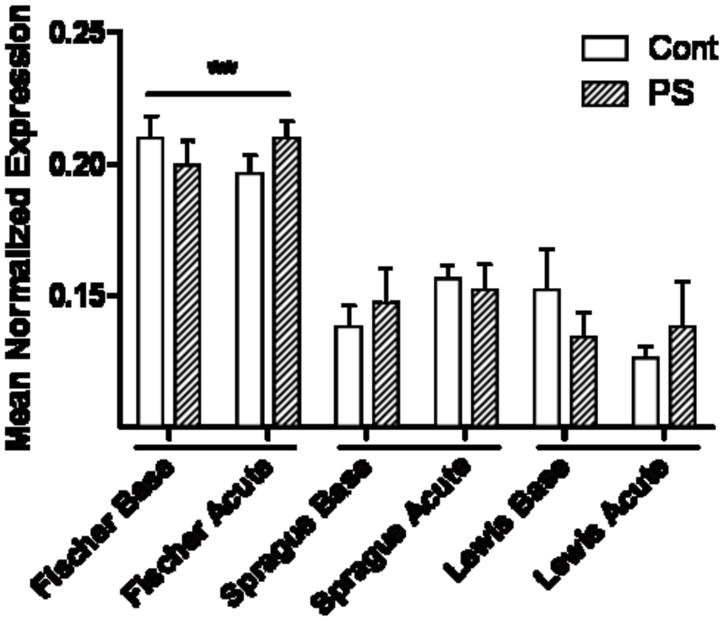
Expression of Chrna7 was significantly lower in Sprague-Dawley and Lewis than in Fischer rats (**, p<0.01).
5. Discussion
The use of six random stressors presented prenatally in the last week of fetal rat gestation results in altered phenotypes that are similar to those seen in schizophrenia [26,22]. We added genetic diversity to this paradigm by studying the effects of the stressors in three rat strains who differ in their stress response. Gestational stress exposure elicits enduring changes in the behavioral phenotype of the adult offspring. While repeated exposure to restraint stress results in a behavioral phenotype related to depression and anxiety, prenatal exposure to repeated mild, variable stressors produces a pattern of behavioral differences closer to those seen in schizophrenia [22]. Studies done by Kinnunen et al. [26] and Bogoch et al. [39] previously identified a number of gene expression differences associated with excitatory or inhibitory synaptic transmission in the frontal cortex and hippocampus. The present study extends these earlier findings by examining gene expression differences across three different animal strains. Since gene-environment interactions contribute prominently to neuropsychiatric illnesses, it is important to identify gene expression differences that can be attributed to genetic differences or environmental insults.
Nr3c1, also known as the glucocorticoid receptor, is important in regulating the negative feedback loop of the HPA-axis, and in its regulatory role of gene transcription. In our study, the Lewis rats had a smaller CORT response to an acute stress than the other strains. In addition, there was an up-regulation of Nr3c1 in Lewis rats compared to Fischer. This suggests that having more receptors available might blunt the CORT response, possibly through increased negative feedback and tighter regulation, which is consistent with the known physiology of the HPA-axis [40]. The negative feedback signal that dampens the stress response may not be as robust in Fischer rats, allowing an enhanced CORT response to an acute stressor. Our results agree with three studies showing lower hippocampal Nr3c1 mRNA expression in Fischer rats, compared to Lewis rats [29,41,42]. However, two studies show lower Nr3c1 mRNA levels in Lewis rats [43,44], and another shows no difference between the two strains [45]. These differences may reflect genetic diversity due to vendor source or different techniques for analysis of mRNA expression. Our study used quantitative real-time PCR to study mRNA levels derived from both dorsal and ventral hippocampus, while others used in situ hybridization, northern blot, or cytosolic receptor binding assays.
Expression of Grin2b was increased in adult male offspring of PS Sprague-Dawley and Lewis rats. NMDA receptors are part of the ionotropic glutamate class of receptors; glutamate is the predominant excitatory neurotransmitter in the brain. The receptors are involved in long-term potentiation (LTP) and long-term depression (LTD), important measures of learning and memory [46]. NMDA receptors are heteromeric complexes with one Grin1 subunit and at least one of four NR2 subunit types: Grin2A, Grin2B, Grin2C, or Grin2D. Grin2B is currently thought to be of particular importance for the induction of CA1 hippocampal LTD [47,48,49]. It is also known that stress can enable and facilitate LTD, and that blocking of Grin2B with specific antagonists can impair hippocampal CA1 LTD caused by acute stress [47]. In this model of PS, the expression of Grin2B was increased in both Sprague-Dawley and Lewis rats, but not in Fischer. Increased Grin2B expression would likely increase LTD and might be involved in the underlying mechanism for decreased cognitive performance in PS Sprague-Dawley rats [22].
Stress is a modifier of overall immune function [50,51] and cytokine transcription in the brain [52,53,54]. Tnfα, which can be down-regulated in the hippocampus by different stressors [55], is a pro-inflammatory cytokine normally involved in systemic inflammation and regulation of immune cells. Here, we see a rapid down-regulation of available transcript immediately following stress, demonstrating the tight regulation imposed due to the PS paradigm on the Lewis and Sprague-Dawley rats. Interestingly, Tnfα affects glutamate transmission, and plays a role in synaptic plasticity and LTP [56]. The cytokine can up-regulate AMPA receptors and simultaneously decrease synaptic inhibition through endocytosis of GABA receptors [57]. A decrease in Tnfα, as seen in our PS animals, may decrease the cognitive performance of these rats. Tnfα can also provide neuroprotective benefits in the brain. Tnfα stimulates activation of the transcription factor nuclear factor κB (Nfκb), which may reduce cell death signals in neurons [58,59]. The reduction in Tnfα expression in the PS Sprague-Dawley and Lewis rats after an acute stress could negatively affect cell survival in these animals. We have previously found aberrant neuronal architecture in PS Sprague-Dawley rats [24]. Effects on brain development in the PS animals may be related to the inability of these animals to maintain Tnfα transcript levels after an acute stress.
Another developmental gene, Bdnf, was also affected by PS in the Sprague-Dawley and Lewis rats, but not in the Fischer. Bdnf is well known as an immediate early gene and its ability to respond quickly to stimuli. Interestingly, the 3′-UTR of Bdnf has the ability to either shorten or prolong the half-life of gene transcripts dependent on membrane depolarization [60]. Bdnf transcripts were increased in the non-PS controls of all three strains after an acute stress. However, the mRNA levels were significantly higher at baseline in Sprague-Dawley and Lewis rats in the PS offspring. Further, there was no up-regulation by acute stress in the PS animals in these strains as was seen in the controls. In the PS Lewis rats, acute stress resulted in a marked decrease in Bdnf expression, while there was no change in the PS Fischer rats. Although it is widely known that chronic stress or chronically elevated glucocorticoids can decrease hippocampal BDNF mRNA expression [61,62,63], the contrary is also true immediately after an acute stress, whereby there is a temporary increase in BDNF transcription as we see in our model [64,65]. Bdnf is initially expressed in a precursor form, proBdnf, which is processed to the mature neurotrophin, mBdnf [66]. mBdnf and proBdnf signal through two different receptors, the TrkB and p75 receptors, respectively. The activation of these receptors results in opposing cellular responses [67,68]. proBDNF binding to p75 causes long-term depression, apoptosis, and synaptic retraction. However, mBDNF activation of the TrkB receptor results in the promotion of synaptic plasticity, cell survival, neuronal differentiation, and is sufficient for late-phase LTP while inhibiting LTD. Because these two differing signaling cascades come from the same Bdnf mRNA, examination of the Bdnf protein in PS animals would be of interest. We hypothesize that mBdnf signaling would be decreased in PS animals, given that a decrease in these responses could contribute to the altered phenotype of the model.
The increased baseline transcript levels of Bdnf in the PS Lewis and Sprague-Dawley rats, as well as a failure of the PS strain to respond to an acute stressor with an increase in Bdnf, suggests that this rat strain may be less protected in inflammatory environments. This possibility is consistent with the differences in expression of Chrna7 between the strains. Chrna7 expression was 50% higher in the Fischer rats than in either the Sprague-Dawley or Lewis rats. The receptors expressed from this gene are protective for inflammation [69,70]. In addition, there is a relationship between stimulation of this receptor and Tnfα responses [71,72]. Thus, the low levels of Chrna7 expression in Sprague-Dawley and Lewis rats may contribute to the differences in gene expression seen in these strains, compared to the Fischer rat.
Genetic diversity is a significant and fundamental obstacle in human studies, as are the effects of environmental factors such as stress on gene expression. Our results suggest that developmental pathways are affected by PS in the hippocampus with consequences that extend into adulthood, and that strain differences play an important role. The genes affected in the PS animals may be related to endophenotypic deficits seen in schizophrenia. Both Chrna7 and Bdnf are replicated candidate genes in schizophrenia with decreased expression [73,7,74]. In addition, all the genes that were found to be altered in this study play a role in synaptic transmission pathways. The impact of PS on altered signaling in the hippocampus is, thus, an important consideration. Further investigation of the effects of PS on gene expression in the hippocampus and the interaction with genetic diversity will provide a better understanding of how brain development is affected by the environment.
Supplementary Material
Acknowledgments
This research was funded by NIH grants DA09457, MH81177, and the Veterans Affairs Medical Research Service to SL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait - Evidence from a meta-analysis of twin studies. Arch Gen Psych. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 2.Owen MJ, Williams HJ, O’Donovan MC. Schizophrenia genetics: advancing on two fronts. Curr Opin Gen Develop. 2009;19:266–270. doi: 10.1016/j.gde.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Freedman R, Leonard S, Olincy A, Kaufmann CA, Malaspina D, Cloninger CR, Svrakic D, Faraone SV, Tsuang MT. Evidence for the multigenic inheritance of schizophrenia. Am J Med Genet. 2001;105:794–800. doi: 10.1002/ajmg.10100. [DOI] [PubMed] [Google Scholar]
- 4.Baron M. Genetics of schizophrenia and the new millennium: Progress and pitfalls. Am J Hum Gen. 2001;68:299–312. doi: 10.1086/318212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 6.Angelucci F, Brene S, Mathe AA. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry. 2005;10:345–352. doi: 10.1038/sj.mp.4001637. [DOI] [PubMed] [Google Scholar]
- 7.Chen DC, Wang J, Wang B, Yang SC, Zhang CX, Zheng YL, Li YL, Wang N, Yang KB, Xiu MH, Kosten TR, Zhang XY. Decreased levels of serum brain-derived neurotrophic factor in drug-naive first-episode schizophrenia: relationship to clinical phenotypes. Psychopharmacol. 2009;207:375–380. doi: 10.1007/s00213-009-1665-6. [DOI] [PubMed] [Google Scholar]
- 8.Coyle JT, Tsai G, Goff D. Converging evidence of NMDA receptor hypofunction in the pathophysiology of schizophrenia. Ann N Y Acad Sci. 2003;1003:318–327. doi: 10.1196/annals.1300.020. [DOI] [PubMed] [Google Scholar]
- 9.Adams CE, Stevens KE. Evidence for a role of nicotinic acetylcholine receptors in schizophrenia. Frontiers Biosci. 2007;12:4755–4772. doi: 10.2741/2424. [DOI] [PubMed] [Google Scholar]
- 10.Leonard S, Adler LE, Benhammou K, Berger R, Breese CR, Drebing C, Gault J, Lee MJ, Logel J, Olincy A, Ross RG, Stevens K, Sullivan B, Vianzon R, Virnich DE, Waldo M, Walton K, Freedman R. Smoking and mental illness. Pharmacol Biochem Behav. 2001;70:561–570. doi: 10.1016/s0091-3057(01)00677-3. [DOI] [PubMed] [Google Scholar]
- 11.Farber NB. The NMDA receptor hypofunction model of psychosis. Ann N Y Acad Sci. 2003;1003:119–130. doi: 10.1196/annals.1300.008. [DOI] [PubMed] [Google Scholar]
- 12.Kumari V, Postma P. Nicotine use in schizophrenia: The self medication hypotheses. Neurosci Biobehav Rev. 2005;29:1021–1034. doi: 10.1016/j.neubiorev.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Leonard S, Mexal S, Freedman R. Smoking, Genetics and Schizophrenia: Evidence for Self Medication. J Dual Diag. 2007;3:43–59. doi: 10.1300/J374v03n03_05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClure RK, Lieberman JA. Neurodevelopmental and neurodegenerative hypotheses of schizophrenia: a review and critique. Curr Opin Psychiatry. 2003;16:S15–S28. [Google Scholar]
- 15.Van Os J, Selten JP. Prenatal exposure to maternal stress and subsequent schizophrenia. The May 1940 invasion of The Netherlands. Br J Psychiatry. 1998;172:324–326. doi: 10.1192/bjp.172.4.324. [DOI] [PubMed] [Google Scholar]
- 16.King S, Laplante DP. The effects of prenatal maternal stress on children’s cognitive development: Project Ice Storm. Stress. 2005;8:35–45. doi: 10.1080/10253890500108391. [DOI] [PubMed] [Google Scholar]
- 17.Mednick SA, Huttunen MO, Machon RA. Prenatal influenza infections and adult schizophrenia. Schizophr Bull. 1994;20:263–267. doi: 10.1093/schbul/20.2.263. [DOI] [PubMed] [Google Scholar]
- 18.Howes OD, McDonald C, Cannon M, Arseneault L, Boydell J, Murray RM. Pathways to schizophrenia: the impact of environmental factors. Int J Neuropsychopharmacol. 2004;7 (Suppl 1):S7–S13. doi: 10.1017/S1461145704004122. [DOI] [PubMed] [Google Scholar]
- 19.Freedman R, Adams CE, Adler LE, Bickford PC, Gault J, Harris JG, Nagamoto HT, Olincy A, Ross RG, Stevens KE, Waldo M, Leonard S. Inhibitory neurophysiological deficit as a phenotype for genetic investigation of schizophrenia. Am J Med Genet. 2000;97:58–64. doi: 10.1002/(sici)1096-8628(200021)97:1<58::aid-ajmg8>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 20.Braff DL, Freedman R, Schork NJ, Gottesman II. Deconstructing schizophrenia: An overview of the use of endophenotypes in order to understand a complex disorder. Schizophren Bull. 2007;33:21–32. doi: 10.1093/schbul/sbl049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dawe GS, Hwang EHJ, Tan CH. Pathophysiology and Animal Models of Schizophrenia. Ann Acad Med Singapore. 2009;38:425–430. [PubMed] [Google Scholar]
- 22.Koenig JI, Elmer GI, Shepard PD, Lee PR, Mayo C, Joy B, Hercher E, Brady DL. Prenatal exposure to a repeated variable stress paradigm elicits behavioral and neuroendocrinological changes in the adult offspring: potential relevance to schizophrenia. Behavior Brain Res. 2005;156:251–261. doi: 10.1016/j.bbr.2004.05.030. [DOI] [PubMed] [Google Scholar]
- 23.Koenig JI, Kirkpatrick B, Lee P. Glucocorticoid hormones and early brain development in schizophrenia. Neuropsychopharmacol. 2002;27:309–318. doi: 10.1016/S0893-133X(01)00396-7. [DOI] [PubMed] [Google Scholar]
- 24.Markham JA, Koenig JI. Prefrontal Neuronal Architecture Is Disrupted in the Rat Prenatal Stress Model of Schizophrenia. Schizophren Bull. 2009;35:137–137. [Google Scholar]
- 25.Lee PR, Brady DL, Shapiro RA, Dorsa DM, Koenig JI. Prenatal stress generates deficits in rat social behavior: Reversal by oxytocin. Brain Res. 2007;1156:152–167. doi: 10.1016/j.brainres.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinnunen AK, Koenig JI, Bilbe G. Repeated variable prenatal stress alters pre- and postsynaptic gene expression in the rat frontal pole. J Neurochem. 2003;86:736–748. doi: 10.1046/j.1471-4159.2003.01873.x. [DOI] [PubMed] [Google Scholar]
- 27.Sternberg EM, Young WS, Bernardini R, Calogero AE, Chrousos GP, Gold PW, Wilder RL. A Central Nervous-System Defect in Biosynthesis of Corticotropin-Releasing Hormone Is Associated with Susceptibility to Streptococcal Cell Wall-Induced Arthritis in Lewis Rats. Proc Nat Acad Sci. 1989;86:4771–4775. doi: 10.1073/pnas.86.12.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhabhar FS, McEwen BS, Spencer RL. Stress-Response, Adrenal-Steroid Receptor Levels and Corticosteroid-Binding Globulin Levels - A Comparison Between Sprague-Dawley, Fischer-344 and Lewis Rats. Brain Res. 1993;616:89–98. doi: 10.1016/0006-8993(93)90196-t. [DOI] [PubMed] [Google Scholar]
- 29.Dhabhar FS, Miller AH, McEwen BS, Spencer RL. Differential Activation of Adrenal-Steroid Receptors in Neural and Immune Tissues of Sprague-Dawley, Fischer-344, and Lewis Rats. J Neuroimmunol. 1995;56:77–90. doi: 10.1016/0165-5728(94)00135-b. [DOI] [PubMed] [Google Scholar]
- 30.Moncek F, Kvetnansky R, Jezova D. Differential responses to stress stimuli of Lewis and Fischer rats at the pituitary and adrenocortical level. Endocr Regul. 2001 [PubMed] [Google Scholar]
- 31.Kosten TA, Ambrosio E. HPA axis function and drug addictive behaviors: insights from studies with Lewis and Fischer 344 inbred rats. Psychoneuroendocrinol. 2002;27:35–69. doi: 10.1016/s0306-4530(01)00035-x. [DOI] [PubMed] [Google Scholar]
- 32.Bogerts B, Meertz E, Schonfeldtbausch R. Basal Ganglia and Limbic System Pathology in Schizophrenia - A Morphometric Study of Brain Volume and Shrinkage. Arch Gen Psych. 1985;42:784–791. doi: 10.1001/archpsyc.1985.01790310046006. [DOI] [PubMed] [Google Scholar]
- 33.Heckers S, Konradi C. Hippocampal neurons in schizophrenia. J Neural Transm. 2002;109:891–905. doi: 10.1007/s007020200073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McEwen BS, Magarinos AM. Stress and hippocampal plasticity: Implications for the pathophysiology of affective disorders. Hum Psychopharmacol. 2001;16:S7–S19. doi: 10.1002/hup.266. [DOI] [PubMed] [Google Scholar]
- 35.Maroun M, Richter-Levin G. Exposure to acute stress blocks the induction of long-term potentiation of the amygdala-prefrontal cortex pathway in vivo. J Neurosci. 2003;23:4406–4409. doi: 10.1523/JNEUROSCI.23-11-04406.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alfonso J, Frasch AC, Flugge G. Chronic stress, depression and antidepressants: Effects on gene transcription in the hippocampus. Rev Neurosci. 2005;16:43–56. doi: 10.1515/revneuro.2005.16.1.43. [DOI] [PubMed] [Google Scholar]
- 37.Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 38.Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19:1439–1440. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- 39.Bogoch Y, Biala YN, Linial M, Weinstock M. Anxiety induced by prenatal stress is associated with suppression of hippocampal genes involved in synaptic function. J Neurochem. 2007;101:1018–1030. doi: 10.1111/j.1471-4159.2006.04402.x. [DOI] [PubMed] [Google Scholar]
- 40.de Kloet ER, Joels M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- 41.Gomez F, Lahmame A, deKloet ER, Armario A. Hypothalamic-pituitary-adrenal response to chronic stress in five inbred rat strains: Differential responses are mainly located at the adrenocortical level. Neuroendocrinol. 1996;63:327–337. doi: 10.1159/000126973. [DOI] [PubMed] [Google Scholar]
- 42.Jongen-Relo AL, Pothuizen HHJ, Feldon J, Pryce CR. Comparison of central corticosteroid receptor expression in male Lewis and Fischer rats. Brain Res. 2002;953:223–231. doi: 10.1016/s0006-8993(02)03293-6. [DOI] [PubMed] [Google Scholar]
- 43.Kabbaj M, Yoshida S, Numachi Y, Matsuoka H, Devine DP, Sato M. Methamphetamine differentially regulates hippocampal glucocorticoid and mineralocorticoid receptor rnRNAs in Fischer and Lewis rats. Mol Brain Res. 2003;117:8–14. doi: 10.1016/s0169-328x(03)00257-2. [DOI] [PubMed] [Google Scholar]
- 44.Smith CC, Omeljaniuk RJ, Whitfield HJ, Jr, Aksentijevich S, Fellows MQ, Zelazowska E, Gold PW, Sternberg EM. Differential mineralocorticoid (type 1) and glucocorticoid (type 2) receptor expression in Lewis and Fischer rats. Neuroimmunomodulation. 1994;1:66–73. doi: 10.1159/000097092. [DOI] [PubMed] [Google Scholar]
- 45.Numachi Y, Yoshida S, Toda S, Matsuoka H, Sato M. Two inbred strains of rats, Fischer 344 and Lewis, showed differential behavior and brain expression of corticosterone receptor mRNA induced by methamphetamine. Ann NY Acad Sci. 2000;914:33–45. doi: 10.1111/j.1749-6632.2000.tb05181.x. [DOI] [PubMed] [Google Scholar]
- 46.Malenka RC, Bear MF. LTP and LTD: An Embarrassment of Riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 47.Yang CH, Huang CC, Hsu KS. Behavioral Stress Enhances Hippocampal CA1 Long-Term Depression through the Blockade of the Glutamate Uptake. J Neurosci. 2005;25:4288–4293. doi: 10.1523/JNEUROSCI.0406-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L, Wong TP, Pozza MF, Lingenhoehl K, Wang Y, Sheng M, Auberson YP, Wang YT. Role of NMDA Receptor Subtypes in Governing the Direction of Hippocampal Synaptic Plasticity. Science. 2004;304:1021–1024. doi: 10.1126/science.1096615. [DOI] [PubMed] [Google Scholar]
- 49.Izumi Y, Auberson YP, Zorumski CF. Zinc Modulates Bidirectional Hippocampal Plasticity by Effects on NMDA Receptors. J Neurosci. 2006;26:7181–7188. doi: 10.1523/JNEUROSCI.1258-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marketon JIW, Glaser R. Stress hormones and immune function. Cell Immunol. 2008;252:16–26. doi: 10.1016/j.cellimm.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 51.Padgett DA, Glaser R. How stress influences the immune response. Trends Immunol. 2003;24:444–448. doi: 10.1016/s1471-4906(03)00173-x. [DOI] [PubMed] [Google Scholar]
- 52.Jacque C, Thurin JM. Stress, immunity and nervous system physiology. M S-Med Sci. 2002;18:1160–1166. [Google Scholar]
- 53.Anisman H, Merali Z. Cytokines, stress and depressive illness: brain-immune interactions. Ann Med. 2003;35:2–11. doi: 10.1080/07853890310004075. [DOI] [PubMed] [Google Scholar]
- 54.Dunn AJ, Wang JP, Ando T. Effects of cytokines on cerebral neurotransmission - Comparison with the effects of stress. Adv Exper Med Biol. 1999;461:117–127. doi: 10.1007/978-0-585-37970-8_8. [DOI] [PubMed] [Google Scholar]
- 55.Bartolomucci A, Palanza P, Parmigiani S, Pederzani T, Merlot E, Neveu PJ, Dantzer R. Chronic psychosocial stress down-regulates central cytokines mRNA. Brain Res Bull. 2003;62:173–178. doi: 10.1016/j.brainresbull.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 56.Pickering M, Cumiskey D, O’Connor JJ. Actions of TNF-alpha on glutamatergic synaptic transmission in the central nervous system. Exper Physiol. 2005;90:663–670. doi: 10.1113/expphysiol.2005.030734. [DOI] [PubMed] [Google Scholar]
- 57.Stellwagen D, Beattie EC, Seo JY, Malenka RC. Differential regulation of AMPA receptor and GABA receptor trafficking by tumor necrosis factor-alpha. J Neurosci. 2005;25:3219–3228. doi: 10.1523/JNEUROSCI.4486-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng B, Christakos S, Mattson MP. Tumor Necrosis Factors Protect Neurons Against Metabolic Excitotoxic Insults and Promote Maintenance of Calcium Homeostasis. Neuron. 1994;12:139–153. doi: 10.1016/0896-6273(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 59.Barger SW, Horster D, Furukawa K, Goodman Y, Krieglstein J, Mattson MP. Tumor-Necrosis-Factor-Alpha and Tumor-Necrosis-Factor-Beta Protect Neurons Against Amyloid Beta-Peptide Toxicity - Evidence for Involvement of A Kappa-B-Binding Factor and Attenuation of Peroxide and Ca2+ Accumulation. Proc Nat Acad Sci. 1995;92:9328–9332. doi: 10.1073/pnas.92.20.9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fukuchi M, Tsuda M. Involvement of the 3′-untranslated region of the brain-derived neurotrophic factor gene in activity-dependent mRNA stabilization. J Neurochem. 2010;115:1222–1233. doi: 10.1111/j.1471-4159.2010.07016.x. [DOI] [PubMed] [Google Scholar]
- 61.Chao HM, Sakai RR, Ma LY, Mcewen BS. Adrenal steroid regulation of neurotrophic factor expression in the rat hippocampus. Endocrinol. 1998;139:3112–3118. doi: 10.1210/endo.139.7.6114. [DOI] [PubMed] [Google Scholar]
- 62.Schaaf MJM, de Jong J, de Kloet ER, Vreugdenhil E. Downregulation of BDNF mRNA and protein in the rat hippocampus by corticosterone. Brain Res. 1998;813:112–120. doi: 10.1016/s0006-8993(98)01010-5. [DOI] [PubMed] [Google Scholar]
- 63.Fuchikami M, Morinobu S, Kurata A, Yamamoto S, Yamawaki S. Single immobilization stress differentially alters the expression profile of transcripts of the brain-derived neurotrophic factor (BDNF) gene and histone acetylation at its promoters in the rat hippocampus. Internat J Neuropsychopharmacol. 2009;12:73–82. doi: 10.1017/S1461145708008997. [DOI] [PubMed] [Google Scholar]
- 64.Molteni R, Calabrese F, Cattaneo A, Mancini M, Gennarelli M, Racagni G, Riva MA. Acute stress responsiveness of the neurotrophin BDNF in the rat hippocampus is modulated by chronic treatment with the antidepressant duloxetine. Neuropsychopharmacol. 2009;34:1523–1532. doi: 10.1038/npp.2008.208. [DOI] [PubMed] [Google Scholar]
- 65.Marmigere F, Givalois L, Rage F, Arancibia S, Tapia-Arancibia L. Rapid induction of BDNF expression in the hippocampus during immobilization stress challenge in adult rats. Hippocampus. 2003;13:646–655. doi: 10.1002/hipo.10109. [DOI] [PubMed] [Google Scholar]
- 66.Lessmann V, Brigadski T. Mechanisms, locations, and kinetics of synaptic BDNF secretion: An update. Neurosci Res. 2009;65:11–22. doi: 10.1016/j.neures.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 67.Pang P, Teng H, Zaitsev E, Woo N, Sakata K, Zhen S, Teng K, Yung W, Hempstead B, Lu B. Cleavage of proBDNF by tPA/Plasmin Is Essential for Long-Term Hippocampal Plasticity. Science. 2004;306:487–491. doi: 10.1126/science.1100135. [DOI] [PubMed] [Google Scholar]
- 68.Schweigreiter R. The dual nature of neurotrophins. Bioessays. 2006;28:583–594. doi: 10.1002/bies.20419. [DOI] [PubMed] [Google Scholar]
- 69.Wang H, Yu M, Ochani M, Amella CA, Tanovic M, Susarla S, Li JH, Wang HC, Yang H, Ulloa L, Al-Abed Y, Czura CJ, Tracey KJ. Nicotinic acetylcholine receptor alpha 7 subunit is an essential regulator of inflammation. Nature. 2003;421:384–388. doi: 10.1038/nature01339. [DOI] [PubMed] [Google Scholar]
- 70.de Jonge WJ, Ulloa L. The alpha7 nicotinic acetylcholine receptor as a pharmacological target for inflammation. Brit J Pharmacol. 2007;151:915–929. doi: 10.1038/sj.bjp.0707264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parrish WR, Rosas-Ballina M, Gallowitsch-Pureta M, Ochani M, Ochani K, Yang LH, Hudson L, Lin X, Patel N, Johnson SM, Chavan S, Goldstein RS, Czura CJ, Miller EJ, Al-Abed Y, Traccy KJ, Pavlov VA. Modulation of TNF release by choline requires alpha 7 subunit nicotinic acetylcholine receptor-mediated signaling. Mol Med. 2008;14:567–574. doi: 10.2119/2008-00079.Parrish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pavlov VA, Parrish WR, Rosas-Ballina M, Ochani M, Puerta M, Qchani K, Chavan S, Al-Abed Y, Tracey KJ. Brain acetylcholinesterase activity controls systemic cytokine levels through the cholinergic anti-inflammatory pathway. Brain Behav Immun. 2009;23:41–45. doi: 10.1016/j.bbi.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 74.Freedman R, Hall M, Adler LE, Leonard S. Evidence in postmortem brain tissue for decreased numbers of hippocampal nicotinic receptors in schizophrenia. Biol Psychiatry. 1995;38:22–33. doi: 10.1016/0006-3223(94)00252-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


