Abstract
Diurnal-nocturnal, or circadian-like, rhythms are 24-h variations in biological processes, evolved for the efficient functioning of living organisms. Such oscillations and their regulation in many peripheral tissues are still unclear. In this study, we used Affymetrix gene chips in a rich time-series experiment involving 54 animals killed at 18 time points within the 24-h cycle to examine light-dark cycle patterns of gene expression in rat lungs. Data mining identified 646 genes (represented by 1,006 probe sets) showing robust oscillations in expression in lung that were parsed into 8 distinct temporal clusters. Surprisingly, more than two-thirds of the probe sets showing cyclic expression peaked during the animal's light/inactive period. Six core clock genes and nine clock-related genes showed rhythmic oscillations in their expression in lung. Many of the genes that peaked during the inactive period included genes related to extracellular matrix, cytoskeleton, and protein processing and trafficking, which appear to be mainly involved in the repair and remodeling of the organ. Genes coding for growth factor ligands and their receptors, which play important roles in maintaining normal lung function, also showed rhythmic expression. In addition, genes involved in the metabolism and transport of endogenous compounds, xenobiotics, and therapeutic drugs, along with genes that are biomarkers or potential therapeutic targets for many lung diseases, also exhibited 24-h cyclic oscillations, suggesting an important role for such rhythms in regulating various aspects of the physiology and pathophysiology of lung.
Keywords: circadian rhythms, microarrays, growth factors, drug metabolism, lung repair
many behavioral, physiological, cellular, and molecular processes exhibit 24-h cyclic variations, often cued by the light-dark cycle. Daily cyclic processes are directly or indirectly controlled by the circadian clock, an internal time-keeping system that has evolved in order for the organism to adapt by efficiently anticipating and responding to changes, thereby maximizing survival of the organism. Circadian rhythms are 24-h variations in any process in an organism controlled by an endogenous clock (44, 68). These rhythms exist even without an external stimulus but are entrained and synchronized to environmental factors, such as the presence or absence of light and food availability. In mammals, the central or master clock is present in the paired suprachiasmatic nuclei in the anterior part of the hypothalamus and receives direct input through the retinohypothalamic tract, which synchronizes the endogenous clock to the environmental light-dark cycle (13, 44). The peripheral clocks are the circadian oscillators in other parts of the brain and other tissues and are, at least in part, controlled, coordinated, and synchronized by the central clock (5, 58).
It is accepted that the molecular mechanism controlling the central clock entails a transcriptional and translational autoregulatory feedback loop involving molecular clock proteins. The Per-Arnt/AhR-Sim (PAS)-basic helix-loop-helix (bHLH) transcription factor brain and muscle Arnt-like protein 1 (BMAL1) heterodimerizes with CLOCK or neuronal PAS domain protein 2 (NPAS2) and drives the expression of Per and Cry genes by binding to the E-box element present in the promoter of these genes (14, 37). The Per and Cry transcription factors, in turn, negatively feed back on the transcriptional activity of the BMAL1:CLOCK/BMAL1:NPAS2 complex. Apart from these core clock transcription factors, other transcription factors, including REV-ERBs (NR1Ds), retinoic acid-related orphan receptors (RORs), PAR b-ZIP, and class B bHLHs (bHLHBs; DECs), are also involved in regulating various aspects of the molecular clock and the circadian oscillations in the expression of the transcriptome (18, 25, 43).
Apart from breathing and respiration, lung plays an important role in many other processes, including the innate immune response against airborne foreign materials and pathogens and metabolism of endogenous and exogenous compounds, and also as a paraendocrine organ producing a wide variety of signaling molecules, including growth factors and cytokines (6, 17, 23, 45). Owing to the structural complexity of the organ, lung is made up of various cell types, including type 1 and type 2 pneumocytes, Clara cells, immune cells (predominantly alveolar macrophages, mast cells, and dendritic cells), fibroblasts, airway smooth muscle cells, vasculature, and nerve cells (7). In addition to these cellular components, lung interstitium is made up of extracellular matrix components that not only provide a structural support and physical barrier but are also involved in determining cell growth, morphology, migration, and functions (15). All these cellular and extracellular components continuously interact to allow a dynamic regulation of various functions, and any disruption of these interactions can lead to disease. Some common lung pathologies, including asthma, chronic obstructive pulmonary disorder (COPD), acute lung injury (ALI), pulmonary hypertension, and cancer, involve disruption of the balance between the above-mentioned components by foreign materials, impaired immune activation, impaired cell cycle regulation, or other factors.
The nature of the processes carried out by lung and the expression of a large number of genes that regulate these processes suggest that many of these genes may be under direct or indirect control of circadian oscillators for the efficient functioning of the organ. Furthermore, disruption of this regulation could be a cause or an effect of lung pathologies. In addition, some symptoms of these pathologies also show rhythmic variations across the light-dark cycle that could be related to variations in the processes carried out by the organ (58). For example, it is a common observation that the symptoms of asthma, including shortness of breath, wheezing, and chest tightness, worsen during the late inactive period (late night in humans) (51, 58). In this study, we use Affymetrix gene chips to characterize the diurnal-nocturnal oscillations in gene expression in lungs of Wistar rats to understand the role of circadian oscillators in coordinating and controlling the functioning of lung. We also compared the data with data from our previous studies, where we characterized the 24-h cyclic oscillation in gene expression from liver, skeletal muscle, and white adipose tissue from this same set of animals, along with relevant physiological parameters, to understand the tissue-specific functions and variation in the circadian oscillators between these peripheral tissues (2, 3, 59).
MATERIALS AND METHODS
Animals.
An extensive description of the animal experiment can be found in our previously published reports (2, 59). Our research protocol adhered to the “Principles of Laboratory Animal Care” (National Institutes of Health Publication 85-23, revised 1985) and were approved by the State University of New York at Buffalo Institutional Animal Care and Use Committee. Briefly, 54 normal male Wistar rats purchased in 2 batches of 27 animals each from Harlan Laboratories (Indianapolis, IN) were allowed to acclimatize in a constant-temperature environment (22°C) and a 12:12-h light-dark cycle with free access to standard rat chow and drinking water. Animals were anesthetized with a mixture of ketamine and xylazine (80 and 10 mg/kg, respectively) and killed by exsanguination on 3 successive days at 0.25, 1, 2, 4, 6, 8, 10, 11, and 11.75 h after lights on for time points in the light period and at 12.25, 13, 14, 16, 18, 20, 22, 23, and 23.75 h after lights on for time points in the dark period. Time points were designed with extra equal weight given to light-dark transition periods, as it is likely that more dynamic changes would occur during such transitions. Animals killed at the same time on the 3 successive days were treated as triplicate measurements. Blood was drawn from the abdominal aortic arteries into syringes that contained EDTA (4 mM final concentration) as anticoagulant. Plasma was prepared from blood by centrifugation (2,000 g, 4°C, 15 min), divided into aliquots, and stored at −80°C. Both lungs, along with trachea, were harvested, flash-frozen in liquid nitrogen, and stored at −80°C.
Microarrays.
A mortar and pestle cooled by liquid nitrogen were used to grind whole frozen lung samples into a fine powder, and 100 mg of ground tissue were added to 1 ml of QIAzol lysis reagent (Qiagen Sciences, Germantown, MD). Total RNAs were extracted according to the manufacturer's instructions and were further purified using RNeasy mini columns (RNeasy Mini Kit, Qiagen Sciences). Final RNA preparations were resuspended in RNase-free water and stored at −80°C. RNAs were quantified spectrophotometrically, and purity and integrity were assessed by agarose gel electrophoresis. All samples used for arrays exhibited 260-nm-to-280-nm absorbance ratios of ∼2.0, and all showed intact ribosomal 28S and 18S RNA bands in an approximate ratio of 2:1, as visualized by ethidium bromide staining. Isolated RNA from each sample was used to prepare the target according to the manufacturer's protocols. The biotinylated cRNAs were hybridized to 54 individual Affymetrix GeneChips Rat Genome 230_2 (Affymetrix, Santa Clara, CA). The 230_2 chips contain >31,000 different probe sets. The high reproducibility of in situ synthesis of oligonucleotide chips allows accurate comparison of signals generated by samples hybridized to separate arrays. This data set has been submitted to the Gene Expression Omnibus database (GSE25612).
Data set construction and mining.
Affymetrix Microarray Suite 5.0 was used for initial data acquisition and analysis. A distribution of all genes around the 50th percentile was used to normalize the signal intensities for each chip. Animals that were killed at the same time on different days were considered replicate measurements (n = 3) for that time and were used for construction of the time series. In addition, for better visualization and analysis of the data for the light-dark and dark-light transitions, two 24-h cycles were concatenated to form a 48-h time series. A nonlinear least-squares fitting of the replicate data was conducted using MATLAB (Mathworks, Natick, MA), which utilized a regular sinusoidal function [A × sin(B × t + C) + mean], where A, B, and C reflect the amplitude, period, and phase of the oscillations, t represents the circadian time, and mean is the average of intensities across the 48-h time series for the given probe set. Genes that could be curve-fitted with Pearson's correlation (R2) values >0.75 were kept for further analysis. This particular approach is only viable because of our rich data set. The data set was then loaded into the data-mining program GeneSpring 7.3.1 (Silicon Genetics, Redwood City, CA), where we normalized the value of each probe set on each chip to the median of that probe set on all chips. To identify genes that show similar expression patterns within the light-dark cycle, we used a quality threshold (QT) clustering algorithm in GeneSpring software, with R2 as the similarity measurement. One limitation of this procedure is that genes that do not follow the conventional sinusoidal pattern in expression but, rather, show a different rhythmic pattern will not be identified by this analysis. Another limitation is that such a stringent selection criterion (R2 > 0.75) will result in some false negatives, where genes that show a 24-h pattern might be missed.
Functional clustering.
Gene accession numbers corresponding to each of the probe sets showing rhythmic oscillations were analyzed using various online tools and databases, including National Center for Biotechnology Information Basic Local Alignment Search Tool and GeneCards to confirm the identity and the annotations of the probe sets provided by Affymetrix and to obtain alternate names and symbols for the genes corresponding to these probe sets. From this information, extensive literature searches were performed to identify the lung-specific functions and other relevant information for these probe sets. The use of currently available pathway analysis tools was avoided, as the databases for these tools are not complete (i.e., they do not represent all the functional genes identified) and do not take into account the tissue-specific physiological functions of the genes.
Quantitative real-time RT-PCR.
The quantities of lung glucocorticoid receptor and glutamine synthase mRNA, along with gene-specific in vitro-transcribed cRNA standards, were determined by real-time quantitative RT-PCR (qRT-PCR) using TaqMan-based probes. Primer and probe sequences were designed using PrimerExpress software (Applied Biosystems, Foster City, CA) and custom-synthesized by Biosearch Technologies (Novato, CA). The qRT-PCR was performed using Brilliant qRT-PCR Core Reagent Kit, 1-Step (Stratagene, La Jolla, CA) in a Stratagene MX3005P thermocycler according to the manufacturer's instructions. A standard curve was generated using in vitro-transcribed sense cRNA standards. Primer and probe sequences are as follows: 5′ AACATGTTAGGTGGGCGTCAA 3′ (forward primer), 5′ GGTGTAAGTTTCTCAAGCCTAGTATCG 3′ (reverse primer), and 5′ TGATTGCAGCAGTGAAATGGGCAAAG 3′ (FAM-labeled probe) for glucocorticoid receptor; 5′ CGCCCGCCGTCTGA 3′ (forward primer), 5′ TCTCCTGGCCGACAATCC 3′ (reverse primer), and 5′ TCCACGAAACCTCCAACATCAACGACTTT 3′ (FAM-labeled probe) for glutamine synthase. Samples were run in triplicate and standards in duplicate. Additional minus-RT controls were run for each RNA sample analyzed to check for genomic DNA contamination; all controls exhibited lack of amplification in minus-RT controls. Intra- and interassay coefficients of variation were <15%.
RESULTS
Data mining and QT clustering of the data.
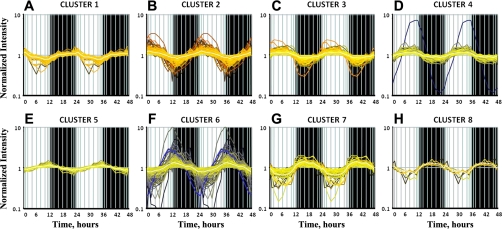
The sinusoidal model applied to the data set identified 1,006 probe sets showing cyclic oscillations that satisfied the selection criterion of R2 > 0.75, as described in our previous publication (59). The values of the parameters of the sine function that were estimated for all 1,006 probe sets are given in Supplemental Table S1 (see Supplemental Material for this article, available online at the Journal website). Parameter B, which is a measure of periodicity of the rhythmic oscillations, was estimated to be ∼0.260 (2π/24) for all these probe sets, which further confirms that the oscillations follow a circadian pattern. The richness of the time-series data set allowed us to apply a QT algorithm to cluster the probe sets based on similarities in the expression patterns within the light-dark cycle. This procedure yielded eight distinct temporal clusters, with the probe sets in each of these clusters showing R2 ≥ 0.75 with the centroid curve of its assigned cluster. As shown in Fig. 1 and Table 1, 50 probe sets (5%) peaked during the dark-light transition (cluster 1). What is very interesting and strikingly different from the other peripheral tissues is that 652 probe sets (65%) peaked during the light period (clusters 2, 3, and 4), which is the inactive and nonfeeding period in rodents. Twenty-seven probe sets (3%) peaked during the light-dark transition (cluster 5). Two hundred sixty-one probe sets (26%) peaked during the dark period (clusters 6, 7, and 8); of these, 201 probe sets peaked during the very early dark period (cluster 6).
Fig. 1.
Quality threshold (QT) clustering of genes showing circadian-like oscillations in expression in lung. For each probe set, Pearson's correlation (R2) >0.75; centroid curve of the cluster (average curve) is shown in white. Shaded areas indicate dark periods; unshaded areas indicate light periods.
Table 1.
Clustering of rhythmic oscillations in expression in lung
| Cluster No. | No. of Probe Sets | Acrophase, h |
|---|---|---|
| 1 | 50 | 0 |
| 2 | 201 | 2 |
| 3 | 304 | 5 |
| 4 | 147 | 8 |
| 5 | 27 | 11 |
| 6 | 201 | 14 |
| 7 | 53 | 17 |
| 8 | 7 | 19 |
Expression of clock and clock-related genes.
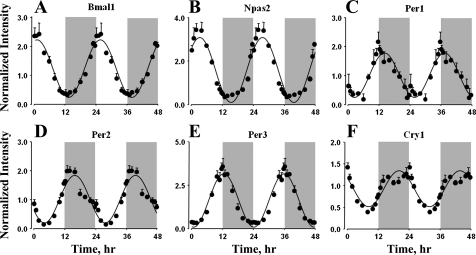
All the core clock genes that showed rhythmic oscillations in other peripheral tissues from the same animals also showed similar oscillations in lungs (Fig. 2). The PAS-bHLH transcription factors Bmal1 (Arntl) and Npas2 showed maximal expression during the early light period, with peaks at zeitgeber time (ZT) 1 and ZT 3. The mRNA expression of Period genes (Per1, Per2, and Per3), which are transcriptionally controlled by the BMAL1:CLOCK/BMAL1:NPAS2 transcription factor complex, peaked during the early dark period, with maxima at ZT 14, ZT 16, and ZT 13, respectively. The expression of the other core clock gene, Cry1, peaked during the late dark period at ZT 20. Consistent with other peripheral tissues, signal intensities for Clock and Cry2 genes were very low, which might have prevented the identification of oscillations in the mRNA expression of these genes.
Fig. 2.
Expression patterns of core clock transcription factors in lung as a function of circadian time. ●, Mean data; error bars, SD; solid lines, curves fitted to a sine function.
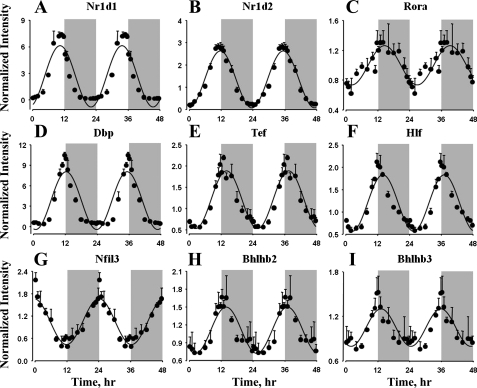
Many of the clock-related and clock-controlled transcription factors also showed circadian-like oscillations in gene expression in lung (Fig. 3). The orphan nuclear receptors Nr1d1 (Rev-Erbα) and Nr1d2 (Rev-Erbβ) peaked during the late light period and the light-dark transition, with peaks at ZT 11 and ZT 12. In contrast to other peripheral tissues from these animals, the nuclear receptor RORα did show circadian-like oscillations in lung, with peak expression during the early dark period at ZT 14. The clock-controlled PAR b-Zip transcription factors, including Dbp, Tef, and Hlf, peaked during the early dark period, with maximum expression at ZT 13 for Dbp and ZT 14 for Tef and Hlf. However, the mRNA expression of Nfil3, which is a repressor of genes containing D-box-binding elements, showed opposite oscillations, with a peak during the dark-light transition at ZT 0 (or ZT 24). Transcription repressor proteins Bhlhb2 and Bhlhb3 (Dec1 and Dec2) peaked during the early dark period at ZT 13. Furthermore, it is interesting to observe ∼30- and 60-fold differences between the peak and the nadir mRNA expression of Dbp and Nr1d1 in lung. Signal intensities for RORβ were very low, which might have prevented the identification of oscillations in its mRNA expression.
Fig. 3.
Expression patterns of clock-related and clock-controlled transcription factors in lung as a function of circadian time. ●, Mean data; error bars, SD; solid lines, curves fitted to a sine function.
Functional clustering of the data.
From the data mining and extensive literature searches, functions for 719 probe sets corresponding to 646 genes (as some of the genes contained multiple probe sets) were identified. These genes were further classified into 11 groups on the basis of the physiological and molecular function of the gene (see relevant information in Supplemental Table S2). Signal Transduction corresponds to the largest functional group, represented by 132 genes (153 probe sets). These include genes coding for proteins that act as ligands, receptors, or other components in signal transduction cascades involved in a variety of signaling pathways, including pathways involved in growth factor signaling, Wnt and Tgf pathways, angiogenesis, vasoconstriction and vasodilation, cell adhesion and migration, and lung development. Genes Involved in Regulating Transcription and Translation form the second-largest functional group, containing 104 genes (122 probe sets). This group includes genes that act as transcription factors, enhancers, activators, and repressors and genes that are involved in histone and chromatin modification, thereby regulating gene expression. All the core clock, clock-related, and clock-controlled transcription factors are members of this functional group. Genes that are involved in regulating translation are also grouped into this category. Metabolism forms the next-largest functional group, with 65 genes (69 probe sets) that are involved in the metabolism of energy substrates, endogenous compounds, and xenobiotics. Protein Processing contains 58 genes (61 probe sets). This group includes genes that are involved in posttranslational protein modifications, protein folding, protein degradation through ubiquitination, and also the endopeptidases and proteases. As the name suggests, Cell Cycle/Apoptosis consists of 41 genes (44 probe sets) that are involved in the regulation of cell cycle progression and apoptosis and, hence, regulate the turnover of different cell types in the organ. Extracellular Matrix and Cytoskeleton form two interesting functional groups, containing 40 genes (47 probe sets) and 20 genes (21 probe sets), respectively. Transport consists of 39 genes (41 probe sets) that play important roles in the transportation of a variety of endogenous and exogenous compounds, metabolites, and metal ions. The genes encoding for the members of the solute carrier family are included in this category. Vesicular Transport contains 32 genes (34 probe sets) that code for proteins that regulate and are involved in retrograde and anterograde trafficking and also the secretion of proteins. The mRNA Processing category includes 33 genes (35 probe sets) that are involved in posttranscriptional modification and turnover of mRNA in the cell. Immune Regulation forms the last functional group consisting of 26 genes (30 probe sets) that are involved in regulating natural immune responses and inflammation associated with the entry of foreign materials, which includes the soluble immune regulating factors, such as chemokine ligand family proteins.
Lung homeostasis and repair.
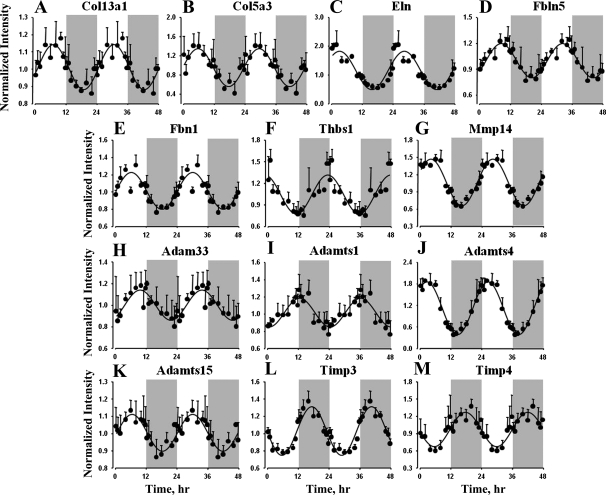
Data mining identified many genes coding for proteins that are involved in regulation, maintenance, and repair of the lung tissues, interstitium, and vasculature that showed oscillations in expression, and most of these genes peak during the light/inactive period. The genes that are directly related to these processes are primarily categorized into the following functional groups: Extracellular Matrix, Cytoskeleton, Protein Processing, Vesicular Transport, and Signal Transduction. Extracellular matrix forms an important part of the lung interstitium and vasculature and is important for maintenance and functioning (15). Of the 40 genes that are classified as extracellular matrix and associated proteins, 27 are extensively expressed in lung tissue and play an important role not only in maintaining homeostasis and repair but also in remodeling and development of the organ (47). Table 2 provides specific details and the functions of these genes with respect to lung physiology. Genes coding for extracellular matrix components, such as collagen family members (Col13a1 and Col5a1), elastin (Eln), fibulin 5 (Fbln5), fibrillin 1 (Fbn1), and thrombospondin 1 (Thbs1), showed circadian-like oscillations in expression (Table 2 and Fig. 4). It is also interesting that all these genes showed maximum expression during the light/inactive period, with the expression of Col13a1, Col5a1, Eln, and Fbn1 peaking in the middle of the light period (cluster 3), while the expression of Thbs1 peaked during the dark-light transition (cluster 1) and Fbln5 during the late light period (cluster 4). This list also includes the metallopeptidases that are involved in extracellular matrix turnover, including matrix metallopeptidase 14 (Mmp14), A disintegrin and metalloproteinase (ADAM) metallopeptidase 33 (Adam33), ADAM metallopeptidase with thrombospondin motif family members (Adamts1, Adamts4, and Adamts15), and its inhibitors, including tissue inhibitor of metalloproteinase (Timp3 and Timp4). Adamts4, Mmp14, and Adamts15 showed maximal expression during the light period (clusters 2 and 3), while Adamts1, Timp3, and Timp4 expression peaked during the dark period (clusters 6 and 7). Similar to the extracellular matrix, many cytoskeleton proteins, including microtubule-actin cross-linking factor 1 (Macf1), septin 4 (Sept4), vasodilator-stimulated phosphoprotein (Vasp), and keratin (Krt19 and Krt80), also showed circadian-like oscillations in their gene expression, with peak expression during the light period.
Table 2.
Extracellular matrix components and processing proteins showing circadian-like oscillations in gene expression
| Symbol | Cluster No. | Gene Name | Gene Function |
|---|---|---|---|
| Adamtsl2 | 1 | ADAMTS-like 2 | Glycoprotein; cell surface and ECM binding |
| Thbs1 | 1 | Thrombospondin 1 | Cell attachment, invasion; cell-ECM interactions |
| Adamts4 | 2 | ADAM metallopeptidase 4 | ECM protease; proteoglycan cleavage |
| Itgb6 | 2 | Integrin-β6 | Receptor: fibronectin and cytotactin |
| Selp | 2 | Selectin P | Receptor for carbohydrates in immune cells |
| Adamts15 | 3 | ADAM metallopeptidase 15 | ECM protease; interacts with integrins |
| Dag1 | 3 | Dystroglycan 1 | ECM receptor; CM-cytoskeleton interactions |
| Eln | 3 | Elastin | Structural component of elastic fibers |
| Emilin1 | 3 | Elastin microfibril interfacer 1 | Formation of elastic fiber |
| Fbn1 | 3 | Fibrillin 1 | Extracellular Ca2+ binding; structural support |
| Itga4 | 3 | Integrin-α4 | Receptor for fibronectin |
| Lox | 3 | Lysyl oxidase | Cross-linking of collagen and elastin |
| Mmp14 | 3 | Matrix metallopeptidase 14 | Breakdown of ECM components |
| Ndst1 | 3 | N-deacetylase/N-sulfotransferase 1 | Biosynthesis of heparan sulfate |
| Spon2 | 3 | Spondin 2 | Pattern recognition for microbes in ECM |
| Xpnpep2 | 3 | X-prolyl aminopeptidase 2 | Hydrolase specific for ECM degradation components |
| Col13a1 | 3 | Collagen, type XIII, α1 | Nonfibrillar collagen; branching morphogenesis of lung |
| Col5a3 | 3 | Collagen, type V, α3 | Interaction with heparan sulfate and thrombospondin |
| Fbln5 | 4 | Fibulin 5 | Interactions with integrins; vascular development |
| Gpc4 | 4 | Glypican 4 | Cell surface proteoglycan; heparan sulfate |
| Tnc | 4 | Tenascin C | Fiber-associated structural protein |
| Adam33 | 5 | ADAM metallopeptidase 33 | ECM protease-cell matrix interactions |
| Adamts1 | 6 | ADAM metallopeptidase 1 | ECM protease-proteoglycan cleavage |
| Itga6 | 6 | Integrin, α6 | Receptor for laminin |
| Mgp | 6 | Matrix Gla protein | Inhibitory morphogen; vascular branching |
| Timp3 | 7 | TIMP metallopeptidase inhibitor 3 | Inhibits MMP |
| Timp4 | 7 | TIMP metallopeptidase inhibitor 4 | Inhibits MMP |
ADAMS, A disintegrin and metalloproteinase with thrombospondin motifs; ECM, extracellular matrix; CM, calmodulin; MMP, matrix metalloproteinase; TIMP, tissue inhibitor of metalloproteinase.
Fig. 4.
Expression patterns of selected extracellular matrix components and processing proteins in lung as a function of circadian time. ●, Mean data; error bars, SD; solid lines, curves fitted to a sine function.
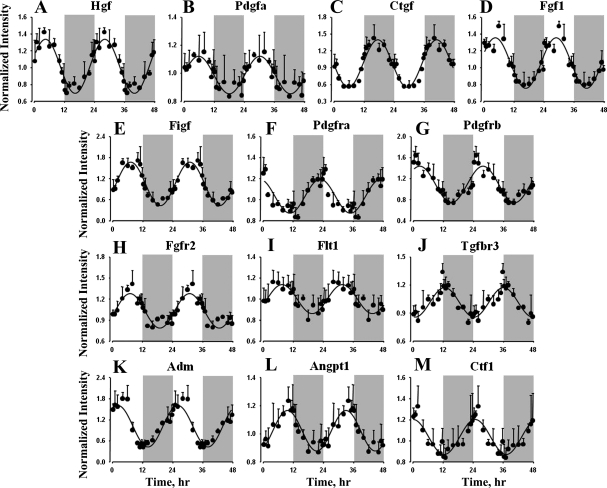
Signal Transduction, which forms the largest functional category, contains many genes coding for proteins that play important roles in regulating the processes involved in the maintenance, repair, and remodeling of the organ. Table 3 lists the 18 protein ligands and 25 cell surface receptors that show rhythmic oscillations in expression. This list includes growth factor family proteins, including hepatocyte growth factor (Hgf), platelet-derived growth factor-α (Pdgfa), connective tissue growth factor (Ctgf), fibroblast growth factor 1 (Fgf1), c-Fos-induced growth factor (Figf), and receptors for growth factor proteins, including platelet-derived growth factor receptors (Pdgfra and Pdgfrb), fibroblast growth factor receptor 2 (Fgfr2), Fms-related tyrosine kinase 1 (Flt1), and transforming growth factor-β receptor 3 (Tgfbr3), all of which showed circadian oscillations, but with peak expressions at different points in the 24-h light-dark cycle (Fig. 5). Maxima for Hgf, Pdgfa, Fgf1, and Figf occurred in the middle of the light period (cluster 3), while Pdgfra (cluster 1), Pdgfrb (cluster 2), Fgfr2 (cluster 4), and Flt1 (cluster 4) peaked during the dark-light transition, early light period, and late light period, respectively. Tgfbr3 peaked during the early dark period and Ctgf in the middle of the dark period. These growth factors are not only involved in cell growth, cell migration, and remodeling of various components of the organ but are also very important for the morphogenesis and development of the organ (12, 19, 21, 32, 63). This list also includes other signaling ligands, including adrenomedullin (Adm), angiopoietin 1 (Angpt1), and cardiotrophin 1 (Ctf1), which, in addition to the above-mentioned growth factors, are involved in maintaining the normal functioning of the organ (10, 28, 69). Adm and Ctf1 peaked during the early light period (cluster 2), and Angpt1 peaked during the late light period (cluster 4). Furthermore, most of these proteins are also involved in regulating angiogenesis, which is important in the remodeling of the pulmonary vasculature (34).
Table 3.
Signaling ligand molecules and receptors showing circadian-like oscillations in gene expression
| Symbol | Cluster No. | Gene Name | Gene Function |
|---|---|---|---|
| Ligands | |||
| Adm | 2 | Adrenomedullin | Hypotensive and vasodilatory agent |
| Fgg | 2 | Fibrinogen γ-chain | Platelet aggregation, blood clotting |
| Ctf1 | 2 | Cardiotrophin 1 | Angiogenesis, anti-inflammatory cytokine |
| Inhbb | 2 | Inhibin-βB | Inhibits cellular proliferation |
| Wisp2 | 2 | Wnt1 inducible signaling 2 | Wnt signaling |
| Hgf | 3 | Hepatocyte growth factor | Mitogenesis, matrix invasion, angiogenesis |
| Pdgfa | 3 | Platelet-derived growth factor- α | Mitogenesis, wound healing, tissue repair |
| Wnt2 | 3 | Wingless-type MMTV integration 2 | Wnt signaling, cell proliferation and differentation |
| Fgf1 | 3 | Fibroblast growth factor 1 | Mitogenesis, tissue repair, angiogenesis |
| Apln | 3 | Apelin | Control of body fluid homeostasis |
| Figf | 3 | c-Fos-induced growth factor | Angiogenesis and lymphangiogenesis |
| Tfpi | 4 | Tissue factor pathway inhibitor | Inhibition of TF-dependent blood coagulation |
| Angpt1 | 4 | Angiopoietin 1 | Vascular development and angiogenesis |
| Slit2 | 4 | Slit homolog 2 | Angiogenesis and cell migration |
| Gas6 | 4 | Growth arrest-specific 6 | Thrombosis and cell adhesion, migration, proliferation |
| Ctgf | 7 | Connective tissue growth factor | Connective tissue mitoattractant |
| Areg | 7 | Amphiregulin | Fibroblast proliferation and mitogenesis |
| Nrg4 | 7 | Neuregulin 4 | Activation of type 1 growth factor receptors |
| Receptors | |||
| Pdgfra | 1 | Platelet-derived growth factor receptor-α | Mitogenesis, wound healing, tissue repair |
| Tnfrsf12a | 1 | TNF receptor superfamily, 12A | Angiogenesis, endothelial cell proliferation, cell adhesion |
| Adora2b | 1 | Adenosine A2b receptor | Activates adenylyl cyclase |
| Pdgfrb | 2 | Platelet-derived growth factor receptor-β | Vascular and tissue remodeling |
| Thra | 2 | Thyroid hormone receptor-α | Growth and metabolism |
| Nmur1 | 2 | Neuromedin U receptor 1 | Vasoconstriction, SM contraction |
| Met | 2 | Met protooncogene | HGF receptor: cell proliferation and survival |
| Adora2a | 3 | Adenosine A2a receptor | Activates adenylyl cyclase |
| Antxr1 | 3 | Anthrax toxin receptor 1 | Cell adhesion and migration |
| Antxr2 | 3 | Anthrax toxin receptor 2 | Cell adhesion and migration |
| Plxnd1 | 3 | Plexin D1 | Vascular development and angiogenesis |
| Epha4 | 3 | EPH receptor A4 | Cell adhesion, migration, survival |
| P2rx1 | 3 | Purinergic receptor P2X1 | Ca2+ transport, vasoconstriction |
| Ednra | 3 | Endothelin receptor A | Vasoconstriction, cell proliferation |
| Acvr2a | 3 | Activin A receptor, type IIA | Activates Smad transcriptional regulators |
| Fgfr2 | 4 | Fibroblast growth factor receptor 2 | Mitogenesis, tissue repair, angiogenesis |
| Ptch1 | 4 | Patched homolog 1 | Hedgehog signaling; lung development |
| Flt1 | 4 | Fms-related tyrosine kinase 1 | Vascular development and angiogenesis |
| Adrb2 | 6 | Adrenergic-β2 receptor | Catecholamine-induced activation of adenylate cyclase |
| Lifr | 6 | Leukemia inhibitory factor receptor-α | Cell proliferation, differentiation, survival |
| Vipr1 | 6 | Vasoactive intestinal peptide receptor 1 | SM relaxation; fluid homeostasis |
| Tgfbr3 | 6 | Transforming growth factor-β receptor 3 | Facilitates TGFβ binding |
| Lpar1 | 6 | Lysophosphatidic acid receptor 1 | Cell proliferation, platelet aggregation, SM contraction |
| Robo2 | 6 | Roundabout homolog 2 | Receptor for Slit2; cell migration |
| Gria3 | 6 | Glutamate receptor, AMPA 3 | Opening of cation channels |
MMTV, mouse mammary tumor virus; TF, tissue factor; SM, smooth muscle; EPH, ephrin; HGF, heaptocyte growth factor; TGF, transforming growth factor; AMPA, 3-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid.
Fig. 5.
Expression patterns of selected signaling ligand molecules and receptors in lung as a function of circadian time. ●, Mean data; error bars, SD; solid lines, curves fitted to a sine function.
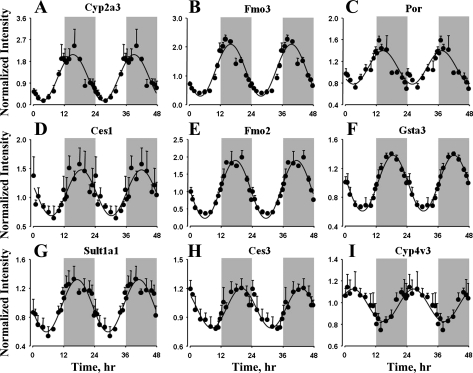
Metabolism and transport of small molecules.
The genes that are classified under the functional groups Metabolism and Transport are involved in the production, processing, and degradation of endogenous compounds and the subsequent transport and movement of these compounds between different compartments of the organ. This includes genes involved in metabolism of carbohydrates and lipids, which are not only important energy substrates but are also important in forming structural and functional components of the organ. Circadian-like oscillations in the expression of these genes may help ensure the proper maintenance of homeostasis and efficient functioning of the system. The lung is an important organ that is involved in the metabolism of not only the endogenous compounds but also toxic xenobiotics and therapeutic drugs (6, 17). Genes that play an important role in the metabolism of xenobiotics, along with the transporters involved, are listed in Table 4. This list includes genes coding for phase 1 and phase 2 enzymes, which are involved in the functionalization and conjugation of many drugs. Figure 6 shows the cyclic oscillations in the expression of these genes. Cytochrome P-450, family 2, subfamily A, polypeptide 3 (Cyp2a3), flavin-containing monooxygenase 3 (Fmo3), and P-450 (cytochrome) oxidoreductase (Por) expression peaked during the early dark period (cluster 6), the expression of carboxylesterase 1 (Ces1), flavin-containing monooxygenase 2 (Fmo2), glutathione S-transferase-α3 (Gsta3), and sulfotransferase family 1A (Sult1a1) peaked in the middle of the dark period (cluster 7), and the expression of carboxylesterase 3 (Ces3) peaked during the late dark period. In contrast, cytochrome P-450, family 4, subfamily V, polypeptide 2 (Cyp4v3) expression peaked during the early light period (cluster 2). Of the 17 solute carrier family protein coding genes that showed circadian oscillations in expression in lung, 8 are relevant to the transport of therapeutic drugs (Table 4).
Table 4.
Therapeutic drug and xenobiotic metabolizing enzymes and transporters showing circadian-like oscillations in gene expression
| Symbol | Cluster No. | Gene Name | Gene Function |
|---|---|---|---|
| Drug metabolism | |||
| Ces3 | 1 | Carboxylesterase 3 | Hydrolysis of amide and ester bonds |
| Cyp4v3 | 2 | Cytochrome P-450 4V3 | Oxidation of fatty acid and steroid moieties |
| Por | 6 | P-450 oxidoreductase | Electron transfer: NADP to cytochrome P-450 |
| Fmo3 | 6 | Flavin-containing monooxygenase 3 | Oxidation of xenobiotics |
| Cyp2a3 | 6 | Cytochrome P-450 2A3 | Hydroxylation of xenobiotics |
| Ces1 | 7 | Carboxylesterase 1 | Hydrolysis of amide and ester bonds |
| Fmo2 | 7 | Flavin-containing monooxygenase 2 | Oxidation of xenobiotics |
| Sult1a1 | 7 | Sulfotransferase family, cytosolic, 1A1 | Sulfate conjugation of xenobiotics |
| Gsta3 | 7 | Glutathione S-transferase-α3 | Glutathione conjugation of xenobiotics |
| Drug transport | |||
| Slc15a2 | 1 | Solute carrier family 15A2 | Oligopeptide transport |
| Slc3a2 | 2 | Solute carrier family 3A2 | Neutral amino acid transport |
| Slc25a35 | 2 | Solute carrier family 25A35 | Mitochondrial transport of xenobiotics |
| Slc7a7 | 2 | Solute carrier family 7A7 | Dibasic and neutral amino acid transport |
| Slc28a3 | 2 | Solute carrier family 28A3 | Nucleoside transport |
| Abca8 | 3 | ATP-binding cassette (ABC1) 8 | ATP-dependent lipophilic drug transport |
| Slc22a5 | 3 | Solute carrier family 22A5 | Organic cation transport |
| Slc15a3 | 3 | Solute carrier family 15A3 | Proton oligopeptide cotransport |
| Abca5 | 6 | ATP-binding cassette (ABC1) 5 | Intracellular sterol and steroid transport |
| Slc43a2 | 6 | Solute carrier family 43A2 | Neutral amino acid transport |
Fig. 6.
Expression patterns of genes coding for enzymes involved in endogenous and exogenous compound metabolism in lung as a function of circadian time. ●, Mean data; error bars, SD; solid lines, curves fitted to a sine function.
Lung diseases, disorders, microbial infection, and potential drug targets.
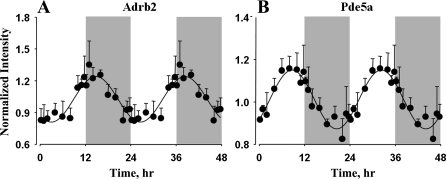
Another interesting set of genes that showed 24-h cyclic oscillations in expression in lung and are highly relevant to its physiology are the genes that are differentially regulated and/or involved in pathogenesis of many lung diseases and disorders. Several genes that are involved in the sensing, binding, killing, and clearance of pathogenic microbes and viruses also showed such oscillations in expression in lung. Furthermore, many of these genes that are involved in the pathophysiology of lung diseases and disorders are also important drug targets that can be exploited to treat and cure these diseases. Genes associated with lung diseases that showed circadian-like oscillations in expression, along with their related diseases, are listed in Table 5 (for more detailed information about these genes and their role in lung diseases, see Supplemental Table S3). These genes are associated with lung diseases and disorders, including asthma, COPD (which includes lung emphysema and chronic bronchitis), ALI, pulmonary hypertension, fibrosis (pulmonary and cystic), pneumonia, endotoxemia, and cancer. Furthermore, genes that are potential therapeutic targets for drugs or the drugs themselves that are used for the treatment of these diseases that are currently in use or in clinical trials or preclinical development are also listed in Table 5. Figure 7 shows the oscillations in the gene expression of two important drug targets in the treatment of lung diseases. β2-Adrenergic receptors (Adrb2) are cell surface receptors that mediate the catecholamine-induced activation of adenylate cyclase and promote bronchodilation (26). As shown in Fig. 7 and Table 5, Adrb2 is a member of cluster 6 and showed peak expression during the early dark/active period and reached its nadir during the early light period. The protein encoded by this gene is an important therapeutic target for a class of drugs called β2-adrenergic agonists, which are used in the treatment of symptoms of asthma and COPD. Phosphodiesterase 5A (Pde5a) is an enzyme that is highly expressed in lung tissue and is involved in the hydrolysis of cGMP (16). This gene is a member of cluster 4 and showed circadian-like oscillations in expression, with peak at the late light period and nadir at the late dark period. Inhibition of this gene by compounds such as sildenafil causes an increase in cGMP levels, resulting in dilation of the pulmonary vasculature, and, hence, can be used to treat pulmonary hypertension. There were 271 genes (see Supplemental Table S4) showing rhythmic oscillations in expression in lung that are differentially regulated or involved in the pathogenesis of different types of tumors, and some of these are potential therapeutic targets and biomarkers for lung cancer.
Table 5.
Genes associated with lung diseases showing circadian-like oscillations in expression
| Symbol | Cluster No. | Gene Name | Drug Target |
|---|---|---|---|
| Asthma | |||
| *Pdgfra | 1 | Platelet-derived growth factor receptor-α | |
| Eif4e | 1 | Eukaryotic translation initiation factor 4E | |
| *Vasp | 2 | Vasodilator-stimulated phosphoprotein | |
| Selp | 2 | Selectin P | Bimosiamose, rPSGL-1 |
| Cxcl3 | 2 | Chemokine (C-X-C motif) ligand 3 | |
| *Irf1 | 2 | Interferon regulatory factor 1 | |
| Tgm2 | 2 | Transglutaminase 2 | |
| *Adm | 2 | Adrenomedullin | |
| *Rhoa | 2 | Ras homolog gene A | |
| Pdgfrb | 2 | Platelet-derived growth factor receptor-β | |
| Nfil3 | 2 | Nuclear factor, IL-3-regulated | |
| Aqp1 | 2 | Aquaporin 1 | |
| *Itga4 | 3 | Integrin-α4 | |
| Tnfsf10 | 3 | TNF superfamily, member 10 | |
| Emr1 | 3 | EGF-like module containing, 1 | |
| Cxcl12 | 3 | Chemokine (C-X-C motif) ligand 12 | |
| *Adora2a | 3 | Adenosine A2a receptor | Binodenoson |
| Hgf | 3 | Hepatocyte growth factor | Recombinant HGF |
| Pdgfa | 3 | Platelet-derived growth factor-α | |
| *Tnc | 4 | Tenascin C | |
| Ptprd | 4 | Protein tyrosine phosphatase, receptor D | |
| Fyn | 4 | FYN oncogene related to SRC, FGR, YES | |
| Angpt1 | 4 | Angiopoietin 1 | |
| Flt1 | 4 | Fms-related tyrosine kinase 1 | |
| *Marcks | 4 | Myristoylated Ala-rich PKC substrate | Marcks inhibitor peptide |
| *Adam33 | 5 | ADAM metallopeptidase 33 | |
| *Adrb2 | 6 | Adrenergic-β2 receptor | β-Agonists |
| Ppargc1a | 6 | PPARγ coactivator 1α | |
| Cd163 | 7 | CD163 molecule | |
| *Ctgf | 7 | Connective tissue growth factor | |
| *Mrc1 | 7 | Mannose receptor, C1 | |
| *Hspb1 | 8 | Heat shock 27-kDa protein 1 | |
| COPD | |||
| *Pdgfra | 1 | Platelet-derived growth factor receptor α | |
| *Adm | 2 | Adrenomedullin | |
| *Cx3cl1 | 2 | Chemokine (C-X3-C motif) ligand 1 | |
| Mmp14 | 3 | Matrix metallopeptidase 14 | |
| Eln | 3 | Elastin | |
| Fgf1 | 3 | Fibroblast growth factor 1 | |
| Fbn1 | 3 | Fibrillin 1 | |
| *Pde5a | 4 | Phosphodiesterase 5A, cGMP-specific | Sildenafil (Viagra) |
| *Marcks | 4 | Myristoylated Ala-rich PKC substrate | Marcks inhibitor peptide |
| *Adam33 | 5 | ADAM metallopeptidase 33 | |
| *Adrb2 | 6 | Adrenergic β2 receptor | β2-Agonist |
| Tgfbr3 | 6 | Transforming growth factor-β receptor 3 | |
| Hey1 | 6 | Hairy/enhancer-of-split related with YRPW 1 | |
| *Ctgf | 7 | Connective tissue growth factor | |
| *Timp3 | 7 | TIMP metallopeptidase inhibitor 3 | |
| Lung injury | |||
| *Vasp | 2 | Vasodilator-stimulated phosphoprotein | |
| *Ctf1 | 2 | Cardiotrophin 1 | |
| *Itga4 | 3 | Integrin-α4 | |
| Ampd3 | 3 | Adenosine monophosphate deaminase 3 | |
| *Adora2a | 3 | Adenosine A2a receptor | A2a receptor agonist |
| *Tnc | 4 | Tenascin C | |
| *Angpt1 | 4 | Angiopoietin 1 | COMP-Ang1 |
| Mapk1 | 4 | Mitogen-activated protein kinase 1 | PD98059 |
| Mt2A | 6 | Metallothionein 2A | |
| Ddit4 | 6 | DNA damage-inducible transcript 4 | |
| Lifr | 6 | Leukemia inhibitory factor receptor-α | |
| *Rock2 | 6 | Rho-associated, coiled-coil containing PK2 | Y-27632 |
| Map3k2 | 6 | Mitogen-activated protein 3K2 | |
| Aoc3 | 6 | Amine oxidase, copper containing 3 | SZE 5302 |
| *Timp3 | 7 | TIMP metallopeptidase inhibitor 3 | |
| *Hspb1 | 8 | Heat shock 27-kDa protein 1 | |
| Microbial infection | |||
| *Cx3cl1 | 2 | Chemokine (C-X3-C motif) ligand 1 | |
| *Irf1 | 2 | Interferon regulatory factor 1 | |
| Spon2 | 3 | Spondin 2 | |
| Ncf4 | 3 | Neutrophil cytosolic factor 4 | |
| Antxr1/TEM8 | 3 | Anthrax toxin receptor 1 | |
| Colec12 | 4 | Collectin subfamily member 12 | |
| Tbk1 | 4 | TANK-binding kinase 1 | |
| Dhx36 | 4 | DEAH box polypeptide 36 | |
| Ivns1abp | 6 | Influenza virus NS1A binding protein | |
| *Mrc1 | 7 | Mannose receptor, C type 1 | |
| Pulmonary and cystic fibrosis | |||
| *Pdgfra | 1 | Platelet-derived growth factor receptor-α | Imatinib |
| Atf4 | 1 | Activating transcription factor 4 | |
| *Adm | 2 | Adrenomedullin | |
| Grk5 | 2 | G protein-coupled receptor kinase 5 | |
| *Cxcl12 | 3 | Chemokine (C-X-C motif) ligand 12 | Neutralizing antibody |
| Nox4 | 3 | NADPH oxidase 4 | |
| Ednra | 3 | Endothelin receptor type A | |
| LRP6 | 6 | LDLR-related protein 6 | |
| Pulmonary hypertension | |||
| *Thbs1 | 1 | Thrombospondin 1 | |
| *Adm | 2 | Adrenomedullin | |
| *Ctf1 | 2 | Cardiotrophin 1 | Recombinant CTF1 |
| *Pdgfrb | 2 | Platelet-derived growth factor receptor-β | |
| *Rhoa | 2 | Ras homolog gene family, member A | Fasudil |
| Fbln5 | 4 | Fibulin 5 | |
| *Pde5a | 4 | Phosphodiesterase 5A, cGMP-specific | Sildenafil (Viagra) |
| *Tfpi | 4 | Tissue factor pathway inhibitor | |
| Alas1 | 6 | Aminolevulinate-δ synthase 1 | |
| *Rock2 | 6 | Rho-associated, coiled-coil containing PK2 | Hydroxyfasudil |
| Others (idiopathic interstitial pneumonia, community-acquired pneumonia, endotoxemia) | |||
| *Thbs1 | 1 | Thrombospondin 1 | |
| *Irf1 | 2 | Interferon regulatory factor 1 | |
| Neurl3 | 3 | Neuralized homolog 3 | |
| *Tfpi | 4 | Tissue factor pathway inhibitor | Tifacogin |
| Cancer (selected genes) | |||
| *Pdgfrb | 2 | Platelet-derived growth factor receptor-β | Telatinib |
| Tnfsf10 | 3 | TNF superfamily, member 10 | Dulanermin |
| *Cxcl12 | 3 | Chemokine (C-X-C motif) ligand 12 | AMD 3100 and CTCE-9908 |
| Hgf | 3 | Hepatocyte growth factor | AMG 102 |
| Fyn | 4 | FYN oncogene related to SRC, FGR, YES | Dasatinib |
| *Angpt1 | 4 | Angiopoietin 1 | AMG 386 |
| Flt1 | 4 | Fms-related tyrosine kinase 1 | Cediranib, angiozyme |
COPD, chronic obstructive pulmonary disorder; EGF, epidermal growth factor; rPSGL, recombinant P-selectin glycoprotein ligand; ADAM, A disintegrin and metalloproteinase; PPAR, peroxisome proliferator-activated receptor; LDLR, LDL receptor.
Genes associated with multiple lung diseases.
Fig. 7.
Expression patterns of Adrb2 and Pde5a in lung as a function of circadian time. ●, Mean data; error bars, SD; solid lines, curves fitted to a sine function.
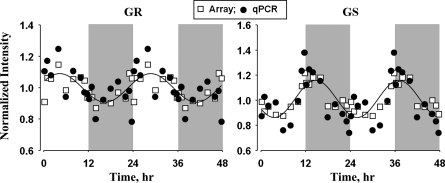
Confirmation of array data for glutamine synthase and glucocorticoid receptor circadian-like expression by quantitative PCR.
Oscillations in the expression of glucocorticoid receptor and glucocorticoid-regulated glutamine synthase gene expression were probed using qRT-PCR to confirm and validate the data obtained from gene chips. Glucocorticoid receptor is a cytosolic receptor for endogenous and exogenous glucocorticoids and is involved in the transcriptional regulation of a wide range of genes involved in immune and metabolic processes. As shown in Fig. 8, the peak expression of glucocorticoid receptor mRNA occurred at around ZT 4 in the early light period, and the nadir occurred in the early dark period. Cyclic oscillation in the expression of this receptor in lung is very similar to that in skeletal muscle but is very different from the expression in liver and adipose tissue from the same animals, where the peak expression occurred at the light-dark transition. The expression of glutamine synthase mRNA peaked during the light-dark transition (ZT 12), and the nadir occurred at the dark-light transition. The circadian-like oscillation in glutamine synthase was very similar to the 24-h oscillation in endogenous corticosterone, which also peaked during the light-dark transition (3).
Fig. 8.
Cyclic pattern of expression of glucocorticoid receptor (GR) and glutamine synthase (GS) quantified by quantitative real-time PCR and Affymetrix gene chips. Solid lines, curves fitted to a sine function.
DISCUSSION
In the present study, we describe for the first time the genome-wide analysis of diurnal/nocturnal patterns in mRNA expression from lungs using Affymetrix gene chips, which was validated by qRT-PCR for selected genes. This study was carried out in intact rats acclimated to a tightly controlled 12:12-h light-dark cycle, but with the animals otherwise unperturbed. We use the term “circadian-like” to describe oscillating patterns of expression that occur with a periodicity of ∼24 h in the normal animal. The richness of our data set (54 animals killed across 18 time points), which is one of the salient features of this study, allowed us to apply a nonlinear fitting of a sinusoidal model to the expression data to identify these oscillations using the same filtering criteria used in our previous studies (59). However, a limitation of this approach is that this model is designed to select expression patterns that exhibit a traditional sine-cosine oscillatory behavior and will not capture other cyclic patterns that potentially could occur in the data. Because our animals were maintained in regular 12:12-h light-dark cycles (as opposed to constant darkness or constant light) with food and water ad libitum, we cannot distinguish primary endogenous rhythms from those entrained by secondary exogenous factors such as food or light. An additional consideration in analyzing such oscillations in gene expression from organs such as lung is that it is possible that the expression of a gene in different cell types may differ (i.e., genes that are rhythmic in one cell type may not be rhythmic in another or may have a different phase).
However, the richness of this data set allowed us to cluster circadian-like oscillations in the mRNA expression based on the similarity in their temporal expression using a QT algorithm. We identified 1,006 probe sets coding for 646 genes of known function (as some genes were represented by multiple probe sets and some probe sets corresponded to expressed sequence tags or genes of unknown functions) that showed oscillations in expression; these genes were further parsed into 8 distinct temporal clusters. One of the striking observations from the clustering analysis is that two-thirds of the genes showing circadian-like oscillations in lung expression peaked during the light/inactive period. This is in contrast to studies in all other tissues performed in nocturnal rodents from our laboratory and others, where more genes that showed circadian-like expression peak during the dark/active period (2–4, 59). In addition, most genes that peaked during the light period in lung did not show cyclic oscillations in expression in other peripheral tissues.
Studies of the central clock in the suprachiasmatic nuclei and peripheral clocks in other tissues showed that a number of transcription factors are involved in the maintenance and regulation of the clock. In this study, we demonstrate that the mRNA expression of six core clock genes (Bmal1, Npas2, Per1, Per2, Per3, and Cry1) shows circadian oscillations, which agrees with their expression in the central clock and peripheral clocks in other tissues (61). Furthermore, nine clock-related and clock-controlled transcription factors (Nr1d1, Nr1d2, Rorα, Dbp, Nfil3, Tef, Hlf, Bhlhb2, and Bhlhb3) that are involved in the regulation of the molecular clock and the control of the circadian oscillations in the expression of the transcriptome also showed robust oscillations in expression in lung. Of these nine genes, the phases of the oscillations in the expression of seven are in agreement with other peripheral tissues. The exceptions were RORα and Bhlhb2 (Dec1). Neither of these genes showed cyclic oscillations in white adipose, liver, and muscle tissue from the same animals; yet both genes showed robust circadian-like oscillations in expression in lung. This suggests that although most components of the molecular clock are conserved in lung, a few tissue-specific differences are also observed compared with other peripheral tissues. Apart from these transcription factors, the F-box protein Fbxl3, which is a component of the SKP1-CUL1-F-box-protein (SCF)-E3 ubiquitin ligase complex, also showed oscillations in its expression in lung (see Supplemental Fig. S1). Forward genetic approaches identified Fbxl3 as an important protein involved in posttranslational control of clock proteins (49). Fbxl3 interacts with Cry proteins and leads to an increase in the proteasome-mediated degradation of the Cry proteins. Mutation in this gene was found to cause a longer circadian period (∼26 h) in mouse and results in global transcriptional repression of Per and Cry genes. It is interesting to observe that the expression of Fbxl3 peaked during the late light period, which is exactly opposite to Cry1 gene expression, which showed its nadir at the same time. The other component of the SCF complex, Cul1, also showed circadian-like oscillations in its expression in lung, with peak expression in the middle of the light period (see Supplemental Fig. S1).
Lung is one of the organs that constantly interacts with the environment and is highly exposed to foreign materials (45). Hence, to maintain the normal functioning and homeostasis of the organ, constant repair and turnover of its components, ranging from extracellular and intracellular components to whole cells, are essential. Cyclic oscillations in the expression of genes associated with extracellular matrix, cytoskeleton, cell cycle, and apoptosis suggest that the repair and turnover of these different components in lung are directly or indirectly under the regulation of the molecular clock. Furthermore, many of the growth factor ligands and receptors that are involved in the direct regulation of these processes and the maintenance of the homeostasis in lung also showed circadian-like oscillations in expression. These growth factor ligands and receptors are directly involved in controlling cell growth, motility, angiogenesis, and tissue repair (12, 19, 32). In addition to these functions, many of these proteins are also involved in regulating wound healing, extracellular matrix invasion, immune response, and production of inflammatory cytokines (21, 34, 64). These processes, in turn, can regulate repair and turnover of different components of the organ (36, 53, 56).
In addition to the cyclic oscillations in the above-mentioned components, many of the genes involved in processing of proteins, ranging from posttranslational modification (e.g., glycosylation and proteolytic modifications) to ubiquitin-mediated proteasomal degradation, also showed circadian-like oscillations in expression in lung (see Supplemental Table S2). Furthermore, genes that are involved in protein sorting and trafficking to different organelles and the cell surface, as well as secretion to the extracellular space and systemic circulation, also showed rhythmic expression in lung. Posttranslational modification of proteins, their degradation, and their transport are essential for the repair and turnover processes of different components of the organ; hence, 24-h rhythms in the expression of the genes involved in protein processing and trafficking processes further confirm that repair and turnover processes in lung are controlled and coordinated, directly or indirectly, by the endogenous clock, allowing for maximum efficiency in the functioning of lung (29, 50). Another interesting observation is that most of the genes involved in all the above-mentioned processes mainly peaked during the light period, which is also the inactive period in these nocturnal rodents. This suggests that the repair and turnover processes mainly occur during the period when the organism is sleeping/inactive and, therefore, has lower pulmonary demands, as the organism's requirement for oxygen is at its minimum. Studies in humans show that pulmonary function and breathing show cyclic rhythms, with the nadir during the sleeping period (1, 54, 55).
The 24-h oscillations in the expression of genes involved in the regulation of other aspects of lung function were also observed in this analysis. Genes coding for inflammatory molecules, including chemokine ligands (Cx3cl1, Cxcl3, and Cxcl12), tumor necrosis factor ligand member 10 (Tnfsf10), and others, showed circadian-like oscillations in expression in lung, suggesting that the molecular clock could directly or indirectly regulate the immune response in the organ. Lung has a very well-developed innate immune system, as it is constantly exposed to pathogenic microorganisms, viruses, and other foreign materials (8, 45). Apart from the resident alveolar macrophages, dendritic cells, and mast cells in the organ, granulocytes, such as eosinophils and neutrophils, can also migrate into lung from the systemic circulation if necessary, and these cell types largely contribute to the inflammatory and immune-regulating molecules produced by the organ (7, 60). Furthermore, genes that are involved in the metabolism of xenobiotics also showed circadian-like oscillations in expression in lung (Table 4, Fig. 6). The entry of xenobiotics into the organism is mainly through the breathing process, which is more active during the dark/active period, or through ingestion, which also occurs mostly during the dark/active period in nocturnal rodents. Liver, which receives blood through the hepatic portal vein, is the organ that predominantly encounters and metabolizes xenobiotics, which gain entry through ingestion, while the lung predominantly encounters and metabolizes airborne xenobiotics (52). It is interesting to note that cyclic expression of most of these genes coding for metabolizing enzymes in lungs peaked during the dark/active period, as the probability of entry of these agents is higher during the active period. For example, Cyp2a3, which is a cytochrome P-450 enzyme that is predominantly expressed in lung, peaked during the early dark period and is involved in the metabolism of many airborne carcinogens and xenobiotics, including compounds produced by tobacco smoking (e.g., nicotine and nitrosoamine compounds) (27, 62). Similarly, the phase 2 metabolizing enzyme Sult1a1, which peaked in the middle of the dark period, is also involved in the metabolism of cigarette smoke toxicants (67).
Along with liver, lung plays an important role in the metabolism of drugs, both inhaled drugs and those from the systemic circulation, and, hence, can affect their pharmacokinetics (6). The same enzyme systems that are involved in metabolizing endogenous compounds and xenobiotics are also involved in metabolizing therapeutic drugs. Studies have shown that the pharmacokinetics of some drugs used for treating diseases including asthma, lung cancer, and many others show circadian-like variations based on the time of dosing, which could partially be affected by the oscillations in the metabolism of drugs (31). Pharmacokinetics and the subsequent action of a drug are not just affected by metabolism, but also by the absorption, transport, and distribution of drugs between different components of an organ (58). Proteins from the solute carrier family play an important role in these processes, and many of these proteins show circadian-like oscillations in their mRNA expression in lung (Table 4; see Supplemental Table S2). For example, ipratropium and tiotropium are anticholinergic drugs used for the treatment of pulmonary diseases such as asthma and COPD (38). Absorption of these drugs after inhalation occurs through the solute carrier protein coded by Slc22a5 (Octn2), which showed cyclic oscillations in expression in lung (Table 4; see Supplemental Fig. S2), suggesting that the absorption rate of these drugs could vary depending on the time of dosing. Apart from genes coding for metabolizing enzymes and transporters, biomarkers that are used for tissue-specific targeting of therapeutic drugs also showed circadian-like oscillations in expression. For example, preclinical studies have shown that aminopeptidase P, which is enriched in pulmonary vasculature, can be used as a target biomarker for tissue-specific delivery of imaging agents and drugs for treating diseases such as lung cancer, lung fibrosis, and pulmonary hypertension (11). Similarly, Pdgfrb can be targeted for the delivery of anticancer agents, specifically to tumor stromal cells to increase efficacy, and at the same time to decrease systemic toxicity (42). Both of these proteins show 24-h cyclic oscillations in their mRNA expression; hence, the efficiency of targeting could depend on the dosing time.
In this study, rhythmic oscillations were observed in the mRNA expression of glucocorticoid receptor, which was further confirmed by quantitative PCR. It is interesting to see two different time-dependent patterns in the expression of glucocorticoid receptor in different peripheral tissues, with expression peaking at ZT 4 in lung and skeletal muscle and at ZT 12 in white adipose tissue and liver from the same animals (22, 59, 66). This later time corresponds to the peak in corticosterone concentrations in plasma (the endogenous glucocorticoid in rats) in these animals. This suggests that transcriptional regulation of glucocorticoid receptor is tissue-specific and that the cyclic expression could be negatively regulated by the oscillations in concentrations of its own ligand, corticosterone (as the nadir of the receptor expression occurs a few hours after the peak corticosterone), in lung and skeletal muscle, but not in liver and white adipose tissue. Furthermore, oscillations in the expression of glucocorticoid receptor in lung could affect the therapeutic efficacy of synthetic corticosteroids, which are widely used as treatment for asthma and other lung inflammation-related disorders (35). The expression of the glucocorticoid-regulated gene for glutamine synthase peaked at the same time as expression of the corticosterone gene, suggesting that expression of glutamine synthase could be driven by oscillations in the concentrations of plasma corticosterone, as observed in skeletal muscle (66).
The analysis of genes showing circadian-like oscillations in expression in lung and their association with the pathophysiology of lung diseases identified many genes that are biomarkers, potential therapeutic targets, or differentially regulated in many lung diseases. These genes are involved in regulating diverse functions and are classified across different functional categories (see Supplemental Table S2). It is not surprising that some of these genes, which are directly or indirectly involved in regulating immune responses, are associated with more than one lung disease. One common factor between many lung diseases, such as asthma, COPD, fibrosis, ALI, pneumonia, and endotoxemia, is excessive and uncontrolled inflammation and innate immune response. Furthermore, stimulation of immune response is also associated with invasion by foreign pathogens and, in a controlled manner, is required for elimination of pathogens. For example, chemokine ligand (CXCL12) levels are elevated and are involved in the pathogenesis of asthma and pulmonary fibrosis (39, 48, 57). Similarly, chemokine ligand (CX3CL1) expression is not only elevated in cigarette smoke-induced emphysema but also during viral infection as part of normal immune response (24, 33). Studies have shown that extracellular matrix plays an important role in the pathophysiology of many lung diseases, including asthma, COPD, fibrosis, and ALI, where changes in extracellular matrix turnover and its remodeling are evident in the diseased state (46, 53). Furthermore, extracellular matrix can influence the recruitment, activation, and survival of immune cells, and in turn immune response and inflammation can influence the remodeling of extracellular matrix (15, 47). In addition, many signaling molecules (such as the growth factors discussed previously) can regulate inflammation and extracellular matrix remodeling, and the complex interaction between all these processes results in the pathophysiology of lung diseases (12, 21). This suggests that some of these signaling molecules will be of therapeutic importance as drugs and drug targets for treating lung diseases, and some preclinical and clinical studies show promising results (32). For example, recombinant hepatocyte growth factor treatment attenuated allergic airway inflammation, recombinant Angpt1 treatment attenuated ALI, and recombinant Ctf1 treatment attenuated pulmonary hypertension in rodent models (30, 40, 41). Similarly, some of the targets for agents to treat lung cancer that are currently in various stages of clinical development also show 24-h oscillations in expression in lung (Table 5) (9, 20, 65). Circadian-like oscillations in expression of these potential therapeutic target genes should be taken into account to obtain optimal dosing times, to maximize efficacy, and, at the same time, to reduce side effects. Caution should be taken while translating gene expression data obtained from normal conditions to diseased states, since the pathophysiology of the disease might cause changes in the cyclic expression pattern of some of the associated genes. Hence, characterizing the rhythmic expression of the gene of interest under disease states, along with normal conditions, will be helpful in understanding the causal relationship of the disease with circadian regulation and also in obtaining better insight into the optimal dosing time for the drugs targeting the disease.
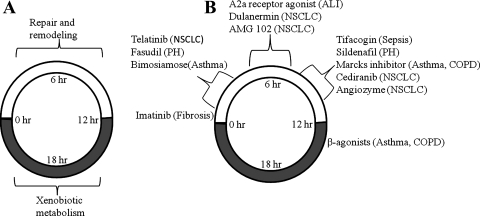
In summary, this study provides a temporal and functional categorization of genes showing 24-h oscillations in expression in lung and its implication in the physiology and pathophysiology of this dynamic organ. A large number of genes showed cyclic expression in lung that adequately fit our model for circadian-like oscillations. Consistent with other peripheral tissues, the molecular clock machinery was conserved in lung. As summarized in Fig. 9, analysis of diurnal-nocturnal gene expression suggests that repair and remodeling of the organ occurs primarily during the light/inactive period, when the animals are at rest, and metabolism of xenobiotic compounds occurs primarily during the dark period, when rats are active. Furthermore, many of the genes showing circadian-like oscillations in expression are drug targets and/or biomarkers involved in the pathogenesis of lung diseases. Figure 9 also presents some important drugs used for the treatment of lung diseases and the circadian timing in nocturnal animals at which the lung will be more susceptible to the action of these drugs based on the gene expression data. However, study of the oscillations in the protein concentrations and activity will be required to confirm the observations based on transcription profiling. The information obtained from rodent models may serve as a starting point for designing studies in humans to exploit the knowledge of chronopharmacology in maximizing efficacy and, at the same time, minimizing adverse effects for therapeutic drugs used for treating lung diseases.
Fig. 9.
Summary of select physiological functions (A) and targets of drug action (B) in relation to circadian time. PH, pulmonary hypertension; NSCLC, non-small cell lung carcinoma; COPD, chronic obstructive pulmonary disorder; ALI, acute lung injury.
GRANTS
This work was supported by National Institute of General Medical Sciences Grant GM-24211.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
Supplementary Material
REFERENCES
- 1. Adamczyk W, Tafil-Klawe M, Siekierka M, Zlomanczuk P, Weber P, Klawe JJ. Daily pattern of breathing in healthy young men. J Physiol Pharmacol 59 Suppl 6: 115–122, 2008 [PubMed] [Google Scholar]
- 2. Almon RR, Yang E, Lai W, Androulakis IP, DuBois DC, Jusko WJ. Circadian variations in rat liver gene expression: relationships to drug actions. J Pharmacol Exp Ther 326: 700–716, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Almon RR, Yang E, Lai W, Androulakis IP, Ghimbovschi S, Hoffman EP, Jusko WJ, DuBois DC. Relationships between circadian rhythms and modulation of gene expression by glucocorticoids in skeletal muscle. Am J Physiol Regul Integr Comp Physiol 295: R1031–R1047, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bailey MJ, Coon SL, Carter DA, Humphries A, Kim JS, Shi Q, Gaildrat P, Morin F, Ganguly S, Hogenesch JB, Weller JL, Rath MF, Moller M, Baler R, Sugden D, Rangel ZG, Munson PJ, Klein DC. Night/day changes in pineal expression of >600 genes: central role of adrenergic/cAMP signaling. J Biol Chem 284: 7606–7622, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartness TJ, Song CK, Demas GE. SCN efferents to peripheral tissues: implications for biological rhythms. J Biol Rhythms 16: 196–204, 2001 [DOI] [PubMed] [Google Scholar]
- 6. Boer F. Drug handling by the lungs. Br J Anaesth 91: 50–60, 2003 [DOI] [PubMed] [Google Scholar]
- 7. Bowden DH. Cell turnover in the lung. Am Rev Respir Dis 128: S46–S48, 1983 [DOI] [PubMed] [Google Scholar]
- 8. Boyton RJ, Openshaw PJ. Pulmonary defences to acute respiratory infection. Br Med Bull 61: 1–12, 2002 [DOI] [PubMed] [Google Scholar]
- 9. Burger JA, Stewart DJ. CXCR4 chemokine receptor antagonists: perspectives in SCLC. Expert Opin Investig Drugs 18: 481–490, 2009 [DOI] [PubMed] [Google Scholar]
- 10. Chinoy MR, Graybill MM, Miller SA, Lang CM, Kauffman GL. Angiopoietin-1 and VEGF in vascular development and angiogenesis in hypoplastic lungs. Am J Physiol Lung Cell Mol Physiol 283: L60–L66, 2002 [DOI] [PubMed] [Google Scholar]
- 11. Chrastina A, Valadon P, Massey KA, Schnitzer JE. Lung vascular targeting using antibody to aminopeptidase P: CT-SPECT imaging, biodistribution and pharmacokinetic analysis. J Vasc Res 47: 531–543, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Desai TJ, Cardoso WV. Growth factors in lung development and disease: friends or foe? Respir Res 3: 2, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duffy JF, Czeisler CA. Effect of light on human circadian physiology. Sleep Med Clin 4: 165–177, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dunlap JC. Molecular bases for circadian clocks. Cell 96: 271–290, 1999 [DOI] [PubMed] [Google Scholar]
- 15. Dunsmore SE, Rannels DE. Extracellular matrix biology in the lung. Am J Physiol Lung Cell Mol Physiol 270: L3–L27, 1996 [DOI] [PubMed] [Google Scholar]
- 16. Fan Chung K. Phosphodiesterase inhibitors in airways disease. Eur J Pharmacol 533: 110–117, 2006 [DOI] [PubMed] [Google Scholar]
- 17. Foth H. Role of the lung in accumulation and metabolism of xenobiotic compounds—implications for chemically induced toxicity. Crit Rev Toxicol 25: 165–205, 1995 [DOI] [PubMed] [Google Scholar]
- 18. Gachon F. Physiological function of PARbZip circadian clock-controlled transcription factors. Ann Med 39: 562–571, 2007 [DOI] [PubMed] [Google Scholar]
- 19. Gibbons GH, Dzau VJ. The emerging concept of vascular remodeling. N Engl J Med 330: 1431–1438, 1994 [DOI] [PubMed] [Google Scholar]
- 20. Gordon MS, Sweeney CS, Mendelson DS, Eckhardt SG, Anderson A, Beaupre DM, Branstetter D, Burgess TL, Coxon A, Deng H, Kaplan-Lefko P, Leitch IM, Oliner KS, Yan L, Zhu M, Gore L. Safety, pharmacokinetics, and pharmacodynamics of AMG 102, a fully human hepatocyte growth factor-neutralizing monoclonal antibody, in a first-in-human study of patients with advanced solid tumors. Clin Cancer Res 16: 699–710, 2010 [DOI] [PubMed] [Google Scholar]
- 21. Hassoun PM, Mouthon L, Barbera JA, Eddahibi S, Flores SC, Grimminger F, Jones PL, Maitland ML, Michelakis ED, Morrell NW, Newman JH, Rabinovitch M, Schermuly R, Stenmark KR, Voelkel NF, Yuan JX, Humbert M. Inflammation, growth factors, and pulmonary vascular remodeling. J Am Coll Cardiol 54: S10–S19, 2009 [DOI] [PubMed] [Google Scholar]
- 22. Hazra A, Pyszczynski N, DuBois DC, Almon RR, Jusko WJ. Modeling receptor/gene-mediated effects of corticosteroids on hepatic tyrosine aminotransferase dynamics in rats: dual regulation by endogenous and exogenous corticosteroids. J Pharmacokinet Pharmacodyn 34: 643–667, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heinemann HO, Ryan JW, Ryan US. Is the lung a para-endocrine organ? Am J Med 63: 595–603, 1977 [DOI] [PubMed] [Google Scholar]
- 24. Herd KA, Nelson M, Mahalingam S, Tindle RW. Pulmonary infection of mice with human metapneumovirus induces local cytotoxic T-cell and immunoregulatory cytokine responses similar to those seen with human respiratory syncytial virus. J Gen Virol 91: 1302–1310, 2010 [DOI] [PubMed] [Google Scholar]
- 25. Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal 7: e003, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson M. The β-adrenoceptor. Am J Respir Crit Care Med 158: S146–S153, 1998 [DOI] [PubMed] [Google Scholar]
- 27. Kalscheuer S, Zhang X, Zeng Y, Upadhyaya P. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis 29: 2394–2399, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keith IM. The role of endogenous lung neuropeptides in regulation of the pulmonary circulation. Physiol Res 49: 519–537, 2000 [PubMed] [Google Scholar]
- 29. Kim KJ, Malik AB. Protein transport across the lung epithelial barrier. Am J Physiol Lung Cell Mol Physiol 284: L247–L259, 2003 [DOI] [PubMed] [Google Scholar]
- 30. Kim SR, Lee KS, Park SJ, Min KH, Lee KY, Choe YH, Hong SH, Koh GY, Lee YC. Angiopoietin-1 variant, COMP-Ang1 attenuates hydrogen peroxide-induced acute lung injury. Exp Mol Med 40: 320–331, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lemmer B. The clinical relevance of chronopharmacology in therapeutics. Pharmacol Res 33: 107–115, 1996 [DOI] [PubMed] [Google Scholar]
- 32. Lindsay CD. Novel therapeutic strategies for acute lung injury induced by lung damaging agents: the potential role of growth factors as treatment options. Hum Exp Toxicol. In press [DOI] [PubMed] [Google Scholar]
- 33. McComb JG, Ranganathan M, Liu XH, Pilewski JM, Ray P, Watkins SC, Choi AM, Lee JS. CX3CL1 up-regulation is associated with recruitment of CX3CR1+ mononuclear phagocytes and T lymphocytes in the lungs during cigarette smoke-induced emphysema. Am J Pathol 173: 949–961, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McDonald DM. Angiogenesis and remodeling of airway vasculature in chronic inflammation. Am J Respir Crit Care Med 164: S39–S45, 2001 [DOI] [PubMed] [Google Scholar]
- 35. McFadden ER., Jr Inhaled glucocorticoids and acute asthma: therapeutic breakthrough or nonspecific effect? Am J Respir Crit Care Med 157: 677–678, 1998 [DOI] [PubMed] [Google Scholar]
- 36. McGowan SE. Extracellular matrix and the regulation of lung development and repair. FASEB J 6: 2895–2904, 1992 [PubMed] [Google Scholar]
- 37. Mirsky HP, Liu AC, Welsh DK, Kay SA, Doyle FJ., 3rd A model of the cell-autonomous mammalian circadian clock. Proc Natl Acad Sci USA 106: 11107–11112, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakamura T, Nakanishi T, Haruta T, Shirasaka Y, Keogh JP, Tamai I. Transport of ipratropium, an anti-chronic obstructive pulmonary disease drug, is mediated by organic cation/carnitine transporters in human bronchial epithelial cells: implications for carrier-mediated pulmonary absorption. Mol Pharm 7: 187–195, 2010 [DOI] [PubMed] [Google Scholar]
- 39. Negrete-Garcia MC, Velazquez JR, Popoca-Coyotl A, Montes-Vizuet AR, Juarez-Carvajal E, Teran LM. Chemokine (C-X-C motif) ligand 12/stromal cell-derived factor-1 is associated with leukocyte recruitment in asthma. Chest 138: 100–106, 2010 [DOI] [PubMed] [Google Scholar]
- 40. Nomura N, Asano M, Saito T, Sasaki S, Suzuki H, Manabe T, Mishima A. Cardiotrophin-1 is a prophylactic against the development of chronic hypoxic pulmonary hypertension in rats. Ann Thorac Surg 76: 237–243, 2003 [DOI] [PubMed] [Google Scholar]
- 41. Okunishi K, Sasaki O, Okasora T, Nakagome K, Imamura M, Harada H, Matsumoto T, Tanaka R, Yamamoto K, Tabata Y, Dohi M. Intratracheal delivery of hepatocyte growth factor directly attenuates allergic airway inflammation in mice. Int Arch Allergy Immunol 149 Suppl 1: 14–20, 2009 [DOI] [PubMed] [Google Scholar]
- 42. Prakash J, de Jong E, Post E, Gouw AS, Beljaars L, Poelstra K. A novel approach to deliver anticancer drugs to key cell types in tumors using a PDGF receptor-binding cyclic peptide containing carrier. J Control Release 145: 91–101, 2010 [DOI] [PubMed] [Google Scholar]
- 43. Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251–260, 2002 [DOI] [PubMed] [Google Scholar]
- 44. Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature 418: 935–941, 2002 [DOI] [PubMed] [Google Scholar]
- 45. Reynolds HY. Lung host defenses: a status report. Chest 75: 239–242, 1979 [PubMed] [Google Scholar]
- 46. Roman J. Extracellular matrix and lung inflammation. Immunol Res 15: 163–178, 1996 [DOI] [PubMed] [Google Scholar]
- 47. Roman J, Limper AH, McDonald JA. Lung extracellular matrix: physiology and pathophysiology. Hosp Pract (Off Ed) 25: 125–128, 131,–125, 139–140, 1990 [DOI] [PubMed] [Google Scholar]
- 48. Scotton CJ, Chambers RC. Molecular targets in pulmonary fibrosis: the myofibroblast in focus. Chest 132: 1311–1321, 2007 [DOI] [PubMed] [Google Scholar]
- 49. Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129: 1011–1023, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sixt SU, Peters J. Extracellular alveolar proteasome: possible role in lung injury and repair. Proc Am Thorac Soc 7: 91–96, 2010 [DOI] [PubMed] [Google Scholar]
- 51. Smolensky MH, Reinberg AE, Martin RJ, Haus E. Clinical chronobiology and chronotherapeutics with applications to asthma. Chronobiol Int 16: 539–563, 1999 [DOI] [PubMed] [Google Scholar]
- 52. Somers GI, Lindsay N, Lowdon BM, Jones AE, Freathy C, Ho S, Woodrooffe AJ, Bayliss MK, Manchee GR. A comparison of the expression and metabolizing activities of phase I and II enzymes in freshly isolated human lung parenchymal cells and cryopreserved human hepatocytes. Drug Metab Dispos 35: 1797–1805, 2007 [DOI] [PubMed] [Google Scholar]
- 53. Sorokin L. The impact of the extracellular matrix on inflammation. Nat Rev Immunol 10: 712–723, 2010 [DOI] [PubMed] [Google Scholar]
- 54. Spengler CM, Czeisler CA, Shea SA. An endogenous circadian rhythm of respiratory control in humans. J Physiol 526: 683–694, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Spengler CM, Shea SA. Endogenous circadian rhythm of pulmonary function in healthy humans. Am J Respir Crit Care Med 162: 1038–1046, 2000 [DOI] [PubMed] [Google Scholar]
- 56. Spurzem JR. Function at the junction: dynamic interactions between lung cells and extracellular matrix. Thorax 51: 956–958, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Strieter RM, Keeley EC, Hughes MA, Burdick MD, Mehrad B. The role of circulating mesenchymal progenitor cells (fibrocytes) in the pathogenesis of pulmonary fibrosis. J Leukoc Biol 86: 1111–1118, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sukumaran S, Almon RR, DuBois DC, Jusko WJ. Circadian rhythms in gene expression: relationship to physiology, disease, drug disposition and drug action. Adv Drug Deliv Rev 62: 904–917, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sukumaran S, Xue B, Jusko WJ, DuBois DC, Almon RR. Circadian variations in gene expression in rat abdominal adipose tissue and relationship to physiology. Physiol Genomics 42A: 141–152, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Suzuki T, Chow CW, Downey GP. Role of innate immune cells and their products in lung immunopathology. Int J Biochem Cell Biol 40: 1348–1361, 2008 [DOI] [PubMed] [Google Scholar]
- 61. Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet 37: 187–192, 2005 [DOI] [PubMed] [Google Scholar]
- 62. Visoni S, Meireles N, Monteiro L, Rossini A, Pinto LF. Different modes of inhibition of mouse Cyp2a5 and rat CYP2A3 by the food-derived 8-methoxypsoralen. Food Chem Toxicol 46: 1190–1195, 2008 [DOI] [PubMed] [Google Scholar]
- 63. Warburton D, Bellusci S, Del Moral PM, Kaartinen V, Lee M, Tefft D, Shi W. Growth factor signaling in lung morphogenetic centers: automaticity, stereotypy and symmetry. Respir Res 4: 5, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ware LB, Matthay MA. Keratinocyte and hepatocyte growth factors in the lung: roles in lung development, inflammation, and repair. Am J Physiol Lung Cell Mol Physiol 282: L924–L940, 2002 [DOI] [PubMed] [Google Scholar]
- 65. Weng DE, Usman N. Angiozyme: a novel angiogenesis inhibitor. Curr Oncol Rep 3: 141–146, 2001 [DOI] [PubMed] [Google Scholar]
- 66. Yao Z, DuBois DC, Almon RR, Jusko WJ. Modeling circadian rhythms of glucocorticoid receptor and glutamine synthetase expression in rat skeletal muscle. Pharm Res 23: 670–679, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yasuda S, Idell S, Fu J, Carter G, Snow R, Liu MC. Cigarette smoke toxicants as substrates and inhibitors for human cytosolic SULTs. Toxicol Appl Pharmacol 221: 13–20, 2007 [DOI] [PubMed] [Google Scholar]
- 68. Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol 11: 764–776 [DOI] [PubMed] [Google Scholar]
- 69. Zhou D, Zheng X, Wang L, Stelmack G, Halayko AJ, Dorscheid D, Bai TR. Expression and effects of cardiotrophin-1 (CT-1) in human airway smooth muscle cells. Br J Pharmacol 140: 1237–1244, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.