Abstract
Demyelination occurs in multiple inherited mitochondrial diseases. We studied which genes were induced as a consequence of differentiation in rodent and human oligodendroglia. Cholesterol, myelin and mitochondrial genes were significantly increased with oligodendroglial differentiation. Mitochondrial DNA content per cell and acetyl CoA-related transcripts increased significantly; thus, the large buildup of cholesterol necessary for myelination appears to require mitochondrial production of acetyl-CoA. Oligodendroglia were treated with low doses of the mitochondrial inhibitor rotenone to test the dependence of differentiation on mitochondrial function. Undifferentiated cells were resistant to rotenone, whereas differentiating cells were much more sensitive. Very low doses of rotenone that did not affect viability or ATP synthesis still inhibited differentiation, as measured by reduced levels of the myelin transcripts 2′,3′-Cyclic Nucleotide-3′-Phosphodiesterase and Myelin Basic Protein. Thus, mitochondrial transcripts and mtDNA are amplified during oligodendroglial differentiation, and differentiating oligodendroglia are especially sensitive to mitochondrial inhibition, suggesting mechanisms for demyelination observed in mitochondrial disease.
Keywords: Oligodendroglia, Mitochondria, Myelination, Cholesterol, Rotenone
1. Introduction
Oligodendrocytes are responsible for myelination of the axons in the Central Nervous System (CNS). Axons are initially unmyelinated and become myelinated during development. Myelination buffers the dissipation of the action potential along the axons, and provides trophic support for neurons. Oligodendroglial precursor cells grow into immature/galactosylceramide-positive oligodendrocytes, and finally into mature, myelin-producing oligodendrocytes (Baumann and Pham-Dinh, 2001).
Crosstalk between neurons, astrocytes and oligodendrocytes is necessary for proper oligodendrocyte differentiation and myelination. Growth factors such as PDGF, FGF-2, IGF-1, NT-3 and CNTF, released by neurons and astrocytes, function to promote oligodendrocyte differentiation. Neuregulin promotes differentiation via interaction with the ErbB receptor on oligodendrocytes (reviewed in (Simons and Trajkovic, 2006)). The interaction between F3/contactin and notch also promotes oligodendrocyte maturation (Hu et al., 2003). The electrical activity of neurons and the release of leukemia inhibitory factor from astrocytes have been shown to be important for initiating myelination (Ishibashi et al., 2006).
Demyelination occurs in multiple mitochondrial diseases, including Leber’s Hereditary Optic Neuropathy (Kovacs et al., 2005), Friedreich’s ataxia (Carelli et al., 2002), MELAS (Rusanen et al., 1995), Charcot-Marie Tooth 2a, caused by a defect in mitofusin 2 (Niemann et al., 2006), and Dominant Optic Atrophy, caused by a defect in opa1 (Johnston et al., 1979). Demyelination also occurs in Periventricular Leukomalacia (PVL), as a consequence of hypoxia/ischemia (Volpe, 2008). Consistent with the observation of demyelination in the mitochondrial genetic diseases named above, a recent microarray study of five mitochondrial diseases produced the unexpected result of significant down-regulation of several transcripts involved in myelination (Cortopassi et al., 2006). These results suggest that mitochondrial functions might be required for proper oligodendrocyte differentiation and myelination. To test this idea, we microarrayed undifferentiated and differentiated rat and human oligodendroglia. We also tested the effects of mitochondrial inhibition on oligodendroglial differentiation and found differentiating cells to be particularly sensitive to rotenone.
2. Materials and methods
Biochemical reagents were purchased from Sigma (St. Louis, MO), Invitrogen (Carlsbad, CA) or Bio-Rad (Hercules, CA). Micro-array chips and reagents were purchased from Affymetrix (Santa Clara, CA).
2.1. Cell culture
Purified primary cultures of oligodendroglial lineage from 0 to 2 day-old rats were prepared by serial immunopanning procedure as reported and characterized in detail elsewhere (Itoh et al., 2002, 2009; Horiuchi et al., 2006). Briefly, purified oligodendroglial cultures contained a more than 90% A2B5-positive O4-negative and glial fibrillary acidic protein-negative cell population which corresponded to the developmental stage of OPC. More importantly, the OPC cultures were virtually free of microglia as determined immunocytochemically by expression of CD11b antigen. The OPC cultures were expanded by up to four passages in the culture medium containing B104 neuroblastoma conditioned medium, bovine FGF2, and human recombinant PDGFAA.
To induce differentiation of OPC to oligodendrocytes, the culture medium was switched to a defined medium free of the mitogens and B104 neuroblastoma conditioned medium. Based on the immunocytochemical data at 4 days after induction of differentiation, these oligodendroglial cultures contained >80% cells positive for Myelin Basic Protein, and could be used as cultures of mature oligodendrocytes.
HOG cells, an oligodendroglioma cell line (a kind gift from Dr. G. Dawson, University of Chicago, Chicago, IL) and MO3.13 cells, an oligodendrocyte – rhabdomyosarcoma line, were grown in DMEM supplemented with 10% FBS. Differentiation into more mature oligodendrocytes was performed as described (Buntinx et al., 2003). Briefly, cells were allowed to sit overnight in growth media and replaced with differentiation media (DMEM, 0.05% FBS, 30 nM triiodothyronine, 30 nM selenium, 0.5 µg/ml insulin, and 50 µg/ml transferrin) the following day. Cells were differentiated for 10 days, replacing the differentiation media every 3 days. Cell viability was determined by the trypan blue exclusion method in the ViCell (Beckman-Coulter), a unit that objectively counts cells based on 100 images. Student’s t-test was used to determine significance.
2.2. Microarray
Total RNA was extracted using RNeasy columns (Qiagen, Valencia, CA). Ten micrograms of RNA was labeled for hybridization to the microarray chips according to the manufacturer’s recommendations. Rat samples were analyzed using the Affymetrix RAE230A arrays and human samples with U133A arrays.
Four sets of microarray data were compared, two from primary rat oligodendrocytes and two from human oligodendroglial cell lines HOG and MO3.13. Data set 1 was generated using total RNA from three preparations of primary rat oligodendroglial precursors and differentiated cells (seven chips) while data set 2 was previously published data from similar cells (nine chips) (Nielsen et al., 2006). Data sets 3 and 4 were generated using total RNA from undifferentiated and differentiated samples of Human HOG and MO3.13 cells, respectively (four chips per cell line). Each of the four data sets was analyzed with dChip, then filtered for transcripts altered at p < 0.10 and pCall > 0.
Data was analyzed individually for each sample type using dChip v2005 (Li and Hung Wong, 2001). Within each group, samples were normalized to the median intensity chip and fluorescence values generated using the perfect match/mismatch difference model. Probe sets with a pCall > 0% and p < .10 were considered significantly altered. Comparisons were made by comparing all undifferentiated to all differentiated and by parsing the group into subsets.
Updated annotations and orthologs were obtained from the NetAffx database (Liu et al., 2003) and multi-sample analysis performed by combining the dChip lists using Excel. Significantly altered probesets present in three or more groups were categorized using Onto-Express (Draghici et al., 2003). The false discovery rate method was used to correct for errors generated by multiple t-tests.
2.3. Quantitative reverse transcriptase PCR (QRT-PCR)
Total RNA was extracted from cells and QRT-PCR performed as previously described (Tan et al., 2001). The Roche Lightcycler 480 (Roche Diagnostics, Indianapolis, IN) was used to amplify the genes of interest with the following conditions, initial denaturation at 94 °C for 5 min, followed by 40 cycles of denaturation at 94 °C for 10 s, annealing at 60 °C for 20 s and extension at 72 °C for 5 s. The primer sequences for MBP and CNPase were obtained from Buntinx et al., and the primer sequences for vimentin were as follows: forward, 5′-GTCAGCAATATGAAAGTGTGGC-3′, reverse, 5′-GGTAGTTAGCAGCTTCAACGG-3′; a standard curve was amplified for each gene to quantify the samples. MBP and CNPase transcripts were normalized to vimentin expression.
Rat samples were amplified on the Roche Lightcycler 1.2, with the following conditions: 40 cycles of denaturation at 94 °C for 0 s, annealing at 60 °C for 10 s and extension at 72 °C for 20 s. The primers used were as follows: GAPDH forward, 5′-TCAAGAAGGTGGTGAAGCAGG-3′, reverse, 5′-GGTCTGGGATGGAATTGTGAG-3′; DHCR24 forward, 5′-ATGCTGGTACCCATGAAATGC-3′, reverse, 5′-CGAGATCTTGTCATACACCTC-3′; HMGCS1 forward, 5′-TCAAGGCTTGACTCAAGAACG-3′, reverse, 5′-GGAATATGCTCTGTAGCTGTG-3′; IDI1 forward, 5′-ACAGGTTCAGCTTCTAGCAGA-3′, reverse, 5′-CCTTTAAGCGCTTCTGTGCTG-3′; PDK2 forward, 5′-CATCAAAATGAGTGACCGAGG-3′, reverse, 5′-AGGCAGACTTGTTGTAGACAG-3′; SOD2 forward, 5′-CTCCCTGACCTGCCTTAC-3′, reverse, 5′-CGACCTTGCTCCTTATTG-3′; and HADHSC forward, 5′-CACCAG ACAAGACCGATTTGC-3′, reverse, 5′-ATTCATGGAAGGACTGGGCTG-3′. Rat samples were normalized to GAPDH, which was found to be unchanged between differentiated and undifferentiated samples.
2.4. Mitochondrial/nuclear copy number analysis
Human mitochondrial and nuclear copy number analysis was performed as previously described (Wong et al., 2002). Briefly, genomic DNA preparations (also containing mitochondrial DNA) were quantitatively PCR’d in a Lightcycler using ND1 (mitochondrial) and CFTR (nuclear) primers. Calculations assumed 300,000 mitochondrial copies and 150 nuclear copies per ng input DNA. ND2 and CFTR were used for rat mitochondrial and nuclear copy number analysis, respectively. The primers used were: ND2 forward, 5′-CTGAGGAGGACTTAACCAGAC-3′, reverse, 5′-GTGATAGGAGGATGATGGATG-3′ and CFTR forward, 5′-TGCTTTTAGTGCACGGATCTG-3′, reverse, 5′-GGCTTCACATGTGATCATCA-3′.
2.5. ATP synthesis assay
ATP synthesis was measured as described by Atorino et al. (2003). Briefly, cells were permeabilized with digitonin and incubated in buffer containing 0.25 M sucrose, 20 mM MOPS, 1 mM EDTA, 5 mM inorganic phosphate, 0.1% BSA fatty acid free, and 1 mM ADP, pH 7.4. To measure complexes I–V activity, 5 mM glutamate and 1 mM malate were added to permeabilized cells. Oligomycin (10 µg/ml) was added to determine the amount of non-mitochondrial ATP production in each sample. Non-mitochondrial ATP production was subtracted from the overall production. Cellular ATP levels were determined by a luminometric assay (Roche) following the manufacturer’s instructions and the ATP synthesis rate determined by calculating the slope of ATP level over time. Student’s t-test was performed to determine the significance values for untreated and treated cells.
3. Results
3.1. Microarray analysis
We carried out four independent microarray comparisons of undifferentiated to differentiated oligodendroglia, two from primary rat cells and two from human cells lines (HOG and MO3.13), using 4–9 chips per group. We then counted those genes that were significantly altered in the same direction in at least three of the four groups. These included 559 activated and 535 inhibited genes. Thus, differentiation induced and inhibited similar numbers of transcripts (Supplementary Fig. 1).
3.2. Oligodendroglial differentiation induces mitochondrial transcripts and inhibits cell cycle transcripts
We carried out gene analysis of the 559 activated and 535 repressed genes to determine if oligodendroglial differentiation engages different cellular compartments by using Onto-Express (Draghici et al., 2003) (Table 1). The two most active cellular compartments with oligodendroglial differentiation were the plasma membrane and Golgi, as expected of a cell converting to massive secretion of cholesterol-containing membrane. Surprisingly, transcripts assigned to the mitochondrial compartment were also significantly induced (Table 1), including the categories mitochondrion, mitochondrial inner membrane, and mitochondrial envelope, which are described in more detail below. Genes involved in cell cycle and sorting to the spindle and nuclear pore were the most significantly down-regulated, as expected because differentiating oligodendroglia repress proliferation-specific genes in order to stop division and in preparation for differentiation (Baumann and Pham-Dinh, 2001).
Table 1.
Cell compartment analysis of 559 significantly activated and 535 inhibited genes altered transcripts, rank-ordered by significant p-value. Genes assorting to membrane, secretory and mitochondrial compartments are activated, whereas nuclear genes are inhibited after oligodendrocyte differentiation.
| Cell compartments | Genes | p-value |
|---|---|---|
| Up-regulated | ||
| Integral to membrane | 100 | 1.E–05 |
| Golgi apparatus | 15 | 2.E–05 |
| Mitochondrion | 34 | 5.E–05 |
| Cytoplasm | 48 | 1.E–04 |
| Mitochondrial inner membrane | 15 | 5.E–04 |
| Membrane | 71 | 5.E–04 |
| Late endosome | 5 | 6.E–04 |
| Mitochondrial envelope | 5 | 6.E–04 |
| Lysosome | 10 | 1.E–03 |
| Endoplasmic reticulum | 22 | 3.E–03 |
| Down-regulated | ||
| Nucleus | 98 | <1.E–10 |
| Spindle | 9 | 3.E–08 |
| Nuclear pore | 8 | 9.E–07 |
3.3. Multiple mitochondrially-expressed genes are induced as a consequence of oligodendroglial differentiation
The surprising result of the microarray was that so many mitochondrially-targeted genes were induced as a consequence of oligodendroglial differentiation in both rat and human oligodendroglia, with examples shown in Table 2. Several mitochondrial fatty acid oxidation transcripts were activated (Echs1, Hadhsc, Hadh2, Hadha), as were mitochondrial electron transport and ATP synthesis transcripts (Ndufs2, Ndufc2, Cox7a2, Cox4i1, ATP5c1). The mitochondrial morphology genes Opa1 and Fis1 were also induced, suggesting that differentiated oligodendroglia require altered mitochondrial morphology. Two genes that protect mitochondria specifically from oxidative stress, mitochondrial superoxide dismutase (MnSOD) and mitochondrial rhodanese, which repairs oxidatively damaged iron–sulfur clusters, were induced 3-fold and 2-fold, respectively with differentiation. Finally, multiple apoptosis genes were induced (Bnip3, Bcl2l1, Mtch, Psen1), suggesting that the process of differentiation produces a state of vulnerability, as explored further in following paragraphs.
Table 2.
Mitochondrial transcripts significantly induced in three or more experiments after oligodendrocyte differentiation.
| Mitochondrially-targeted genes induced by differentiation |
Symbol | Fold Induction |
|---|---|---|
| Lipid oxidation/acetyl CoA/cholesterol production | ||
| Hydroxyacyl-coenzyme A dehydrogenase | HADHA | 1.4 |
| L-3 hydroxyacyl-coenzyme A dehydrogenase, short chain | HADHSC | 1.7 |
| Hydroxyacyl-coenzyme A dehydrogenase type II | HADH2 | 1.4 |
| Enoyl coenzyme A hydratase | ECHS1 | 2.5 |
| Diazepam binding inhibitor/cholesterol transport protein | DBI | 1.4 |
| Pyruvate dehydrogenase kinase, isoenzyme 2 | PDK2 | 3.4 |
| Electron transport/OXPHOS genes | ||
| NADH dehydrogenase (ubiquinone) Fe–S protein 2 | NDUFS2 | 1.7 |
| NADH dehydrogenase (ubiquinone) 1, 2 | NDUFC2 | 1.3 |
| Succinate dehydrogenase, subunit C | SDHC | 1.3 |
| Cytochrome c oxidase, subunit VIIa 2 | COX7A2 | 1.5 |
| Cytochrome c oxidase, subunit IV isoform 1 | COX4I1 | 1.4 |
| ATP synthase | ATP5C1 | 1.3 |
| Myelination genes | ||
| Myelin-associated oligodendrocytic basic protein | MOBP | 4 |
| Fyn proto-oncogene | FYN | 2.1 |
| Mitochondrial morphology genes | ||
| Fission 1 | FIS1 | 1.9 |
| Optic atrophy 1 | OPA1 | 1.4 |
| Iron–sulfur and oxidative stress genes | ||
| Rhodanese | TST | 2.2 |
| Superoxide dismutase 2, mitochondrial | SOD2 | 2.9 |
| Apoptosis-related genes | ||
| BCL2/adenovirus E1B interacting protein 3 | BNIP3 | 4 |
| Bcl2-like 1 | BCL2L1 | 3.4 |
| Mitochondrial carrier homolog 1/presenilin interactor | MTCH1 | 2.4 |
| Presenilin 1 | PSEN1 | 1.3 |
| Other genes | ||
| Dihydrolipoamide branched chain transacylase E2 | DBT | 2.3 |
| Glycine amidinotransferase (l-arginine:glycine amidinotransferase) | GATM | 1.9 |
| Translocase of inner mitochondrial membrane | TIMM8B | 1.9 |
| Adenine nucleotide translocator 4 | SLC25A4 | 1.7 |
3.4. Up-regulation of cholesterol biosynthesis requires mitochondrial beta-oxidation and mitochondrial cholesterol transporters
We also used Onto Express to analyze differentiation-dependent biological processes in our oligodendroglial samples (Table 3). Cholesterol metabolism and myelination categories were very strongly induced, consistent with expectations of myelinating oligodendrocytes (Nielsen et al., 2006; Saher et al., 2005). In addition to the very strong induction of genes encoding cytoplasmic cholesterol, DBI, a mitochondrial cholesterol transporter, was also induced.
Table 3.
Pathway analysis of significantly altered transcripts in the same direction in at least three of four groups by oligodendrocytic differentiation. Transcripts involved in sterol and myelination metabolism are induced, those involved in replication and cell cycle are inhibited.
| Bioprocess | Genes | p-value |
|---|---|---|
| Up-regulated | ||
| Cholesterol biosynthesis | 11 | 1.E–10 |
| Sterol biosynthesis | 6 | 1.E–08 |
| Adult locomotory behavior | 5 | 9.E–06 |
| Isoprenoid biosynthesis | 4 | 3.E–05 |
| Lipid metabolism | 14 | 2.E–03 |
| Proton transport | 6 | 3.E–03 |
| Protein transport | 13 | 6.E–03 |
| Steroid biosynthesis | 4 | 8.E–03 |
| Dendrite morphogenesis | 4 | 8.E–03 |
| Myelination | 4 | 8.E–03 |
| Response to oxidative stress | 5 | 2.E–02 |
| Down-regulated | ||
| DNA replication | 13 | 8.E–10 |
| Cell cycle | 19 | 3.E–08 |
| Mitosis | 10 | 7.E–07 |
| DNA repair | 12 | 1.E–06 |
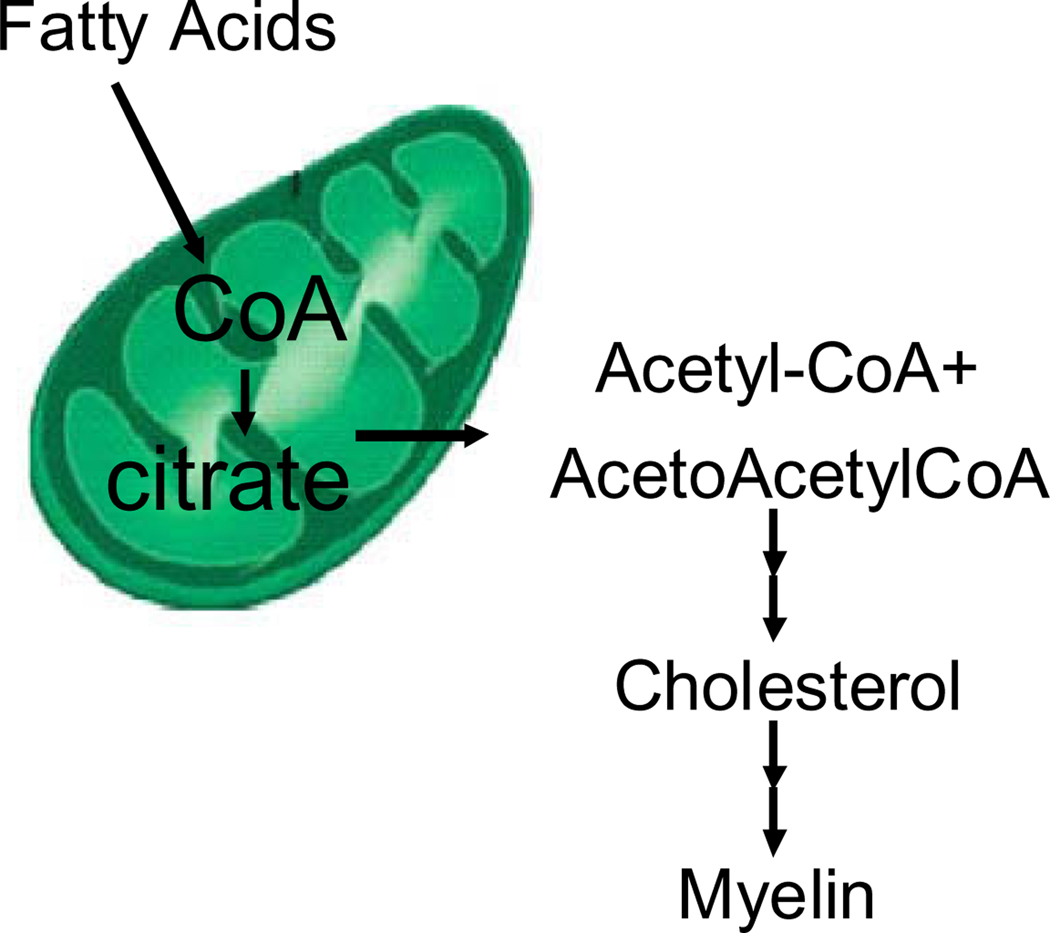
The large induction of the cholesterol pathway in any cell type requires acetyl-CoA generated in the mitochondria, which is produced by oxidation of fatty acids and then transferred to the cytoplasm as citrate for cholesterol synthesis (Fig. 1) (Giudetti et al., 2003). We observed the induction of multiple genes of mitochondrial fatty acid oxidation necessary to support the intense cholesterologenic process of the oligodendrocyte (Table 2, described above). Consistent with this mitochondrial result, cytoplasmic transcripts that produce acetyl-CoA were also induced in all four experimental groups, including acetoacetyl CoA synthetase and acyl-CoA synthetase long-chain family member 3 (Supplemental Fig. 1).
Fig. 1.
Mitochondrial origin of acetyl-CoA for cholesterol synthesis in oligodendrocytes.
3.5. Mitochondrial copy number increases in differentiated oligodendroglia
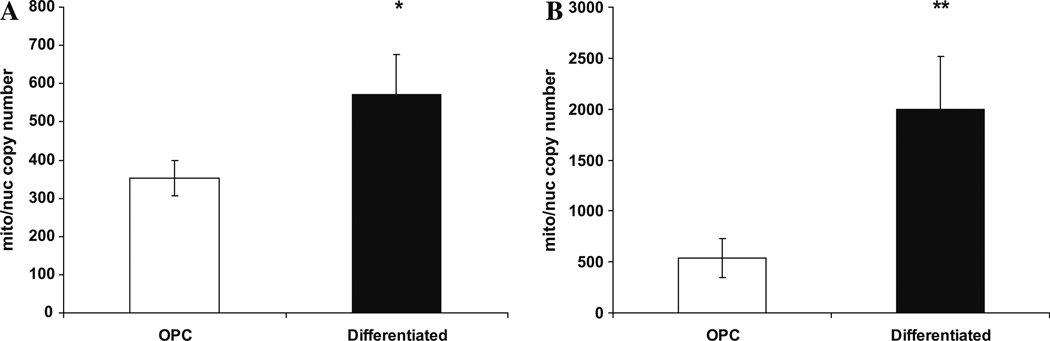
Since oligodendroglial differentiation triggered transcription of several mitochondrial genes, we investigated whether differentiation induces mitochondrial proliferation by assaying the mtDNA/nDNA ratio. mtDNA/nDNA ratio number significantly increased in differentiated oligodendroglia, by 4-fold in HOG cells (p < .0005) and by 1.6-fold in rat cells (p < .001, Fig. 2). The results agreed in general with the 30–400% differentiation-dependent induction of mitochondrial transcripts observed in Table 2.
Fig. 2.
Mitochondrial/nuclear copy number in undifferentiated and differentiated: (A) rat oligos and (B) Human HOG cells. White bars = undifferentiated; black bars = differentiated; * = p < .05; ** = p < .0005. Error bars indicate 2 SEM.
3.6. QRT-PCR verification of microarray results
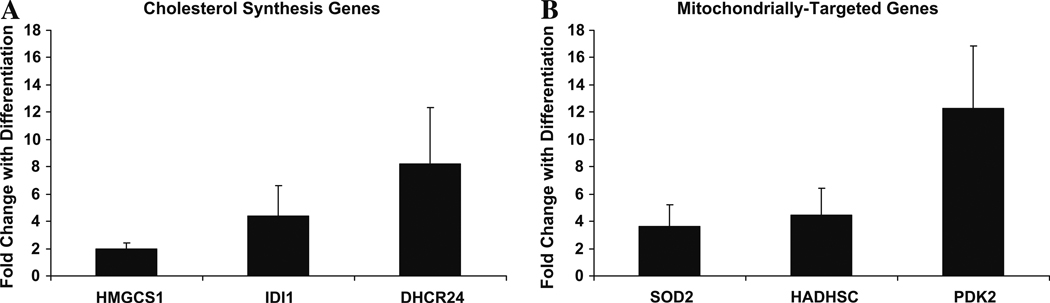
Transcript levels of several genes identified by microarray and categorized in the cholesterol and mitochondrial categories were measured by quantitative RT-PCR using rat oligodendroglial RNA. As predicted by the microarray, we observed that steady-state transcripts of the rate-limiting step of cholesterol biosynthesis HMGCS1, as well as IDI1 and DHCR24 increased with oligodendroglial differentiation (Fig. 3A), as did the nuclear-encoded mitochondrially-targeted transcripts SOD2, HADHSC, and PDK2 (Fig. 3B), at p < 0.05.
Fig. 3.
Quantitative RT-PCR of cholesterol biosynthesis and mitochondrial genes in rat cells confirms significant fold induction with differentiation. Four undifferentiated and 4 differentiated samples were tested in triplicate. (A) cholesterol synthesis genes; (B) mitochondrially-targeted genes. Analysis with student’s t-test indicated up-regulation at p < 0.05 for all samples. Error bars = 2 SEM.
3.7. Differentiated and undifferentiated oligodendroglia are relatively resistant to mitochondrial toxicity
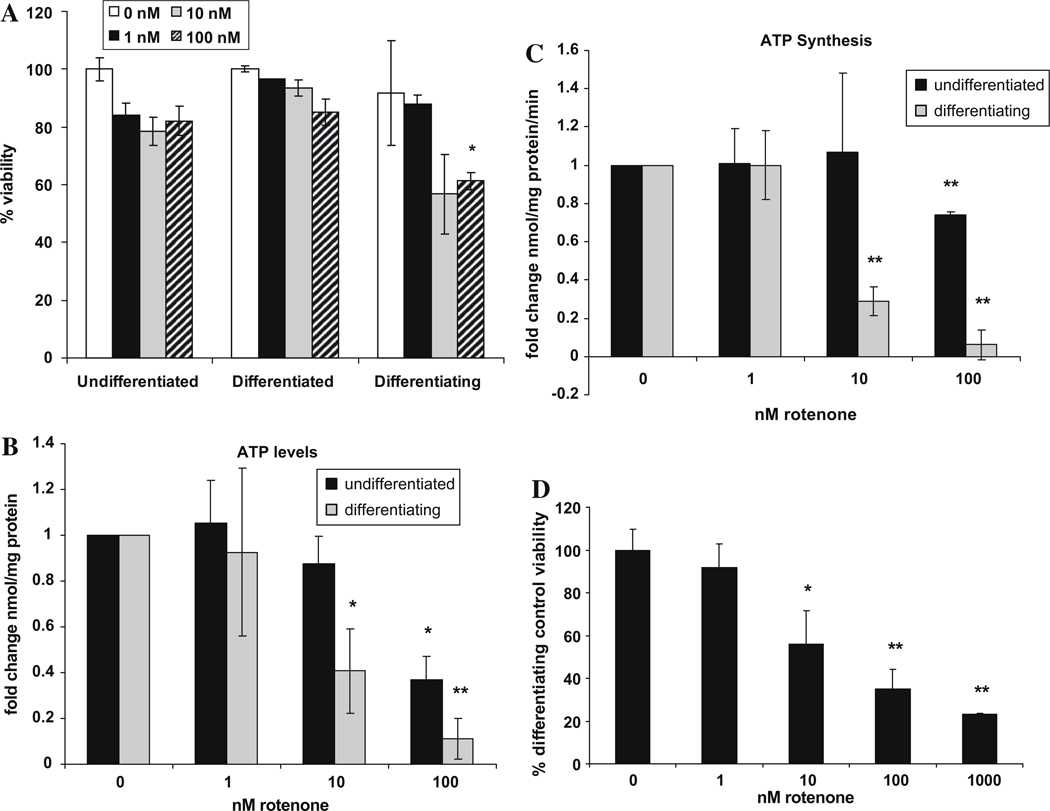
We tested the effects of mitochondrial inhibition before, during and after the differentiation process by treating the human oligodendroglial cells with rotenone for 24 h. Rotenone is a specific inhibitor of mitochondrial complex I, the first step in the electron transport chain. We found that 1 – 100 nM rotenone had little effect on cell viability in undifferentiated and 10 day-differentiated cells, in that their viability remained >80% (Fig. 4A). By contrast, differentiating oligodendroglia treated with 10 and 100 nM rotenone for just 24 h were much more sensitive to mitochondrial inhibition (Fig. 4A). Thus, differentiating oligodendroglial cells were more sensitive to mitochondrial dysfunction than were undifferentiated and differentiated oligodendroglia.
Fig. 4.
(A) Differentiating Human HOG cells are more sensitive to rotenone than undifferentiated and differentiated cells. Viability was measured in Human-ODCs (undifferentiated, differentiating for 1 day and 10-day differentiated) treated with rotenone for 24 h. (B) Differentiating Human HOG cells contain lower ATP levels than do undifferentiated cells. Differentiating cells were exposed to rotenone for 10 days, undifferentiated cells for 4 h. Total cellular ATP (µmol/mg protein) was measured in 3 independent experiments, and normalized to untreated cells. (C) Differentiating Human HOG cells have lower ATP synthesis rates (complexes I–V, µmol/min/mg protein) than do undifferentiated cells (cells were treated as in Fig. 4B). (D) One nM rotenone does not decrease viability in 10-day rotenone-differentiation of oligodendrocytes, however 10–1000 nM produces a dose-dependent decrease in viability. Means of three independent experiments are expressed as viable cells/untreated. Error bars represent SEM, *p < 0.05, **p < 0.005.
3.8. Differentiating oligodendroglia are especially sensitive to mitochondrial inhibition
Given that differentiating oligodendroglia were more rotenone-sensitive than undifferentiated or differentiated oligodendroglia (Fig. 4A), we investigated the mechanism of this sensitivity and whether inhibition of mitochondrial function would inhibit the differentiation process itself. Human oligodendroglial cells differentiating in the presence of 10 nM rotenone demonstrated a 60% decrease in both ATP concentration (Fig. 4B) and ATP synthesis rate (Fig. 4C), with a 40% decrease in cell viability at this same dose and no change in viability at 1 nM rotenone (Fig. 4D). In comparison, undifferentiated cells required 100 nM rotenone (i.e. 10-fold higher inhibition) before decreased ATP levels and synthesis rate were observed (Fig. 4B and C) and required 1 µM rotenone (i.e. a 100-fold higher dose) before a reduction in viability was observed (Silva et al., 2009). Thus, differentiating oligodendroglial cells were more sensitive to lower doses of rotenone than were undifferentiated cells, both in terms of viability, ATP levels and ATP synthesis.
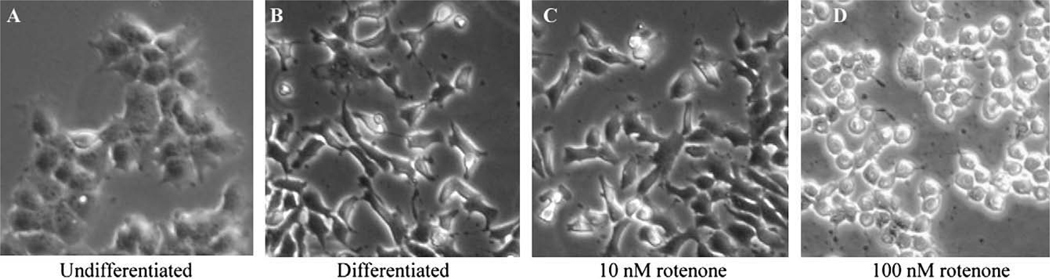
The morphology of differentiating HOG cells was similar between untreated cells and cells treated with 10 nM rotenone for 4 days (Fig. 5B and C). Cells treated with 100 nM rotenone, however, did not have the elongated shape of a differentiated cell, were more rounded, and dead cells could be seen (Fig. 5D).
Fig. 5.
Differentiation of Human HOG cells after 4 days of rotenone treatment. Human-ODCs were differentiated for 10 days in the absence (B) or presence of 10 or 100 nM rotenone (C and D).
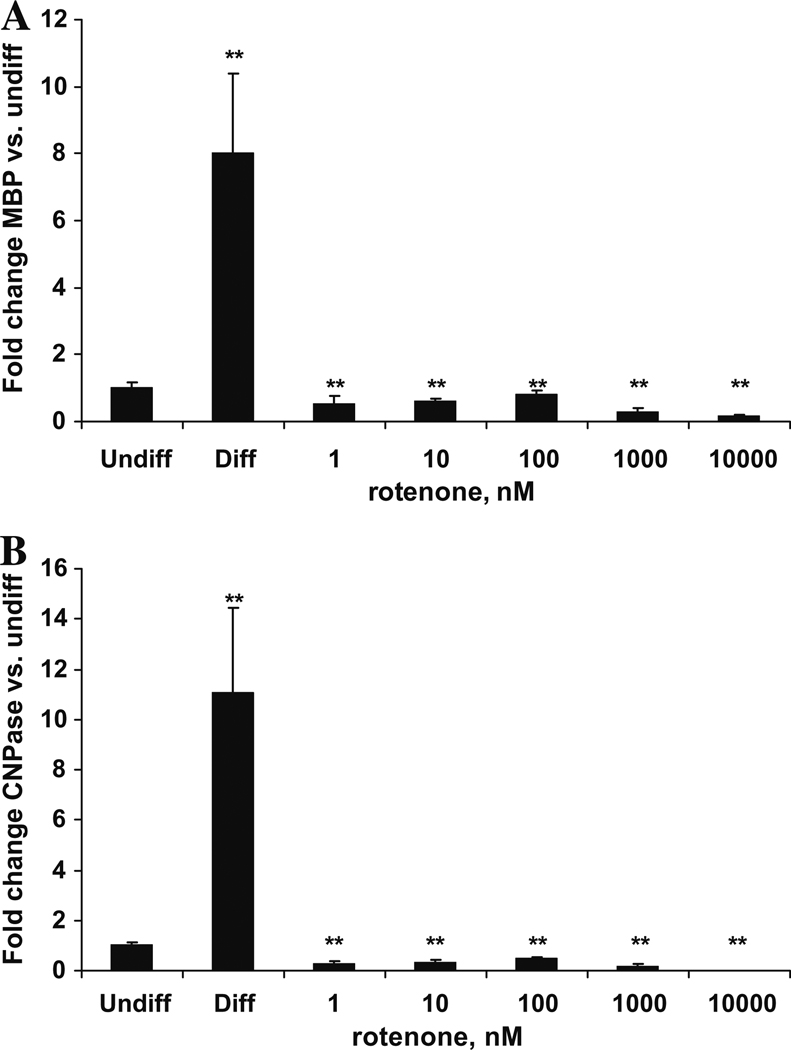
3.9. Differentiation-dependent induction of MBP and CNPase transcripts is blocked by very low doses of rotenone
Two myelination-specific markers, Myelin Basic Protein (MBP) and 2′,3′-Cyclic Nucleotide-3′-Phosphodiesterase (CNPase), are hugely increased by oligodendroglial differentiation in vitro (Buntinx et al., 2003), and we investigated whether mitochondrial inhibition with rotenone would affect these markers. Upon differentiation in the absence of mitochondrial inhibitor, transcript levels of MBP and CNPase increased 8-fold and 11-fold respectively, relative to undifferentiated human oligodendroglial cells. By contrast, all doses of rotenone, including the 1 nM dose, which does not affect viability, completely blocked the differentiation-specific induction of MBP and CNPase transcripts (Fig. 6A and B). Thus, mild mitochondrial inhibition at levels that do not decrease viability blocks transcripts of oligodendroglial differentiation.
Fig. 6.
Differentiated Human HOG cells express more Myelin Basic Protein (MBP) and Cyclic Nucleotide Phosphodiesterase CNPase transcript than undifferentiated cells, but not when differentiated with >1 nM rotenone. Human HOG cells were differentiated for 10 days with or without rotenone, transcript levels were examined by quantitative RT-PCR. Transcripts were normalized to vimentin and expressed per undifferentiated or untreated samples. Error bars represent SEM, **p < 0.005.
4. Discussion
4.1. Oligodendroglial differentiation induces cholesterologenic and mitochondrial transcripts
We microarrayed and performed gene ontology analysis on oligodendroglial precursor cells and differentiated oligodendroglia. A strong and significant induction of cholesterol-related, mitochondrial and myelination transcripts was observed with differentiation, demonstrating an intense activation of the cholesterol pathway and a requirement for mitochondrial functions. In contrast, many important cell proliferation genes, including DNA ligase 1, Cyclins B1 and D1 and cell division cycle 20 and 25 were significantly inhibited with differentiation. Thus, the three major transcriptional features of oligodendroglial differentiation we observed by microarray were the induction of cholesterologenic and mitochondrial transcripts, and the inhibition of cell proliferation transcripts.
4.2. Intense cholesterol biosynthesis requires induction of mitochondrial functions
Cholesterol biosynthesis is required for myelination (Pleasure and Kim, 1976; Vance et al., 2000). Nearly all oligodendrocytic cholesterol is self-generated rather than imported (Bjorkhem and Meaney, 2004; Morell and Jurevics, 1996; Saher et al., 2005). In turn, cholesterol biosynthesis requires a large amount of acetyl-CoA, which is provided by mitochondrial beta-oxidation of fatty acids, as in Fig. 1 (Schulz, 1991). This explains why there is induction of several transcripts of mitochondrial beta-oxidation (Echs1, Hadhsc, Hadh2, Hadha) in a cell whose cytoplasm is actively synthesizing cholesterol lipid. The mitochondrial acetyl-CoA produced by beta-oxidation is transported as citrate through mitochondrial transporters to the cytosol, where it is cleaved into oxaloacetate and acetyl-CoA, to be used for cholesterologenesis. This also explains the induction of the mitochondrial cholesterol transporter DBI, and the induction of PDK2, which inhibits flux through pyruvate dehydrogenase, because mitochondria have a very sufficient acetyl-CoA source, i.e. from beta-oxidation. Presumably some of this acetyl-CoA also runs through the Krebs cycle and electron transport chain as well, consistent with the induction of multiple complexes I and IV electron transport chain transcripts observed (Ndufs2, Ndufc2, Cox7a2, Cox4i1, ATP5c1).
The need for mitochondrial antioxidant protection in oligodendrocytes is highlighted by the 3-fold induction of mitochondrial superoxide dismutase (MnSOD) which scavenges superoxide, and the 2-fold induction of mitochondrial rhodanese, which repairs oxidant-sensitive iron–sulfur clusters. A further demonstration of the relevance of mitochondrial function in oligodendroglial differentiation is the strong and significant induction of mitochondrial copy number with oligodendroglial differentiation.
4.3. A sublethal dose of mitochondrial inhibitor represses differentiation-specific transcripts
We observed that differentiating oligodendrocytes were much more sensitive to mitochondrial inhibition with rotenone than either undifferentiated or differentiated oligodendroglia. Differentiating HOG cells exposed to rotenone for 10 days were sensitive to 10 and 100 nM rotenone in terms of both viability and ATP synthesis. In contrast, undifferentiated cells were resistant to 10 nM and 100 nM rotenone treatment in terms of viability and required 100 nM rotenone before a decrease in ATP synthesis was observed. This suggests that differentiating oligodendroglia have an altered mitochondrial structure and/or are more vulnerable to mitochondrial inhibition.
With respect to this latter point, treatment with 1 nM rotenone, a dose at least 10-fold lower than that which affects viability or ATP synthesis, completely abolished the differentiation-dependent induction of MBP and CNPase expression (p < 0.005). Thus, differentiating oligodendroglia are much more sensitive to mitochondrial inhibition than undifferentiated or differentiated oligodendrocytes, and very low doses of mitochondrial inhibition completely abolish the differentiation program, in the absence of toxicity. This suggests that differentiating oligodendroglia pass through a ‘window’ requiring high mitochondrial function, and even slight inhibition of that function interrupts the differentiation program completely.
These in vitro results are consistent with in vivo observations that formation of the myelin sheath is reduced by mitochondrial inhibitors (Bizzozero et al., 1999). They are also consistent with those of Ingram, who observed that a mitochondrial uncoupler selectively inactivated myelin protein kinases (Luo and Ingram, 2001). Additionally, mitochondrial inhibitors produce demyelination in the optic nerve (Hirano et al., 1967, 1968a,b; Lessell and Kuwabara, 1974; Levine et al., 1967).
4.4. Demyelination in mitochondrial disease
The data presented here support the view that differentiating oligodendroglia are especially sensitive to mitochondrial dysfunction, which could be relevant to demyelination in those who inherit defects of mitochondrial function. Demyelination occurs in the mitochondrial disease Leber’s Hereditary Optic Neuropathy (LHON), LHON patients have 100-fold increased risk of developing multiple sclerosis (Harding, 1992; Vanopdenbosch, 2000). Demyelination also occurs in the mitochondrial disease Friedreich’s ataxia (FRDA) (Al-Mahdawi et al., 2006; Machkhas et al., 1998); in MELAS, caused by a mitochondrial tRNA mutation (Ohara et al., 1988); in Charcot-Marie Tooth 2a and in Dominant Optic Atrophy, caused by mutations in the mitofusins mitofusin 2 and Opa1, respectively (Niemann et al., 2006; Johnston et al., 1979).
Periventricular Leukomalacia (PVL) is caused by hypoxic-ischemic brain injury, primarily affecting pre-oligodendrocytes in cerebral white matter (Volpe, 2003, 2008; Deng et al., 2008). These cells have been shown to be energy-sensitive, in that cultured pre-oligodendrocytes are more sensitive to oxygen-glucose deprivation and glutamate-dependent excitotoxic cell death (Deng et al., 2003). Additionally, this sensitivity can be enhanced in the presence of mitochondrial toxins (Deng et al., 2006). Thus, mitochondrial dysfunction in differentiating oligodendrocytes may play a role in the pathology of PVL.
We have demonstrated here that: (1) mitochondrial transcripts and copy number are induced by oligodendroglial differentiation, (2) that slight mitochondrial inhibition inhibits differentiation, and (3) stronger mitochondrial inhibition selectively decreases viability of differentiating oligodendroglia but not undifferentiated cells. These results suggest that oligodendroglial differentiation is a ‘window’ of selective vulnerability to mitochondrial inhibition, and could be relevant to the mechanisms of demyelination in inherited mitochondrial disease and in birth ischemia.
Supplementary Material
Acknowledgements
This work was supported by grants from the USPHS – EY12245, AG11967, AG16719, AG23311 (to G.A.C.), NS25044 (to D.P.) and National Multiple Sclerosis Society award RG3419A1/1T (to T.I.). We thank J. Nielsen and L. Hudson for sharing microarray data.
Footnotes
Appendix A. Supplementary material
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.mito.2009.12.141.
References
- Al-Mahdawi S, Pinto RM, Varshney D, Lawrence L, Lowrie MB, Hughes S, et al. GAA repeat expansion mutation mouse models of Friedreich ataxia exhibit oxidative stress leading to progressive neuronal and cardiac pathology. Genomics. 2006;88:580–590. doi: 10.1016/j.ygeno.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atorino L, Silvestri L, Koppen M, Cassina L, Ballabio A, Marconi R, et al. Loss of m-AAA protease in mitochondria causes complex I deficiency and increased sensitivity to oxidative stress in hereditary spastic paraplegia. J. Cell Biol. 2003;163:777–787. doi: 10.1083/jcb.200304112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol. Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- Bizzozero OA, Sanchez P, Tetzloff SU. Effect of ATP depletion on the palmitoylation of myelin proteolipid protein in young and adult rats. J. Neurochem. 1999;72:2610–2616. doi: 10.1046/j.1471-4159.1999.0722610.x. [DOI] [PubMed] [Google Scholar]
- Bjorkhem I, Meaney S. Brain cholesterol: long secret life behind a barrier. Arterioscler. Thromb. Vasc. Biol. 2004;24:806–815. doi: 10.1161/01.ATV.0000120374.59826.1b. [DOI] [PubMed] [Google Scholar]
- Buntinx M, Vanderlocht J, Hellings N, Vandenabeele F, Lambrichts I, Raus J, et al. Characterization of three human oligodendroglial cell lines as a model to study oligodendrocyte injury: morphology and oligodendrocyte-specific gene expression. J. Neurocytol. 2003;32:25–38. doi: 10.1023/a:1027324230923. [DOI] [PubMed] [Google Scholar]
- Carelli V, Ross-Cisneros FN, Sadun AA. Optic nerve degeneration and mitochondrial dysfunction: genetic and acquired optic neuropathies. Neurochem. Int. 2002;40:573–584. doi: 10.1016/s0197-0186(01)00129-2. [DOI] [PubMed] [Google Scholar]
- Cortopassi G, Danielson S, Alemi M, Zhan SS, Tong W, Carelli V, Martinuzzi A, Marzuki S, Majamaa K, Wong A. Mitochondrial disease activates transcripts of the unfolded protein response and cell cycle and inhibits vesicular secretion and oligodendrocyte-specific transcripts. Mitochondrion. 2006;6:161–175. doi: 10.1016/j.mito.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Deng W, Rosenberg PA, Volpe JJ, Jensen FE. Calcium-permeable AMPA/kainate receptors mediate toxicity and preconditioning by oxygen-glucose deprivation in oligodendrocyte precursors. Proc. Natl. Acad. Sci. USA. 2003;100:6801–6806. doi: 10.1073/pnas.1136624100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Yue Q, Rosenberg PA, Volpe JJ, Jensen FE. Oligodendrocyte excitotoxicity determined by local glutamate accumulation and mitochondrial function. J. Neurochem. 2006;98:213–222. doi: 10.1111/j.1471-4159.2006.03861.x. [DOI] [PubMed] [Google Scholar]
- Deng W, Pleasure J, Pleasure D. Progress in periventricular leukomalacia. Arch. Neurol. 2008;65:1291–1295. doi: 10.1001/archneur.65.10.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz SA, Tainsky MA. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudetti AM, Sabetta S, di Summa R, Leo M, Damiano F, Siculella L, et al. Differential effects of coconut oil- and fish oil-enriched diets on tricarboxylate carrier in rat liver mitochondria. J. Lipid Res. 2003;44:2135–2141. doi: 10.1194/jlr.M300237-JLR200. [DOI] [PubMed] [Google Scholar]
- Harding AE, Sweeney MG, Miller DH, Mumford CJ, Kellar-Wood H, Menard D, McDonald WI, Compston DA. Occurrence of a multiple sclerosis-like illness in women who have a Leber's hereditary optic neuropathy mitochondrial DNA mutation. Brain. 1992;115:979–989. doi: 10.1093/brain/115.4.979. [DOI] [PubMed] [Google Scholar]
- Hirano A, Levine S, Zimmerman HM. Experimental cyanide encephalopathy: electron microscopic observations of early lesions in white matter. J. Neuropathol. Exp. Neurol. 1967;26:200–213. doi: 10.1097/00005072-196704000-00002. [DOI] [PubMed] [Google Scholar]
- Hirano A, Levine S, Zimmerman HM. Remyelination in the central nervous system after cyanide intoxication. J. Neuropathol. Exp. Neurol. 1968a;27:234–245. doi: 10.1097/00005072-196804000-00005. [DOI] [PubMed] [Google Scholar]
- Hirano A, Zimmerman HM, Levine S. Remyelination in the central nervous system after cyanide intoxication. J. Neuropathol. Exp. Neurol. 1968b;27:144–145. [PubMed] [Google Scholar]
- Horiuchi M, Itoh A, Pleasure D, Itoh T. MEK-ERK signaling is involved in interferon-gamma-induced death of oligodendroglial progenitor cells. J. Biol. Chem. 2006;281:20095–20106. doi: 10.1074/jbc.M603179200. [DOI] [PubMed] [Google Scholar]
- Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, et al. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell. 2003;115:163–175. doi: 10.1016/s0092-8674(03)00810-9. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Dakin KA, Stevens B, Lee PR, Kozlov SV, Stewart CL, et al. Astrocytes promote myelination in response to electrical impulses. Neuron. 2006;49:823–832. doi: 10.1016/j.neuron.2006.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Beesley J, Itoh A, Cohen AS, Kavanaugh B, Coulter DA, et al. AMPA glutamate receptor-mediated calcium signaling is transiently enhanced during development of oligodendrocytes. J. Neurochem. 2002;81:390–402. doi: 10.1046/j.1471-4159.2002.00866.x. [DOI] [PubMed] [Google Scholar]
- Itoh A, Horiuchi M, Bannerman P, Pleasure D, Itoh T. Impaired regenerative response of primary sensory neurons in ZPK/DLK gene-trap mice. Biochem. Biophys. Res. Commun. 2009;383:258–262. doi: 10.1016/j.bbrc.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Johnston PB, Gaster RN, Smith VC, Tripathi RC. A clinicopathologic study of autosomal dominant optic atrophy. Am. J. Ophthalmol. 1979;88:868–875. doi: 10.1016/0002-9394(79)90565-8. [DOI] [PubMed] [Google Scholar]
- Kovacs GG, Hoftberger R, Majtenyi K, Horvath R, Barsi P, Komoly S, et al. Neuropathology of white matter disease in Leber's hereditary optic neuropathy. Brain. 2005;128:35–41. doi: 10.1093/brain/awh310. [DOI] [PubMed] [Google Scholar]
- Lessell S, Kuwabara T. Fine structure of experimental cyanide optic neuropathy. Invest. Ophthalmol. 1974;13:748–756. [PubMed] [Google Scholar]
- Levine S, Hirano A, Zimmerman HM. Experimental cyanide encephalopathy: electron microscopic observations of early lesions in white matter. J. Neuropathol. Exp. Neurol. 1967;26:172–174. [PubMed] [Google Scholar]
- Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Loraine AE, Shigeta R, Cline M, Cheng J, Valmeekam V, et al. NetAffx: affymetrix probesets and annotations. Nucleic Acids Res. 2003;31:82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Ingram VM. Uncoupling of mitochondria activates protein phosphatases and inactivates MBP protein kinases. J. Alzheimers Dis. 2001;3:593–598. doi: 10.3233/jad-2001-3610. [DOI] [PubMed] [Google Scholar]
- Machkhas H, Bidichandani SI, Patel PI, Harati Y. A mild case of Friedreich ataxia: lymphocyte and sural nerve analysis for GAA repeat length reveals somatic mosaicism. Muscle Nerve. 1998;21:390–393. doi: 10.1002/(sici)1097-4598(199803)21:3<390::aid-mus13>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Morell P, Jurevics H. Origin of cholesterol in myelin. Neurochem. Res. 1996;21:463–470. doi: 10.1007/BF02527711. [DOI] [PubMed] [Google Scholar]
- Nielsen JA, Maric D, Lau P, Barker JL, Hudson LD. Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling. J. Neurosci. 2006;26:9881–9891. doi: 10.1523/JNEUROSCI.2246-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann A, Berger P, Suter U. Pathomechanisms of mutant proteins in Charcot-Marie-Tooth disease. Neuromolecular Med. 2006;8:217–242. doi: 10.1385/nmm:8:1-2:217. [DOI] [PubMed] [Google Scholar]
- Ohara S, Ohama E, Takahashi H, Ikuta F, Nishizawa M, Tanaka K, et al. Alterations of oligodendrocytes and demyelination in the spinal cord of patients with mitochondrial encephalomyopathy. J. Neurol. Sci. 1988;86:19–29. doi: 10.1016/0022-510x(88)90004-4. [DOI] [PubMed] [Google Scholar]
- Pleasure D, Kim SU. Sterol synthesis by myelinating cultures of mouse spinal cord. Brain Res. 1976;103:117–126. doi: 10.1016/0006-8993(76)90691-0. [DOI] [PubMed] [Google Scholar]
- Rusanen H, Majamaa K, Tolonen U, Remes AM, Myllyla R, Hassinen IE. Demyelinating polyneuropathy in a patient with the tRNA(Leu) (UUR) mutation at base pair 3243 of the mitochondrial DNA. Neurology. 1995;45:1188–1192. doi: 10.1212/wnl.45.6.1188. [DOI] [PubMed] [Google Scholar]
- Saher G, Brugger B, Lappe-Siefke C, Mobius W, Tozawa R, Wehr MC, et al. High cholesterol level is essential for myelin membrane growth. Nat. Neurosci. 2005;8:468–475. doi: 10.1038/nn1426. [DOI] [PubMed] [Google Scholar]
- Schulz H. Beta oxidation of fatty acids. Biochim. Biophys. Acta. 1991;1081:109–120. doi: 10.1016/0005-2760(91)90015-a. [DOI] [PubMed] [Google Scholar]
- Silva JM, Wong A, Carelli V, Cortopassi GA. Inhibition of mitochondrial function induces an integrated stress response in oligodendroglia. Neurobiol. Dis. 2009;34:357–365. doi: 10.1016/j.nbd.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Simons M, Trajkovic K. Neuron-glia communication in the control of oligodendrocyte function and myelin biogenesis. J. Cell Sci. 2006;119:4381–4389. doi: 10.1242/jcs.03242. [DOI] [PubMed] [Google Scholar]
- Tan G, Chen LS, Lonnerdal B, Gellera C, Taroni FA, Cortopassi G. Frataxin expression rescues mitochondrial dysfunctions in FRDA cells. Hum. Mol. Genet. 2001;10:2099–2107. doi: 10.1093/hmg/10.19.2099. [DOI] [PubMed] [Google Scholar]
- Vance JE, Campenot RB, Vance DE. The synthesis and transport of lipids for axonal growth and nerve regeneration. Biochim. Biophys. Acta. 2000;1486:84–96. doi: 10.1016/s1388-1981(00)00050-0. [DOI] [PubMed] [Google Scholar]
- Vanopdenbosch L, Dubois B, D'Hooghe MB, Meire F, Carton H. Mitochondrial mutations of Leber's hereditary optic neuropathy: a risk factor for multiple sclerosis. J. Neurol. 2000;247:535–543. doi: 10.1007/s004150070153. [DOI] [PubMed] [Google Scholar]
- Volpe JJ. Cerebral white matter injury of the premature infant-more common than you think. Pediatrics. 2003;112:176–180. doi: 10.1542/peds.112.1.176. [DOI] [PubMed] [Google Scholar]
- Volpe JJ. Neurology of the Newborn. Philadelphia: W.B. Saunders; 2008. [Google Scholar]
- Wong A, Cavelier L, Collins-Schramm HE, Seldin MF, McGrogan M, Savontaus ML, et al. Differentiation-specific effects of LHON mutations introduced into neuronal NT2 cells. Hum. Mol. Genet. 2002;11:431–438. doi: 10.1093/hmg/11.4.431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.