Abstract
Diverse histone modifications play important roles in transcriptional regulation throughout eukaryotes, and recent studies have implicated histone H2B ubiquitylation in active transcription. The necessity of at least three enzymes (E1, E2 and E3), as well as ongoing transcription events, for efficient H2B ubiquitylation complicates mechanistic studies of H2B ubiquitylation relative to other histone modifications. Here we describe experimental protocols for preparation of human H2B ubiquitylation factors, ubiquitylation substrates and transcription factors, as well as the use of these factors to establish H2B ubiquitylation mechanisms during transcription. The methods include reliable protein interaction and E3 ubiquitylation assays that can be widely applied to confirm cognate E2–E3 pairs in other protein ubiquitylation systems, optimized in vitro ubiquitylation assays for various histone substrates, and a transcription-coupled H2B ubiquitylation assay in a highly purified transcription system. These comprehensive analyses have revealed (i) that RAD6 serves as the cognate E2 for the BRE1 complex in human cells, as previously established in yeast, (ii) that RAD6, through direct interaction with the BRE1 complex, ubiquitylates chromatinized H2B at lysine 120 and (iii) that PAF1 complex-mediated transcription is required for efficient H2B ubiquitylation. This experimental system permits detailed mechanistic analyses of H2B ubiquitylation during transcription by providing information concerning both precise enzyme functions and physical interactions between the transcription and histone modification machineries.
Keywords: Histone H2B ubiquitylation, RAD6, BRE1 complex, PAF1 complex, Chromatin, Transcription
1. Introduction
Protein ubiquitylation is mediated by the sequential action of three classes of enzymes – the ubiquitin activating enzyme (E1), ubiquitin conjugating enzymes (E2s) and ubiquitin ligases (E3s). An E2 ubiquitylates its target protein in conjunction with its cognate RING finger-type E3, which specifically interacts with the protein substrate and the E2 [1]. Despite the discovery of histone H2B (H2B) ubiquitylation about 30 years ago, identification of enzymes responsible for H2B ubiquitylation was relatively recent. Thus, yeast genetic studies showed that Rad6 [2] and Bre1 [3,4] serve as E2 and E3 enzymes, respectively, and their intrinsic enzyme activities were confirmed by recent biochemical analysis [5].
In addition to basic ubiquitylation enzymes Rad6 and Bre1, a requirement of the RNA polymerase II-associated Paf1 complex for efficient H2B ubiquitylation in yeast implicated H2B ubiquitylation in transcription-related events [6,7]. In support of this idea, studies in yeast further showed, first, that the level of ubiquitylated H2B (ubH2B) is transiently increased around the core promoter region upon activator-dependent transcription of some genes [8,9] and, second, that H2B ubiquitylation is a prerequisite for the downstream H3K4 methylation that marks actively transcribed genes [10,11]. Since H2B ubiquitylation occurs at a lysine residue (K123 in yeast and K120 in human) that is embedded within the C-terminal α-helix, it is plausible that changes in nucleosome structure during transcription allow Paf1 complex-recruited H2B ubiquitylation factors easier access to the ubiquitylation site [12].
Although H2B ubiquitylation systems are highly conserved from yeast to human, clear demonstrations of factors and mechanistic explanations for H2B ubiquitylation in human cells have been more recent. This reflects in part the limitations on genetic approaches in mammalian cells, which in turn led to an emphasis on the establishment of in vitro H2B ubiquitylation assays with purified factors. In the establishment of a reliable in vitro mammalian H2B ubiquitylation assay, several important issues need to be considered. As shown for several E2s [13,14], uncontrolled accessibility of E2s to highly basic histones results in non-specific ubiquitylation in purified systems in the absence of E3. This complicates establishment of an E3 requirement for histone ubiquitylation. Moreover, the in vitro histone ubiquitylation reaction generates a large number of protein-ubiquitin conjugates that include a ubiquitin dimer, ubiquitylated E2, ubiquitylated E3 and ubiquitylated histones. These issues have made it almost impossible to detect H2B that is specifically ubiquitylated at the single physiological site with anti-ubiquitin or anti-H2B antibodies. Importantly, however, the recent development of an anti-ubH2B monoclonal antibody [15] that specifically recognizes ubiquitylated H2B at lysine 120 enabled us to overcome these problems. The robust assays that have been developed clearly demonstrate requirements for RAD6 and the BRE1 complex for H2B ubiquitylation at lysine 120 in human cells and, in addition, reveal a mechanism of PAF1 complex-dependent transcription-coupled H2B ubiquitylation that involves a direct interaction between PAF1 and BRE1 complexes [12].
Here we describe methods for the purification of H2B ubiquitylation factors and histone/chromatin substrates, comprehensive in vitro H2B ubiquitylation assays with various histone substrates, a transcription-coupled chromatin ubiquitylation assay and protein interaction assays that are important to establish cognate E2–E3 pairs. Although we here describe the assays with human proteins, these methods have been used to analyze H2B ubiquitylation factors from other organisms [5] and are applicable for analyses of other ubiquitylation systems.
2. Description of Methods
2.1. Materials
For recombinant protein preparations, cDNAs were obtained from ATCC and subcloned into appropriate plasmids. All purified protein concentrations are determined by Coomassie Blue staining with BSA standards.
2.1.1. Preparation of recombinant histones, and histone octamers and natural oligonucleosomes
Recombinant Xenopus histones are prepared according to the protocol described by Luger et al. [16]. Briefly, individual histones are expressed in E. coli, and purified by Sephacryl S-200 (GE Healthcare) gel filtration and SP Sepharose (GE Healthcare) ion exchange chromatography under denaturing conditions. Pure individual histones are then reconstituted into an octamer that is further purified and separated through Superdex 200 (GE Healthcare) gel filtration at high salt. Prepared histones are stored at −80°C in 20 mM Tris, pH 8.0, 0.25 mM EDTA, 2 M NaCl, 30% glycerol, 1 mM 2-mercaptoethanol (Fig. 1A). Purification of natural oligonucleosomes from HeLa cells is as follows. Chromatin is extracted by homogenization in high salt buffer (20 mM HEPES, pH 7.5, 0.65 M NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol and 0.34 M sucrose) from nuclear pellet prepared by the Dignam method [17] and then equilibrated stepwise to 20 mM HEPES, pH 7.5, 0.1 M NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol. Cleared extracts are incubated with 10 U/ml micrococcal nuclease (Sigma) in the presence of 3 mM CaCl2. Digested oligonucleosomes are purified and separated through Sepharose CL-6B (GE Healthcare) gel filtration at 0.6 M NaCl. Fractions containing oligonucleosomes that are composed mainly of 10–20 nucleosomes are pooled and stored at −80°C in 20 mM HEPES, pH 7.5, 1 mM EDTA, 1 mM 2-mercaptoethanol (Fig. 1A). Note that natural oligonucleosomes contain pre-existing post-translational modifications and are likely present in various states of chromatin structure, which in turn can influence recognition/recruitment and/or function of the ubiquitylation enzymes in an in vitro assay.
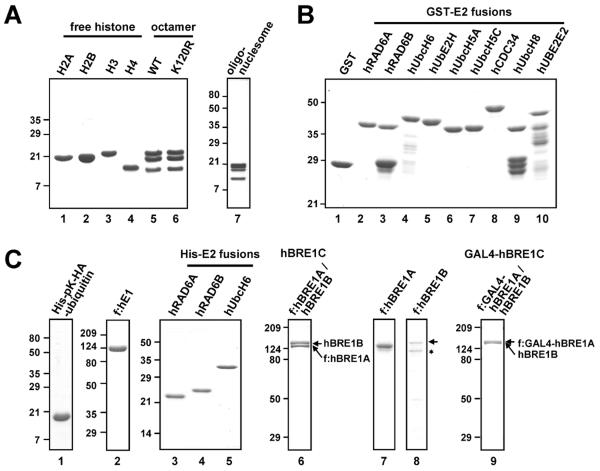
Fig. 1.
Analysis of purified factors by SDS-PAGE with Coomassie Blue staining. (A) Histone ubiquitylation substrates: recombinant histones, reconstituted histone octamers and oligonucleosomes. (B) GST and GST-E2 proteins. (C) Ubiquitin and ubiquitylation enzymes: His-pK-HA-ubiquitin, FLAG-hE1, His-E2 proteins, hBRE1C (composed of FLAG-hBRE1A and untagged hBRE1B), FLAG-hBRE1A, FLAG-hBRE1B and GAL4-hBRE1C (composed of FLAG-GAL4-hBRE1A and untagged hBRE1B). Asterisk, degradation product. Part of this figure is reproduced, with permission, from Ref. [12].
2.1.2. Preparation of recombinant ubiquitin, human E1, human RAD6 and the human BRE1 complex
For GST-tagged E2 proteins, cDNAs are subcloned into pGEX4T (GE Healthcare), expressed in E. coli, and purified by binding to glutathione-Sepharose 4B beads (GE Healthcare) in BC buffer (20 mM Tris, pH 7.9, 0.2 mM EDTA, 20% glycerol) containing 1000 mM KCl (BC1000; hereafter, the number indicates the KCl concentration added to the BC buffer) and 0.1% NP40. After extensive washing with BC1000/0.1% NP40, GST-proteins are eluted with 10 mM glutathione (Sigma) in BC1000/0.1% NP40 and stored at −80°C in BC100/0.01% NP40 (Fig. 1B). His-pKHA-ubiquitin [18; pK indicates a protein kinase recognition site] and His-tagged E2 proteins (cDNAs subcloned into pET28 (Novagen)) are expressed in E. coli and purified by binding to Ni-NTA agarose (Qiagen) in BC500/0.1% NP40 (with exclusion of EDTA) containing 5 mM imidazole (Sigma). After extensive washing with BC500/0.1% NP40 (with exclusion of EDTA) containing 30 mM imidazole, His-proteins are eluted with 400 mM imidazole in BC500/0.1% NP40 and stored at −80°C in BC100/0.01% NP40 (Fig. 1C).
For hE1, hBRE1A and hBRE1B, cDNAs are subcloned in pFASTBAC1 (Invitrogen) with or without an epitope tag and corresponding baculoviruses are generated according to the manufacturer's instruction (Invitrogen). Sf9 cells are infected either with individual baculoviruses or combinations of baculoviruses and incubated for 72 h. Cell extracts are then prepared in BC300/0.1% NP-40 supplemented with 1 mM DTT, 0.5 mM PMSF and protease inhibitor cocktail (Roche). Clarified extracts are subjected to affinity purification on M2 agarose beads (Sigma). After extensive washing with BC150/0.1% NP40, FLAG-proteins/complexes are eluted with BC150/0.1% NP40 containing 0.25 mg/ml FLAG peptide and stored at −80 °C (Fig. 1C). Note that the FLAG-hBRE1B polypeptide is easily degraded when expressed alone but stable when coexpressed with hBRE1A, suggesting an associated stabilization of hBRE1B by hBRE1A in the complex.
2.1.3. Preparation of chromatin assembly factors Acf1, ISW1 and NAP1
Baculoviruses for recombinant Drosophila Acf1 and ISW1 were obtained from the Kadonaga lab [19]. These factors are independently expressed and purified as FLAG-tagged proteins in Sf9 cells as described above. His-tagged mouse NAP-1 is expressed in E. coli, followed by purification through Ni-NTA and Q-Sepharose (GE Healthcare) chromatography. Preparations of these proteins are shown in our earlier publications [12,20].
2.1.4. Preparation of transcription factors
Comprehensive purification protocols for most of these factors are essentially as described [21–24]. Briefly, p53, individual TFIIA subunits (p55 and p12), TFIIB, and TFIIE subunits (α and β) are expressed in E. coli as FLAG-tagged proteins and affinity purified on M2 agarose. TFIIF subunits (RAP30 and RAP74) are independently expressed in E. coli and purified on Ni-NTA agarose. TFIIA and TFIIF are then reconstituted from individual subunits following denaturation and renaturation. Non-tagged PC4 is expressed in E. coli and purified by heparin-Sepharose (GE Healthcare) and phosphocellulose (P11, Whatman) chromatography. The multisubunit TFIID, TFIIH, Mediator and RNA polymerase II (Pol II) complexes are purified from HeLa cell lines expressing complex-specific epitope-tagged subunits (FLAG-TBP for TFIID; FLAG-ERCC3 for TFIIH; FLAG-TRAP220/MED1 (1–670 amino acids) for Mediator; FLAG-RPB9 for Pol II). TFIID, TFIIH and the Mediator complexes are purified from nuclear extracts, prepared as described [17], using a combination of conventional chromatography on phosphocellulose and DEAE cellulose (DE52, Whatman) and affinity purification on M2 agarose as the final step. Pol II is purified from a high salt-solubilized nuclear pellet fraction, prepared as described [25], by conventional ion exchange chromatography followed by affinity purification on M2 agarose. His-tagged SII/TFIIS is expressed in E. coli and purified by Ni–NTA and HiTrap SP (GE Healthcare) chromatography. The multisubunit human PAF1 complex (hPAF1C) is reconstituted in Sf9 cells by coinfection with baculoviruses that individually express hCTR9, hLEO1, FLAG-hPAF1, hRTF1, hCDC73 and hSKI8, and affinity purified on M2 agarose. These procedures provide essentially homogenous preparations of these components as shown by SDS–PAGE with Coomassie blue staining or silver staining in our earlier publications [12,23,24].
2.1.5. Antibodies
The following antibodies are obtained commercially: anti-H2A (Millipore 07-146), anti-H2B (Abcam ab1790), anti-H3 (Abcam ab1791), anti-H4 (Abcam ab7311), anti-HA (Abcam ab9110), and anti-FLAG (HRP-conjugated, Sigma A8592). Polyclonal anti-hBRE1A [26] and anti-hBRE1B [12] antibodies were developed against His-tagged N-terminal fragments and affinity purified. Anti-BRE1A (Bethyl Laboratories A300-714A) and anti-hBRE1B (Bethyl Laboratories A300-720A) antibodies are also commercially available. Mouse monoclonal anti-ubH2B antibody was obtained from the Oren lab [15] and is now commercially available at Millipore (05-1312) and Medimabs (MM0029). In our recent test, rabbit monoclonal anti-ubH2B antibody from Cell Signaling Technology (5546) also showed very good results.
2.2. Analysis of physical and functional interactions of cognate E2–E3 pairs
About two dozen E2 and several hundred E3s are present in mammalian cells, making it important to identify correct cognate E2–E3 pairs in order to establish relevant in vitro protein ubiquitylation assays. Each RING finger-containing E3 specifically and directly binds to its cognate E2 [1] and this direct interaction results in both substrate and E3 ubiquitylation by E2 [27]. From these principles and in order to identify a cognate E2 for the human BRE1 complex (hBRE1C), we set up in vitro assays to screen for an E2 that specifically interacts with and ubiquitylates hBRE1C.
2.2.1. In vitro protein-protein interaction assays
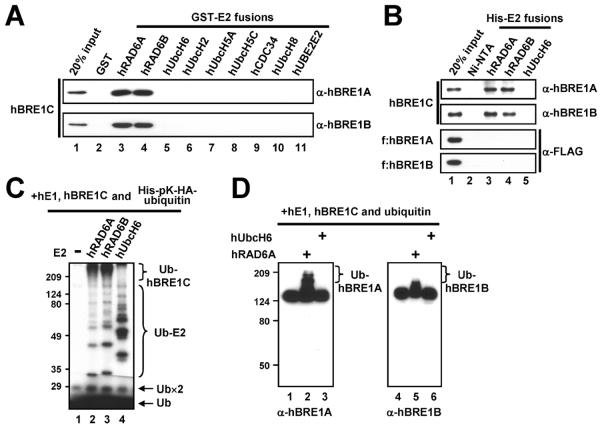
To examine direct interactions between E2 and hBRE1C, two in vitro binding assays are employed. For GST-pull down assays, 2 μg of purified GST or GST-fused E2s immobilized on glutathione-Sepharose 4B beads are incubated with 200 ng of purified hBRE1C in the binding buffer A (20 mM Tris, pH 7.5, 200 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.1% NP-40, 0.5 mg/ml BSA and 0.5 mM PMSF) at 4°C for 3 h and then beads are extensively washed with binding buffer A without BSA. Bound proteins are analyzed by immunoblotting. Fig. 2A illustrates that, among the nine tested E2s, only hRAD6A and hRAD6B directly bind to hBRE1C. This interaction is confirmed by an alternative assay. For His-pull down assays, Ni-NTA agarose preparations previously coupled with 2 μg purified His-tagged E2s are incubated with 200 ng purified hBRE1C or 100 ng individual hBRE1A or hBRE1B polypeptides in binding buffer B (20 mM Tris, pH 7.9, 150 mM KCl, 20% glycerol, 0.05% NP-40, 0.2 mg/ml BSA and 0.5 mM PMSF) at 4 °C for 3 h and then beads are extensively washed with wash buffer (20 mM Tris, pH 7.9, 150 mM KCl, 20% glycerol, 0.05% NP-40, 0.5 mM PMSF and 50 mM imidazole). Bound proteins are analyzed by immunoblotting. The results show that hRAD6 interacts with hBRE1C but not with individual hBRE1A or hBRE1B polypeptides (Fig. 2B), indicating that formation of the heteromeric hBRE1C is critical for direct interaction with hRAD6. This result accounts for the failure to identify a cognate E2 for hBRE1C using only hBRE1A as bait [26].
Fig. 2.
The human BRE1 complex physically and functionally interacts with human RAD6. (A and B) Direct binding of purified hBRE1C to GST-E2 versus GST (A). Direct binding of purified hBRE1C, FLAG-hBRE1A and FLAG-hBRE1B to His-hRAD6 versus His-hUbcH6 (B). Bound proteins were scored by immunoblotting with indicated antibodies. (C and D) Ubiquitylation of hBRE1C by hRAD6. Purified hBRE1C was analyzed in an E3 ubiquitylation assay with indicated E2 enzymes in the presence of either 32P-labeled (C) or unlabeled (D) ubiquitin, and ubiquitylation of hBRE1C was monitored by autoradiography (C) or immunoblot (D), respectively. Free ubiquitin (Ub), ubiquitin dimer (Ub×2), ubiquitylated E2 (Ub-E2) and poly-ubiquitylated hBRE1 proteins (Ub-hBRE1A, Ub-hBRE1B or Ub-hBRE1C) are indicated. Part of this figure is reproduced, with permission, from Ref. [12].
2.2.2. In vitro E3 ubiquitylation assay
To further confirm the functional relevance of the physical interaction between hRAD6 and hBRE1C, an in vitro E3 ubiquitylation assay is performed. Reactions containing 100 ng hE1, 200 ng His-tagged E2, 150 ng hBRE1C, 1.3 μg unlabeled ubiquitin or 32P-labeled His-pK-HA-ubiquitin, 50 mM Tris, pH 7.9, 5 mM MgCl2, 2 mM NaF, 0.4 mM DTT and 4 mM ATP in 20 μl are incubated at 37°C for 1 h, resolved by SDS-PAGE and subjected to immunoblots with anti-hBRE1 antibodies (when unlabeled ubiquitin is used) or auto-radiography (when 32P-labeled ubiquitin is used). 32P-labeled ubiquitin is prepared as follows. A reaction mixture (20 μl) containing 25 μg His-pK-HA-ubiquitin, 1 U cAMP kinase (Sigma), 20 mM Tris, pH 7.4, 12 mM MgCl2, 2 mM NaF, 50 mM NaCl, 25 μM ATP, 5 μCi of [μ-32P] ATP and 0.1 mg/ml BSA is incubated at 37°C for 30 min and then heated at 70°C for 3 min to inactivate the kinase. As described below, the immunoblotting method is useful for monitoring ubiquitylation of E3 but is not informative for measuring E2 activities. Instead, the auto-radiography method allows a direct comparison of the ubiquitin-conjugating activities of E2s. This assay reveals hRAD6-dependent poly-ubiquitylation of hBRE1C that is visualized by autoradiography (Fig. 2C, lanes 2 and 3) and immunoblot (Fig. 2D, lanes 2 and 5). As a control, unrelated hUbcH6 E2 that fails to interact with hBRE1C (Fig. 2A, lane 5; Fig. 2B, lane 5) does not mediate any detectable poly-ubiquitylation of hBRE1C (Fig. 2D, lanes 3 and 6) although it exhibits a strong ubiquitin conjugating activity (Fig. 2C, lane 4).
2.3. In vitro recombinant chromatin assembly
Recombinant chromatins are used for conventional and transcription-coupled chromatin ubiquitylation assays (below). Procedures for chromatin assembly with the recombinant ACF system are adapted from Ito et al. [19]. For transcription assays, the reaction containing core histone octamer (350 ng) and NAP1 (2.5 μg) in 55 μl HEG buffer (25 mM HEPES, pH 7.6, 0.1 mM EDTA and 10% glycerol) is incubated on ice for 30 min. After the further addition of ACF1 (30 ng), ISW1 (40 ng) and plasmid (350 ng), the reaction is adjusted to 25 mM HEPES, pH 7.6, 0.1 mM EDTA, 10% glycerol, 50 mM KCl, 3.4 mM ATP and 4.8 mM MgCl2 in a final volume of 70 μl and incubated at 27°C for 3 h. For ubiquitylation assays, reactions contain two-fold higher amounts of the plasmid and all proteins under the same reaction conditions and in the same (70 μl) volume. The DNA templates employed in these assays are the p53ML [20] and the pG5ML [28] plasmids that contain five p53 and five GAL4 binding sites, respectively, along with the adenovirus major late promoter followed by a 390-nucleotide G-less cassette.
2.4. In vitro histone H2B ubiquitylation assay
To clearly demonstrate direct E2 and E3 functions of hRAD6 and hBRE1C for H2B ubiquitylation, respectively, in vitro chromatin ubiquitylation assays employing oligonucleosomes derived from HeLa cells are used. It is crucial to use epitope-tagged ubiquitin (His-pK-HA-ubiquitin in our assay), especially with natural oligonucleosome substrates, because newly ubiquitylated H2B (modified by His- and HA-tagged ubiquitin) can be distinguished from the naturally ubiquitylated H2B that is present in the oligonucleosome substrate. Note also that, in our experience, GST-tagged hRAD6 proteins exhibit much weaker H2B ubiquitylation activities than do His- or FLAG-tagged hRAD6 proteins -- probably because the relatively large epitope at the N-terminus of hRAD6 interferes its E2 activity. In addition, we also found that any kind of C-terminal epitope tag on hRAD6 completely abrogates its enzyme activity. Importantly, as documented below, the ubiquitylation of nucleosomal H2B (in the absence of transcription) is much less efficient than the ubiquitylation of free H2B. Appropriate methods for H2B ubiquitylation assays with various histone substrates are described below.
2.4.1. Ubiquitylation assay with recombinant histones, histone octamers and oligonucleosome
Reactions containing 300 ng recombinant free histone, 1.2 μg recombinant histone octamer or 5 μg HeLa cell-derived oligonucleosomes and 100 ng hE1, 200 ng His-tagged E2, 600 ng hBRE1C and 2.8 μg His-pK-HA-ubiquitin in 20 μl reaction buffer (50 mM Tris, pH 7.9, 5 mM MgCl2, 2 mM NaF, 0.4 mM DTT and 4 mM ATP) are incubated at 37°C for 1 h. Proteins are resolved by SDS-PAGE and subjected to immunoblotting. Although reactions were incubated for 6–10 h in our earlier publications [5,12], the H2B ubiquitylation level is now known to be maximal at 1 h.
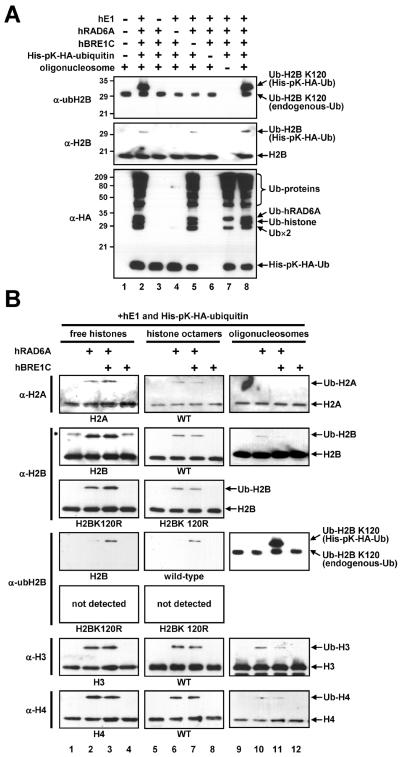
The results show that complete reactions containing purified hE1, hRAD6A, hBRE1C, ubiquitin and oligonucleosomes generate a lysine 120-ubiquitylated H2B (Fig. 3A, top panel, lanes 2 and 8), whereas reactions lacking any of these components do not (top panel, lanes 3–7). Although a significant level of H2B ubiquitylation (probed by an anti-H2B antibody) is detected in the absence of hBRE1C, this clearly represents H2B ubiquitylation at a site(s) other than lysine 120 (compare top panel, lane 5 versus middle panel, lane 5).
Fig. 3.
The hBRE1 complex is required for H2B ubiquitylation at lysine 120. (A) Collective requirement of factors for H2B ubiquitylation. Reactions containing the indicated combinations of purified hE1, hRAD6A, hBRE1C, ubiquitin and oligonucleosome were subjected to immunoblot with indicated antibodies. (B) Requirement of hBRE1C for H2B ubiquitylation at lysine 120. Reactions containing free histone, histone octamer or oligonucleosome substrates with hE1, hRAD6A, hBRE1C and ubiquitin, where indicated, were subjected to in vitro ubiquitylation assays. Ubiquitylated histones were scored by immunoblot with the indicated antibodies. Asterisk, non-specific band. Part of this figure is reproduced, with permission, from Ref. [12].
The detection of a significant level of hBRE1C-independent H2B ubiquitylation by hRAD6 led us to more detailed analyses of substrate and site specificities of hRAD6 in the presence and in the absence of hBRE1C (Fig. 3B). Consistent with earlier reports of the ability of yeast Rad6 to ubiquitylate free H2A and H2B [14, 29], hRAD6 ubiquitylates all four core histones both in free and octamer forms in the absence of hBRE1C (lanes 2 and 6, in the anti-H2A, -H2B, -H3 and -H4 immunoblots) and the inclusion of hBRE1C does not affect the overall histone ubiquitylation efficiency (lanes 3 and 7 versus lanes 2 and 6, respectively, in the anti-H2A, -H2B, -H3 and -H4 immunoblots). Importantly, the efficiency of histone ubiquitylation with the nucleosomal substrate is significantly lower than that with free histones or histone octamers (lanes 9–11 versus lanes 1–3 or lanes 5–7, in the anti-H2A, -H2B, -H3 and -H4 immunoblots). This suggests that the nucleosome structure restricts non-specific access of ubiquitylation factors to the histone core regions. However, and most importantly, H2B ubiquitylation at lysine 120 is critically dependent upon hBRE1C for all substrates tested (compare lanes 3, 7 and 11 versus lanes 2, 6 and 10, respectively, in the anti-ubH2B immunoblots). This indicates that hBRE1C directs the hRAD6 ubiquitin-conjugating activity toward a specific ubiquitylation site (lysine 120) through recognition of the target substrate. Overall, these results indicate (i) a non-specific RAD6 ubiquitin conjugating activity that is manifested in a purified assay system lacking constraints that normally control accessibility of RAD6 to histones and (ii) an H2B lysine 120-specific hRAD6 ubiquitylation activity that is dependent upon hBRE1C. These results also emphasize the utility and validity of the anti-ubH2B antibody [15] for monitoring precise in vitro H2B ubiquitylation events.
2.4.2. Ubiquitylation assay with recombinant chromatin
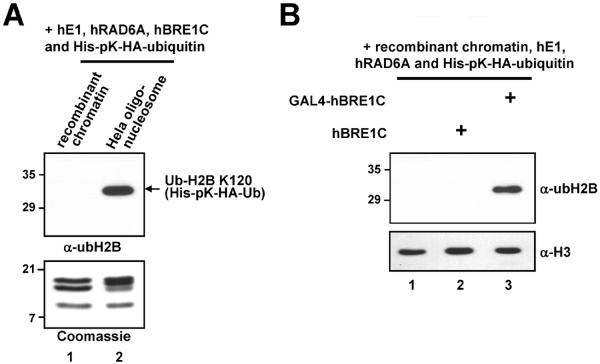
The conventional chromatin ubiquitylation assays described above employ natural oligonucleosome arrays derived from HeLa cells since they have been found to serve as preferential substrates, relative to recombinant chromatin with unmodified histones, for H2B ubiquitylation in vitro (Fig. 4A). These results raise the possibility that pre-existing histone modifications and/or altered chromatin structures (both present in natural oligonucleosome arrays) may provide preferential ubiquitylation enzyme recruitment/recognition conditions for H2B ubiquitylation. To test whether recruitment of the ubiquitylation machinery to chromatin is important for efficient H2B ubiquitylation, an in vitro ubiquitylation assay containing recombinant chromatin assembled with pG5ML plasmid (containing GAL4 binding sites) and GAL4-hBRE1C (composed of FLAG-GAL4-hBRE1A and untagged hBRE1B, Fig. 1C) is employed. The assay is performed as follow: 350 ng (histone amount) recombinant chromatin template (35 μl) is supplemented with 50 ng hE1, 100 ng His-hRAD6A, 200 ng GAL4-hBRE1C or hBRE1C, 1.4 μg His-pK-HA-ubiquitin, 2 mM NaF, 0.4 mM DTT and 4 mM ATP in a final reaction volume of 45 μl. Reactions are incubated at 37°C for 1 h, resolved by SDS-PAGE and subjected to immunoblotting. The result shows that GAL4-directed recruitment of hBRE1C to chromatin greatly enhances H2B ubiquitylation (Fig. 4B, lane 3 versus lane 2). Note that these reactions contain the ATP-dependent remodeling factors employed for chromatin assembly and that previous studies have shown ATP-dependent chromatin remodeling upon binding of GAL4-fusion proteins [30,31]. Thus, these results suggest that proximity of the ubiquitylation machinery to chromatin and/or alteration of chromatin structure mediated by chromatin remodeling factors upon GAL4-hBRE1C binding to chromatin are crucial for efficient H2B ubiquitylation, and that prior histone modifications per se are dispensable for H2B ubiquitylation.
Fig. 4.
Altered chromatin structure and/or human BRE1 complex recruitment to chromatin is crucial for efficient H2B ubiquitylation. (A) The chromatin ubiquitylation assay was performed with the indicated substrates for H2B ubiquitylation (top). Analyses of purified recombinant histone octamers and natural oligonucleosome derived from HeLa cells by SDS-PAGE with Coomassie Blue staining to ensure comparable levels of protein usage for in vitro chromatin ubiquitylation assay (bottom). Note that endogenous ubH2B is not detected in lane 2 because a much smaller amount (350 ng) of nucleosome substrate was used in this assay. (B) hBRE1C recruitment to chromatin is critical for efficient H2B ubiquitylation. A recombinant chromatin template containing the pG5ML plasmid was subjected to the in vitro ubiquitylation assay with hE1, hRAD6A, ubiquitin and either hBRE1C or GAL4-hBRE1C, as indicated, and in the continued presence of the chromatin assembly factors. Reaction products were analyzed by immunoblot with indicated antibodies.
2.4.3. Transcription-coupled ubiquitylation assay with recombinant chromatin
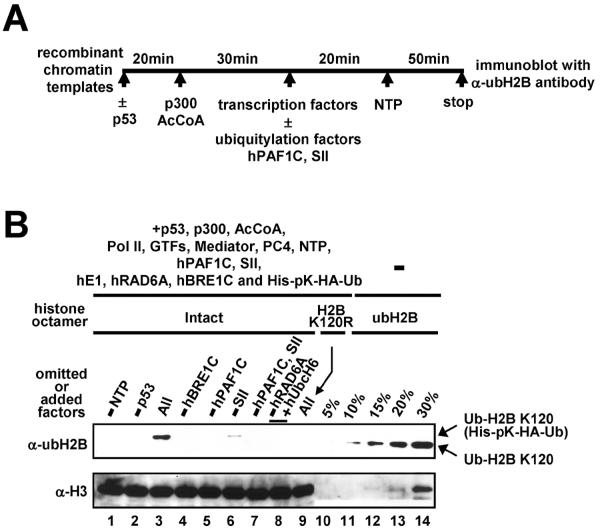
In addition to basic ubiquitylation enzymes, efficient H2B ubiquitylation in vivo requires the PAF1 complex [6,7], an RNA polymerase II-associated [32] transcription elongation factor [33] that directly interacts with the BRE1 complex [5,12]. These results suggest that the PAF1 complex links H2B ubiquitylation and transcription machineries. In addition, the requirement for an altered chromatin structure for H2B ubiquitylation (Fig. 4), along with documented effects of transcription on chromatin structure [34,35] further suggests that H2B ubiquitylation is related to ongoing transcription. In order to test this possibility, an in vitro transcription-coupled ubiquitylation assay that employs purified factors and a chromatinized p53ML array template is performed as follows (schematized in Fig. 5A): (i) the chromatin template (8 μl, 40 ng DNA) is incubated with p53 (10 ng) in 0.5×HAT buffer (10 mM HEPES, pH 7.8, 30 mM KCl, 2.5 mM DTT, 0.25 mM EDTA and 5 mM sodium butyrate) at 30 °C for 20 min in a 20 μl reaction; (ii) the activator-bound chromatin template is incubated with p300 (15 ng) and 20 μM acetyl-CoA at 30 °C for 30 min in a 25 μl reaction; (iii) after addition of 1.25 μl BSA (20 mg/ml), 1 μl DTT (250 mM) and 2.5 μl 20×RB buffer (400 mM HEPES, pH 7.8, 120 mM MgCl2), BC200 buffer is added to adjust the final KCl concentration to 60 mM in a final reaction volume of 50 μl; (iv) highly purified transcription factors (35 ng Pol II, 50 ng TFIID, 20 ng TFIIA, 10 ng TFIIB, 5 ng TFIIEα, 2.5 ng TFIIEβ, 20 ng TFIIF, 20 ng TFIIH, 100 ng Mediator and 55 ng PC4) are added and preinitiation complex (PIC) formation is allowed to proceed at RT for 20 min; (v) purified ubiquitylation factors (25 ng E1, 25 ng E2, 75 ng hBRE1C and 350 ng His-pK-HA-ubiquitin), hPAF1C (240 ng) and/or SII (10 ng) are added and the reaction is further incubated at RT for 5 min; (vi) 2.5 μl 20×nucleotide mixture (10 mM ATP, UTP, GTP and CTP) and 0.25 μl RNasin (40 U/μl) are added to each reaction, along with water to adjust the final volume to 50 μl, and transcription is allowed to proceed for 50 min at 30 °C. (vii) in each case, nine standard transcription reactions are combined, concentrated (G Biosciences) and subjected to immunoblotting.
Fig. 5.
Transcription-dependent H2B ubiquitylation. (A) Schematic representation of the standard in vitro transcription assay. Transcription factors included TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, PC4, Mediator and Pol II. Ubiquitylation factors included E1, E2 (hRAD6A or hUbcH6), hBRE1C and ubiquitin. Chromatin-based assays also contained the components (ACF1, ISWI and NAP1) employed for chromatin assembly. (B) Transcription-coupled H2B ubiquitylation assays were performed with deletions and additions as indicated (lanes 1–9) and reactions were subjected to immunoblot with indicated antibodies. Histone octamers containing fully ubiquitylated H2B (indicated as % of histone octamers present in the transcription assay) were loaded in lanes 10–14. Part of this figure is reproduced, with permission, from Ref. [12].
An assay under transcription conditions with all components present generates a very significant level of H2B ubiquitylation at lysine 120 (Fig. 5B, compare lanes 3 and 9). This corresponds to ubiquitylation of about 15% of the total H2B used for transcription, as judged by comparison with histone octamer standards (compare lane 3 versus lanes 10–14) containing fully ubiquitylated H2B [36]. This analysis also confirms a requirement for hBRE1C (lane 4). Importantly, this efficient H2B ubiquitylation is coupled to activator-dependent transcription since omission of either p53 or nucleoside triphosphates (but with ATP present) reduces ubH2B to an undetectable level (lanes 1 and 2). Similarly, omission of hPAF1C results in an undetectable level of ubH2B (lanes 5 and 7), whereas SII omission significantly decreases the efficiency of H2B ubiquitylation (lane 6). We have recently shown that hPAF1C and SII act independently and cooperatively in chromatin transcription [33]. Hence, the efficiency of H2B ubiquitylation generally correlates with the transcription level. These results thus suggest that hPAF1C facilitates H2B ubiquitylation through an effect on transcription and thereby directly links transcription to H2B ubiquitylation factors. However, given that SII and hPAF1C each stimulate transcription to comparable levels [33], the fact that omission of hPAF1C completely eliminates H2B ubiquitylation, while omission of SII does not, indicates that H2B ubiquitylation is also dependent upon hPAF1C functions other than those directly connected to transcription.
3. Concluding remarks
The increasingly appreciated roles of H2B ubiquitylation in diverse transcription events must ultimately be understood at a mechanistic level, necessitating both genetic and biochemical analyses with properly defined (physiological) components of the ubiquitylation machinery. Here we describe comprehensive protein interaction assays and robust in vitro chromatin ubiquitylation assays with biochemically defined factors that establish bona fide enzymes for H2B ubiquitylation in human cells. Further deployment in biochemically defined transcription-coupled ubiquitylation assays reveals significant mechanistic aspects of H2B ubiquitylation during transcription. These assays can be further applied to define protein domains that are responsible for protein-protein interactions and for the various enzyme activities. As more detailed studies of the role of H2B ubiquitylation and associated histone modifications (e.g. H3K4 and H3K79 methylation) in transcription become necessary, the defined systems described here should prove critical for a further characterization of transcription-related factors that are dependent upon H2B ubiquitylation. In addition, the methods described here might be also used to identify other cognate E2–E3 enzyme pairs and to verify their enzyme activities.
Acknowledgements
We thank Drs. N. Minsky and M. Oren for anti-ubH2B antibody. This work was supported by NIH grants (CA129325 and DK071900), a Leukemia and Lymphoma Society SCOR grant and a Starr Foundation Cancer Consortium grant. J.K. is a senior fellow of the Charles H. Revson Foundation.
Abbreviations
- E1
ubiquitin-activating enzyme
- E2
ubiquitin-conjugating enzyme
- E3
ubiquitin ligase
- Pol II
RNA polymerase II
- WT
wild type
- H2A
histone H2A
- H2B
histone H2B
- H3
histone H3
- H4
histone H4
- ubH2B
lysine 120-ubiquitylated H2B
- GST
glutathione S-transferase
- Ni-NTA
nickel-nitrilotriacetic acid
- HA
hemagglutinin
- PMSF
phenylmethylsulfonyl fluoride
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- BSA
bovine serum albumin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hershko A, Ciechanover A. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- [2].Robzyk K, Recht J, Osley MA. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- [3].Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. Mol. Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- [4].Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. Mol. Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- [5].Kim J, Roeder RG. J. Biol. Chem. 2009;284:20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ng HH, Dole S, Struhl K. J. Biol. Chem. 2003;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- [7].Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. J. Biol. Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- [8].Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- [11].Sun ZW, Allis CD. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- [12].Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TM, Roeder RG. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Goebl MG, Yochem J, Jentsch S, McGrath JP, Varshavsky A, Byers B. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- [14].Sung P, Prakash S, Prakash L. Genes Dev. 1988;2:1476–1485. doi: 10.1101/gad.2.11.1476. [DOI] [PubMed] [Google Scholar]
- [15].Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Nat. Cell. Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- [16].Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. J. Mol. Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- [17].Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Mol. Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- [19].Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].An W, Kim J, Roeder RG. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- [21].Ge H, Martinez E, Chiang CM, Roeder RG. Methods Enzymol. 1996;274:57–71. doi: 10.1016/s0076-6879(96)74008-9. [DOI] [PubMed] [Google Scholar]
- [22].Malik S, Roeder RG. Methods Enzymol. 2003;364:257–284. doi: 10.1016/s0076-6879(03)64015-2. [DOI] [PubMed] [Google Scholar]
- [23].Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG. Cell. 2006;125:275–286. doi: 10.1016/j.cell.2006.01.055. [DOI] [PubMed] [Google Scholar]
- [24].Guermah M, Kim J, Roeder RG. Methods. 2009;48:353–360. doi: 10.1016/j.ymeth.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bitter GA. Anal. Biochem. 1983;128:294–301. doi: 10.1016/0003-2697(83)90378-0. [DOI] [PubMed] [Google Scholar]
- [26].Kim J, Hake SB, Roeder RG. Mol. Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- [27].Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. Proc. Natl. Acad. Sci. USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kundu TK, Palhan VB, Wang Z, An W, Cole PA, Roeder RG. Mol. Cell. 2000;6:551–561. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- [29].Jentsch S, McGrath JP, Varshavsky A. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- [30].Owen-Hughes T, Utley RT, Côté J, Peterson CL, Workman JL. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- [31].Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- [32].Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. Mol. Cell. Biol. 1996;16:669–676. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kim J, Guermah M, Roeder RG. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Workman JL, Kingston RE. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- [35].Li B, Carey M, Workman JL. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- [36].McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]