Abstract
Environmental factors are known to affect the strength and the specificity of interactions between hosts and parasites. However, how this shapes patterns of coevolutionary dynamics is not clear. Here, we construct a simple mathematical model to study the effect of environmental change on host–parasite coevolutionary outcome when interactions are of the matching-alleles or the gene-for-gene type. Environmental changes may effectively alter the selective pressure and the level of specialism in the population. Our results suggest that environmental change altering the specificity of selection in antagonistic interactions can produce alternating time windows of cyclical allele-frequency dynamics and cessation thereof. This type of environmental impact can also explain the maintenance of polymorphism in gene-for-gene interactions without costs. Overall, our study points to the potential consequences of environmental variation in coevolution, and thus the importance of characterizing genotype-by-genotype-by-environment interactions in natural host–parasite systems, especially those that change the direction of selection acting between the two species.
Keywords: host-parasite coevolution, antagonistic interaction, environment, Red Queen dynamics, genotype-by-genotype-by-environment
1. Introduction
Hosts are under selective pressure to resist parasites, and parasites are selected to overcome host defences. This may lead to coevolutionary dynamics, where gene frequency changes in one species trigger gene frequency changes in the other species and vice versa (reviewed in [1,2]). Because rare genotypes are expected to be advantageous in such a scenario, cyclic gene-frequency dynamics may ensue, sometimes referred to as Red Queen (RQ) dynamics. Although RQ dynamics have been documented in some study systems [3–5], they remain poorly understood. This gap in our knowledge is particularly noteworthy since antagonistic coevolution between hosts and parasites has far-reaching implications for many topics in biology, including local adaptation [6], maintenance of genetic polymorphism in populations [7,8], molecular evolution [9], deployment of resistance genes in agriculture [10], evolution of pathogen virulence [11,12], emergence and spread of infectious diseases [2] and the evolution of sex [13–15].
It is commonly accepted that the genetics of the host and the parasite are a major determinant of infection success. A number of studies have shown that one parasite genotype may be more infective than another parasite genotype on a given host, but on another host this hierarchy is reversed—a pattern known as genotype-by-genotype (G × G) interaction ([16–19]; see [20] for review). Modelling has demonstrated that the exact type of genetic interaction is decisive for the coevolutionary dynamics expected to occur, and in particular for whether RQ dynamics ensue or polymorphism at the interaction loci is lost (e.g. [21–24]). On the other hand, there is increasing evidence that the outcome of host–parasite interactions can be substantially affected by environmental factors, e.g. temperature or availability of nutrients (reviewed in [25–28]). Thus, it is uncertain how stable G × G interactions are in the presence of environmental fluctuations, and how the coevolutionary dynamics of hosts and parasites are affected by such fluctuations.
How can a host–parasite G × G interaction responds to environmental conditions? First, environmental variation could have an equally strong influence on the fitness values of all genotypes involved. Within a population genetic framework (i.e. when there is no density-dependent selection), this type of impact is not expected to affect the coevolutionary dynamics because the relative fitness of each genotype is not affected by the environment. Second, fitness values of different genotypes could be affected differently by environmental factors, but still in a way that the ranking of fitness values remains the same. Finally, the specificity of the G × G could be changed through environmental variation. Assuming only two host and two parasite genotypes, this means that on each host, the fittest parasite in one environment is the least fit in a different environment (cf. box 2 in [28]). The latter two situations are often referred to as G × G × E interactions, and cases of environmental impact on both the strength and the specificity of selection have been reported (B. Sadd, 2010, Submitted; [29,30]; see also §4).
In this study, we examine the impact of the second and third kind of environmental change—where the specificity and the strength of the G × G are changed—on host–parasite coevolutionary dynamics by means of a simple mathematical model. The impact of temporal environmental heterogeneity has previously been studied in the context of the geographical mosaic theory [31], where the authors considered an environment that alters the quality of interaction between species (antagonism versus mutualism). Here, we investigate G × G × E effects in strictly antagonistic interactions. Although we focus on situations where both the strength and the specificity are affected by the environment, our model also covers cases where only the strength of selection is environment dependent. As the mechanistic basis of host–parasite interactions remains poorly understood, we assume that the genetic basis of interaction is mediated by two standards, biologically documented interaction models, namely the matching-alleles (MA) and the gene-for-gene (GFG) models (see §2 for a brief discussion of these models). Our results show that if the specificity of the interaction is changed through environmental variation, the resulting coevolutionary dynamics can be qualitatively affected, even to the extent that RQ dynamics disappear completely where they would occur in a constant environment, or emerge where they would not.
2. The model
(a). General construction
To investigate the impact of environment on host–parasite RQ dynamics, we consider a standard, discrete-time, population-genetic model of host–parasite dynamics (e.g. [32,33]). For the sake of simplicity, both species are assumed to be haploid and to reproduce asexually. Each species carries a single, biallelic locus. The allele frequencies in the next generation are determined by the fitness values, given by the interaction model (table 1), as well as frequencies of host and parasite alleles in the current generation. First, each species undergoes selection, which operates as follows. If fiH denotes the frequency of allele i of the host and fiP denotes the frequency of allele i of the parasite, then the frequencies after selection will be given by
| 2.1 |
where the vector wis denotes the fitness conferred by allele i in species s and 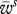 denotes the mean fitness of species s (host H or parasite P). These fitness values are given by
denotes the mean fitness of species s (host H or parasite P). These fitness values are given by
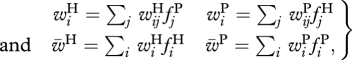 |
2.2 |
where the matrix ws = (wijs)2×2 denotes the fitness values of an individual of species s with allele i when encountering an individual of the second species with allele j. These fitness values are given by the interaction model (cf. table 1). In order to avoid extinction of one of the alleles, selection is followed by reproduction during which mutation between the two alleles can occur at a rate μ = 10−5. Both host and parasite population are assumed to be infinitely large and are started with random allele frequencies. Simulation are started with a burn-in phase of at least 6000 generations, followed by 2000 generations during which the actual dynamics are recorded.
Table 1.
Interaction models in one locus, two-allele models. The matching-alleles (MA) model is thought to represent interactions between hosts with the immune system and antigenic parasites, which have to specifically match the host in order to infect it. The gene-for-gene (GFG) type of interaction, inspired by interactions of plants with their pathogens, represents the situation where host needs to recognize specific ‘effectors’ of the parasite in order to launch its defence, hence here matching is equivalent to resistance. Mutations both in the host and the parasite would lead to the lack of such recognition, and hence to infection. Therefore, the parasite population consists of specialists (parasite allele A can only infect host A) and generalists (parasite B can infect any host allele). Above, sH denotes the relative fitness cost of the host owing to parasitic infection, sP denotes the relative fitness cost of the parasite for the inability to infect the host. We assume that 0 < sH < 1 and 0 < sP < 1.
| host fitness | host A | host B | parasite fitness | host A | host B |
|---|---|---|---|---|---|
| MA model | |||||
| parasite A | 1 − sH | 1 | parasite A | 1 | 1 − sP |
| parasite B | 1 | 1 − sH | parasite B | 1 − sP | 1 |
| GFG model | |||||
| parasite A | 1 | 1 − sH | parasite A | 1 − sP | 1 |
| parasite B | 1 − sH | 1 − sH | parasite B | 1 | 1 |
(b). Environment
We allow the abiotic environment to affect the interaction between the host and the parasite. In particular, the environment is given by a parameter E, which varies continuously between two extreme environments, E1 and E2. The interaction model between the two species is given by the interaction matrix ws,E1 in environment E1 and ws,E2 in environment E2, respectively, where s stands for the host (H) or parasite (P) species. The fitness matrix for an arbitrary environment E ∈ [0,1] is then given by the equation
| 2.3 |
Here, E = 1 yields the interaction model in environment E1 and E = 0 in E2. There are at least two interpretations of such environmental impact on the fitness values assumed. The first interpretation is that the fitness values of individuals in the population depend linearly on the parameter E, which can vary on a continuum between 0 and 1. Here, all individuals are assumed to be equally affected by an environmental factor (a good example might be temperature), and the fitness of each genotype is an E-weighted mean of the fitness values in the extreme environments. A second interpretation is that environment is discrete (e.g. presence or absence of a certain nutrient). Thus, only environments E1 and E2 occur, but these two environments are distributed spatially in the habitat of the population. In each generation, a randomly chosen fraction E of genotypes then undergoes an interaction defined by ws,E1 and the remaining fraction 1 − E undergoes an interaction defined by ws,E2. This interpretation of the model requires global competition and complete mixing of the population during reproduction. Finally, we generally assume the environment to be abiotic, but a biotic interpretation is also possible as long as the environmental fluctuations are independent of the coevolutionary dynamics.
By default, we assume E to be time dependent, oscillating between the extreme values according to
Here, both the mean value of the environment and the amplitude of oscillations are equal to 1/2 and T is the period of oscillations. We also investigated the impact of other types of environmental change, which are discussed in §3.
(c). Environment-dependent interactions
We examine two classes of interaction models: the MA class of interactions and the GFG class of interactions (table 1). The main difference between the two models is the degree of specialism/generalism. The MA model represents a full degree of specialism where a given parasite is better than any other parasite in infecting a given host but is worse off on any different host. This type of a lock–key mechanism (match versus non-match) is thought to emulate animal self-/non-self-recognition systems [34]. The GFG model, on the other hand, allows for both specialist and generalist parasites, the latter being equally effective in infecting all hosts. Although initially introduced to describe plant–fungal interactions [35,36], the general context of this model is beginning to be recognized [37–40]. Both models can produce persistant RQ dynamics, although these dynamics differ in some respects [21,36].
When MA interactions are considered, we assume that the ‘matching’ is environment dependent, where ‘matching’ refers to the interaction between the host allele and the parasite allele that result in infection. In particular, in environment E1 allele, A matches allele A and B matches B, whereas in environment E2, allele A matches B, and B matches A. The interaction matrices then take the following form:
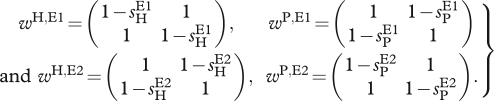 |
2.4 |
The notation we use is the following: sHE1 and sPE1 are the selection coefficients for the hosts and parasites, respectively, in environment E1 and sHE2 and sPE2 are the selection coefficients in environment E2. (Unless noted otherwise, we assume sHE1 > 0, sHE2 > 0, sPE1 > 0, sPE2 > 0.) For any given environment, E, the effective interaction matrices are obtained by the use of equation (2.3).
When GFG interactions are considered, we assume that the ‘matching’, which in this context results in host resistance, occurs solely between one pair of loci: in environment E1 only host allele A matches parasite allele A, and in environment E2 only host allele B matches parasite allele B. Therefore, in the host, allele A confers resistance in environment E1 and susceptibility to parasite in E2, whereas the converse is true for allele B. In the parasites, allele B is a generalist in E1 because it allows infecting any host (universal virulence) and is a specialist in E2 because it allows infecting only host B (avirulence); the converse is true for allele A. The interaction matrices are given by
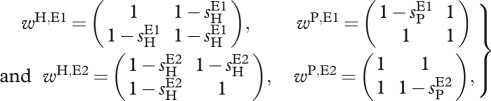 |
2.5 |
where the notation is identical to the one in the case of the MA model, and the effective interaction matrices are also obtained by the use of equation (2.3).
3. Results
(a). Matching-alleles interactions
In order to examine the impact of environment on the host–parasite dynamics defined by the MA-type of interaction, we examined the robustness of RQ dynamics when the environment is assumed to alter the specificity of the interaction between the two species. In particular, we assumed that the ‘matching’ of alleles depends on the environment in which the interaction takes place (see §2). Given the interaction matrices (2.4) for the two extreme environments, we first calculated the effective interaction model in any given environment E ∈ [0,1] from equation (2.3). One can show that this effective interaction model is again exactly an MA model (see table 1) with the original coefficients sH and sP replaced by the following ‘effective selection coefficients’ ξH and ξP:
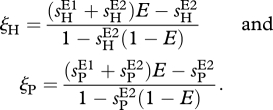 |
3.1 |
Thus, the environment-dependent model behaves like a regular MA model for any fixed value of E, although now the effective selection coefficients need not be positive.
As we were interested in the conditions for the occurrence of oscillatory behaviour, we next developed a general framework in order to derive such conditions given an arbitrary, but constant fitness interaction model, which is given in electronic supplementary material, part I. Using inequalities (S6), we first derived the conditions for the occurrence of RQ dynamics given a static environment, i.e. when E = const., and then used these to examine the case of a slowly changing environment. It can be shown that the condition (S6) in the context of the interaction model (2.4) is equivalent to
| 3.2 |
This means that for RQ dynamics to occur, the genetic interaction must be ‘antagonistic’, defined as an interaction where a host allele that is optimal for the host, given interaction with a particular parasite, does not maximize parasite fitness, and vice versa. By contrast, when the optimal allele for the host is also optimal for the parasite, a synergistic genetic interaction occurs (see electronic supplementary material).
A graphical representation of condition (3.2) is shown in figure 1a. One can see that as E increases from 0, both ξH and ξP increase from negative to positive values. The point where ξH and ξP change sign marks a switch in the specificity in the interaction for each species, but importantly, this switch will in general occur for a different value of E in hosts and parasites. The result of this partial change in specificity is that there will be a range of E (between sHE2/(sHE1 + sHE2) and sPE2/(sPE1 + sPE2)), where negative frequency-dependent selection (FDS) changes into positive FDS, leading to fixation of one host allele and one parasite allele. Negative FDS results from an antagonistic genetic interaction between the two species, whereas positive FDS stems from synergistic interaction. Positive FDS occurs only in models where the specificity of interaction varies. When the environment changes solely the strength of selection, synergism of genetic interaction never occurs and hence only the speed of cycles is affected. This can be seen from the fact that condition (3.2) is fulfilled for any value of E when sHE2< 0 and sPE2< 0.
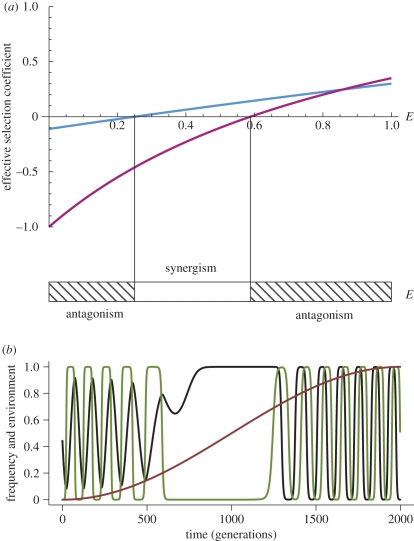
Figure 1.
Impact of environment on host–parasite coevolutionary dynamics with MA interactions. (a) Theoretical predictions for the persistence of RQ dynamics. The horizontal axis shows environmental parameter E and the vertical axis shows effective selection coefficients. The blue and the purple curves show the effective selection coefficient of the host and the parasite, respectively. The horizontal bar shows the parameter areas of E where the interaction is antagonistic (ξHξP>0; hatched areas), and where it is synergistic (ξH ξP<0; white area). Blue line, ξH; purple line, ξP. (b) The coevolutionary dynamics for the situation in (a) when the environment changes slowly. RQ dynamics occur only for the values of E where ξH ξP>0. Black lines, host A; green lines, parasite A; brown line, environment. Values used in (a) and (b) are sHE1 = 0.3, sPE1 = 0.35, sHE2 = 0.1, sPE2 = 0.5. Panel (b) further assumes T = 4000.
The results can now be applied to understand the impact of environmental change on the coevolutionary dynamics, i.e. the case when E = E(t) and hence ξH = ξH(t) and ξP = ξP(t). When the environment changes slowly (T ≫1), ξH and ξP will stay approximately constant relative to the velocity of allele-frequency change, and thus this case is readily understood from the analytical predictions illustrated in figure 1a. Figure 1b shows an example of the resulting coevolutionary dynamics. It can be seen that, as expected, for intermediate values of E (when ξH and ξP are of opposite sign), a time window of allele fixation emerges. In this time window, antagonistic interaction changes into a synergistic interaction, leading to positive FDS. These results are qualitatively the same for other selection coefficients and other (non-zero) mutation rates. Examples for allele-frequency dynamics when the environment affects only the strength, but not the specificity of the interaction (i.e. the case of sHE2<0 and sPE2<0), are shown in electronic supplementary material, figure S5.
As the environment changes more swiftly, the ‘constant E’ approximation is not valid anymore. For this case, the geometric mean of effective selection coefficients over many generations determines the dynamics of the system. Defining
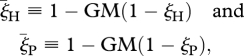 |
3.3 |
where GM denotes the geometric mean, it can be shown (see electronic supplementary material) that the condition for the maintenance of RQ dynamics under temporal environmental variation (S11) reads
| 3.4 |
Note that condition (3.4) is a generalization of the condition (3.2).
In the extreme case of maximally rapid environmental change (T = 2), we have
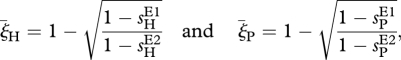 |
3.5 |
and condition (3.4) implies that either sHE2> sHE1 and sPE2> sPE1, or sHE2< sHE1 and sPE2< sPE1 must hold for RQ dynamics to occur. If not, the antagonism will effectively change into synergism and polymorphism will not be maintained (see electronic supplementary material, figure S2b). Extensive numerical screenings of the parameter space have confirmed the analytical predictions (results not shown).
If condition (3.4) is fulfilled, a phenomenon is sometimes observed that may on first sight be counterintuitive: RQ dynamics can proceed slower in systems with larger selection coefficients than in systems with smaller selection coefficients (see figure 2 for an example). This effect can again be understood with the help of the geometric mean effective selection coefficients. Equation (3.5) shows that the effective selection acting on the populations can be small even if the selection coefficients measured in environments E1 and E2 are large, provided they are of comparable magnitude. As a result, the environmental fluctuations may effectively weaken the average selection acting on the population. Such long-term impact of environment stands in contrast to a short-term impact, which stems from the changes of the magnitude and the direction of selection from one generation to another. These changes will lead to rapid allele fluctuations, which in turn may contribute to shaping temporal patterns of genetic variation in the population (see small subplots in figure 2a,b). Such clear distinction between a long-term and a short-term effect becomes blurry as T becomes larger: the short-term effect and the long-term effect will gradually merge into the regular cycles observed for E ≈ const.
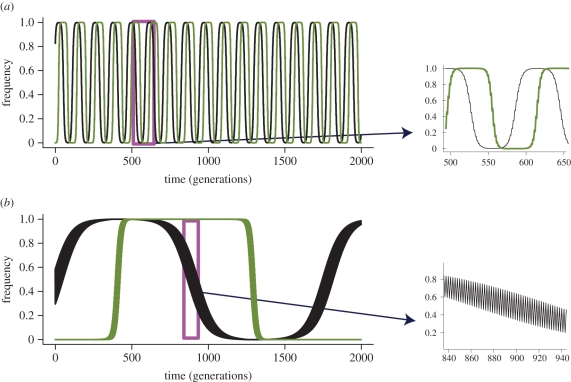
Figure 2.
Impact of the rapidly changing environment on host–parasite coevolutionary dynamics with MA interactions. Both panels show the simulation results of the model. (a) Rapid RQ dynamics in spite of comparatively weak selection. (b) Slow RQ dynamics in spite of comparatively strong selection. Allele-frequency changes proceed faster in (a) than in (b) because the long term, average selection acting on the population (effective selection) in (a) is stronger than such long-term selection in (b). In both panels, subplots show rapid allele fluctuations from one generation to another caused by rapidly altering direction of selection. Importantly, in both panels, the condition 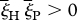 is fulfilled; its violation would lead to a permanent synergistic interaction, and hence allele fixation. Values used in (a) are sHE1 = 0.4, sPE1 = 0.6, sHE2 = 0.01, sPE2 = 0.1, and values used in (b) are sHE1 = 0.71, sPE1 = 0.95, sHE2 = 0.7, sPE2 = 0.94. All simulations use T = 2 (environment not shown). Black lines, host A; green lines, parasite A.
is fulfilled; its violation would lead to a permanent synergistic interaction, and hence allele fixation. Values used in (a) are sHE1 = 0.4, sPE1 = 0.6, sHE2 = 0.01, sPE2 = 0.1, and values used in (b) are sHE1 = 0.71, sPE1 = 0.95, sHE2 = 0.7, sPE2 = 0.94. All simulations use T = 2 (environment not shown). Black lines, host A; green lines, parasite A.
Finally, we also studied the impact of two other types of environmental change. First, we examined the impact of discrete switches between the extreme environments (i.e. E takes only the values 0 and 1 for n generations). In the case of maximally short persistence (n = 1), this situation is equivalent to the earlier discussed situation of T = 2. In the case of long persistence (n ≫1), the coevolutionary cycles observed in both environments are only briefly interrupted following a one-generation switch from E1 to E2 (or vice versa); otherwise the cycles are defined by the interaction model for a given environment (either E1 or E2; see electronic supplementary material, figure S2). Second, we examined the effect of stochastic environmental change (see electronic supplementary material, figure S3). For weak selection, the geometric mean condition for oscillatory allele-frequency dynamics can in this case be approximated by
| 3.6 |
where the derivation is based on the one given in Nuismer et al. [31], and < x > denotes the expected value of x. As anticipated, the long-term/short-term distinction can again be observed in a rapidly changing environment, whereas for more steady environments, the E ≈ const. approximation is informative of the coevolutionary dynamics.
(b). Gene-for-gene interactions
In this section, we analyse the impact of environmental change on the host–parasite interactions defined by a class of GFG models. Specifically, we consider here a situation where the environment changes specificity in both the host and the parasite (§2). This means that the two alternative phenotypes of the host interaction locus (resistance and susceptibility) as well as two alternative phenotypes of the parasite interaction locus (avirulence and virulence) are interchanged under the influence of the environment. Under these circumstances, the interaction model is given by equation (2.5). Based on the general results given in electronic supplementary material, part I, one can show that with fixed E coevolutionary cycles will ensue if and only if
| 3.7 |
Figure 3 shows simulation results for slowly and rapidly changing environments. Figure 3a shows cycling of allele frequencies in time when environment changes slowly. E can therefore be considered as approximately constant in each time point, which explains the resulting lack of coevolutionary dynamics for E = 0 and E = 1. As the environment changes between the two extremes, the antagonism and specialism, and hence the cycles, resume. Interestingly, as the environment changes between E1 and E2, the interaction model changes from the GFG into an MA-like model. In particular, for E = sHE2/(sHE1 + sHE2), the host exactly undergoes the MA interaction given in table 1 with
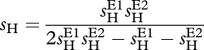 |
and the same reasoning applies to the parasite. Hence, a changing environment that affects the specificity of a GFG interaction can induce negative FDS, thus explaining the maintenance of polymorphism in the population.
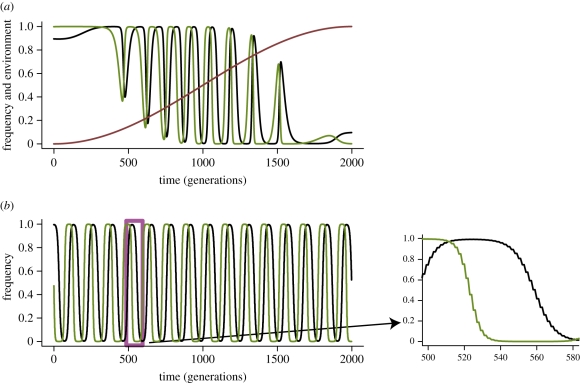
Figure 3.
Impact of slowly and rapidly changing environment on host–parasite coevolutionary dynamics with GFG interactions. (a) Impact of a slowly changing environment. Coevolutionary cycles emerge for E ∈ (0,1). As E changes between E1 and E2, the interaction model changes from the GFG into the MA-like interaction. (b) Impact of rapidly changing environment. The impact of environment is again subdivided into a long-term and a short-term effect. The long-term effect is determined by the geometric mean effective selection coefficient. The short-term effect yields step-like allele fluctuations owing to an inherent asymmetry of the GFG model (selection acting on an allele in one environment is much stronger than the selection acting in the other environment). Values used in (a) and (b), are sHE1 = 0.35, sPE1 = 0.48, sHE2 = 0.3, sPE2 = 0.45, and furthermore T = 4000 in (a) and T = 2 in (b). Black lines, host A; green lines, parasite A; brown lines, environment (for sake of clarity, not shown in panel b).
Figure 3b shows the dynamics for the case when environmental change is rapid (i.e. changes every generation). Similar to MA interactions, we can see that the impact of environment can be subdivided into a short-term effect and a long-term effect. The long-term effect is determined by the geometric mean selection coefficients over many generations, which again can be derived in analogy to the MA model case (although now there are two coefficients per species; see electronic supplementary material). The short-term effect stems from the selection coefficients acting on each allele in each of the extreme environments, thus changing every generation. In the case considered here, it can be seen that the rapid allele fluctuations observed for the MA model are replaced by the step-like fluctuations, suggesting that the short-term selection acting on them is uni-directional (see the small sub-figure in figure 3b). Such qualitatively different dynamics are the consequence of the inherent asymmetry of the GFG model, where one observes recurrent sweeps of the resistant and the virulent alleles. Since here, virulence and resistance are phenotypes expressed solely in one of the environments, the periods of increase are interrupted by periods of allele-frequency stagnation. Finally, the condition for the maintenance of polymorphism in a stochastic environment can be derived in the weak selection limit in analogy to condition (3.6); see the electronic supplementary material, part II.
We analysed different impacts of environment on the GFG model and found that polymorphism persists only if the specificity of interaction is altered in both species. Similar conclusions can be drawn if one ‘inverts’ the GFG model by assuming that the matching does not yield resistance but infectivity. Such model typically represents pathogens that possess receptors mediating their entrance into their host [38] but also in the interaction of plants with their necrotrophic parasites [41]. In this case, a switch in specificity of selection in both species is again required in order to yield evolutionary cycles. We also extended our analysis to switches between different interaction models (see electronic supplementary material, figure S4). Even though the results depend on the interaction model considered, in each case, the analysis presented here can be repeated in order to examine the underlying dynamics. Overall, we have seen that if the environment affects the specificity of interaction between the host and the parasite, the antagonism and hence coevolutionary dynamics can be affected in a way that is not to be expected from interactions obtained in a constant environment.
4. Discussion
Using a simple mathematical model, we studied the impact of environmental changes on coevolutionary dynamics between hosts and parasites. Our results show that when environmental factors influence the specificity of G × G host–parasite interactions, this can have profound effects on the coevolutionary dynamics. Most importantly, temporal environmental changes can inhibit RQ dynamics where they would occur in a stable environment (MA interactions) and trigger RQ dynamics where they would not occur (GFG interactions). These effects can be understood through the notion of effective selection coefficients, which clarify how both specificity and antagonism in the host–parasite interaction can be affected by environmental change.
An important factor is the velocity of environmental change. If the environment changes rapidly between the two extremes (e.g. every generation), then the impact of the environment can be subdivided into a long-term and a short-term effect. The long-term effect stems from the average of host and parasite selection coefficients over many generations. This long-term selection acting on the populations is typically weaker than the selection defined in a single, constant environment. On the other hand, the short-term effect stems from changes in selection coefficients from one generation to another. This can produce rapid allele fluctuations, the amplitude and direction of which depend on the difference in the relative selective pressure between the two interacting species. As environmental changes become slower, the short-term and the long-term effects will gradually merge together producing dynamics increasingly similar to those observed for a constant environment (cf. electronic supplementary material, figure S1). When the environmental change is slow relative to the generation time, the coevolutionary dynamics are governed by an approximately constant interaction model at each time point (figure 1).
Our results have a number of interesting implications for studies of host–parasite interactions. First, in populations that undergo G × G × E interactions with change in the specificity of selection, the effective selection acting in a population may be weaker than selection measured in constant environments. In particular, if strong selection coefficients in a host–parasite interaction are measured in two different environmental states, this does not necessarily mean that rapid coevolutionary dynamics—or any dynamics—are to be expected when the environment changes between these two states. There may be periods with slow allele-frequency oscillations (owing to weak effective selection) or fixations of alleles (owing to temporary loss of antagonism or specificity) that alternate with periods of rapid allele-frequency change.
Our findings can also be viewed within the framework of the geographical mosaic theory of coevolution [42,43]. This theory states that three processes are primary drivers of coevolutionary dynamics: (i) a selection mosaic mediated by G × G × E interactions, (ii) the existence of communities where selection may or may not be reciprocal, yielding evolutionary hot and cold spots, respectively, and (iii) a dynamic genetic structure of the coevolving species affected by gene flow, random genetic drift and other factors. Mathematical models studying the geographical mosaic have mainly focused on the impact of spatial environmental heterogeneity on the coevolutionary process (e.g. [44–47]; but see also Nuismer et al. [31] for an investigation of temporal environmental variability). Here, we have assumed that the sign and the magnitude of selection in the host–parasite interaction change with temporal environmental variation within a coevolutionary hot spot. We have shown that in spite of an inherent antagonistic interaction, temporal environmental variation can remove the negative FDS where it would occur in a constant environment (MA) and produce such selection where it would not occur (costless GFG). As a result, the environmental change affecting the direction of G × G interactions within a coevolutionary hot spot may qualitatively affect the coevolutionary host–parasite dynamics. It would be interesting to embed our model into a spatial context and examine the impact of environmental change affecting the specificity of host–parasite interactions in local coevolutionary hot spots on the global coevolutionary dynamics in a metapopulation.
Even though a sole change in the strength of selection can substantially affect the speed of allele-frequency change (see electronic supplementary material, figure S5), our results show that it is the change in specificity of selection that leads to the most dramatic impact on RQ dynamics. Although the latter form of environmental impact might not be as empirically common as assumed in this model, our study points to the importance of extensively characterizing these interactions in natural host–parasite populations. At present, the scale of occurrence of this latter type of environmental impact remains not well understood. To our knowledge, only three studies have provided direct evidence for G × G × E interactions (B. Sadd, 2010, Submitted; [29,30]). However, many studies have demonstrated strong G × E interactions, i.e. environmentally induced switches in the specificity of infection success of several parasite genotypes on a single host or vice versa (e.g. [48–55]; reviewed in [28]). Importantly, none of these studies could reject the presence of G × G × E interactions because either only a single host or only a single parasite genotype was tested. Taking the available evidence for both G × G and G × E interactions together and also taking into consideration that only a small fraction of genotypes and environmental conditions can be tested in experiments, it is to be expected that G × G × E interactions are common in natural systems. Our study points to the importance of extensively characterizing such interactions in natural host–parasite populations.
Environmental change as studied here may also have implications for the maintenance of polymorphism in host–parasite systems that undergo the GFG type of interaction. One important property of the GFG model is that in the absence of costs associated with the resistance allele in the host and the virulence allele in the parasite, the virulence allele will become fixed in the parasite population and coevolutionary dynamics cease. In the context of the GFG interactions, the notion of costly resistance/virulence has been a subject of debate [56–58], and a number of alternative explanations have been put forward [41,59,60]. Recently, it has been suggested that heterogeneous environments affecting selection in host–parasite systems undergoing GFG interactions may serve as yet another explanation for the persistence of polymorphism in natural populations [39]. Here, we show that persistent coevolutionary cycles can indeed emerge in the absence of costs and spatial structure, provided that the environment affects the specificity of the GFG interaction in both species.
We have deliberately kept the model as simple as possible, as this enabled us to obtain analytical solutions and provide intuitive interpretations of the simulation results. We realize that this model is too simple to fit any experimental case of G × G × E interactions in host–parasite systems. However, we believe its simplicity may reveal basic patterns of environmental impact on coevolutionary dynamics, which might underlie the outcome of interactions in real host–parasite systems with complex interaction networks. Extending our model to include multiple loci, recombination, population dynamics and life history of both species, epistatic effects and potentially diverging impact of different environmental factors on different loci would be a valuable future task.
Acknowledgements
We wish to thank Bruce McDonald, Dominik Refardt, Ben Sadd and two anonymous referees for helpful comments on the manuscript, as well as Sebastian Bonhoeffer and Megan McDonald for stimulating discussions. This work was supported by the Swiss National Science Foundation.
References
- 1.Little T. J. 2002. The evolutionary significance of parasitism: do parasite-driven genetic dynamics occur ex silico? J. Evol. Biol. 15, 1–9 10.1046/j.1420-9101.2002.00366.x (doi:10.1046/j.1420-9101.2002.00366.x) [DOI] [Google Scholar]
- 2.Woolhouse M. E. J., Webster J. P., Domingo E., Charlesworth B., Levin B. R. 2002. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 32, 569–577 10.1038/ng1202-569 (doi:10.1038/ng1202-569) [DOI] [PubMed] [Google Scholar]
- 3.Barrett J. A. 1988. Frequency-dependent selection in plant–fungal interactions. Phil. Trans. R. Soc. Lond. B 319, 473–483 10.1098/rstb.1988.0060 (doi:10.1098/rstb.1988.0060) [DOI] [Google Scholar]
- 4.Decaestecker E., Gaba S., Raeymaekers J. A. M., Stoks R., Van Kerckhoven L., Ebert D., De Meester L. 2007. Host–parasite Red Queen dynamics archived in pond sediment. Nature 450, 870–873 [DOI] [PubMed] [Google Scholar]
- 5.Jokela J., Dybdahl M. F., Lively C. M. 2009. The maintenance of sex, clonal dynamics, and host–parasite coevolution in a mixed population of sexual and asexual snails. Am. Nat. 174, S43–S53 10.1086/599080 (doi:10.1086/599080) [DOI] [PubMed] [Google Scholar]
- 6.Kaltz O., Shykoff J. A. 1998. Local adaptation in host–parasite systems. Heredity 81, 361–370 10.1046/j.1365-2540.1998.00435.x (doi:10.1046/j.1365-2540.1998.00435.x) [DOI] [Google Scholar]
- 7.Roff D. A. 1997. Evolutionary quantitative genetics, 1st edn. New York, NY: Chapman & Hall [Google Scholar]
- 8.Tellier A., Brown J. K. 2007. Stability of genetic polymorphism in host–parasite interactions. Proc. R. Soc. B 274, 809–817 10.1098/rspb.2006.0281 (doi:10.1098/rspb.2006.0281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paterson S., et al. 2010. Antagonistic coevolution accelerates molecular evolution. Nature 464, 275–278 10.1038/nature08798 (doi:10.1038/nature08798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rausher M. D. 2001. Co-evolution and plant resistance to natural enemies. Nature 411, 857–864 10.1038/35081193 (doi:10.1038/35081193) [DOI] [PubMed] [Google Scholar]
- 11.Day T., Burns J. G. 2003. A consideration of patterns of virulence arising from host–parasite coevolution. Evolution 57, 671–676 [DOI] [PubMed] [Google Scholar]
- 12.Read A. F. 1994. The evolution of virulence. Trends Microbiol. 2, 73–76 10.1016/0966-842X(94)90537-1 (doi:10.1016/0966-842X(94)90537-1) [DOI] [PubMed] [Google Scholar]
- 13.Hamilton W. D. 1980. Sex versus non-sex versus parasite. Oikos 35, 282–290 10.2307/3544435 (doi:10.2307/3544435) [DOI] [Google Scholar]
- 14.Jaenike J. 1978. An hypothesis to account for the maintenance of sex within populations. Evol. Theory 3, 191–194 [Google Scholar]
- 15.Salathé M., Kouyos R. D., Bonhoeffer S. 2008. The state of affairs in the kingdom of the Red Queen. Trends Ecol. Evol. 23, 439–445 [DOI] [PubMed] [Google Scholar]
- 16.Carius H. J., Little T. J., Ebert D. 2001. Genetic variation in a host–parasite association: potential for coevolution and frequency-dependent selection. Evolution 55, 1136–1145 [DOI] [PubMed] [Google Scholar]
- 17.Ebert D., Zschokke-Rohringer C. D., Carius H. J. 1998. Within- and between-population variation for resistance of Daphnia magna to the bacterial endoparasite Pasteuria ramosa. Proc. R. Soc. Lond. B 265, 2127–2134 10.1098/rspb.1998.0549 (doi:10.1098/rspb.1998.0549) [DOI] [Google Scholar]
- 18.Lively C. M. 1989. Adaptation by a parasitic trematode to local populations of its snail host. Evolution 43, 1663–1671 10.2307/2409382 (doi:10.2307/2409382) [DOI] [PubMed] [Google Scholar]
- 19.Luijckx P., Ben-Ami F., Mouton L., Du Pasquier L., Ebert D. In press. Cloning of the unculturable parasite Pasteuria ramosa and its Daphnia host reveals extreme genotype–genotype interactions. Ecol. Lett. (doi:10.1111/j.1461-0248.2010.01561) [DOI] [PubMed] [Google Scholar]
- 20.Lambrechts L., Fellous S., Koella J. C. 2006. Coevolutionary interactions between host and parasite genotypes. Trends Parasitol. 22, 12–16 10.1016/j.pt.2005.11.008 (doi:10.1016/j.pt.2005.11.008) [DOI] [PubMed] [Google Scholar]
- 21.Agrawal A., Lively C. M. 2002. Infection genetics: gene-for-gene versus matching-alleles models and all points in between. Evol. Ecol. Res. 4, 79–90 [Google Scholar]
- 22.Engelstädter J., Bonhoeffer S. 2009. Red Queen dynamics with non-standard fitness interactions. PLoS Comp. Biol. 5, 140–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayakar S. D. 1970. A mathematical model for interaction of gene frequencies in a parasite and its host. Theor. Popul. Biol. 1, 140–164 10.1016/0040-5809(70)90032-8 (doi:10.1016/0040-5809(70)90032-8) [DOI] [PubMed] [Google Scholar]
- 24.Mode C. J. 1958. A mathematical model for the co-evolution of obligate parasites and their hosts. Evolution 12, 158–165 10.2307/2406026 (doi:10.2307/2406026) [DOI] [Google Scholar]
- 25.Laine A.-L. 2009. Role of coevolution in generating biological diversity: spatially divergent selection trajectories. J. Exp. Bot. 60, 2957–2970 10.1093/jxb/erp168 (doi:10.1093/jxb/erp168) [DOI] [PubMed] [Google Scholar]
- 26.Lazzaro B. P., Little T. J. 2009. Immunity in a variable world. Phil. Trans. R. Soc. B 364, 15–26 10.1098/rstb.2008.0141 (doi:10.1098/rstb.2008.0141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wade M. J. 2007. The co-evolutionary genetics of ecological communities. Nat. Rev. Genet. 8, 185–195 10.1038/nrg2031 (doi:10.1038/nrg2031) [DOI] [PubMed] [Google Scholar]
- 28.Wolinska J., King K. C. 2009. Environment can alter selection in host–parasite interactions. Trends parasitol. 25, 236–244 10.1016/j.pt.2009.02.004 (doi:10.1016/j.pt.2009.02.004) [DOI] [PubMed] [Google Scholar]
- 29.Tétard-Jones C., Kertesz M. A., Gallois P., Preziosi R. F. 2007. Genotype-by-genotype interactions modified by a third species in a plant–insect system. Am. Nat. 170, 492–499 [DOI] [PubMed] [Google Scholar]
- 30.Bryner S. F., Rigling D. 2011. Temperature dependent genotype-by-genotype interaction between a pathogenic fungus and its hyperparasitic virus. Am. Nat. 177, 65–74 10.1086/657620 (doi:10.1086/657620) [DOI] [PubMed] [Google Scholar]
- 31.Nuismer S. L., Gomulkiewicz R., Morgan M. T. 2003. Coevolution in temporally variable environments. Am. Nat. 162, 195–204 10.1086/376582 (doi:10.1086/376582) [DOI] [PubMed] [Google Scholar]
- 32.Nee S. 1989. Antagonistic co-evolution and the evolution of genotypic randomization. J. Theor. Biol. 140, 499–518 10.1016/S0022-5193(89)80111-0 (doi:10.1016/S0022-5193(89)80111-0) [DOI] [PubMed] [Google Scholar]
- 33.Otto S. P., Nuismer S. L. 2004. Species interactions and the evolution of sex. Science 304, 1018–1020 10.1126/science.1094072 (doi:10.1126/science.1094072) [DOI] [PubMed] [Google Scholar]
- 34.Grosberg R. K., Hart M. W. 2000. Mate selection and the evolution of highly polymorphic self/nonself recognition genes. Science 289, 2111–2114 10.1126/science.289.5487.2111 (doi:10.1126/science.289.5487.2111) [DOI] [PubMed] [Google Scholar]
- 35.Flor H. 1955. Host–parasite interaction in flax rust—its genetics and other implications. Phytopathology 45, 680–685 [Google Scholar]
- 36.Parker M. A. 1994. Pathogens and sex in plants. Evol. Ecol. 8, 560–584 10.1007/BF01238258 (doi:10.1007/BF01238258) [DOI] [Google Scholar]
- 37.Dangl J. L., Jones J. D. 2001. Plant pathogens and integrated defence responses to infection. Nature 411, 826–833 10.1038/35081161 (doi:10.1038/35081161) [DOI] [PubMed] [Google Scholar]
- 38.Fenton A., Antonovics J., Brockhurst M. A. 2009. Inverse-gene-for-gene infection genetics and coevolutionary dynamics. Am. Nat. 174, E230–E242 10.1086/645087 (doi:10.1086/645087) [DOI] [PubMed] [Google Scholar]
- 39.Laine A.-L., Tellier A. 2008. Heterogeneous selection promotes maintenance of polymorphism in host–parasite interactions. Oikos 117, 1281–1288 10.1111/j.0030-1299.2008.16563.x (doi:10.1111/j.0030-1299.2008.16563.x) [DOI] [Google Scholar]
- 40.Nürnberger T., Brunner F., Kemmerling B., Piater L. 2004. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266 [DOI] [PubMed] [Google Scholar]
- 41.Stukenbrock E. H., McDonald B. A. 2009. Population genetics of fungal and oomycete effectors involved in gene-for-gene interactions. MPMI 22, 371–380 10.1094/MPMI-22-4-0371 (doi:10.1094/MPMI-22-4-0371) [DOI] [PubMed] [Google Scholar]
- 42.Thompson J. N. 1994. The coevolutionary process. Chicago, IL: University of Chicago Press [Google Scholar]
- 43.Thompson J. N. 2005. The geographic mosaic of coevolution.1st edn Chicago, IL: University of Chicago Press [Google Scholar]
- 44.Gavrilets S., Michalakis Y. 2008. Effects of environmental heterogeneity on victim–exploiter coevolution. Evolution 62, 3100–3116 10.1111/j.1558-5646.2008.00513.x (doi:10.1111/j.1558-5646.2008.00513.x) [DOI] [PubMed] [Google Scholar]
- 45.Gomulkiewicz R., Thompson J. N., Holt R. D., Nuismer S. L., Hochberg M. E. 2000. Hot spots, cold spots, and the geographic mosaic theory of coevolution. Am. Nat. 156, 156–174 10.1086/303382 (doi:10.1086/303382) [DOI] [PubMed] [Google Scholar]
- 46.Nuismer S. L., Thompson J. N., Gomulkiewicz R. 1999. Gene flow and geographically structured coevolution. Proc. R. Soc. Lond. B 266, 605–609 10.1098/rspb.1999.0679 (doi:10.1098/rspb.1999.0679) [DOI] [Google Scholar]
- 47.Nuismer S. L., Thompson J. N., Gomulkiewicz R. 2000. Coevolutionary clines across selection mosaics. Evolution 54, 1102–1115 [DOI] [PubMed] [Google Scholar]
- 48.Blanford S., Thomas M. B., Pugh C., Pell J. K. 2003. Temperature checks the Red Queen? Resistance and virulence in a fluctuating environment. Ecol. Lett. 6, 2–5 10.1046/j.1461-0248.2003.00387.x (doi:10.1046/j.1461-0248.2003.00387.x) [DOI] [Google Scholar]
- 49.Fels D., Kaltz O. 2006. Temperature-dependent transmission and latency of Holospora undulata, a micronucleus-specific parasite of the ciliate Paramecium caudatum. Proc. R. Soc. B 273, 1031–1038 10.1098/rspb.2005.3404 (doi:10.1098/rspb.2005.3404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laine A. L. 2007. Pathogen fitness components and genotypes differ in their sensitivity to nutrient and temperature variation in a wild plant–pathogen association. J. Evol. Biol. 20, 2371–2378 10.1111/j.1420-9101.2007.01406.x (doi:10.1111/j.1420-9101.2007.01406.x) [DOI] [PubMed] [Google Scholar]
- 51.Lambrechts L., Chavatte J. M., Snounou G., Koella J. C. 2006. Environmental influence on the genetic basis of mosquito resistance to malaria parasites. Proc. R. Soc. B 273, 1501–1506 10.1098/rspb.2006.3483 (doi:10.1098/rspb.2006.3483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lazzaro B. P., Flores H. A., Lorigan J. G., Yourth C. P. 2008. Genotype-by-environment interactions and adaptation to local temperature affect immunity and fecundity in Drosophila melanogaster. PLoS Pathog. 4, e1000025. 10.1371/journal.ppat.1000025 (doi:10.1371/journal.ppat.1000025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell S. E., Rogers E. S., Little T. J., Read A. F. 2005. Host–parasite and genotype-by-environment interactions: temperature modifies potential for selection by a sterilizing pathogen. Evolution 59, 70–80 [PubMed] [Google Scholar]
- 54.Price J. S., Bever J. D., Clay K. 2004. Genotype, environment, and genotype by environment interactions determine quantitative resistance to leaf rust (Coleosporium asterum) in Euthamia graminifolia (Asteraceae). New Phytol. 162, 729–743 10.1111/j.1469-8137.2004.01082.x (doi:10.1111/j.1469-8137.2004.01082.x) [DOI] [PubMed] [Google Scholar]
- 55.Vale P. F., Stjernman M., Little T. J. 2008. Temperature-dependent costs of parasitism and maintenance of polymorphism under genotype-by-environment interactions. J. Evol. Biol. 21, 1418–1427 10.1111/j.1420-9101.2008.01555.x (doi:10.1111/j.1420-9101.2008.01555.x) [DOI] [PubMed] [Google Scholar]
- 56.Bergelson J., Purrington C. B. 1996. Surveying patterns in the cost of resistance in plants. Am. Nat. 148, 536–558 10.1086/285938 (doi:10.1086/285938) [DOI] [Google Scholar]
- 57.Sacristán S., García-Arenal F. 2008. The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9, 369–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vila-Aiub M. M., Neve P., Powles S. B. 2009. Fitness costs associated with evolved herbicide resistance alleles in plants. New Phytol. 184, 751–767 10.1111/j.1469-8137.2009.03055.x (doi:10.1111/j.1469-8137.2009.03055.x) [DOI] [PubMed] [Google Scholar]
- 59.Damgaard C. 1999. Coevolution of a plant host-pathogen gene-for-gene system in a metapopulation model without cost of resistance or cost of virulence. J. Theor. Biol. 201, 1–12 10.1006/jtbi.1999.1007 (doi:10.1006/jtbi.1999.1007) [DOI] [PubMed] [Google Scholar]
- 60.Salathé M., Scherer A., Bonhoeffer S. 2005. Neutral drift and polymorphism in gene-for-gene systems. Ecol. Lett. 8, 925–932 [DOI] [PubMed] [Google Scholar]