Abstract
Adequate expression of surfactant protein-B (SP-B) is critical in the function of pulmonary surfactant to reduce alveolar surface tension. Expression of SP-B mRNA is restricted to specific lung-airway epithelial cells, and human SP-B mRNA stability is increased in the presence of the synthetic glucocorticoid dexamethasone (DEX). Although the mechanism of SP-B mRNA stabilization by DEX is unknown, studies suggest involvement of the glucocorticoid receptor (GR). We developed a dual-cistronic plasmid-based expression assay in which steady-state levels of SP-B mRNA, determined by Northern analysis, reproducibly reflect changes in SP-B mRNA stability. Using this assay, we found that steady-state levels of SP-B mRNA increased greater than twofold in transfected human-airway epithelial cells (A549) incubated with DEX (10−7 M). DEX-mediated changes in SP-B mRNA levels required the presence of the SP-B mRNA 3′-untranslated region but did not require ongoing protein synthesis. The effect of DEX on SP-B mRNA levels was dose dependent, with maximal effect at 10−7 M. DEX increased levels of SP-B mRNA in cells lacking GR, and the presence of the GR antagonist RU486 did not interfere with the effect of DEX. Surprisingly, other steroid hormones (progesterone, estradiol, and vitamin D; 10−7 M) significantly increased SP-B mRNA levels, suggesting a common pathway of steroid hormone action on SP-B mRNA stability. These results indicate that the effect of DEX to increase SP-B mRNA stability is independent of activated GR and suggests that the mechanism is mediated by posttranscriptional or nongenomic effects of glucocorticoids.
Keywords: steady-state mRNA, 3′-untranslated region, intrinsic mRNA stability, hormone
pulmonary surfactant, a lipoprotein complex composed of 90% lipid and 10% protein, plays a central role in reducing alveolar surface tension and in innate immunity of the lung (7, 23, 56). Surfactant is synthesized by lung alveolar type II epithelial cells during the terminal sac stage of human embryonic lung development and coats the lining of the alveolus (40). The protein component of surfactant consists of four major proteins, surfactant protein-A (SP-A), SP-B, SP-C, and SP-D (35). SP-A and SP-D are hydrophilic oligomeric proteins that enhance the clearance of invasive bacteria and viruses (56). SP-B and SP-C are small hydrophobic proteins associated with surfactant lipids and function to accelerate surfactant film formation and stabilize the surfactant monolayer (43). Although the phospholipid portion of surfactant, comprised largely of dipalmitoylphosphatidylcholine, acts directly to reduce surface tension, SP-B is also essential in this function, and the lack of sufficient mature active SP-B leads to respiratory failure (17).
Mature SP-B is a highly processed, positively charged 8-kDa dimeric protein exclusively expressed by alveolar type II epithelial cells, but SP-B mRNA is present in both type II and nonciliated bronchioalveolar (Clara) cells (55). SP-B mRNA is detectable as early as 13-wk gestation in human lung tissue (28). However, mature SP-B protein is not detectable until 31 wk of gestation in amniotic fluid, and protein levels increase exponentially thereafter (36). Inadequate levels of surfactant attributable to premature birth can lead to respiratory distress syndrome (RDS), the leading cause of neonatal morbidity and mortality in developed countries (1). In clinical situations where a premature delivery is imminent, antenatal glucocorticoids are given to accelerate fetal lung development and augment expression of pulmonary surfactant (14).
Glucocorticoids have drastic and complex effects on SP-B gene expression by increasing both SP-B gene transcription and mRNA stability. Glucocorticoids have been shown to increase transcription of SP-B mRNA of several mammalian species through the actions of the glucocorticoid receptor (GR) (3, 28). This steroid hormone also increases the stability of human SP-B mRNA in vitro and in vivo. In explants from midtrimester human fetal lung, dexamethasone (DEX) increased SP-B mRNA levels in a dose-dependent manner (53). DEX increased SP-B mRNA levels rapidly (within 12–24 h) and increased SP-B mRNA stability 2.6-fold (half-life of 7.5 h to 18 h), and the increase was not affected by the protein synthesis inhibitor cycloheximide, indicating that the factors involved in glucocorticoid regulation of SP-B gene expression do not require new protein synthesis (50). These characteristics, and the fact that half-maximal stimulation of SP-B mRNA levels in response to glucocorticoids occurs at 1 nM, suggest the involvement of the GR in regulation of SP-B gene expression.
The regulatory mechanisms of mRNA stability are complex and highly variable (10, 13, 15). Steroid hormones have been shown to regulate mRNA stability by a variety of mechanisms (21, 45). Although we have shown that the effect of glucocorticoids to enhance human SP-B mRNA stability requires segments of the SP-B mRNA 3′-untranslated region (UTR), the mechanism by which this stabilization occurs remains unclear (20). We hypothesized that regulation of human SP-B mRNA stability by glucocorticoids requires the action of the GR and characterized hormonal regulation of human SP-B mRNA stability using a reproducible, easily assayable method in which steady-state levels of SP-B mRNA reflect changes in mRNA stability. In the course of these investigations, we have found that DEX regulation of human SP-B mRNA stability does not involve activated GR.
MATERIALS AND METHODS
Materials.
Steroid hormones were purchased from Sigma Chemical (St. Louis, MO) and diluted in ethanol, including DEX (no. D1756), betamethasone (BMZ, no. B7005), hydrocortisone (HCT, no. H0888), β-estradiol (E2, no. E3725), progesterone (PROG, no. P0130), cholecalciferol (vitamin D3, VitD, no. C9756) and mifepristone (RU486, no. M8046).
Plasmid description.
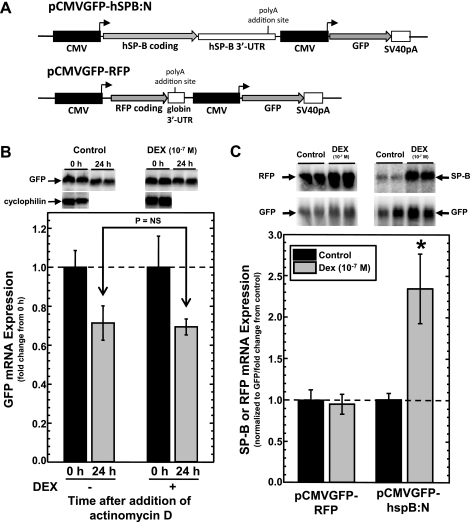
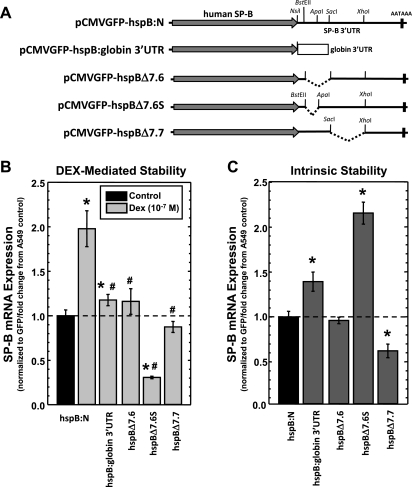
Standard cloning, PCR techniques and bacterial manipulations were used in this study. The expression cassette that permits expression of human SP-B mRNA has been described previously (20). In this cassette, the full-length human SP-B cDNA with the SP-B gene 3′-flanking genomic DNA was placed under transcriptional control of the cytomegalovirus (CMV) E1 promoter of pcDNA3.1 (Invitrogen, Carlsbad, CA). This expression cassette was placed in pShuttle-CMV (18), and the resulting plasmid (pCMVGFP-hspB:N), shown in Fig. 1A, is a dual-cistronic vector in which two identical CMV promoters independently drive expression of the full-length SP-B cDNA with its bona fide polyadenylation site and expression of the gene-encoding green fluorescent protein (GFP). pCMVGFP- red fluorescent protein (RFP), shown in Fig. 1A, is essentially identical to pCMVGFP-hspB:N with the coding sequence and 3′-UTR of the human SP-B cDNA replaced with the coding sequence for the RFP (derived from pIRES-dsRED2; Clontech Laboratories, Mountain View, CA) and the rabbit β-globin 3′-UTR derived from pTET-BBB (58). Plasmids containing various deletions or replacement of the SP-B mRNA 3′-UTR are shown in Fig. 3A. In pCMVGFP-hspB:globin3′UTR, the entire SP-B 3′-UTR region was replaced with the rabbit β-globin 3′-UTR. pCMVGFP-hspBΔ7.6, pCMVGFP-hspBΔ7.6S, and pCMVGFP-hspBΔ7.7 have 236 bp, 126 bp, and 313 bp deletions of the SP-B mRNA 3′-UTR, respectively.
Fig. 1.
Analysis of dexamethasone (DEX)-induced changes in steady-state levels of surfactant protein-B (SP-B) or red fluorescent protein (RFP) mRNA in A549 cells transfected with pCMVGFP-hspB:N or pCMV-GFP-RFP, where CMV is cytomegalovirus and GFP is green fluorescent protein. A: schematic of pCMVGFP-hspB:N and pCMV-GFP-RFP plasmid shown in scale. Black boxes represent identical, independent CMV E1 promoters; arrows denote the direction of transcription. Gray arrows represent the coding regions for human SP-B (hSP-B), RFP, and GFP. Hatched boxes represent the SP-B mRNA 3′-untranslated region (hSP-B 3′-UTR), rabbit β-globin 3′-UTR (globin 3′-UTR) and the SV40 large T antigen mRNA 3′-UTR (SV40pA). The positions of the polyadenylation signals are indicated. B: analysis of GFP mRNA stability in A549 cells. Cells were transfected with pCMVGFP-hspB:N and incubated in the absence or presence of DEX (10−7 M) and then treated with actinomycin D (10 μg/ml) for 24 h. RNA was isolated at time = 0 and 24 h and subjected to Northern analysis for the presence of GFP mRNA and cyclophilin mRNA (for use as a loading control, only at t = 0 h). Shown is a typical image of the analysis generated by phosphorimaging. Levels of GFP mRNA at t = 0 h were set as 1 and normalized GFP levels (means ± SE) at 24 h relative to levels at t = 0 h are shown (N = 6, 2 independent experiments). C: effect of DEX on steady-state levels of SP-B and RFP mRNA in A549 cells. Cells were transfected with pCMVGFP-RFP or pCMVGFP-hspB:N and incubated in the absence or presence of DEX (10−7 M). RNA was isolated and subjected to Northern analysis for the presence for SP-B, RFP, or GFP mRNA. Shown is a typical image of the analysis generated by phosphorimaging. The RFP mRNA/GFP mRNA or SP-B mRNA/GFP mRNA ratio was determined in each sample. The average ratio in untreated samples was set as 1; the ratio from DEX-treated samples was normalized to this average. Shown are normalized SP-B levels (means ± SE) in DEX-treated samples relative to levels in untreated samples (N ≥ 10, 3 independent experiments; *P < 0.01).
Fig. 3.
DEX-induced changes in steady-state levels of SP-B mRNA requires portions of the human SP-B mRNA 3′-UTR. A: schematics of the various expression cassettes harboring deletions of the SP-B 3′-UTR shown in scale. The gray arrow denotes the coding region for human surfactant protein-B, whereas the thin black lines represent the 3′-UTR. The hatched box represents the rabbit β-globin 3′-UTR. The positions of restriction endonuclease cleavage sites used to generate deletions of the SP-B 3′-UTR (shown by dashed lines) are indicated. AATAAA indicates the position of the polyadenylation signal of SP-B mRNA. B: effect of DEX on steady-state levels of SP-B mRNA in A549 cells transfected with plasmids shown above. The SP-B mRNA/GFP mRNA ratio was determined in each sample. The average ratio in untreated samples was set as 1; the ratio from DEX-treated samples was normalized to this average. Shown are normalized SP-B levels in DEX-treated samples relative to levels in untreated samples (means ± SE; *P < 0.01 relative to untreated controls, #P < 0.01 relative to SP-B mRNA levels from pCMVGFP-hspB:N treated with DEX). C: effect of deletions of the SP-B mRNA 3′-UTR on intrinsic stability of SP-B mRNA expressed from A549 cells transfected with plasmids shown above. The SP-B mRNA/GFP mRNA ratio was determined in each sample. The average ratio in pCMVGFP-hspB:N samples was set as 1; the ratio from other samples was normalized to this average. Shown are the normalized SP-B levels (means ± SE; N ≥ 7, 2 independent experiments; *P < 0.01 relative to SP-B mRNA levels from pCMVGFP-hspB:N).
Cell culture.
Human lung epithelial A549 cells (ATCC CCL-185), human epithelial kidney (HEK)293 cells (ATCC CRL-1573), human umbilical vein endothelial ECV-304 cells (48), and human cervical epithelial HeLa cells (ATCC CCL-2) were cultured in Weymouth's MB 752/1 medium (no. 11220, Invitrogen), containing FBS (10% vol/vol) in a humidified incubator at 37°C with 5% CO2.
Cell transfection and assay design.
Transfection of cells with plasmid DNA was performed using the protocol prescribed by the lipofectamine plus reagent (no. 11514, Invitrogen) with slight modifications; 4 μg of plasmid combined with 12 μl of lipofectamine reagent and 12 μl of the plus reagent were utilized to transfect 60-mm plates. A typical assay was performed as follows: cells were transfected with plasmid DNA for 4 h and allowed to recover overnight in media containing 2% charcoal-stripped serum (no. 12676, Invitrogen) to prevent undue influence of serum-derived steroid hormones. Steroids were added to the cells 18 h after transfection, and incubation continued for 36 h, at which time RNA was isolated for analysis.
Isolation of RNA.
RNA was isolated and purified from the cells using Trizol reagent (no. 15596–026, Invitrogen). The concentration of the RNA was determined by measuring absorbance at 260 nm.
RT-PCR.
RT-PCR of mRNA was performed using the Superscript one-step RT-PCR kit (no. 10928, Invitrogen). To isolate a 542-bp cDNA fragment of the human serum/glucocorticoid regulated kinase 1 (sgk1) gene for use in Northern analysis, RT-PCR was performed on RNA derived from A549 cells using hsgk1 forward (5′-GCATACGCCGAGCCGGTCTT-3′) and sgk1 reverse (5′-GAAGGCCCACCAGGAAAGGG-3′) primers. The reaction was performed for 30 cycles at a hybridization temperature of 55°C. To detect the presence of mRNA encoding human GR, RT-PCR analysis of mRNA isolated from A549, HEK293, ECV-304, and HeLa cells was performed using primers described previously (FGRα: 5′-GGCAATACCAGGTTTCAGGAACTTACA-3′, RGRα: 5′-ATTTCACCATCTACTCTCCCATCACTG-3′) that produces a DNA fragment of 824 bp (37). The reaction was performed for 35 cycles at a hybridization temperature of 58°C.
Northern analysis of mRNA.
Northern analysis of sgk1, cyclophilin, RFP, GFP, and SP-B mRNA expression was performed as described in detail previously (4). Total RNA (20 μg) was electrophoresed, transferred to nylon membrane (Zeta-Probe, no. 162-0165; Bio-Rad Laboratories, Hercules, CA), and probed using a radiolabeled DNA probes derived from pCMVGFP-hspB:N (GFP, SP-B), pCMVGFP-RFP (RFP), sgk1 cDNA (described above), or rabbit cyclophilin cDNA (a gift from Dr. Miles Wilkenson). Signal was visualized and quantified using a Storm 840 phosphorimager (Amersham Biosciences, Piscataway, NJ).
Data analysis.
In this study, at least two independent experiments were performed in each analysis. The data were analyzed by with SigmaPlot (ver 10; Systat Software, San Jose, CA) software. Differences between groups were assessed by the student's t-test. The results are expressed as means ± SE. Statistical differences were considered significant when P ≤ 0.05.
RESULTS
Description and justification of the steady-state mRNA assay system that reflects mRNA stability.
Previously, we reported the use of a plasmid-based expression system in which the full-length SP-B cDNA under transcriptional control of the ubiquitously-expressed CMV E1 promoter and SP-B mRNA maturity is attained by inclusion of the bona fide SP-B polyadenylation signal in the context of SP-B genomic DNA sequences (20). We found that a lung epithelial cell line, A549 (27), can be used to assay SP-B mRNA stability in vivo when transfected with these plasmids. Although this approach was used successfully to assay DEX-induced changes in stability of SP-B mRNA deleted in various regions of the 3′-UTR, there were drawbacks. It proved difficult to normalize the results of several experiments. It was impossible to compare the analysis of one type of SP-B construct to another because of plasmid-specific quality affecting transfection (only DEX-induced changes could be determined using a single construct type). The potential adverse effects of the transcription inhibitor used in the assay on mRNA stability had to be considered; actinomycin D is toxic to cells and is known to affect the stability of various mRNAs (58). In the strategy used here, we reasoned that expression of two different genes under transcriptional control of two identical but independent promoters should result in relatively consistent transcription rates of two different mRNAs. The presence of agents that alter transcriptional activity of the CMV promoters would affect the overall rate of transcription, but the relative ratio of the steady-state levels of both transcripts should remain the same if the agents had no effect on mRNA stability of transcripts under control of the promoters. However, the ratio of the steady-state levels of the two mRNAs would change if the agent alters the stability of only one mRNA species. In addition, mutagenesis of the sequences that affect mRNA stability in one of the mRNAs would also be reflected in the ratio of the steady-state levels. The use of this strategy would exclude the need for actinomycin D, eliminating potential adverse effects of this transcription initiation inhibitor in the analysis. We then reasoned that, if we put both promoters on a single plasmid independently driving the expression of two expression cassettes, then the need for cotransfection of a plasmid used for normalization is eliminated, as are the potential adverse effects of plasmid quality. If there are any differences in quality or transfection efficiency between plasmids, then expression of the common gene can be used as a normalizing factor among the samples.
In Fig. 1A, the schematics of pCMVGFP-hspB:N, one of the dual cistronic plasmids used in the assay, are shown. The cDNA coding for the GFP is under transcriptional control of one of the CMV promoters, and the resulting normalizing mRNA is under maturation control of the SV40 large T 3′-UTR. The other CMV promoter drives expression of the previously described human SP-B cDNA expression cassette. An artificial but unique NsiI was placed immediately downstream of the stop codon for the SP-B protein and provides a convenient restriction endonuclease site for subsequent manipulations of the SP-B DNA. There are several assumptions that underlie interpretation of the results of this steady-state assay, such as the fact that DEX has no effect on GFP mRNA stability. This possibility was tested in a manner described previously where the stability of SP-B mRNA in the absence or presence of DEX was determined using actinomycin D (20). A549 cells were transfected with pCMVGFP-hspB:N and incubated in the absence or presence of DEX (10−7 M). Twenty-four hours after transfection the cells were treated with actinomycin D (10 μg/ml). RNA was isolated at 0 and 24 h after addition of actinomycin D, GFP mRNA levels were visualized by Northern analysis, and the signal was quantified as described in materials and methods. The levels of cyclophilin mRNA was determined as loading control at 0 h, but cyclophilin levels were not used after addition of actinomycin D, as these levels reflect the stability of cyclophilin mRNA and confound the results. Levels of GFP mRNA remaining after addition of actinomycin D were normalized to GFP mRNA levels (± DEX) at 0 h. The results of the assay are shown in Fig. 1B. As can be seen, there is no significant difference between the change in GFP mRNA levels incubated in the absence or presence of DEX, indicating that GFP mRNA stability is not affected by the presence of the steroid hormone.
Another assumption is that transcriptional rate responses are identical for both CMV promoters. Although the sequences of the CMV promoters upstream and immediately around the TATA are the same, there are differences in sequences near the transcription start sites in the SP-B and GFP expression cassettes. These regions may differentially influence promoter activity after steroid treatment, and thus SP-B mRNA responses may reflect differences in transcriptional activity rather than in mRNA stability. To demonstrate the lack of consequence of these potential confounding events in the assay, we tested the plasmid pCMVGFP-RFP, shown in Fig. 1A, which was generated during the course of other investigations. In this plasmid, the gene-encoding RFP replaces the SP-B cDNA and is under control of one CMV promoter and GFP under control of the other. We assayed the effect of DEX on RFP mRNA stability in the manner described above using actinomycin D for GFP mRNA stability and found no effect (data not shown). A549 cells were transfected with pCMVGFP-RFP, incubated in the absence or presence of DEX (10−7 M) and levels of RFP and GFP mRNA in the cells determined by the steady-state assay as described in materials and methods. A typical image of Northern analysis of RFP and GFP mRNA is shown in Fig. 1C. DEX appears to increase the signals of both RFP and GFP mRNA in the analysis, presumably attributable to increases in DEX-induced transcriptional activity of the CMV promoters. In each individual sample, the RFP mRNA signal relative to the GFP mRNA signal was determined as an arbitrary number. In samples incubated in the absence of DEX (control), the average RFP mRNA/GFP mRNA ratio was determined for each independent experiment. This value was used to normalize the RFP mRNA/GFP mRNA value for each individual sample, both control and DEX treated. In this manner, the average of all control samples in the individual experiment should be 1, and the effect of DEX treatment is expressed as a fold change from 1. The same procedure is repeated in each individual experiment. Because all RFP mRNA/GFP mRNA values are expressed as fold change from control, samples from individual experiments could be grouped and provide large N values from multiple experiments, and small changes in stability can be determined that are statistically significant. As can be seen in Fig. 1C, when the RFP mRNA levels are normalized to GFP mRNA levels and the ratio in the absence of DEX is set as 1, there is no change in the ratio in cells incubated in the presence of DEX. These results suggest that there are no promoter-specific differences between the two CMV promoters in the plasmid or that the presence of promoter-specific differences has no effect on interpretation of the results of the assay.
DEX-induced changes in steady-state levels of SP-B mRNA expressed from pCMVGFP-hspb:N reflect posttranscriptional regulation of mRNA stability.
The steady-state assay system was used to determine the effect of DEX on stability of SP-B mRNA. A549 cells were transfected with pCMVGFP-hspB:N and incubated in the absence or presence of DEX (10−7 M), and steady-state levels of SP-B and GFP mRNA in the cells were determined as described in materials and methods. Transfection of cells with pCMVGFP-hspB:N resulted in two transcripts: GFP mRNA (∼800 nt) and SP-B mRNA (∼2150 nt). A typical image of Northern analysis of SP-B and GFP mRNA is shown in Fig. 1C. As noted with pCMVGFP-RFP, DEX increases the signals of both SP-B and GFP mRNA in the analysis, presumably attributable to increases in DEX-induced transcriptional activity of the CMV promoters. However, the change in the SP-B mRNA signal appears to be greater than the change in GFP mRNA signal. These differences became apparent when the SP-B mRNA/GFP mRNA ratios were calculated for each sample. In Fig. 1C is shown the graphical representation of DEX-induced changes in SP-B mRNA levels. As can be seen, the presence of DEX increases steady-state levels of SP-B mRNA ∼2.3-fold compared with untreated samples. These results are in agreement with our previously published results in isolated human alveolar epithelial type II cells in primary culture and with others who have shown that DEX increases SP-B mRNA stability greater than 2.6-fold in human fetal lung tissue in organ culture (20, 50). However, it is important to realize that, although increases in steady-state SP-B mRNA levels reflect changes in stability of the mRNA in this assay, the assay does not directly measure mRNA half-life.
DEX-induced changes in steady-state levels of SP-B mRNA require at least 24 h of exposure.
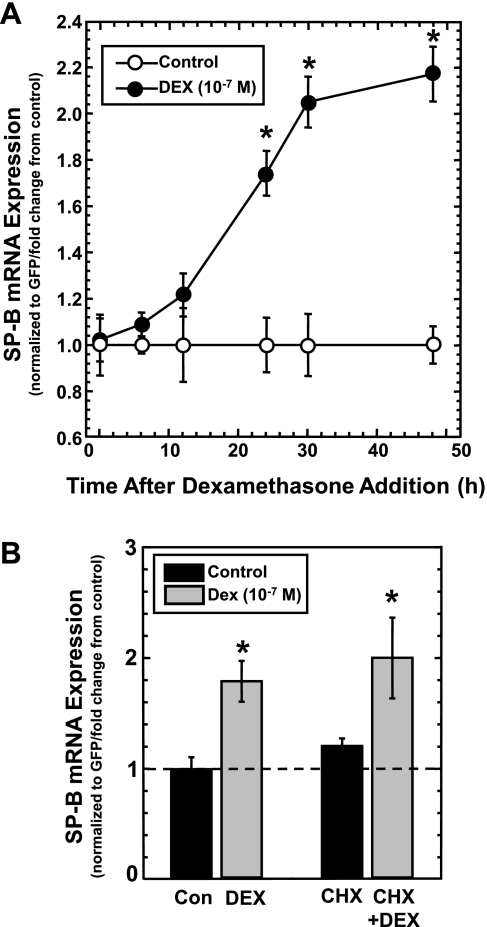
In the results described above, the steady-state levels of SP-B mRNA were determined 36 h after transfection of A549 cells with pCMVGFP-hSPB:N. Although it was assumed that this time frame allows for changes in the steady-state levels of SP-B mRNA to manifest (because of changes in mRNA stability), the rate of change is completely unknown, as is the length of time required for a new steady-state equilibrium between synthesis and decay to be reached. To ascertain the appropriate exposure time, the change in steady-state levels of SP-B as a function of time after addition of DEX was determined. A549 cells were transfected with pCMVGFP-hspB:N and incubated for 24 h. DEX (10−7 M) was added to a portion of the plates, and the steady-state levels of SP-B and GFP mRNA in the cells incubated in the absence or presence of DEX were determined at various time points as described in materials and methods. The DEX-induced SP-B mRNA/GFP mRNA ratio relative to the SP-B mRNA/GFP mRNA ratio in untreated cells was determined. As seen in Fig. 2A, the ratio of SP-B mRNA/GFP mRNA in DEX-treated cells changes rapidly following a lag period, after which a plateau was reached by 30 h of exposure. Overall, this curve indicates that a significant change in SP-B steady-state levels requires a minimum of 24 h of exposure to DEX. These results do not suggest that the mechanism of activation of SP-B mRNA stability by DEX requires at least 24 h; these results indicate that the use of this assay to measure changes that reflect changes in mRNA stability requires at least 24 h.
Fig. 2.
Analysis of DEX-induced changes in steady-state levels of SP-B mRNA as a function of time and in the presence of a protein synthesis inhibitor. A: changes in steady-state SP-B mRNA levels in the absence or presence of DEX as a function of time. A549 cells were transfected with pCMVGFP-hspB:N and incubated in the absence or presence of DEX (10−7 M). RNA was isolated from the samples at the indicated times and subjected to Northern analysis for the presence of SP-B and GFP mRNA. Shown is a line graph of DEX-induced SP-B mRNA/GFP mRNA levels (means ± SE) relative to untreated samples, normalized as 1 (N ≥ 7, 2 independent experiments; *P < 0.01). B: effect of DEX on steady-state levels of SP-B mRNA in A549 cells transfected with pCMVGFP-hspB:N in the absence or presence of cyclohexamide (CHX). A549 cells were transfected with pCMVGFP-hspB:N and incubated in the absence or presence of DEX (10−7 M) and in the absence or presence of cyclohexamide (100 μg/ml) for 24 h. RNA was isolated and subjected to Northern analysis for the presence of SP-B and GFP mRNA, and the SP-B mRNA/GFP mRNA ratio was determined in each sample. The average ratio in untreated samples was set as 1; the ratio from DEX-treated samples was normalized to this average. Shown are normalized SP-B mRNA levels in DEX-treated samples relative to levels in untreated samples (means ± SE; N ≥ 7, 2 independent experiments; *P < 0.03 relative to controls without DEX).
DEX-induced changes in steady-state levels of SP-B mRNA do not require ongoing protein synthesis.
DEX-mediated changes in biological processes generally involve the GR (42). Binding of DEX to GR and activation of GR is a posttranslational mechanism that does not require synthesis of new protein. To determine whether the mechanism by which DEX stabilizes SP-B mRNA requires ongoing protein synthesis, the steady-state assay system was performed in the presence of cyclohexamide, a protein synthesis inhibitor (2). A549 cells were transfected with pCMVGFP-hspB:N and incubated overnight. Cells were then incubated in the absence or presence of cyclohexamide (100 μg/ml) and in the absence or presence of DEX (10−7 M) for 24 h. Steady-state levels of SP-B and GFP mRNA in the cells were determined as described in materials and methods. Shown in Fig. 2B are the results of the assay. As can be seen, cyclohexamide had no effect on the ability of DEX to significantly increase the steady-state levels of SP-B. These results indicate that new protein synthesis is not required for stabilization of SP-B mRNA by DEX, suggesting mechanisms involving activated GR or posttranslational modification of other proteins.
The absence of specific segments of the SP-B mRNA 3′-UTR alters DEX-induced and intrinsic SP-B mRNA stability.
Our previous studies using an assay involving inhibition of transcription indicated that the SP-B mRNA sequences necessary for DEX-induced stabilization of mRNA stability are localized to the 3′-UTR (20). Those findings were verified using the dual cistronic plasmid steady-state assay described here. The plasmids containing expression cassettes resulting in SP-B mRNA deleted in various regions of the 3′-UTR are described in materials and methods and are shown in Fig. 3A. To determine the necessity of sequences of the human SP-B 3′-UTR for DEX-induced stabilization of SP-B mRNA, A549 cells were transfected with pCMVGFP-hspB:N and pCMVGFP-hspB:globin3′UTR, incubated in the absence or presence of DEX (10−7 M) for 36 h, and steady-state levels of SP-B and GFP mRNA in the cells were determined as described in materials and methods. As seen in Fig. 3B, the presence of DEX did significantly increase steady-state levels of SP-B mRNA expressed from pCMVGFP-hspB:globin3′UTR, albeit only 1.2-fold compared with control levels, suggesting that DEX can induce stabilization of this hybrid mRNA. However, replacement of the SP-B 3′-UTR with the rabbit β-globin 3′-UTR significantly reduced the ability of DEX to stabilize the hybrid mRNA relative to DEX-induced levels of SP-B mRNA expressed from pCMVGFP-hspB:N, suggesting that the 3′-UTR of the human SP-B mRNA is necessary for DEX regulation of mRNA stability and that coding regions of the human SP-B mRNA are not involved.
In the previous study, the 837-nt-long SP-B mRNA 3′-UTR was dissected into three distinct segments and used in vitro analysis in the formation of specific mRNA:protein complexes, which was seen using the 126-nt-long 7.6S fragment (20). Using the assay system described here, the necessity of segments of the SP-B mRNA 3′-UTR for mediating DEX-stabilization of SP-B mRNA was tested. A549 cells were transfected with pCMVGFP-hspB:N, pCMVGFP-hspB:Δ7.6, pCMVGFP-hspB:Δ7.6S, or pCMVGFP-hspB:Δ7.7 (Fig. 3A) and incubated in the absence or presence of DEX for 36 h, and steady-state levels of SP-B and GFP mRNA in the cells were determined as described in materials and methods. The results, shown in Fig. 3B, indicate that deletion of large segments of the SP-B mRNA 3′-UTR significantly reduced the ability of DEX to stabilize the mRNA compared with the unaltered DEX-induced SP-B transcript, reproducing the results described previously using a different assay (20). In fact, DEX significantly decreased stability of an SP-B transcript deleted in a 126-nt-long segment (7.6S) compared with levels in cells incubated in the absence of DEX. Interestingly, a larger 313-nt-long segment (7.7) was also found to be necessary for DEX-induced stabilization. These results suggest that separate segments of the entire SP-B mRNA 3′-UTR are necessary to elicit sufficient DEX-stabilization of SP-B transcripts and are suggestive that interactions of multiple regions or elements in the SP-B mRNA 3′UTR are a mechanism by which DEX stabilizes SP-B transcripts.
In addition to investigations to determine sequences necessary for hormone-induced changes in SP-B mRNA stability, this assay system allows investigation of the effect of mRNA segments on intrinsic mRNA stability in the absence of DEX treatment. Intrinsic stability is determined by normalizing the expression of altered mRNA species to the unaltered SP-B mRNA (expressed from pCMVGFP-hspB:N). The results of this type of analysis are shown in Fig. 3C. Substitution of the human SP-B mRNA 3′-UTR with the rabbit β-globin 3′-UTR resulted in a significant increase in intrinsic stability, about 1.4-fold. Because the SP-B mRNA half-life is ∼6 h and the rabbit β-globin mRNA stability is ∼9.5 h, the change in SP-B mRNA/GFP mRNA ratios between these plasmids reflects this difference in intrinsic stability (8, 50). When the 7.6 region is deleted, no change in intrinsic stability is observed. However, a more restricted deletion of this region (7.6S) resulted in a significant increase in intrinsic mRNA stability (∼2.3-fold). These results suggest the presence of a destabilizing element in the 7.6S regions whose activity is repressed by sequences of the 7.6 region that do not include 7.6S. Deletion of the 7.7 region significantly reduced intrinsic stability of SP-B mRNA, suggesting the presence of stabilizing elements in this region. These results portend a complex regulatory mechanism of SP-B mRNA stability in which multiple segments of the 3′-UTR participate.
Changes in steady-state levels of SP-B mRNA exposed to various concentrations of DEX.
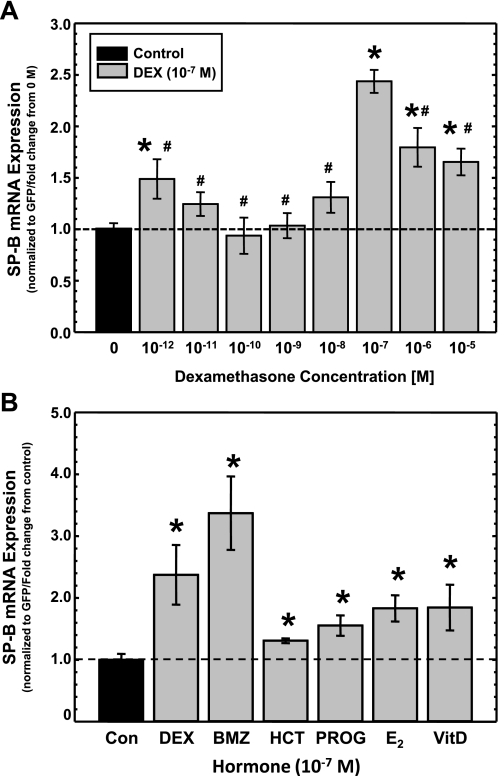
The ease and reproducibility of this dual-cistronic assay system allows more efficient characterization of the effect of DEX on human SP-B mRNA stability. Cellular processes affected by glucocorticoids generally involve the GR (41). When the GR is involved, the magnitude of the response generally follows a typical sigmoidal dose-response curve in which half-maximal response occurs at ∼1.5 × 10−9 M DEX (42). The goal of this assay was to define the dose-response curve of SP-B mRNA stability in response to DEX. A549 cells, which express GR (51), were transfected with pCMVGFP-hspB:N and incubated in the absence or presence of DEX at various concentrations ranging from 10−12 to 10−5 M for 36 h, and steady-state levels of SP-B and GFP mRNA in the cells were determined as described in materials and methods. The data from all concentrations were normalized against the control (no DEX exposure). As can be seen from the results in Fig. 4A, statistically significant differences in the levels of SP-B mRNA were observed in DEX concentrations of 10−12, 10−7, 10−6, and 10−5 M compared with control. Surprisingly, the magnitude of the response at these concentrations was not distributed in the typical sigmoidal dose-response curve of glucocorticoid activation of the GR, but the curve is biphasic. In fact there is a peak of maximal SP-B mRNA stabilization at 10−7 M DEX, and the stability of SP-B mRNA decreases significantly at 10−6 and 10−5 M DEX compared with 10−7 M DEX. In addition, we calculated the half-maximal activation constant for the effect of DEX on steady-state levels of SP-B mRNA from the curve, Kd = ∼10−8 M. This value is about 10–50 times higher than the typical Kd of GR in the presence of DEX, ∼2 × 10−9 M (41). These results suggest an effect of DEX on SP-B mRNA stability that does not reflect normal activation of the GR by DEX.
Fig. 4.
Effects of DEX concentration and other steroid hormones on steady-state levels of SP-B mRNA. A: changes in steady-state levels of SP-B mRNA in response to various concentrations of DEX. A549 cells were transfected with pCMVGFP-hspB:N and incubated in the absence or presence of indicated concentrations of DEX. RNA was isolated and subjected to Northern analysis for the presence of SP-B and GFP mRNA, and the SP-B mRNA/GFP mRNA ratio was determined in each sample. The average ratio in untreated samples was set as 1; the ratio from DEX-treated samples was normalized to this average. Shown are normalized SP-B levels in DEX-treated samples relative to levels in untreated samples (means ± SE; N ≥ 7, 2 independent experiments; *P < 0.02 relative to untreated controls; #P < 0.02 relative to levels at 10−7 M DEX). B: changes in steady-state levels of SP-B mRNA in response to other steroid hormones. A549 cells were transfected with pCMVGFP-hspB:N and incubated for 36 h in the absence (Con) or presence of various steroid hormones (10−7 M), including DEX, betamethasone (BMZ), hydrocortisone (HCT), progesterone (PROG), β-estradiol (E2), and vitamin D (VitD). RNA was isolated and subjected to Northern analysis. The levels of SP-B and GFP were quantified, and the SP-B mRNA/GFP mRNA ratio was determined in each sample. The average ratio in untreated samples was set as 1; the ratio from the hormone-treated samples was normalized to this average. Shown are normalized SP-B levels in treated samples relative to levels in untreated samples (means ± SE; N ≥ 7, 2 independent experiments; *P < 0.03 relative to untreated controls).
Steady-state levels of SP-B mRNA are increased in the presence of other steroid hormones.
The activity of the GR is affected by the type of glucocorticoid that binds the receptor; HCT is generally less effective than DEX, whereas BMZ is generally more effective than DEX (11). To determine whether this response pattern is repeated with regard to SP-B mRNA stability and to determine whether the effect of steroid hormones of SP-B mRNA stability is restricted to glucocorticoids, A549 cells were transfected with pCMVGFP-hspB:N and incubated in the absence or presence of DEX, BMZ, HCT, PROG, E2 and VitD (all at 10−7 M) for 36 h, and steady-state levels of SP-B and GFP mRNA in the cells were determined as described in materials and methods. The SP-B mRNA/GFP mRNA ratio in the treated cells was determined relative to the SP-B mRNA/GFP mRNA ratio in untreated cells, and the results are shown in Fig. 4B. Exposure of the cells to any of the glucocorticoids (DEX, BMZ, HCT) resulted in a significant increase is steady-state levels of SP-B mRNA compared with control. These results recapitulated the expected potency of the hormones towards the GR; BMZ > DEX > HCT. Surprisingly, exposure of the transfected cells to other steroid hormones significantly increased the steady-state levels of SP-B mRNA compared with untreated cells, albeit at lower levels than that attained by exposure to DEX. These results suggest that the pathway of action of steroid hormones to increase SP-B mRNA stability is not specific to glucocorticoids and may involve a common component.
The presence of GR in cells has little correlation with DEX-induced stabilization of SP-B mRNA stability.
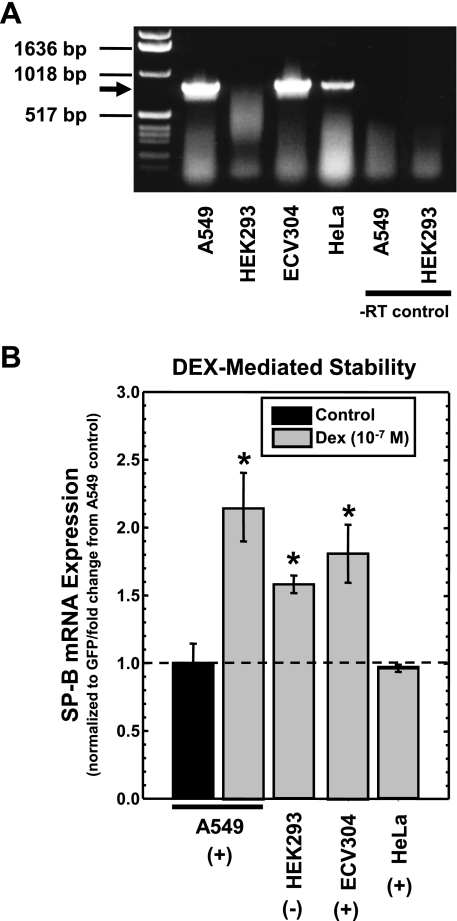
In the previous study, we reported that the ability of DEX to increase SP-B mRNA stability is not specific for the human pulmonary epithelial cells that express SP-B (20). The use of these cells in conjunction with the steady-state assay described here provides an unambiguous means of determining involvement of the GR in the ability of DEX to increase SP-B mRNA stability. Whereas human pulmonary epithelial cell line (A549), endothelial cell line (ECV304), and cervical epithelial cell line (HeLa) express GR, the human kidney epithelial cell line (HEK293) lacks detectable GR (47, 49, 51, 52). The reported absence or presence of GR in these cell lines was verified by RT-PCR as described in materials and methods with primers used previously to quantify GR mRNA in A549 cells (37). As can be seen in Fig. 5A, GR mRNA is detectable in all cells lines except HEK293 cells. These cells were transfected with pCMVGFP-hspB:N and incubated in the absence or presence of DEX (10−7 M) for 36 h, and steady-state levels of SP-B and GFP mRNA in the cells were determined as described in materials and methods. The SP-B mRNA/GFP mRNA ratio in the treated cells was determined relative to the SP-B mRNA/GFP mRNA ratio in untreated cells, and the results are shown in Fig. 5B. As can be seen, DEX significantly increased steady-state levels of SP-B mRNA in cells that express GR (A549 and ECV304) as well as cells that do not express GR (HEK293). Interestingly, DEX does not induce SP-B mRNA stability in HeLa cells, which express GR, suggesting that these cells lack a component of the regulatory mechanism that the other cells possess. These results suggest that the absence or presence of GR has no correlation with the mechanism by which DEX stabilizes SP-B mRNA.
Fig. 5.
DEX-induced changes in steady-state levels of SP-B mRNA in various cells that lack or possess expression of the glucocorticoid receptor (GR). A: detection of GR mRNA in the cell lines. RT-PCR analysis was performed on RNA isolated from A549, human epithelial kidney (HEK)293, ECV304, and HeLa cells as described in materials and methods. The size of the expected 825-bp band is indicated by the arrow. RT control indicates reaction performed without reverse transcriptase. B: effect of DEX on steady-state levels of SP-B mRNA in various cells. A549, HEK293, ECV304 and HeLa cells were transfected with pCMVGFP-hspB:N and incubated in the absence or presence of DEX (10−7 M). Cells that express GR are indicated by +, whereas cells that lack expression are indicated by -. RNA was isolated, subjected to Northern analysis for the presence of SP-B and GFP mRNA, and the SP-B mRNA/GFP mRNA ratio was determined in each sample. The average SP-B mRNA/GFP mRNA ratio in untreated samples was set as 1; the ratio from DEX-treated samples was normalized to this average. Shown are normalized SP-B levels in DEX-treated samples relative to levels in untreated samples (means ± SE; N ≥ 7, 2 independent experiments; *P < 0.01 relative to untreated controls).
The presence of the potent GR antagonist RU486 does not reduce DEX-induced stabilization of SP-B mRNA.
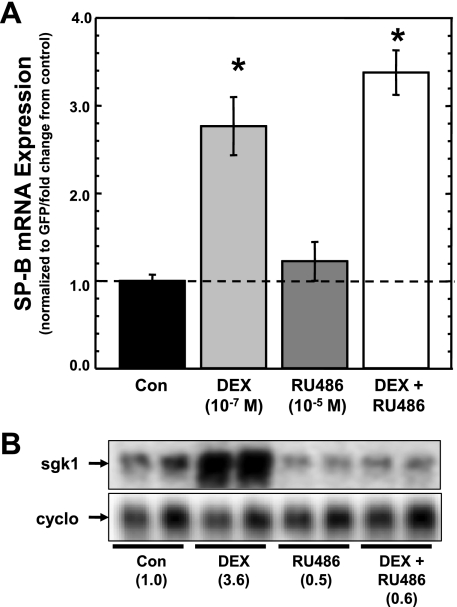
Whereas all types of glucocorticoids act as agonists of GR activity, mifepristone (RU486) is recognized as a potent antagonist of GR activity (5). These assays were undertaken to unambiguously show that GR is not involved in DEX-induced stabilization of SP-B mRNA. A549 cells were transfected with pCMVGFP-hspB:N and incubated in the absence or presence of DEX (10−7 M) and in the absence or presence of RU486 (10−5 M) for 36 h, and steady-state levels of SP-B and GFP mRNA in the cells determined as described in materials and methods. Because RU486 is 100× more concentrated than DEX, RU486 should compete the agonist. The SP-B mRNA/GFP mRNA ratio in the treated cells was determined relative to the SP-B mRNA/GFP mRNA ratio in untreated cells, and the results are shown in Fig. 6A. As can be seen, DEX alone elicited an increase is steady-state levels of SP-B mRNA, whereas the presence of RU486 alone had no effect. However, the steady-state levels of SP-B mRNA in cells incubated with the agents in combination were not significantly different from levels in cells incubated with DEX alone, and the levels were significantly higher than in untreated cells. On the other hand, as seen in Fig. 6B, RU486 was able to suppress the expression of the GR-responsive sgk1 gene in these cells, as reported previously for A549 cells (22). These results indicate that activated GR is not necessary for DEX-induced stabilization of SP-B mRNA.
Fig. 6.
DEX-induced changes in steady-state levels of SP-B mRNA in the presence of RU486. A: A549 cells were transfected with pCMVGFP-hspB:N and incubated in the absence or presence of indicated concentrations of DEX and/or the GR antagonist RU486 for 36 h. RNA was isolated subjected to Northern analysis. The levels of SP-B and GFP were quantified, and the SP-B mRNA/GFP mRNA ratio was determined in each sample. The average ratio in untreated samples was set as 1; the ratio from treated samples was normalized to this average. Shown are normalized SP-B levels in DEX-treated samples relative to levels in untreated samples (means ± SE; N ≥ 7, 2 independent experiments; *P < 0.02 relative to untreated controls). B: analysis of the GR-responsive human serum/glucocorticoid regulated kinase 1 (sgk1) gene in A549 cells in the absence or presence of DEX and/or RU486. Northern analysis of RNA from the cells used in A was performed for detection of sgk1 and cyclophilin (cyclo) RNA. Shown is a typical image of the analysis generated by phosphorimaging. Numbers in parenthesis indicate the fold change from control of the ratio of sgk1/cyclophilin signal.
DISCUSSION
In these investigations, we sought to describe and understand the molecular mechanisms involved in DEX-induced stabilization of SP-B mRNA. To accomplish this goal, a reproducible, easily assayable method in which steady-state levels of SP-B mRNA reflect changes in mRNA stability was developed and employed. This assay method was found to recapitulate the change in SP-B mRNA stability induced by DEX, and that stabilization requires elements of the SP-B mRNA 3′-UTR as described in previous reports (20, 50). It was also determined that large segments of the SP-B mRNA 3′-UTR necessary for hormonal regulation of SP-B mRNA stability also act as stabilization or destabilization elements of intrinsic SP-B stability. The stabilization of SP-B mRNA by DEX is dose dependent and does not require new protein synthesis. The DEX dose-response curve, although biphasic, does not resemble the typical sigmoidal curve expected for responses mediated by hormone receptors, with a peak in activity at 10−7 M DEX. Because DEX-induced stabilization of SP-B mRNA is a phenomenon not specific to lung epithelial cells, the direct involvement of GR in the response was tested in cells that do not express GR, such as kidney-derived HEK293 cells. In these cells, DEX was able to significantly increase SP-B mRNA stability. Additionally, the presence of excess GR antagonist (RU486) does not affect the ability of DEX to stabilize SP-B mRNA. Also, the presence of other steroid hormones (10−7 M) significantly increases SP-B mRNA stability. These findings indicate that DEX-activated GR is not required in DEX regulation of SP-B mRNA stability and suggest that hormones act through a common pathway that involves the SP-B mRNA 3′-UTR to alter SP-B mRNA stability.
Regulation of SP-B mRNA stability by DEX is not restricted to human tissue; rabbit lung tissue treated with DEX resulted in a 2.5-fold increase in SP-B mRNA stability (29). An increase in SP-B mRNA stability by DEX was reported in a mouse-lung epithelial cell line when added in concert with keratinocyte growth factor (32). On the other hand, we have reported that DEX alone does not increase the stability of SP-B mRNA expressed from a mouse type II epithelial cell line (MLE12) (20). In regard to other hormones, their effects on SP-B mRNA stability varies; insulin has no effect on human SP-B mRNA stability (30), whereas retinoic acid increases human SP-B mRNA stability (12). The fact that retinoic acid increases SP-B mRNA stability is in agreement with the findings reported here; hormones other than the glucocorticoids increase SP-B mRNA stability. However, the mechanisms of posttranscriptional regulation of human SP-B mRNA stability by glucocorticoids (or other factors) are still unknown.
The importance of posttranscriptional mechanisms has become recognized as a major point of regulation because the levels of expression of a particular protein depend on the levels of its mRNA. Differential regulation of mRNA stability and mRNA turnover is primarily determined by interactions between specific sequences within mRNA (cis-acting elements) and cellular RNA-binding proteins (trans-acting factors) modulating nuclear export, stabilization, and translation of transcripts, as well as ribonuclease degradation of mRNA (19). The mechanisms involved in these interactions are varied and complex (54), but the fate of an mRNA depends on its rate of degradation. Nucleotide patterns or motifs located in 3′-UTRs can interact with specific RNA-binding proteins, but the biological activity of regulatory motifs at the RNA level relies on a combination of primary and secondary structure (31), leading to mRNA stabilization or degradation.
Regulation of mRNA stability is an important control point for the action of steroid hormones in a variety of biological schemes, and there are several mechanisms that are presently studied to understand this emerging area (33). Perhaps the best studied is the estrogen-mediated stabilization of vitellogenin mRNA in Xenopus liver. Activation of the estrogen receptor by estrogen induces a protein that binds to a particular segment of the 3′-UTR of the vitellogenin mRNA containing the sequence ACUGUA, increasing its stability 30-fold (9). The binding of this protein, vigilin, prevents site-specific endonucleolytic cleavage of the mRNA. A review concerning the role of steroid hormones and posttranscriptional regulation of mRNA stability indicates that, in general, glucocorticoids destabilize mRNAs, except for the mRNAs for fatty-acid synthase and growth hormone (21). It seems that the role of glucocorticoids is more closely associated with the destabilization of the mRNAs for inflammatory response proteins (25). Although these studies described here indicate that hormonal regulation of SP-B mRNA stability does not require activated GR, this study does not allow delineation of the mechanism(s) involved. It is interesting to note that the regions of the SP-B mRNA 3′-UTR involved DEX-mediated regulation of stability are also involved in regulation of intrinsic mRNA stability. Many RNA-binding proteins involved in the cytoplasmic posttranscriptional regulation of gene expression also participate in other regulatory processes within the nucleus (57), and the connection between posttranscriptional events in the nucleus and in the cytoplasm can affect its cytoplasmic fate (24). Clearly, hormonal control of mRNA stability is complex, and the mechanisms and components involved vary for each mRNA.
The results of our investigations with cyclohexamide indicate that the mechanism by which SP-B mRNA is stabilized by DEX does not require protein synthesis. In addition, the results using a GR antagonist (RU486) suggest a mechanism of stabilization of SP-B mRNA by glucocorticoids that is not mediated via the classical genomic mechanism of activation of the GR. These findings suggest nongenomic effects of glucocorticoids on GR activity or posttranslational modification of proteins that may be involved in the stabilization. Increasing evidence suggests that glucocorticoids can affect cell function through nongenomic mechanisms, which are insensitive to protein synthesis, and/or transcription and/or receptor dissociation blockade (16). These effects are exerted by actions of the steroids on membrane lipids (which can alter membrane fluidity), membrane proteins such as ion channels, and cytoplasmic proteins, such as kinases and phospholipases (44, 46). These actions can be mediated directly by glucocorticoids or by the proteins dissociated from the liganded GR complex. These mechanisms are not restricted to glucocorticoids, as other steroid hormones mediate rapid changes of regulation of cellular processes that do not involve classical steroid receptor activation of transcription.
Steroid hormones are known to mediate rapid cellular responses involving signal transduction cascades, and insensitivity to steroid antagonists is a characteristic of rapid, nongenomic regulation of signal pathways by steroids (6). These rapid effects are varied, ranging from activation of MAPKs, adenylyl cyclase, PKC, and heterotrimeric guanosine triphosphate-binding proteins (G proteins). Potential targets for these effects are G protein-coupled receptors because they activate several signal transduction pathways. Glucocorticoids are known to affect p38, JNK, and ERK1/2 activation, and these changes occur in a short time span (26, 38). With regard to the findings described here, these kinases are known to regulate mammalian mRNA stability (39). Although the response time of SP-B mRNA stabilization by DEX does not appear to be rapid (or rapid changes cannot be determined by our assay), the characteristics of the effect of DEX to increase SP-B mRNA stability suggest a posttranscriptional mechanism. Further investigation into this phenomenon should include the use of specific inhibitors of the signaling pathways that may be involved in regulation of mRNA stability.
There are two important caveats to consider upon interpretation of the results of this study. Although the results of these investigations using cell lines that do not express GR indicate that the receptor may not be directly involved in the mechanism of DEX to increase SP-B mRNA stability, other results of these investigations do strongly suggest commonalities with previous reports that indicated GR involvement. The DEX dose response and potency of other hormone steroids (10−7 M) in mediating changes in SP-B expression levels reported here are, in general, consistent with GR mediation (41, 42). Biphasic dose responses have been described for human SP-B and SP-A mRNA expression in cells incubated in the presence of steroid hormones. In the case of DEX, the complexity of these responses cannot be ignored, as expression is a composite of transcriptional activation of the promoters by GR as well as the effect of the hormone on mRNA stability (3, 34, 50). The findings of this study do not suggest that GR has no role in the regulation of SP-B mRNA expression in response to DEX. The other caveat to consider is the artificial nature of the assay. Although A459 cells are human alveolar type II in origin, differences in the physiology of cells after prolonged passage are well documented. Such changes may alter regulatory mechanisms such that the present data relate only to A549 cells and may not necessarily apply to primary lung cells. However, until the technology to allow facile manipulation of lung epithelial cells in primary culture is available, lung cell lines do provide an avenue to address mechanistic questions.
The goal of this study is to define the complex molecular mechanisms by which SP-B mRNA stability is regulated by glucocorticoids. Posttranscriptional regulation of mRNA is recognized as a major mechanism for modulating levels of cellular proteins. Premature infants with inadequate surfactant production are predisposed to develop RDS, and glucocorticoids are used clinically in treatment of these infants. It is important to understand the molecular mechanism(s) by which glucocorticoids act. Ultimately, understanding these mechanisms may lead to a common motif of the regulation of surfactant protein gene expression, a motif that can be exploited clinically and applied to general regulation of eukaryotic gene expression.
GRANTS
This research was supported in part by the Richard W. Mithoff Professorship Research Funds at the Neonatal Division of the Department of Pediatrics at the University of Texas Medical School at Houston and by NIH-NHLBI (R01-068116) to J. Alcorn.
DISCLOSURES
No conflicts of interest, financial or otherwise are declared by the authors.
REFERENCES
- 1. Avery ME, Mead J. Surface properties in relation to atelectasis and hyaline membrane disease. Am J Dis Child 97: 517–523, 1959 [DOI] [PubMed] [Google Scholar]
- 2. Baliga BS, Pronczuk AW, Munro HN. Mechanism of cycloheximide inhibition of protein synthesis in a cell-free system prepared from rat liver. J Biol Chem 244: 4480–4489, 1969 [PubMed] [Google Scholar]
- 3. Ballard PL, Ertsey R, Gonzales LW, Gonzales J. Transcriptional regulation of human pulmonary surfactant proteins SP-B and SP-C by glucocorticoids. Am J Respir Cell Mol Biol 14: 599–607, 1996 [DOI] [PubMed] [Google Scholar]
- 4. Boggaram V, Qing K, Mendelson CR. The major apoprotein of rabbit pulmonary surfactant. Elucidation of primary sequence and cyclic AMP and developmental regulation. J Biol Chem 263: 2939–2947, 1988 [PubMed] [Google Scholar]
- 5. Cadepond F, Ulmann A, Baulieu EE. RU486 (mifepristone): mechanisms of action and clinical uses. Annu Rev Med 48: 129–156, 1997 [DOI] [PubMed] [Google Scholar]
- 6. Cato AC, Nestl A, Mink S. Rapid actions of steroid receptors in cellular signaling pathways. Sci STKE 2002: re9, 2002 [DOI] [PubMed] [Google Scholar]
- 7. Clements JA, King RJ. Composition of surface active material. In: The Biochemical Basis of Pulmonary Function, edited by Crystal RG. New York: Marcel Dekker, 1976, p. 363–387 [Google Scholar]
- 8. Curtis PJ. Globin mRNA in Friend cells: its structure, function and synthesis. Biochim Biophys Acta 605: 347–364, 1980 [DOI] [PubMed] [Google Scholar]
- 9. Dodson RE, Shapiro DJ. Vigilin, a ubiquitous protein with 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region-binding protein. J Biol Chem 272: 12249–12252, 1997 [DOI] [PubMed] [Google Scholar]
- 10. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114, 2008 [DOI] [PubMed] [Google Scholar]
- 11. Fruchter O, Kino T, Zoumakis E, Alesci S, De Martino M, Chrousos G, Hochberg Z. The human glucocorticoid receptor (GR) isoform [beta] differentially suppresses GR[alpha]-induced transactivation stimulated by synthetic glucocorticoids. J Clin Endocrinol Metab 90: 3505–3509, 2005 [DOI] [PubMed] [Google Scholar]
- 12. George TN, Miakotina OL, Goss KL, Snyder JM. Mechanism of all trans-retinoic acid and glucocorticoid regulation of surfactant protein mRNA. Am J Physiol Lung Cell Mol Physiol 274: L560–L566, 1998 [DOI] [PubMed] [Google Scholar]
- 13. Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582: 1977–1986, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grier DG, Halliday HL. Effects of glucocorticoids on fetal and neonatal lung development. Treat Respir Med 3: 295–306, 2004 [DOI] [PubMed] [Google Scholar]
- 15. Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene 265: 11–23, 2001 [DOI] [PubMed] [Google Scholar]
- 16. Haller J, Mikics E, Makara GB. The effects of non-genomic glucocorticoid mechanisms on bodily functions and the central neural system. A critical evaluation of findings. Front Neuroendocrinol 29: 273–291, 2008 [DOI] [PubMed] [Google Scholar]
- 17. Hawgood S. Surfactant protein B: structure and function. Biol Neonate 85: 285–289, 2004 [DOI] [PubMed] [Google Scholar]
- 18. He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA 95: 2509–2514, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hollams EM, Giles KM, Thomson AM, Leedman PJ. MRNA stability and the control of gene expression: implications for human disease. Neurochem Res 27: 957–980, 2002 [DOI] [PubMed] [Google Scholar]
- 20. Huang HW, Bi W, Jenkins GN, Alcorn JL. Glucocorticoid regulation of human pulmonary surfactant protein-B mRNA stability involves the 3′-untranslated region. Am J Respir Cell Mol Biol 38: 473–482, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ing NH. Steroid hormones regulate gene expression posttranscriptionally by altering the stabilities of messenger RNAs. Biol Reprod 72: 1290–1296, 2005 [DOI] [PubMed] [Google Scholar]
- 22. Itani OA, Liu KZ, Cornish KL, Campbell JR, Thomas CP. Glucocorticoids stimulate human sgk1 gene expression by activation of a GRE in its 5′-flanking region. Am J Physiol Endocrinol Metab 283: E971–E979, 2002 [DOI] [PubMed] [Google Scholar]
- 23. Johansson J, Curstedt T. Molecular structures and interactions of pulmonary surfactant components. Eur J Biochem 244: 675–693, 1997 [DOI] [PubMed] [Google Scholar]
- 24. Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell 6: 673–682, 2000 [DOI] [PubMed] [Google Scholar]
- 25. Kracht M, Saklatvala J. Transcriptional and post-transcriptional control of gene expression in inflammation. Cytokine 20: 91–106, 2002 [DOI] [PubMed] [Google Scholar]
- 26. Li X, Qiu J, Wang J, Zhong Y, Zhu J, Chen Y. Corticosterone-induced rapid phosphorylation of p38 and JNK mitogen-activated protein kinases in PC12 cells. FEBS Lett 492: 210–214, 2001 [DOI] [PubMed] [Google Scholar]
- 27. Lieber M, Smith B, Szakal A, Nelson-Rees W, Todaro G. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer 17: 62–70, 1976 [DOI] [PubMed] [Google Scholar]
- 28. Liley HG, White RT, Warr RG, Benson BJ, Hawgood S, Ballard PL. Regulation of messenger RNAs for the hydrophobic surfactant proteins in human lung. J Clin Invest 83: 1191–1197, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Margana RK, Boggaram V. Transcription and mRNA stability regulate developmental and hormonal expression of rabbit surfactant protein B gene. Am J Physiol Lung Cell Mol Physiol 268: L481–L490, 1995 [DOI] [PubMed] [Google Scholar]
- 30. Miakotina OL, Dekowski SA, Snyder JM. Insulin inhibits surfactant protein A and B gene expression in the H441 cell line. Biochim Biophys Acta 1442: 60–70, 1998 [DOI] [PubMed] [Google Scholar]
- 31. Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol 3: REVIEWS0004, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mouhieddine-Gueddiche OB, Pinteur C, Chailley-Heu B, Barlier-Mur AM, Clement A, Bourbon JR. Dexamethasone potentiates keratinocyte growth factor-stimulated SP-A and SP-B gene expression in alveolar epithelial cells. Pediatr Res 53: 231–239, 2003 [DOI] [PubMed] [Google Scholar]
- 33. Nielsen DA, Shapiro DJ. Insights into hormonal control of messenger RNA stability. Mol Endocrinol 4: 953–957, 1990 [DOI] [PubMed] [Google Scholar]
- 34. O′Reilly MA, Clark JC, Whitsett JA. Glucocorticoid enhances pulmonary surfactant protein B gene transcription. Am J Physiol Lung Cell Mol Physiol 260: L37–L43, 1991 [DOI] [PubMed] [Google Scholar]
- 35. Orgeig S, Hiemstra PS, Veldhuizen EJ, Casals C, Clark HW, Haczku A, Knudsen L, Possmayer F. Recent advances in alveolar biology: evolution and function of alveolar proteins. Respir Physiol Neurobiol 173, Suppl: S43–S54, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pryhuber GS, Hull WM, Fink I, McMahan MJ, Whitsett JA. Ontogeny of surfactant proteins A and B in human amniotic fluid as indices of fetal lung maturity. Pediatr Res 30: 597–605, 1991 [DOI] [PubMed] [Google Scholar]
- 37. Pujols L, Mullol J, Perez M, Roca-Ferrer J, Juan M, Xaubet A, Cidlowski JA, Picado C. Expression of the human glucocorticoid receptor alpha and beta isoforms in human respiratory epithelial cells and their regulation by dexamethasone. Am J Respir Cell Mol Biol 24: 49–57, 2001 [DOI] [PubMed] [Google Scholar]
- 38. Qiu J, Wang P, Jing Q, Zhang W, Li X, Zhong Y, Sun G, Pei G, Chen Y. Rapid activation of ERK1/2 mitogen-activated protein kinase by corticosterone in PC12 cells. Biochem Biophys Res Commun 287: 1017–1024, 2001 [DOI] [PubMed] [Google Scholar]
- 39. Rodriguez-Gabriel MA, Russell P. Control of mRNA stability by SAPKs. In: Topics in Current Genetics, edited by Posas F, Nebreda AR. Berlin, Germany: Springer-Verlag, 2008, p. 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rooney SA, Young SL, Mendelson CR. Molecular and cellular processing of lung surfactant. FASEB J 8: 957–967, 1994 [DOI] [PubMed] [Google Scholar]
- 41. Rousseau GG. Structure and regulation of the glucocorticoid hormone receptor. Mol Cell Endocrinol 38: 1–11, 1984 [DOI] [PubMed] [Google Scholar]
- 42. Rousseau GG, Baxter JD, Tomkins GM. Glucocorticoid receptors: relations between steroid binding and biological effects. J Mol Biol 67: 99–115, 1972 [DOI] [PubMed] [Google Scholar]
- 43. Serrano AG, Perez-Gil J. Protein-lipid interactions and surface activity in the pulmonary surfactant system. Chem Phys Lipids 141: 105–118, 2006 [DOI] [PubMed] [Google Scholar]
- 44. Stahn C, Lowenberg M, Hommes DW, Buttgereit F. Molecular mechanisms of glucocorticoid action and selective glucocorticoid receptor agonists. Mol Cell Endocrinol 275: 71–78, 2007 [DOI] [PubMed] [Google Scholar]
- 45. Staton JM, Thomson AM, Leedman PJ. Hormonal regulation of mRNA stability and RNA-protein interactions in the pituitary. J Mol Endocrinol 25: 17–34, 2000 [DOI] [PubMed] [Google Scholar]
- 46. Stellato C. Glucocorticoid actions on airway epithelial responses in immunity: functional outcomes and molecular targets. J Allergy Clin Immunol 120: 1247–1263; quiz 1264–1245, 2007 [DOI] [PubMed] [Google Scholar]
- 47. Sun H, Xu B, Inoue H, Chen QM. P38 MAPK mediates COX-2 gene expression by corticosterone in cardiomyocytes. Cell Signal 20: 1952–1959, 2008 [DOI] [PubMed] [Google Scholar]
- 48. Takahashi K, Sawasaki Y, Hata J, Mukai K, Goto T. Spontaneous transformation and immortalization of human endothelial cells. In Vitro Cell Dev Biol 26: 265–274, 1990 [DOI] [PubMed] [Google Scholar]
- 49. Thompson EB, Tomkins GM, Curran JF. Induction of tyrosine alpha-ketoglutarate transaminase by steroid hormones in a newly established tissue culture cell line. Proc Natl Acad Sci USA 56: 296–303, 1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Venkatesh VC, Iannuzzi DM, Ertsey R, Ballard PL. Differential glucocorticoid regulation of the pulmonary hydrophobic surfactant proteins SP-B and SP-C. Am J Respir Cell Mol Biol 8: 222–228, 1993 [DOI] [PubMed] [Google Scholar]
- 51. Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C, Yamamoto KR. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci USA 101: 15603–15608, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wheller SK, Perretti M. Dexamethasone inhibits cytokine-induced intercellular adhesion molecule-1 up-regulation on endothelial cell lines. Eur J Pharmacol 331: 65–71, 1997 [DOI] [PubMed] [Google Scholar]
- 53. Whitsett JA, Weaver TE, Clark JC, Sawtell N, Glasser SW, Korfhagen TR, Hull WM. Glucocorticoid enhances surfactant proteolipid Phe and pVal synthesis and RNA in fetal lung. J Biol Chem 262: 15618–15623, 1987 [PubMed] [Google Scholar]
- 54. Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet 20: 491–497, 2004 [DOI] [PubMed] [Google Scholar]
- 55. Wohlford-Lenane CL, Snyder JM. Localization of surfactant-associated proteins SP-A and SP-B mRNA in rabbit fetal lung tissue by in situ hybridization. Am J Respir Cell Mol Biol 7: 335–343, 1992 [DOI] [PubMed] [Google Scholar]
- 56. Wright JR. Immunomodulatory functions of surfactant. Physiol Rev 77: 931–962, 1997 [DOI] [PubMed] [Google Scholar]
- 57. Xu N, Chen CY, Shyu AB. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol Cell Biol 21: 6960–6971, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu N, Loflin P, Chen CY, Shyu AB. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res 26: 558–565, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]