Abstract
Meiotic homologous recombination in Saccharomyces cerevisiae involves formation of nucleoprotein filaments of Rad51 and Dmc1 that mediate DNA strand exchange between homologous chromosomes. The Mei5-Sae3 protein complex functions as a recombination mediator to promote nucleation of the Dmc1 recombinase onto replication protein A-coated single-stranded DNA. Here, we have expressed and purified the Mei5 protein, Sae3 protein and the Mei5-Sae3 complex for biochemical studies. We show the Mei5-Sae3 complex preferentially binds a fork-like DNA substrate to 3′ overhanging DNA, single-stranded DNA or double-stranded DNA. We demonstrate that Mei5 confers DNA binding activity to the Mei5-Sae3 complex. We determined Mei5-Sae3 interacts with the Rad51 recombinase through the N-terminal domain of Mei5. Unlike Rad52, Mei5-Sae3 lacks recombination mediator activity for Rad51. Importantly, we find that the Mei5-Sae3 complex does not harbor single-strand DNA annealing activity. These properties of the Mei5-Sae3 complex distinguishes it from the Rad52 protein, which serves as the mediator of Rad51 and is involved in the single-strand DNA annealing pathway of homologous recombination.
Keywords: Mei5-Sae3, meiotic recombination, recombination mediator, Rad51
1. Introduction
Homologous recombination (HR) that occurs in meiotic cells is critical for chromosome segregation in meiosis I and is initiated by the introduction of programmed DNA double strand breaks (DSBs) by the Spo11 protein [1]. DSBs are nucleolytically processed to expose 3′ single-strand DNA (ssDNA) tails on which recombinases nucleate to form nucleoprotein filaments. These nucleoprotein filaments, known as presynaptic filaments, catalyze invasion of the sister chromatid or chromosomal homolog. Most eukaryotic organisms, including humans, possess two orthologs of the Escherichia coli RecA recombinase, namely, Rad51 and Dmc1. While Rad51 is found in both mitotic and meiotic cells, Dmc1 is expressed exclusively during meiosis [2, 3]. The activity of Rad51 and Dmc1 is regulated by a variety of factors. By removing secondary structure in ssDNA and sequestering ssDNA generated during DNA strand exchange, the single-strand DNA binding protein replication protein A (RPA) facilitates presynaptic filament assembly and prevents strand exchange reversal [4–6]. However, RPA also competes with the recombinases for sites on ssDNA that result in an inhibition of presynaptic filament assembly [7]. To overcome the inhibitory effect of RPA, recombination mediator proteins such as Rad52 displace RPA to facilitate the formation of the presynaptic filament [7].
Herein, we focus on the Saccharomyces cerevisiae Mei5-Sae3 complex that is expressed only in meiotic cells. Mutations of MEI5 or SAE3 lead to severe meiotic phenotypes including spore inviability, prophase delay/arrest, accumulation of hyper-resected DNA ends, defective formation of full-length synaptonemal complex, and defective crossover and non-crossover formation [8, 9]. Cytological analysis has shown that localization of Dmc1 to meiotic DSBs is reliant upon Mei5-Sae3 [8, 9]. Biochemical evidence that Mei5-Sae3 facilitates the assembly of the Dmc1 presynaptic filament on RPA-coated ssDNA was recently obtained by Ferrari et al. [10].
In this study, we purified and conducted biochemical analysis of the Mei5-Sae3 complex. Our results show the Mei5-Sae3 complex (herein called Mei5-Sae3) preferentially binds a DNA fork. We also determined that Mei5 is the subunit that confers DNA binding activity to Mei5-Sae3. We show that the DNA binding activity resides within N-terminal half of Mei5. We provide evidence that Mei5-Sae3 interacts with the Rad51 recombinase and that this interaction is mediated through the Mei5 subunit. Unlike Rad52 and the Mei5-Sae3 ortholog in Schizosaccharomyces pombe, Sfr1-Swi5 [11], we find no evidence for mediator activity in Mei5-Sae3 on Rad51. Importantly, we show that Mei5-Sae3 does not harbor single-strand DNA annealing activity, thus further distinguishing Mei5-Sae3 from Rad52.
2. Materials and Methods
2.1. Expression and purification of Mei5-Sae3, MBP-Mei5, Sae3, MBP-Mei5-N, MBP-Mei5-C, Rad51, Rad52, and RPA
Detailed expression and purification protocols for Mei5-Sae3, MBP-Mei5, Sae3-(HIS)6, MBP-Mei5-N, MBP-Mei5-C, Rad51, Rad52, and RPA can be found in the supplementary methods.
2.2. Gel Filtration of the protein complex Mei5-Sae3 and Sae3-(HIS)6
Mei5-Sae3 complex (100 μg) was loaded on a 20 mL Sephacryl S-100 column (0.9 × 30 cm) equilibrated in Buffer A (20 mM KH2PO4 pH 7.5, 10% glycerol, 0.5 mM EDTA and 1 mM dithiothreitol) containing 0.01% Igepal and 150 mM KCl. The proteins were fractionated at 0.25 mL/min in Buffer A containing 150 mM KCl. Fractions were TCA precipitated. The protein samples were resuspended in SDS dye, loaded onto a 12% SDS-PAGE and stained with Coomassie Blue. Protein standards (Bio-Rad; bovine thyroglobulin, 670 kDa; bovine γ-globulin, 158 kDa; chicken ovalbumin, 44 kDa; horse myoglobin, 17 kDa; vitamin B12, 1.35 kDa) were separated on the column and eluted at the indicated positions.
2.3. Spore viability of SAE3-6HIS
Standard PCR-mediated one-step tagging was used to integrate a (HIS)6 tag between the codon encoding the last amino acid of Sae3 and the stop codon [12]. Diploid cells (Table 1) were incubated on sporulation medium plates for 3 days at 30°C. The formed spores were dissected to examine the spore viability for each strain. For the wild type and sae3 null mutant, 20 tetrads were dissected while 40 tetrads were dissected for the SAE3-6HIS strain.
Table 1.
Genotypes of yeast strains used in study.
| Strain | Genotype |
|---|---|
| TBR2065 |
MATa leu2-3, 112 his4-260 ura3-1 ade2-1 thr1-4 trp1-289 lys2 MATα leu2-3, 112 his4-260 ura3-1 ade2-1 thr1-4 trp1-289 lys2 |
| TBR1039 |
MATa leu2-3, 112 his4-260 ura3-1 ade2-1 thr1-4 trp1-289 lys2 sae3::URA3 MATα leu2-3, 112 his4-260 ura3-1 ade2-1 thr1-4 trp1-289 lys2 sae3::URA3 |
| TBR5428 |
MATa leu2-3, 112 his4-260 ura3-1 ade2-1 thr1-4 trp1-289 lys2 SAE3-HIS6-KAN MATα leu2-3, 112 his4-260 ura3-1 ade2-1 thr1-4 trp1-289 lys2 SAE3-HIS6-KAN |
2.4. φX174 DNA mobility shift assay
The indicated amounts of Mei5-Sae3, MBP-Mei5, Sae3-(HIS)6, MBP-Mei5-N, or MBP-Mei5-C were incubated in 12.5 μL Buffer B (50 mM Tris-HCl pH 7.5, 1 mM DTT) containing 100 mM KCl and either φX174 ssDNA (30 μM nucleotides), linearized φX174 dsDNA (30 μM base pairs), or both as indicated at 37°C for 10 min. DNA loading dye (10 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 50% glycerol, 0.1% Orange G) was added to the reaction. The samples were resolved in 0.9% agarose gels and were stained with ethidium bromide. Images were recorded and analyzed using Quantity One (Bio-Rad) software. A control reaction was deproteinized by treatment with SDS (0.5% final) and Proteinase K (0.5 mg/mL) at 37°C for 10 min prior to loading in a 0.9% agarose gel.
2.5. Oligonucleotide DNA mobility shift assay
For the structure specific DNA mobility shift assay, the indicated amounts of Mei5-Sae3, MBP-Mei5, or Sae3-(HIS)6 were incubated for 10 min at 37°C in 10 μL Buffer B containing 0.05 pmol of the indicated structure specific substrates. For the binding site size DNA mobility shift assay, the indicated amount of protein was incubated for 10 min at 37°C in 10 μL Buffer B containing 0.1 pmol of the indicated length of poly dT oligonucleotides. DNA loading dye was added to the reaction. The samples were resolved on 12% non-denaturing TAE polyacrylamide gels. The gels were dried, analyzed with a phosphorimager and quantified with ImageQuant (GE Healthcare) software. A control reaction was deproteinized by treatment with SDS (0.5% final) and Proteinase K (0.5 mg/mL) at 37°C for 10 min prior to loading a 12 % non-denaturing TAE polyacrylamide gel.
2.6. Pull-down assays
Mei5-Sae3 (6 mg), Rad51 (6 mg) or BSA (10 mg) were immobilized on 1 mL of Affi-Gel 15 (Bio-Rad) per the manufacturer’s instructions and stored in Buffer C (50 mM Tris-HCl pH 7.5, 50 mM KCl, 1 mM dithiothreitol, 30% glycerol) at −20°C. The indicated Affi-gel matrices, were equilibrated with Buffer A containing 120 mM KCl. Mei5-Sae3 (6.7 μg), Rad51 (7 μg), Sae3-(HIS)6 (16 μg), or MBP-Mei5 (7.4 μg) was added to the indicated Affi-gel matrix in 30 μL of Buffer A with a final concentration of 120 mM KCl. After agitation for 30 min at 4°C, the supernatant was removed from the beads followed by 3 washes with Buffer A containing 120 mM KCl. Equal volumes of 2xSDS dye was added to the supernatant and wash fractions while 30 μL of 2xSDS loading dye was added to the bead fraction. The supernatant, wash and bead samples were incubated at 95°C prior to loading 8μL of each onto a 15% SDS-PAGE followed by Coomassie Blue staining. For pull-down experiments using amylose resin, MBP-Mei5 (7.4 μg), MBP-Mei5-N truncation (7.4 μg) or MBP-Mei5-C truncation (7.4 μg) was first incubated at 4°C for 30 min with Rad51 (7 μg) or MBP (6.5 μg) in Buffer A containing 120 mM KCl in a final volume of 30 μL. Amylose resin was then added to these reactions and agitated for 30 min at 4°C. The supernatant was removed from the beads followed by 3 washes of the beads with Buffer A containing 120 mM KCl. The supernatant, wash and bead fractions were treated as described for the treatment of Affi-gel matrices and subjected to SDS-PAGE analysis on 15% gels followed by Coomassie Blue staining. For some samples, 6 uL of the indicated fraction was loaded onto SDS-PAGE followed by Western analysis.
2.7. DNA strand exchange assay
In the standard reaction, Rad51 (9.3 μM) was incubated with φX174 (+) virion ssDNA (30 μM nucleotides) in 10 μL of buffer D (50 mM Tris-HCl, pH 7.2, 35 mM KCl, 1 mM dithiothreitol, 2.5 mM ATP, and 3 mM MgCl2) for 7 min at 37°C. After the addition of RPA (1.7 μM) in 0.5 μL, the reaction mixtures were incubated at 37°C for an additional 7 min before the incorporation of linear double-stranded DNA (30 μM nucleotides) in 1 μL followed by 1 μL of 50 mM spermidine hydrochloride (4 mM final concentration). After 120 min of incubation at 37°C, the reaction was terminated by adding an equal volume of 1% SDS containing 1 mg/mL Proteinase K followed by a 20 min incubation at 37°C. The deproteinized samples (12 μL) were run on 0.9% agarose gels in TAE buffer, stained with ethidium bromide for 15 min and then destained for at least 4 h in a large volume of water. Images were recorded and analyzed in a Bio-Rad Quantity One software system. To examine recombination mediator activity, reaction mixtures containing Rad51 and RPA with or without the indicated amounts of Rad52 or Mei5-Sae3 were incubated on ice for 40 min before ssDNA was added. After a 10 min incubation at 37°C, linear double-stranded DNA (30 μM nucleotides) in 1 μL followed by 1 μL of 50 mM spermidine hydrochloride (4 mM final concentration) were incorporated. The reaction mixtures were incubated and analyzed as described above.
2.8. Single-Stranded DNA annealing assay
The 32P-OL83 and unlabeled OL83-c oligonucleotides (0.83 μM nucleotides each, Supp. Table 1) were incubated in separate tubes at 37°C in the presence or absence of RPA (0.12 μM) in Buffer B for 5 min. The annealing reactions were initiated by mixing the oligonucleotides or RPA-coated oligonucleotides with Mei5-Sae3 (0.34 μM) or Rad52 (0.34 μM). The completed reactions (25 μL) were incubated at 37°C. At indicated times, 4 μL of the reaction was removed and quenched by addition of 10-fold excess of unlabeled OL83-c prior to deproteinization by treatment with SDS (0.5% final) and Proteinase K (0.5 mg/mL) for 10 min at 37°C. The samples were subjected to 12% non-denaturing TAE polyacrylamide gel electrophoresis. The gels were dried, analyzed with a Typhoon phosphorimager and quantified with ImageQuant (GE Healthcare) software.
3. Results
3.1. Mei5, Sae3 and Mei5-Sae3 purification
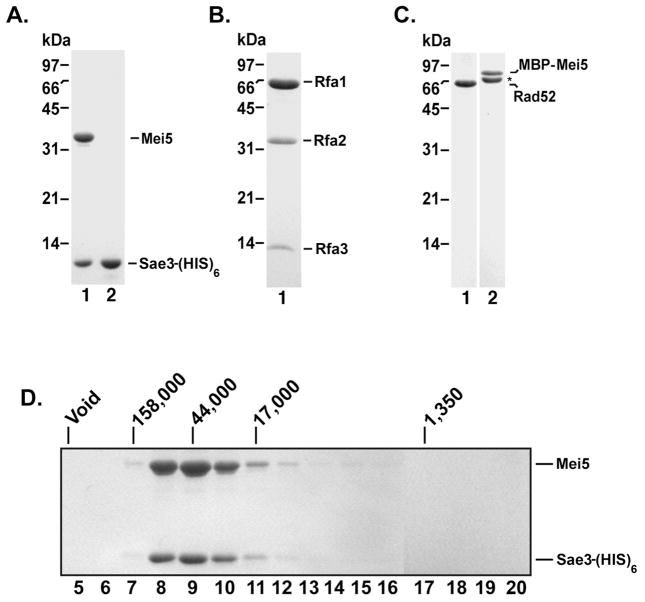
A C-terminal (HIS)6 tag was added to Sae3 to aid in the purification of Sae3 and that of the Mei5-Sae3 complex. The Sae3-(HIS)6 expression plasmid was co-transformed with a compatible expression plasmid harboring an untagged MEI5 gene. The Mei5 and Sae3-(HIS)6 proteins were co-purified from nickel affinity chromatography and remained associated after a 1 M KCl buffer wash. The Mei5-Sae3 complex was purified using Macro-Hydroxyapatite, Mono S and Mono Q columns (Fig. 1A, lane 1; see Supplementary Methods) to greater than 95% homogeneity. We performed gel filtration analysis on Mei5-Sae3 further confirming the two proteins associate as a complex (Fig. 1D). Four independent protein preparations yielded similar results in all of the biochemical experiments.
Figure 1. Purified Mei5-Sae3, Sae3, RPA, Rad52, and MBP-Mei5 proteins and stoichiometry of the Mei5-Sae3 complex.
Mei5-Sae3 complex (1.5 μg; A, lane 1), Sae3-(HIS)6 (1.5 μg; A, lane 2), RPA (3 μg; B), Rad52 (1 μg; C, lane 1) and MBP-Mei5 (1 μg; C, lane 2) were resolved by 12% SDS-PAGE polyacrylamide gel and stained with Coomassie Blue. (*) Indicates the C-terminal truncation product of MBP-Mei5. The Mei5-Sae3 complex (D) was filtered through a 20 mL Sephacryl S-100 column. Fractions were TCA precipitated and subjected to SDS-PAGE on 12% polyacrylamide gels and stained with Coomassie Blue. Protein standards (Bio-Rad) bovine thyroglobulin (670 kDa, Void), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa) and vitamin B12 (1.35 kDa) were filtered on a 20 mL Sephacryl S-100 column and eluted in the indicated fractions.
We also expressed Sae3-(HIS)6 and Mei5 separately in bacteria and devised a procedure to purify each protein. Sae3-(HIS)6 was purified using nickel affinity chromatography, Macro Hydroxyapatite, Mono S, and Mono Q ion exchange chromatography (Fig. 1A, lane 2). To determine whether the (HIS)6 tag fused to the C-terminal end of Sae3 inactivated Sae3, we analyzed spore viability of the SAE3-6HIS strain. Table 2 shows that the SAE3-6HIS strain had the same level of high spore viability as the isogenic wild type strain (98% and 100%, respectively) whereas the sae3 null mutant in the same background showed low spore viability (11%) indicating that the (HIS)6 tag appended to the C-terminus of the Sae3 protein does not interfere with its function.
Table 2.
Spore Viability.
| Strain | Genotype | % Spore viability (viable spores/total spores) |
|---|---|---|
| TBR2055 | wild type | 100% (80/80) |
| TBR1039 | sae3Δ | 11% (9/80) |
| TBR5428 | SAE3-6HIS | 98% (157/160) |
Our initial attempts to purify the Mei5 protein expressed in bacteria resulted in low yields of soluble protein consistent with previous studies [9, 10]. To overcome the poor solubility of Mei5, we fused maltose-binding protein (MBP) to the N-terminal end of Mei5. The presence of MBP improved the solubility of Mei5 and enabled us to purify the fusion protein by amylose affinity chromatography. Expression and purification of MBP-Mei5 from bacteria yields two species (Fig. 1C, lane 2) both of which react with anti-MBP antibodies (Fig. 4B, panel II). By MALDI-TOF analysis, we established that the faster migrating species stemmed from proteolysis of the carboxyl terminus of Mei5 during purification (data not shown).
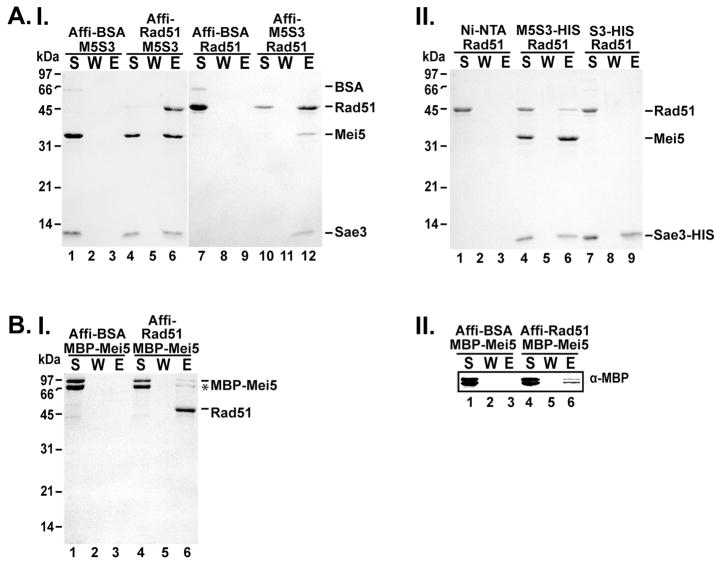
Figure 4. Mei5-Sae3 interacts with Rad51 through Mei5.
(A) Mei5-Sae3 (panel I) was mixed with Affi-Gel containing covalently conjugated BSA or Rad51 (lanes 1–6). Rad51 was mixed with Affi-Gel containing covalently conjugated BSA or Mei5-Sae3 (lanes 7–12). After a wash, the bound protein was eluted with SDS. The supernatant (S), wash (W), and eluate (E) were separated on a SDS-PAGE gel and stained with Coomassie Blue. (A, panel II) Mei5-Sae3 or Sae3-(HIS)6 (panel II) was incubated separately with Rad51. Nickel-NTA beads were added to the reactions and with Rad51 alone with agitation to capture protein complexes. After a wash, the bound protein was eluted and samples were analyzed as described above. (B) MBP-Mei5 (panel I, lanes 1–6) was mixed with Affi-Gel containing covalently conjugated BSA (panel I lanes 1–3) or Rad51 (panel I lanes 4–6). After a wash, the bound protein was eluted and samples were analyzed as described above. Western analysis using anti–MBP antibodies was used to confirm the presence of MBP-Mei5 protein (panel II, lanes 1–6).
3.2. DNA binding activity of Mei5, Sae3 and Mei5-Sae3
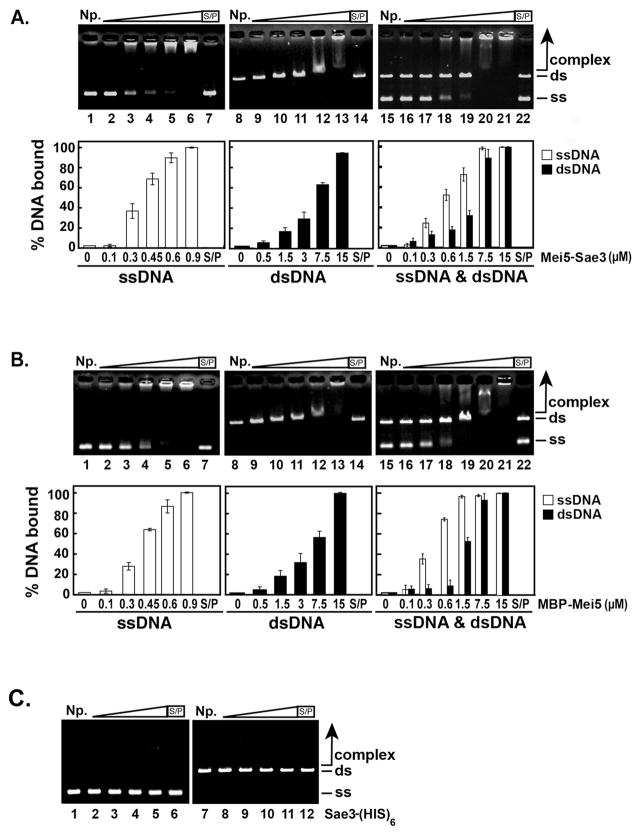
Mei5-Sae3 was reported to possess DNA binding activity with an apparent preference for ssDNA [10]. To determine the role of Mei5 and Sae3 in the DNA binding activity of the Mei5-Sae3 complex, a DNA electrophoretic mobility shift assay with plasmid-length ssDNA and linearized dsDNA was performed. Consistent with a published report [10], increasing concentrations of Mei5-Sae3 shifted both ssDNA (Fig. 2A, lanes 2–6) and dsDNA (Fig. 2A, lanes 9–13) with a preference for ssDNA (Fig. 2A, lanes 16–21). MBP-Mei5 bound both ssDNA (Fig. 2B, lanes 2–6) and dsDNA (Fig. 2B, lanes 9–13) with a preference for ssDNA (Fig. 2B, lanes 16–21) similar to Mei5-Sae3. The nucleoprotein complexes formed by Mei5-Sae3 and MBP-Mei5 both migrate into the agarose gel albeit to a different extent. Sae3-(HIS)6 harbors no detectable DNA binding activity for ssDNA (Fig. 2C, lanes 2–5) or dsDNA (Fig. 2C, lanes 8–11). As expected, we did not detect any mobility shift of either ssDNA or dsDNA substrates by an excess of MBP (data not shown). These results indicate Mei5 is responsible for the DNA binding activity of the Mei5-Sae3 complex.
Figure 2. DNA binding activity of Mei5-Sae3, Mei5 and Sae3 with φX174 DNA.
The indicated concentrations of Mei5-Sae3 (A) or MBP-Mei5 (B) were incubated with φX174 ssDNA (30 μM nucleotides, lanes 2–7), linearized φX174 RF dsDNA (30 μM base pairs, lanes 9–14), or both ssDNA and dsDNA (30 μM nucleotides and 30 μM base pairs, respectively, lanes 16–22) at 37°C for 10 min. (C) Sae3-(HIS)6 protein (1 μM, lanes 2 and 8; 5 μM, lanes 3 and 9; 15 μM, lanes 4 and 10; 30 μM, lanes 5, 6, 11 and 12) was incubated with φX174 ssDNA (30 μM nucleotides, lanes 2–6) or linearized φX174 RF dsDNA (30 μM base pairs, lanes 8–12) at 37°C for 10 min. The reaction products were separated on 0.9 % agarose gels and stained with ethidium bromide. The gels were quantified using Quantity One (Bio-Rad) software. Where indicated, the reaction was treated with SDS (0.5% final) and Proteinase K (0.5 mg/mL) at 37°C for 15 min prior to analysis. S/P, SDS/Proteinase K; NP, no protein control; ss, ssDNA; ds, dsDNA. The average values from 4 experiments were plotted (A and B, bottom panels).
3.3. Substrate preference and minimal length of DNA required for DNA binding of Mei5-Sae3 and Mei5
The plasmid-length ssDNA used in the DNA mobility shift experiments contains a great deal of secondary structure [13]. To determine whether Mei5-Sae3 and MBP-Mei5 have specific affinity for DNA structures, additional DNA mobility shift experiments were performed with 32P-labeled ssDNA, dsDNA, 3′ single-stranded overhang (OH), and fork structure (fork) DNA substrates (Supp. Table 1). We first co-incubated increasing amounts of MBP-Mei5 or Mei5-Sae3 with fork, ssDNA and dsDNA substrates. The DNA-protein complex was separated from free substrate in a non-denaturing polyacrylamide gel.
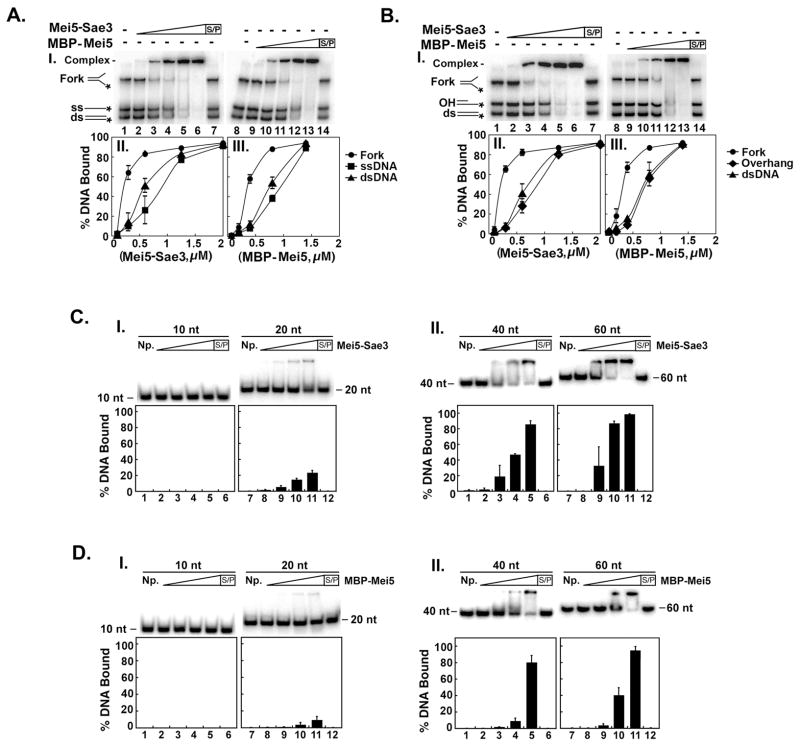
Our results (Fig. 3A) show that Mei5-Sae3 and MBP-Mei5 preferentially bind the fork substrate over ssDNA or dsDNA. When Mei5-Sae3 or MBP-Mei5 was incubated with the mixture of fork, OH and dsDNA, both Mei5-Sae3 and MBP-Mei5 preferentially bound the fork substrate over OH and dsDNA (Fig. 3B). The results in Fig. 3(A and B) reveal that both MBP-Mei5 and Mei5-Sae3 prefer a fork substrate to ssDNA, OH and dsDNA substrates. No DNA binding activity for ssDNA, dsDNA, OH, or fork substrates was detected with increasing concentrations of Sae3-(HIS)6 up to 30 μM or 50 μM MBP (data not shown). In all cases, the DNA-protein complex was trapped within the well. This finding contrasts the results seen when the nucleoprotein complexes were resolved in agarose gels (Fig. 2A and B). It seems likely that in the case of analysis in polyacrylamide gels, nucleoprotein complexes that are near or in the gel well are a result of the high percentage (12 %) acrylamide used.
Figure 3. DNA binding specificity of Mei5-Sae3 and Mei5.
(A and B) Mei5-Sae3 (panel I; 0.1 μM, lane 2; 0.3 μM, lane 3; 0.6 μM, lane 4; 1.25 μM, lane 5; 2.0 μM, lane 6 and 7) and MBP-Mei5 (panel I; 0.07 μM, lane 9; 0.2 μM, lane 10; 0.4 μM, lane 11; 0.83 μM, lane 12; 1.38 μM, lane 13 and 14) were incubated with 0.05 pmol of radiolabeled fork, dsDNA and ssDNA (A, panel I) or radiolabeled fork, dsDNA, and OH (B, panel I) for 10 min at 37°C and subjected to non-denaturing-PAGE. The gels were dried and visualized using a Typhoon phosphorimager. Reactions with Mei5-Sae3 (A and B, panels II) and MBP-Mei5 (A and B, panel III) were quantified using ImageQuant (GE Healthcare) software. (C and D) Mei5-Sae3 and MBP-Mei5, respectively (panel I and II; 0.08 μM, lane 2 and 8; 0.26 μM, lane 3 and 9; 0.52 μM, lane 4 and 10; 1.3 μM, lane 5 and 11; 1.3 μM, lane 6 and 12) were incubated with 0.05 pmol of radiolabeled poly dT 10-mer (panel I, lanes 1–6), poly dT 20-mer (panel I, lanes 7–12), poly dT 40-mer (panel II, lanes 1–6), and poly dT 60-mer (panel II, lanes 7–12) for 10 min at 37°C and treated as described above. Where indicated, the reaction was treated with SDS (0.5% final) and Proteinase K (0.5 mg/mL) at 37°C for 10 min prior to analysis. S/P, SDS/Proteinase K; NP, no protein control; ss, ssDNA; ds, dsDNA.
To determine the minimal length of oligonucleotide necessary for DNA binding by Mei5-Sae3 and MBP-Mei5, DNA mobility shift experiments were performed using 32P-labeled poly dT substrates of 10, 20, 40 or 60 bases in length. Increasing amounts of MBP-Mei5 or Mei5-Sae3 were incubated with the poly dT oligonucletide substrates. Our results show that Mei5-Sae3 (Fig. 3C panel II) and MBP-Mei5 (Fig. 3D panel II) showed similar binding affinity for 40-mer and 60-mer substrates. When a 20-mer substrate was used, only a slight shift of the substrate was detected with Mei5-Sae3 (Fig. 3C panel I) or MBP-Mei5 (Fig. 3C panel I). Neither Mei5-Sae3 nor MBP-Mei5 was able to shift a 10-mer substrate (Fig. 3C and D panel II). The results in Fig. 3(C and D) therefore revealed that Mei5-Sae3 and MBP-Mei5 require a minimum of 20 nucleotides to form a stable nucleoprotein complex, and 40 nucleotides are needed for optimal nucleoprotein complex formation.
3.4. Mei5-Sae3 interacts with Rad51 through Mei5
Genetic and cytological analyses show that Mei5-Sae3 assembly at DSBs is dependent upon Rad51 [8, 9] suggesting Mei5-Sae3 may interact with Rad51. To determine whether Mei5-Sae3 could interact with Rad51, we performed affinity pull-down experiments with Affi-gel matrix conjugated to either Rad51 or Mei5-Sae3. First, we incubated Mei5-Sae3 with Affi-Rad51 matrix. We show that Mei5-Sae3 was retained on the Affi-Rad51 beads and found in the eluate (Fig. 4A panel I, lanes 4–6). As expected, when Mei5-Sae3 was incubated with Affi-matrix conjugated with BSA, Mei5-Sae3 was found in the supernatant indicating no interaction of Mei5-Sae3 with BSA (Fig. 4A panel I, lanes 1–3). We performed the affinity pull-down experiment in the opposite direction by incubating Rad51 with Affi-Mei5-Sae3 matrix. Our results show that Rad51 was retained on Affi-Mei5-Sae3 matrix and was present in the eluate (Fig. 4A panel I, lanes 10–12). Rad51 was not retained on Affi-BSA beads as determined by its presence in the supernatant (Fig. 4A, lanes 7–9). Similar results were observed when we used nickel-NTA matrix to capture the complexes through the (HIS)6 tag on the C-terminal end of Sae3 (Fig. 4A, panel II lane 4–6). We used nickel-NTA resin to perform affinity pull-down experiments to determine whether Sae3 was responsible for interaction with Rad51. Sae3 was incubated with Rad51 and nickel-NTA was used to capture the complexes. We show that Rad51 was found in the supernatant indicating Rad51 failed to interact with Sae3 (Fig. 4A, panel II, lanes 7–9) or nickel-NTA beads (Fig. 4A, panel II, lanes 1–3). We performed the affinity pull-down in the opposite direction by incubating Sae3 with Affi-Rad51 or Affi-BSA. In both pull-downs, Sae3 was found in the supernatant indicating no interaction of Sae3 with Rad51 or BSA (Supp. Fig. 1A). To determine whether Mei5 interacted with Rad51, we performed affinity pull-down experiments using Affi-Rad51 to capture complexes. Our results show that MBP-Mei5 harbored a weak interaction with Rad51 (Fig. 4B, panel 1 and II, lanes 4–6). MBP-Mei5 was not retained on Affi-BSA and was present in the supernatant indicating MBP-Mei5 failed to interact with BSA. We note the interaction between Rad51 and MBP-Mei5 is relatively weak at 120 mM KCl. Similar results were observed when we used amylose resin to capture the protein complexes through the MBP tag on the N-terminal end of Mei5 (Supp. Fig. 1B panel I and II). We cannot rule out the possibility that the MBP fusion to the N-terminal end of Mei5 may interfere with the interaction of Mei5 with Rad51. Taken together, our results reveal a novel interaction of Mei5-Sae3 with Rad51 that is mediated by Mei5.
3.5. The N-terminal domain of Mei5 interacts with the Rad51 recombinase
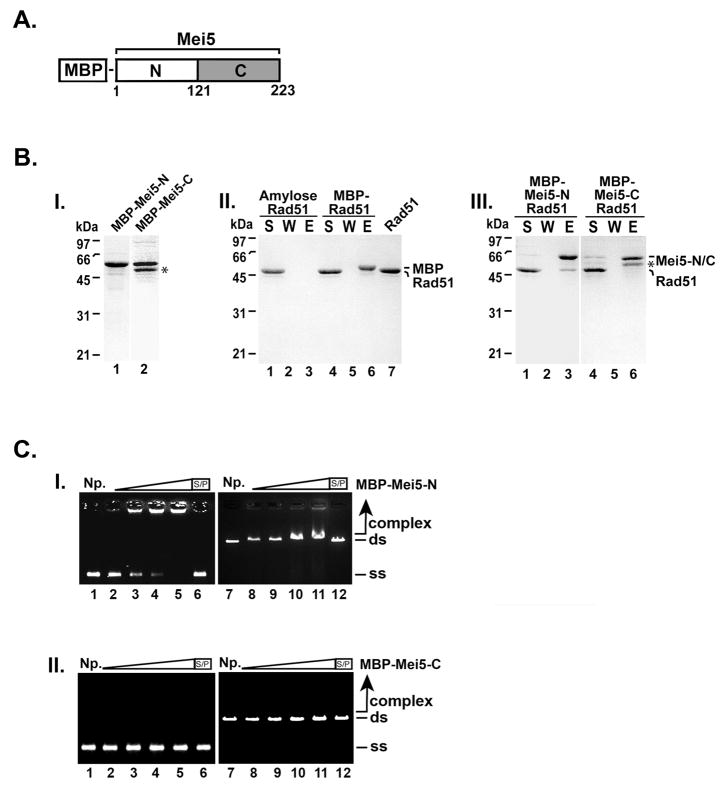
Two-hybrid analysis was used to show Mei5 interacts with the Dmc1 recombinase [9]. In this study, full-length Mei5 was unable to interact with Dmc1, whereas a truncated Mei5 consisting of the N terminal domain (amino acids 1–123) was found to interact with Dmc1 [9]. With this in mind, we wished to delineate the Rad51 interaction domain of Mei5. We constructed an N-terminal and C-terminal truncation of Mei5 fused to MBP and the MBP-Mei5-N and MBP-Mei5-C proteins were purified for use in affinity pull-down experiments. Rad51 was incubated with MBP, MBP-Mei5-N or MBP-Mei5-C to form complexes and were captured using amylose resin. Our results showed Rad51 was retained on MBP-Mei5-N but not MBP-Mei5-C, MBP or amylose resin alone (Fig. 5B). These results indicate the interaction domain of Mei5 for Rad51 resides in the N-terminal domain (amino acids 1–121) of Mei5.
Figure 5. The N-terminal domain of Mei5 interacts with Rad51 and binds DNA.
(A) Domain schematic of Mei5. (B) MBP-Mei5-N (1.3 μg; panel I, lane 1) and MBP-Mei5-C (1.2 μg; panel I, lane 2) were resolved using a 12% SDS-PAGE polyacrylamide gel and stained with Coomassie Blue. Rad51 (7 μg) was incubated with MBP (6.5 μg; panel II, lanes 4–6), MBP-Mei5-N (7.0 μg; panel III, lanes 1–3), MBP-Mei5-C (7.0 μg; panel III, lanes 4–6) for 30 min at 4°C. Amylose beads were added to the reactions and with Rad51 alone (panel II, lanes 1–3) for 30 min at 4°C with agitation to capture protein complexes. The supernatant (S) that contained unbound proteins, wash (W), and SDS eluate (E) were analyzed by SDS-PAGE and stained with Coomassie Blue. (C) MBP-Mei5-N and MBP-Mei5-C (panel I and panel II, respectively; 0.55 μM, lanes 2 and 8; 1.66 μM, lanes 3 and 9; 2.77 μM, lanes 4 and 10; 5.5 μM, lanes 5, 6, 11 and 12) were incubated with φX174 ssDNA (30 μM nucleotides, lanes 2–6) and linearized φX174 RF dsDNA (30 μM base pairs, lanes 7–12) at 37°C for 10 min. The reaction products were separated on 0.9 % agarose gels and stained with ethidium bromide. Where indicated, the reaction was treated with SDS (0.5% final) and Proteinase K (0.5 mg/mL) at 37°C for 15 min prior to analysis. S/P, SDS/Proteinase K; NP, no protein control; ss, ssDNA; ds, dsDNA. (*) Indicates the C-terminal truncation product of MBP-Mei5.
3.5. DNA binding activity resides within the N-terminal half of Mei5
We wished to identify the DNA binding domain of Mei5 using our truncated MBP-Mei5-N and MBP-Mei5-C proteins. Increasing concentrations of MBP-Mei5-N or MBP-Mei5-C was incubated with ssDNA or dsDNA. As shown in Fig. 5C, MBP-Mei5-N preferentially binds ssDNA. These results resemble the DNA binding exhibited by full-length MBP-Mei5 (Fig. 2B) albeit with lower affinity. MBP-Mei5-C failed to shift ssDNA or dsDNA (Fig. 5C). Our data show the N-terminal domain of Mei5 harbors the DNA binding domain.
3.6. Mei5-Sae3 lacks recombination mediator activity for Rad51
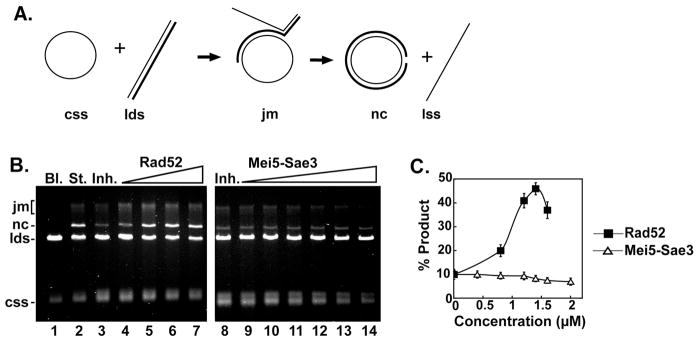
A recent report showed Mei5-Sae3 could overcome RPA inhibition of DNA strand exchange mediated by Dmc1 [10]. Since Mei5-Sae3 interacts with Rad51, we wished to examine the potential of Mei5-Sae3 also functioning as a recombination mediator for Rad51 using a 3 strand plasmid-based DNA strand exchange system [14], [15], [16]. In this system (Fig. 6A), Rad51 is pre-incubated with ssDNA to assemble the presynaptic filament followed by the addition of the ssDNA binding protein, RPA. DNA strand exchange commences upon the addition of dsDNA. When RPA is co-incubated with Rad51, the two compete for access to the ssDNA resulting in greatly reduced strand exchange (Fig. 6B, lanes 3 and 8). The addition of the recombination mediator, Rad52, restores DNA strand exchange (Fig. 6B, lanes 4–7). In contrast, Mei5-Sae3 is unable to overcome the inhibition of RPA in this DNA strand exchange system (Fig. 6B, lanes 9–14) regardless of the order of addition (data not shown). The same results were obtained using two different oligonucleotide based DNA strand exchange systems [17], [18] (data not shown). The results indicate that unlike Rad52 [14] Mei5-Sae3 is unable to aid Rad51 to overcome RPA inhibition. Our data combined with the findings of Ferrari et al. [10] would suggest the recombination mediator activity of Mei5-Sae3 is specific to Dmc1.
Figure 6. Mei5-Sae3 does not mediate Rad51 strand exchange.
(A) Schematic representation of the 3-strand homologous DNA pairing and strand exchange reaction. Homologous pairing between the circular ssDNA (css) and linear duplex DNA (lds) substrates yields a DNA joint molecule (jm), which is converted into a nicked circular duplex molecule (nc) by DNA strand exchange liberating linear ssDNA (lss). (B) The standard reaction (St., lane 2) resulted from pre-incubating the ssDNA with Rad51 (9.3 μM) to allow for the formation of presynaptic filaments before the addition of RPA (1.7 μM). Co-incubation of the ssDNA with Rad51 (9.3 μM) and RPA (1.7 μM) resulted in greatly inhibited DNA strand exchange (Inh., lane 3 and 8). Rad52 (0.8 μM, lane 4; 1.2 μM, lane 5; 1.4 μM, lane 6; 1.6 μM, lane 7) or Mei5-Sae3 (0.4 μM, lane 9, 0.8 μM, lane 10; 1.2 μM, lane 11; 1.6 μM, lane 12; 2.0 μM, lane 13) was included during the incubation of ssDNA with Rad51 and RPA. Bl. indicates no protein was added. (C), the results of (B) were plotted.
3.7. Mei5-Sae3 does not anneal single DNA strands
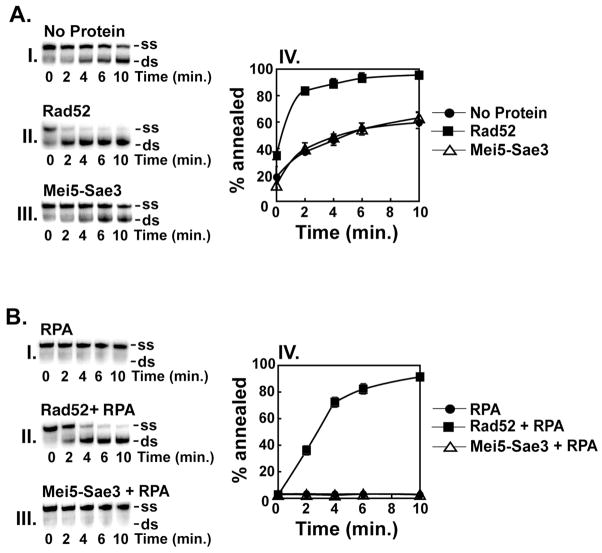
It was of interest to examine whether Mei5-Sae3 possesses single DNA strand annealing activity similar to that of Rad52 [19]. Our data show that Rad52, but not Mei5-Sae3, stimulated annealing of an oligonucleotide to its complement in the absence of RPA (Fig. 7A panel II and III, respectively). Since previous results indicate Mei5-Sae3 binds RPA [10], we wished to investigate whether Mei5-Sae3 could anneal RPA coated-complementary single DNA strands as shown for Rad52 [7]. Our results confirmed that Rad52 anneals RPA coated ssDNA strands (Fig. 7B, panel II) but revealed that Mei5-Sae3 is unable to do so (Fig. 7B, panel III).
Figure 7. Mei5-Sae3 lacks DNA single-strand annealing activity.
(A) The radiolabeled OL83 and unlabeled OL83-c oligonucleotides (0.83 μM nucleotides) were incubated in the absence of protein (A, panel I) or in the presence of Rad52 (0.34 μM, A, panel II) or Mei5-Sae3 (0.34 μM, A, panel III) at 37°C for the indicated times. (B) The same oligonucleotide substrates (0.83 μM nucleotides) were incubated in the presence of RPA (0.12 μM, B, panel I) and either Rad52 (0.34 μM, B, panel II) or Mei5-Sae3 (0.34 μM, B, panel III) at 37°C for the indicated times. For both (A) and (B), the samples were quenched with excess OL83-c and deproteinized by treatment with SDS (0.5% final), Proteinase K (0.5 mg/mL) for 15 min at 37°C. The reaction mixtures were resolved in 12 % non-denaturing TAE polyacrylamide gels. The gels were dried and subjected to phosphorimaging analysis. The position of the ssDNA (ss) and annealed dsDNA (ds) product are indicated on the right in (panels I–III). The results in panels I–III were quantified with ImageQuant (GE Healthcare) and plotted (panel IV).
4. Discussion
In this study, we devised methods for the expression and purification of the S. cerevisiae meiosis-specific recombination mediator complex Mei5-Sae3 and the Mei5 and Sae3 proteins individually. We also examined Mei5-Sae3 and the purified Mei5 and Sae3 proteins for DNA binding activity. In agreement with Ferrari et al., [10] our results showed Mei5-Sae3 preferentially binds plasmid length ssDNA to dsDNA. When defined substrates were used, however, our results revealed that Mei5-Sae3 preferentially binds fork substrates to dsDNA, ssDNA and a 3′ DNA overhang. We showed the minimal length Mei5-Sae3 requires to bind DNA is 20 nucleotides, but 40 nucleotides are needed for optimal DNA engagement. We demonstrated the Mei5 subunit harbors the DNA binding activity of Mei5-Sae3. We reveal the N-terminal 1–121 amino acids of Mei5 harbor the DNA binding domain. Similar to Mei5-Sae3, Mei5 preferentially binds a fork substrate to dsDNA, ssDNA and a 3′ DNA overhang. The specific affinity of Mei5-Sae3 for a DNA fork structure suggests that it nucleates Dmc1 presynaptic filament assembly at regions of ssDNA that harbor similar structures.
Genetic and cytological analysis of mutant yeast strains null for rad51 suggested Mei5-Sae3 interacts with Rad51[8] [9]. We report here that Mei5-Sae3 interacts with Rad51 through the N-terminal domain of Mei5. This region of Mei5 has also been shown to interact with Dmc1 via yeast two-hybrid [9]. Our data are in agreement with a previous report showing Sfr1, the S. pombe ortholog of Mei5, interacts with Rad51 [20]. Recently, murine Swi5 and not Sfr1 was reported to interact with Rad51[21]. This report contrasts our data and that of the Iwasaki laboratory [11, 20].
We examined whether Mei5-Sae3 possessed activities observed for Rad52. While our data confirm the ssDNA annealing activity of Rad52, the lack of ssDNA annealing activity of Mei5-Sae3 distinguishes it from the Rad52 protein. Rad52 also interacts with Rad51 and RPA facilitating its recombination mediator activity on Rad51 [14]. Mei5-Sae3 interacts with Dmc1 and RPA and functions as a recombination mediator of Dmc1 [10]. Although Mei5-Sae3 interacts with Rad51, we find no evidence of a mediator activity in Mei5-Sae3 on Rad51. This differs from a previous report that Sfr1-Swi5, the orthologous complex of Mei5-Sae3 in S. pombe, possesses recombination mediator activity for both Dmc1 and the Rad51 S. pombe ortholog, Rhp51 [11]. Our data supports the hypothesis that Mei5-Sae3 is a recombination mediator specific for Dmc1.
Unlike murine and S. pombe Sfr1-Swi5, Mei5-Sae3 is specifically expressed during meiosis. Taken together with the low sequence conservation of Mei5 to murine and S. pombe Sfr1 (Supp. Fig. 2; 17.6 % and 15.8%, respectively) and Sae3 to murine and S. pombe Swi5 (Supp. Fig. 3; 18% and 22.3%, respectively), it is not surprising that Mei5-Sae3 has properties different from those of Sfr1-Swi5 [11, 21] that likely result in different roles in HR for the two complexes. Overall, our findings provide new information regarding the biochemical properties of the Mei5-Sae3 complex that should prove valuable in guiding future studies on this meiosis-specific mediator and its role in Rad51 mediated recombination.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation grant SCEPSCoR 2004 RII-EPS-0447660 (MGS), Marie Curie Cancer Care Transitional Programme Grant (HT), National Institutes of Health grant GM57814 (PS) and Clemson University. We thank Chau-Wen Chou for mass spectroscopic analysis and William R. Marcotte Jr. for helpful comments on the manuscript.
Abbreviations
- HR
homologous recombination
- ss
single-stranded
- ds
double-stranded
- NTA
nitrilotriacetic acid
- BSA
bovine serum albumin
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
Footnotes
Conflicts of interest
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 2.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 3.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 4.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 5.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 6.Heyer WD, Ehmsen KT, Liu J. Regulation of Homologous Recombination in Eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci U S A. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsubouchi H, Roeder GS. The budding yeast mei5 and sae3 proteins act together with dmc1 during meiotic recombination. Genetics. 2004;168:1219–1230. doi: 10.1534/genetics.103.025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayase A, Takagi M, Miyazaki T, Oshiumi H, Shinohara M, Shinohara A. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell. 2004;119:927–940. doi: 10.1016/j.cell.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 10.Ferrari SR, Grubb J, Bishop DK. The Mei5-Sae3 protein complex mediates Dmc1 activity in Saccharomyces cerevisiae. J Biol Chem. 2009;284:11766–11770. doi: 10.1074/jbc.C900023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y, Iwasaki H. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat Struct Mol Biol. 2006;13:823–830. doi: 10.1038/nsmb1136. [DOI] [PubMed] [Google Scholar]
- 12.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 13.Benevides JM, Stow PL, Ilag LL, Incardona NL, Thomas GJ., Jr Differences in secondary structure between packaged and unpackaged single-stranded DNA of bacteriophage phi X174 determined by Raman spectroscopy: a model for phi X174 DNA packaging. Biochemistry. 1991;30:4855–4863. doi: 10.1021/bi00234a004. [DOI] [PubMed] [Google Scholar]
- 14.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 15.Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- 16.Shi I, Hallwyl SC, Seong C, Mortensen U, Rothstein R, Sung P. Role of the Rad52 amino-terminal DNA binding activity in DNA strand capture in homologous recombination. J Biol Chem. 2009;284:33275–33284. doi: 10.1074/jbc.M109.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 18.San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci U S A. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akamatsu Y, Dziadkowiec D, Ikeguchi M, Shinagawa H, Iwasaki H. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc Natl Acad Sci U S A. 2003;100:15770–15775. doi: 10.1073/pnas.2632890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akamatsu Y, Jasin M. Role for the mammalian Swi5-Sfr1 complex in DNA strand break repair through homologous recombination. PLoS Genet. 2010;6:e1001160. doi: 10.1371/journal.pgen.1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Komen S, Macris M, Sehorn MG, Sung P. Purification and assays of Saccharomyces cerevisiae homologous recombination proteins. Methods Enzymol. 2006;408:445–463. doi: 10.1016/S0076-6879(06)08028-1. [DOI] [PubMed] [Google Scholar]
- 23.Seong C, Sehorn MG, Plate I, Shi I, Song B, Chi P, Mortensen U, Sung P, Krejci L. Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J Biol Chem. 2008;283:12166–12174. doi: 10.1074/jbc.M800763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 25.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.