Abstract
Clostridium cellulovorans uses not only cellulose but also xylan, mannan, pectin, and several other carbon sources for its growth and produces an extracellular multienzyme complex called the cellulosome, which is involved in plant cell wall degradation. Here we report a gene for a cellulosomal subunit, pectate lyase A (PelA), lying downstream of the engY gene, which codes for cellulosomal enzyme EngY. pelA is composed of an ORF of 2,742 bp and encodes a protein of 914 aa with a molecular weight of 94,458. The amino acid sequence derived from pelA revealed a multidomain structure, i.e., an N-terminal domain partially homologous to the C terminus of PelB of Erwinia chrysanthemi belonging to family 1 of pectate lyases, a putative cellulose-binding domain, a catalytic domain homologous to PelL and PelX of E. chrysanthemi that belongs to family 4 of pectate lyases, and a duplicated sequence (or dockerin) at the C terminus that is highly conserved in enzymatic subunits of the C. cellulovorans cellulosome. The recombinant truncated enzyme cleaved polygalacturonic acid to digalacturonic acid (G2) and trigalacturonic acid (G3) but did not act on G2 and G3. There have been no reports available to date on pectate lyase genes from Clostridia.
The plant cell wall is organized as a complex network composed of polysaccharides such as cellulose, hemicellulose, and pectin. In particular, pectin is a heteropolysaccharide composed of α-1,4-linked galacturonate chains with a high percentage of methyl esterification. Pectin degradation requires the combined action of several enzymes that can be classified into two groups, i.e., methylesterases, which remove methoxyl groups from pectin, and depolymerases (hydrolases and lyases), which cleave the bonds between galacturonate units. Pectate lyases (pectate transeliminase, EC 4.2.2.2) are known to play a critical role in the depolymerization process during soft rotting and catalyze random cleavage of internal α-1,4-linkages of polygalacturonate or methylesterified pectins through a trans-eliminative mechanism, thereby generating 4,5-unsaturated oligogalacturonates (1, 2). The breakdown of pectin located in the middle lamella and primary cell wall leads to the maceration of plant tissues (3, 4).
Clostridium cellulovorans, an anaerobic, mesophilic, and spore-forming bacterium, is able to use polysaccharides such as cellulose, pectin, xylan, and several other carbon sources for its growth (5). When C. cellulovorans grows in media containing these carbon sources, the bacterium produces an extracellular multienzyme complex called the cellulosome (6) with a total molecular weight of about 1 × 106 and capable of hydrolyzing crystalline cellulose (7). To elucidate the relationships between the structure and function and the regulatory systems of a variety of enzymes involved in the cellulosome system of C. cellulovorans, we have cloned, sequenced, and characterized several cellulosomal genes (8, 9). The C. cellulovorans cellulosome comprises three major subunits (P170, P100, and P70) (7, 10), i.e., the scaffolding protein CbpA (11), the endoglucanase EngE (12), and the exoglucanase ExgS (13). In addition to these major subunits, we have cloned and sequenced the cellulosomal subunits, including the endoglucanases EngB (14), EngH, EngK, EngL, EngM, and EngY (10, 13, 15), and a mannanase, ManA (15), each containing a duplicated sequence (DS or dockerin) consisting of highly conserved 22-aa repeats. Therefore the C. cellulovorans cellulosomal enzymes identified to date are capable of degrading cellulose, xylan, lichenan, and mannan. However, because we have recently succeeded in converting Arabidopsis and tobacco cells to protoplasts with the C. cellulovorans cellulosomes (Y.T., S. Ui, H. Chan, R.H.D., and B. Liu, unpublished data), and it has been reported that pectin could serve as a carbon source for growth (5), we believed that the C. cellulovorans cellulosome must also have pectinase activity. The present paper provides data that indicate that a cellulosomal gene pelA and the enzyme encoded by this gene can degrade pectin and that pectate lyase A (PelA) contains a DS at its C terminus. Thus the versatility of the C. cellulovorans cellulosome in degrading plant cell wall materials is further established. Furthermore, as far as we are aware, there have been no reports about pectate lyase genes in Clostridium species.
Materials and Methods
Bacterial Strains and Vectors.
C. cellulovorans (ATCC 35296) (5) was used as the source of chromosomal DNA. Escherichia coli XL1-Blue MRF′ and SOLR, λZAPII pBluescript SK (−), and pBluescriptII KS (+) served as cloning hosts and vectors (Stratagene). Plasmids pYT-1 and pYT-2 were derived by using E. coli plasmids XL1-BLUE MRF′ and SOLR. λ plaques were formed after E. coli cells were transfected with λ phages in NZY medium containing 1 mM isopropyl-β-d-thiogalactopyranoside. E. coli was grown at 37°C in LB medium supplemented with ampicillin (100 μg/ml) when required. For agar medium, LB was solidified with 1.5% (wt/vol) agar.
Recombinant DNA Techniques.
The C. cellulovorans genomic library was previously constructed in λZAPII (12) and was immunoscreened with an anti-P70 (cellulase) antiserum (diluted 1:500) previously prepared from C. cellulovorans and alkaline phosphatase-conjugated goat anti-rabbit IgG second antibody (diluted 1:3,000) (Bio-Rad). Immunodetection was carried out with a Western Detection Kit (Bio-Rad), following the manufacturer's instructions. Positive immunocrossreacting plaques were isolated, and clones were obtained by in vivo excision and rescued with the use of ExAssist helper phage (Stratagene). Positive clones were further screened for endoglucanase activity by overlayering with 0.7% soft agar containing 0.3% carboxymethyl cellulose (low viscosity; Sigma). Colonies with carboxymethyl cellulase activity were recognized by the formation of clear haloes on a red background after staining with 0.1% Congo red and destaining with 1 M NaCl (16). Plasmids from E. coli were purified with a Qiagen kit (Qiagen, Chatsworth, CA). Agarose gel electrophoresis, transformation of E. coli, and ligation have been described by Sambrook et al. (17).
Nucleotide Sequence Analysis.
The nucleotide sequence was determined from both strands by the dideoxy chain termination method (18). The 6.7-kb EcoRI fragment (pYT-1) was subcloned with the use of restriction enzymes, and a series of nested deletion mutants from each fragment was constructed with an ExoIII/mung bean nuclease kit (Stratagene). Double-stranded DNA templates were sequenced with sequenase, version 2.0 (United States Biochemical). Both strands were sequenced. Homology searches in the GenBank were carried out with the blast program.
PelA Activity and Protein Assays.
The reaction mixture consisted of 0.1 ml of PelA solution, 1 ml of 0.4% sodium polygalacturonic acid (Sigma), 0.5 ml of 200 mM Tris⋅HCl buffer (pH 8.0), and 0.4 ml of 0.25 mM CaCl2 in a total volume of 2 ml. After reaction at 40°C for 10 min, 2 ml of 50 mM HCl was added to terminate the reaction, and the absorbance in the solution was measured at 235 nm (19). One unit of enzyme activity was defined as the amount of enzyme that liberates 1 μmol of unsaturated oligogalacturonides, equivalent to 1 μmol digalacturonide/min under the above conditions. The molar extinction coefficient value was assumed to be 4,600 M−1⋅cm−1 for unsaturated digalacturonide (19). The influence of Ca2+ and EDTA was investigated by the addition of CaCl2 concentrations ranging from 0 to 1 mM and 1 mM EDTA. The optimum pH was determined by using piperazine-N,N′-bis(2-ethanesulfonic acid); Pipes buffer (pH 6.0 to 8.0), and Tris⋅HCl buffer (pH 7.5–10), each at 50 mM. The activity on substrates with various degrees of methylation was determined by substituting 26%, 67%, and 89% esterified citrus fruit pectins (Sigma), an apple pectin (Sigma), or polygalacturonate.
Protein was measured by the method of Bradford (20) with a protein assay kit from Bio-Rad and BSA as a standard.
SDS/PAGE and Zymogram Analysis.
SDS/PAGE was performed in a 10% polyacrylamide gel by the method of Laemmli (21). After electrophoresis, the gel was stained with Coomassie brilliant blue R. Zymogram analysis was prepared by using 0.2% pectin from apple (Sigma) that was copolymerized with polyacrylamide. Pectinase activity was visualized with 0.05% ruthenium red (Sigma) (22).
Purification of Recombinant PelA.
E. coli harboring pYT-2 was cultured to early stationary phase at 37°C with vigorous shaking. The cells were collected by centrifugation, and the periplasmic fraction was prepared by the osmotic shock method (23). The periplasmic fraction was applied to a Q Sepharose Fast Flow column (2.6 × 22 cm; Amersham Pharmacia) equilibrated with 50 mM Tris⋅HCl buffer (pH 7.5). After being washed with 3 bed volumes of the same buffer, the column was eluted with a linear gradient of NaCl (0 to 0.5 M) at a flow rate of 60 ml/h. The active fractions eluted around 0.2 M NaCl were collected and concentrated by an Amicon concentrator. The concentrated enzyme solution was applied to a Sephacryl S-200 column (2.6 × 75 cm; Amersham Pharmacia) equilibrated with 50 mM Tris⋅HCl buffer (pH 7.5). The active fractions were used as purified enzyme.
TLC.
Oligogalacturonates [galacturonic acid (G1), digalacturonic acid (G2), and trigalacturonic acid (G3)] were purchased from Sigma. Hydrolysis products were separated by TLC on a silica gel 60-plastic sheet (Merck) with a solvent of 1-butanol/water/acetic acid (5:3:2), and the oligosaccharides were visualized by spraying the plate with phosphomolybdic acid (24).
Results
Cloning and Nucleotide Sequence of the pelA Gene.
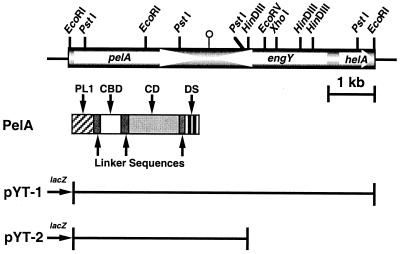
A C. cellulovorans λZAPII library was immunoscreened with anti-P70 (cellulase) antibody and further screened for endoglucanase activity. From ≈2,000 transformants, two positive clones were isolated and further characterized. Both clones contained a common 6.7-kb EcoRI insert. A restriction enzyme map of the cloned gene is shown in Fig. 1. As a result of DNA sequencing, the pYT-1 fragment was found to consist of three ORFs coding for a pectate lyase pelA gene, an endoglucanase engY gene, and a DNA helicase helA gene. As a result of determination of the coding regions, the pelA gene was located on a 3.7-kb EcoRI–HindIII (pYT-2) fragment (Fig. 1).
Figure 1.
Restriction enzyme maps of pYT-1 and domain structure of the PelA polypeptide. Thin lines correspond to vector plasmid DNA. pYT-1 contains the entire 6.7-kb EcoRI fragment. The pectate lyase gene, pelA, was located on a 3.7-kb fragment designated as pYT-2. The white arrows indicate the coding sequences for the PelA, EngY, and HelA polypeptides, respectively. The different domains of PelA are named and filled with different patterns. PL1, pectate lyase family 1; CD, catalytic domain.
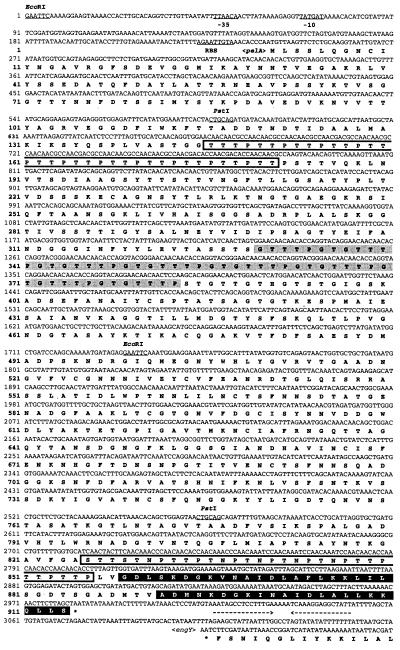
Fig. 2 shows the complete nucleotide sequence of the pelA structural gene along with its flanking regions. The ORF of pelA consists of 2,742 nt encoding a protein of 914 aa with a predicted molecular weight of 94,458. A putative ribosome-binding site, AGAATTGTA, is present at a spacing 8 bp upstream of the translational start codon ATG (25). Upstream of the coding region, possible promoter sequences, TTAACA for the −35 region and TATGAT for the −10 region with a 15-bp spacing between them, were found and showed high homologies to consensus promoter sequences for σ70 or σA factor, TTGACA and TATAAT with a 17-bp spacing, found in E. coli or Bacillus subtilis (26). A possible transcription terminator consisting of a 15-bp palindrome, corresponding to an mRNA hairpin loop with a ΔG of −26.0 kcal/mol (ca. −109 kJ/mol), followed by four Ts after one nucleotide gap, was found downstream of the TAA termination codon. This structure is similar to the ρ factor-independent terminator of E. coli (27). Sequence of the DNA beyond the pelA terminator revealed a convergently oriented ORF that was identical to engY (Fig. 2).
Figure 2.
Nucleotide and deduced amino acid sequences of pelA. The putative promoter (−35 and −10) and ribosome-binding site sequences are underlined. Palindrome sequences are indicated by arrows facing each other. The stop codon is indicated by *. Proline-threonine- or asparagine-serine-threonine-rich regions are boxed. GTTTPGT repeats are shadowed. The DS (or dockerin) is highlighted.
Domain Structure of PelA.
Analysis of the deduced amino acid sequence of PelA revealed a multidomain structure with a unique feature, i.e., a N-terminal domain homologous to pectate lyase family 1, followed by a putative cellulose-binding domain (CBD), a pectate lyase family 4 catalytic domain, and a DS (or dockerin) (Fig. 1). In addition to these domains, there were several linker sequences, i.e., a proline-threonine-rich region downstream of pectate lyase family 1, a region with eight repeats of GTTTPG between the CBD and the catalytic domain, and serine-threonine-asparagine-proline-rich regions between the catalytic domain and the DS (Fig. 2).
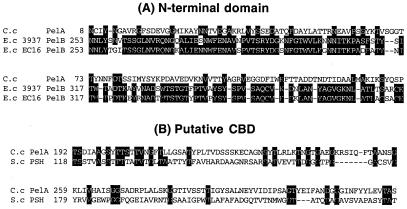
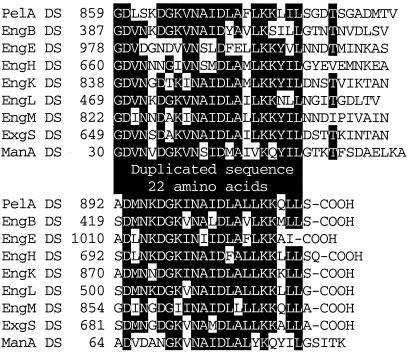
The N-terminal region of PelA (residues 1–145) showed homology with E. chrysanthemi 3937 PelB (24.1% identity) (GenBank accession no. S25262) and EC16 PelB (21.8% identity) (GenBank accession no. M14510) belonging to family 1 of pectate lyase (Fig. 3A). This N-terminal domain coded for a truncated enzyme and did not show any pectate lyase activity. The next domain (residues 186–329) in PelA was homologous to the putative sugar hydrolase of Streptomyces coelicolor (GenBank accession no. AL132662) (Fig. 3B). Interestingly, the region of S. coelicolor putative sugar hydrolase exhibited high sequence similarity with CBDs belonging to family II. Therefore this region of PelA may be able to bind polysaccharides. The catalytic domain (residues 383–825) of PelA showed similarity to catalytic domains of pectate lyases that belong to pectate lyase family 4 (Fig. 4). In particular, the region had high sequence similarity to Bacillus species PelH (49.5% identity) (GenBank accession no. AB028878) and Erwinia chrysanthemi PelL (30.8% identity) (GenBank accession no. S69796) and PelX (30.1% identity) (GenBank accession no. CAB39324). A DS (or dockerin) (residues 849–914) was also found in the C-terminal region of PelA. The DSs consisting of 22-aa repeats are well conserved in cellulosomal enzymatic subunits from C. cellulovorans (Fig. 5).
Figure 3.
Alignment of the N-terminal domain (A) and putative CBD domain (B) of PelA with those of other enzymes. Amino acids that are conserved in three sequences (A) and two sequences (B) are boxed. −, gaps left to improve alignment. Numbers refer to amino acid residues at the start of the respective lines; all sequences are numbered from Met-1 of the peptide. C.c, Clostridium cellulovorans; E.c, Erwinia chrysanthemi; S.c, Streptomyces coelicolor.
Figure 4.
Alignment of pectate lyase family 4 catalytic domains of C. cellulovorans (C.c) PelA, Bacillus species (B.s) PelH, and E. chrysanthemi (E.c) PelL and PelX. Amino acids that are conserved in the four sequences are boxed. −, gaps left to improve alignment. Numbers refer to amino acid residues at the start of the respective lines; all sequences are numbered from Met-1 of the peptide.
Figure 5.
Alignment of the DSs (or dockerins) of cellulosomal subunits of C. cellulovorans. Amino acids that are conserved in at least five of the nine sequences are highlighted. The numbers refer to amino acid residues at the start of the respective lanes; all sequences are numbered from Met-1 of the peptide.
Purification and Characterization of Recombinant PelA (rPelA).
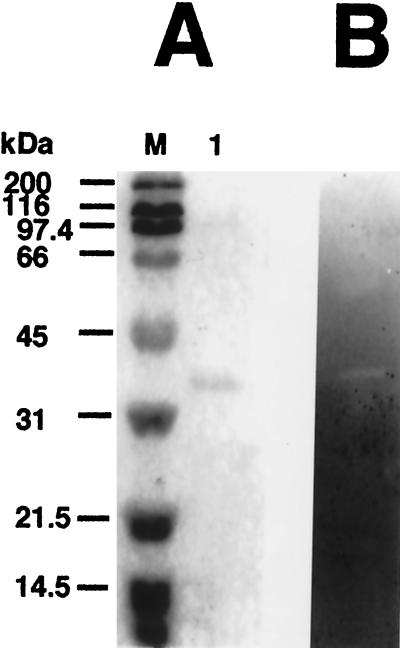
The rPelA was purified from the periplasmic fraction of E. coli harboring pYT-2R, with the use of the Q Sepharose Fast Flow and Sephacryl S-200 columns. After purification, rPelA gave a single band on SDS/PAGE, and the molecular weight of the enzyme was estimated to be around 42,000 (Fig. 6A, lane 1). These results suggested that the purified rPelA had been cleaved by an E. coli protease. Moreover, the estimated molecular weight by SDS/PAGE is in good agreement with the size of the catalytic domain, which is estimated at 42,147, as calculated from its deduced amino acid sequence (Fig. 1). Furthermore, zymogram analysis of the purified enzyme revealed the presence of pectinase activity (Fig. 6B).
Figure 6.
Analysis of the purified rPelA by SDS/PAGE (A) and zymogram (B). Lane M, standard markers [myosin (200 kDa), β-galactosidase (116 kDa), phosphorylase B (97.4 kDa), BSA (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), soybean trypsin inhibitor (21.5 kDa), and lysozyme (14.5 kDa)]; lane A 1, purified rPelA.
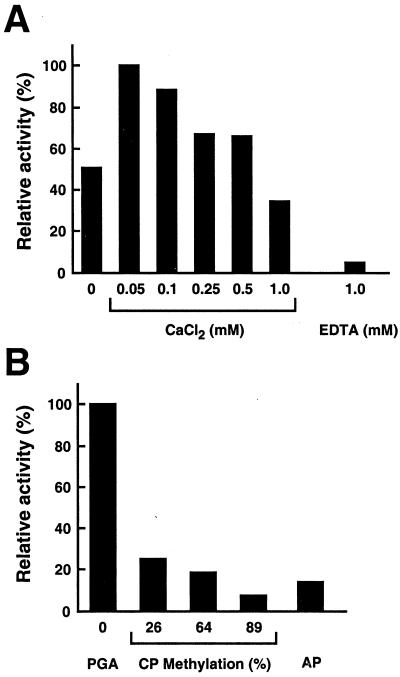
The effect of pH and temperature on the activity of purified rPelA on polygalacturonic acid was determined. The enzyme was most active around pH 8.0. At neutral pH, the activity was less than 30% of the maximum activity. Optimum temperature for rPelA activity was 50°C, whereas more than 50% of maximum activity was found in the temperature range from 40°C to 60°C. Optimal activity was observed throughout a range of CaCl2 concentrations (from 0.05 to 0.5 mM), and the addition of 1 mM CaCl2 reduced activity to less than that with no addition of CaCl2 (Fig. 7A). On the other hand, addition of 1 mM EDTA inhibited activity to less than 5% of maximum. Among pectic substrates, maximum activity was found on polygalacturonic acid, and the purified rPelA showed less than 30% of the maximum activity on citrus and apple pectins (Fig. 7B). Moreover, as the percentage degree of methylation in pectin became higher, the purified rPelA revealed lower activity. As shown in Fig. 8, TLC analysis revealed that the purified enzyme cleaved polygalacturonic acid to form several oligosaccharides, such as digalacturonic acid (G2) and trigalacturonic acid (G3), but did not act on G2 and G3. One must keep in mind that these results are derived with a truncated rPelA.
Figure 7.
Enzymatic properties of PelA. (A) The influence of Ca2+ was tested by using various concentrations of CaCl2 in 50 mM Tris⋅HCl (pH 8) and 0.2% polygalacturonic acid. The cation requirement was confirmed by the addition of 1 mM EDTA. (B) The influence of the degree of pectin methylation on rPelA activity. Activity of rPelA on polygalacturonic acid (PGA), citrus pectins (CP) with a degree of methyl esterification from 26% to 89%, and apple pectin (AP) was determined by the standard assay condition.
Figure 8.
TLC of the hydrolysis products of digalacturonic acid (G2), trigalacturonic acid (G3), and polygalacturonic acid (PG) with the purified rPelA. The reaction mixtures contained 50 mM Tris⋅HCl (pH 8), 0.05 mM CaCl2, 5 units of enzymatic activity/ml, and 1 mg of the substrates/ml. Incubations were performed at 37°C for 12 h after the addition of rPelA or without enzyme.
Discussion
Much knowledge has been obtained about the cellulosomes from several Clostridium species (8, 28), and the previous studies have focused mainly on cellulose degradation. Although the overall features of the cellulosome are reasonably clear, there are several properties of the cellulosome that require further investigation. In particular, the versatility of substrate specificity in the cellulosome is still unknown. An understanding of the mechanism of plant cell wall degradation by the cellulosome may play an important role for biomass recycling and food and energy problems in this century. Our current strategy is to study the properties of the subunits of the C. cellulovorans cellulosome that include not only cellulases but also hemicellulases and pectinases. Our ultimate goal is to apply designer cellulosomes for the utilization of plant biomass as an energy source.
We have cloned and sequenced a 6.7-kb EcoRI fragment consisting of three ORFs, i.e., pelA, engY, and helA (Fig. 1). Both pelA and engY genes have a DS and interestingly possess the same transcription terminator (Fig. 2). In fact, the relationship of gene location between pelA and engY is similar to that for pelL and celZ from E. chrysanthemi (20, 29), although engY belongs to family 9 of cellulases, whereas celZ belongs to family 5. The deduced amino acid sequence of PelA revealed a multidomain structure. The N-terminal domain was homologous to the family 1 of pectate lyases. However, this N-terminal domain coded for a truncated enzyme without a complete catalytic domain, which was followed by a putative CBD, a complete catalytic domain belonging to family 4 of pectate lyases, and a DS. The putative CBD is one of a large group of family 2 CBDs, but the gene banks reveal only two other family II CBDs from Clostridial species. Pectate lyases are classified in five different families according to their primary amino acid sequences (30, 31). The catalytic domain of PelA belongs to family 4 of pectate lyases, which contains PelL and PelX of E. chrysanthemi (20, 29, 31) and PelH of Bacillus species PelH (GenBank accession no. AB028878). In this study, we purified and characterized the rPelA. The molecular weight of the purified rPelA was estimated to be 42,000 by SDS/PAGE (Fig. 6A), and the molecular weight as derived from the DNA sequence was 94,458. It appeared the rPelA had been cleaved by an E. coli protease, but its size was still sufficient to contain the complete catalytic domain (Fig. 1). This type of partial proteolytic degradation had been shown previously for essentially all C. cellulovorans cellulosomal enzyme genes expressed in E. coli. Moreover, the purified enzyme showed pectinase activity on zymogram analysis (Fig. 6B), thus indicating that the specificity and activity of rPelA had been retained in the truncated recombinant enzyme.
Optimum pH and temperature for rPelA activity were pH 8.0 and 50°C, respectively. The purified enzyme revealed the greatest activity with polygalacturonic acid and less activity with citrus and apple pectins (Fig. 7B). Furthermore, the cleavage pattern of rPelA analyzed by TLC showed that the main products from polygalacturonic acid were digalacturonic acid (G2), trigalacturonic acid (G3), and tetragalacturonic acid (G4) (Fig. 8), and those of PelL from E. chrysanthemi showed G3 and larger oligomers (20). On the other hand, the main products from polygalacturonic acid for PelX from E. chrysanthemi were only unsaturated digalacturonic acid (uG2) (31). Furthermore, the purified rPelA did not cleave G2 and G3 (Fig. 8), whereas PelX could act on G3 to form galacturonic acid (G1) and uG2 but not on G2 (31). In addition, G4 is the best substrate for the E. chrysanthemi PelX, and the estimated number of binding subsites for PelX is 4, extending from subsites −2 to +2 (31). Because TLC analysis revealed that rPelA did not act on G2 and G3, the results indicate that rPelA hydrolyzes G4 or longer oligomers. Furthermore, rPelA seems to be an endo-type enzyme, whereas the E. chrysanthemi PelX is an exopoly-galacturonate lyase. Thus, the properties of rPelA of C. cellulovorans differ from those of PelL and PelX of E. chrysanthemi in specific activity, substrates degraded, and the products that are formed, although all of the enzymes belong to family 4 of pectate lyases.
The C. cellulovorans cellulosome has now been shown to be capable of hydrolyzing cellulose, lichenan, mannan, pectin, and xylan. Many of the hydrolytic properties of the whole cell can now be attributed to the enzymatic activities associated with the cellulosome. Therefore, from knowledge of the components of the plant cell wall and by combining the enzymatic subunits in designer C. cellulovorans cellulosomes, we should be able to degrade plant biomass efficiently and convert carbon biomass to much more utilizable forms of energy.
Acknowledgments
This research was supported in part by Grant DE-DDF03-92ER20069 from the U.S. Department of Energy.
Abbreviations
- PelA
pectate lyase A
- rPelA
recombinant PelA
- CBD
cellulose-binding domain
- DS
duplicated sequence
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF105331).
References
- 1.Albersheim P, Killias U. Arch Biochem Biophys. 1962;97:107–115. doi: 10.1016/0003-9861(62)90050-4. [DOI] [PubMed] [Google Scholar]
- 2.Barras F, van Gijsegem F, Chatterjee A K. Annu Rev Phytopathol. 1994;32:201–234. [Google Scholar]
- 3.Collmer A, Keen N T. Annu Rev Phytopathol. 1986;24:383–409. [Google Scholar]
- 4.Lietzke S E, Yoder M D, Keen N T, Jurnak F. Plant Physiol (Rockville) 1994;106:849–862. doi: 10.1104/pp.106.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sleat R, Mah R A, Robinson R. Appl Environ Microbiol. 1984;48:88–93. doi: 10.1128/aem.48.1.88-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamed R, Bayer E A. In: Biochemistry and Genetics of Cellulose Degradation. Aubert J-P, Béguin P, Millet J, editors. San Diego: Academic; 1988. pp. 101–116. [Google Scholar]
- 7.Shoseyov O, Doi R H. Proc Natl Acad Sci USA. 1990;87:2192–2195. doi: 10.1073/pnas.87.6.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doi R H, Park J-S, Liu C-C, Malburg L M, Tamaru Y, Ichi-ishi A, Ibrahim A. Extremophiles. 1998;2:53–60. doi: 10.1007/s007920050042. [DOI] [PubMed] [Google Scholar]
- 9.Tamaru Y, Liu C-C, Malburg L M, Doi R H. In: Genetics, Biochemistry and Ecology of Cellulose Degradation. Ohmiya K, Hayashi K, Sakka K, Kobayashi Y, Karita S, Kimura T, editors. Tokyo: Uni Publishers; 1999. pp. 488–494. [Google Scholar]
- 10.Matano Y, Park J-S, Goldstein M A, Doi R H. J Bacteriol. 1994;176:6952–6956. doi: 10.1128/jb.176.22.6952-6956.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shoseyov O, Takagi M, Goldstein M, Doi R H. Proc Natl Acad Sci USA. 1992;89:3483–3487. doi: 10.1073/pnas.89.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamaru Y, Doi R H. J Bacteriol. 1999;181:3270–3276. doi: 10.1128/jb.181.10.3270-3276.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C-C, Doi R H. Gene. 1998;211:39–47. doi: 10.1016/s0378-1119(98)00081-x. [DOI] [PubMed] [Google Scholar]
- 14.Foong F, Hamamoto T, Shoseyov O, Doi R H. J Gen Microbiol. 1991;137:1729–1736. doi: 10.1099/00221287-137-7-1729. [DOI] [PubMed] [Google Scholar]
- 15.Tamaru Y, Doi R H. J Bacteriol. 2000;182:244–247. doi: 10.1128/jb.182.1.244-247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ali B R S, Romaniec M P M, Hazlewood G P, Freedman R B. Enzyme Microbiol Technol. 1995;17:705–711. doi: 10.1016/0141-0229(94)00118-b. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch J E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasegawa S, Nagel C W. J Food Sci. 1966;31:838–845. [Google Scholar]
- 20.Bradford M M. Anal Biochem. 1976;72:284–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Ried J L, Collmer A. J Bacteriol. 1985;50:615–622. [Google Scholar]
- 23.Manoil C, Beckwith J. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 24.Lojkowska E, Masclaux C, Boccara M, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Mol Microbiol. 1995;16:1183–1195. doi: 10.1111/j.1365-2958.1995.tb02341.x. [DOI] [PubMed] [Google Scholar]
- 25.Tinoco I, Jr, Borer P N, Dengler B, Levine M D, Uhlenbeck O C, Crothers D M, Gralla J. Nat New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein M A, Doi R H. Biotechnol Annu Rev. 1995;1:105–128. doi: 10.1016/s1387-2656(08)70049-8. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberg M, Court D. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 28.Shoham Y, Lamed R, Bayer E A. Trends Microbiol. 1999;7:275–281. doi: 10.1016/s0966-842x(99)01533-4. [DOI] [PubMed] [Google Scholar]
- 29.Alfano J R, Ham J H, Collmer A. J Bacteriol. 1995;177:4553–4556. doi: 10.1128/jb.177.15.4553-4556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henrissat B, Heffron S E, Yoder M D, Lietzke S E, Jurnak F. Plant Physiol. 1995;107:963–976. doi: 10.1104/pp.107.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shevchik V E, Kester H C M, Benen J A E, Visser J, Robeert-Baudouy J, Hugouvieux-Cotte-Pattat N. J Bacteriol. 1999;181:1652–1663. doi: 10.1128/jb.181.5.1652-1663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]