Abstract
The genetic regulation necessary for the formation of a four-chambered heart is tightly regulated by transcription factors such as TBX20, a member of the T-box (TBX) transcription factor family. TBX20 is critical for proper cardiogenesis and is expressed in the heart throughout development. Missense mutations in TBX20 have been found in patients with congenital heart defects (CHD). Characterization of modifiers of TBX20 activity will help elucidate the genetic mechanisms of heart development and CHD. A yeast two-hybrid assay screening an embryonic mouse heart cDNA library with TBX20b as bait was used to identify potential modifiers of TBX20 activity and identified an interaction with muskelin (MKLN1), a primarily cytoplasmic protein with potential roles in signal transduction machinery scaffolding and nucleocytoplasmic protein shuttling. In cellular studies, MKLN1 directly binds to the T-box DNA-binding domain of only the TBX20b isoform by its kelch repeats domain. Immunostaining of mammalian cells transfected with tagged TBX20b and MKLN1 revealed colocalization primarily in the cytoplasm. Immunohistochemistry analysis of embryonic mouse hearts reveals coexpression in the developing endocardial valvular and myocardial interventricular cells. This novel interaction between TBX20b and MKLN1 may help elucidate new regulatory mechanisms within heart development.
Keywords: Yeast two-hybrid, heart development, TBX20, MKLN1, transcription factor, protein-protein interaction
1. Introduction
Cardiogenesis is tightly regulated by networks of different transcription factors. The T-box (TBX) transcription factor family is important for the development of many mesoderm-derived structures, including the heart [1,2]. All TBX transcription factors share a highly conserved DNA-binding domain, designated the T-box [3]. Several TBX factors are directly involved in cardiogenesis including TBX2, TBX3, TBX5, TBX18, and TBX20 [4,5]. Each of these factors can be characterized as activators, repressors, or as having both properties depending on the developmental context. The coexpression of TBX transcription factors, each with different activities, targets, and cofactor binding abilities, adds complexities within the transcriptional networks driving cardiogenesis. Identifying proteins involved in regulating TBX transcription factor activity would provide important information in understanding the transcriptional regulation required for cardiogenesis.
TBX20 is critical for proper cardiogenesis and is expressed throughout heart development. Homozygous Tbx20 null mice die by embryonic day (E) 10.5 due to cardiac insufficiency from an unlooped, un-elongated heart tube that has not differentiated into chamber myocardium [6,7,8,9]. Consistent with mouse models, missense mutations within human TBX20 have been identified in patients with congenital heart defects (CHD) [10,11,12]. TBX20 functions in the early cardiac determination processes, regulates proliferation, and regulates elongation of the heart tube through cell recruitment from the secondary heart field [6,7,8,9]. In later heart development, TBX20 functions in chamber differentiation in part through repression of TBX2 activity [6,7,8]. TBX20 is also important in valvuloseptal formation through regulation of extracellular matrix (ECM) components and epithelial-to-mesenchymal transition (EMT) in the developing atrioventricular canal (AVC) and outflow tract (OFT) regions [13]. Multiple isoforms of TBX20 have been identified with the two major isoforms being TBX20a at 445 amino acids (aa) and TBX20b at 297 aa. Both are identical through the length of TBX20b with TBX20a containing an extended C-terminal region with characterized activation and repression domains [14].
To discover regulators of TBX20 during heart development, a yeast two-hybrid assay was employed to identify novel interacting partners using the human TBX20b isoform as bait. The yeast two-hybrid assay screened a library of cDNAs from E9.5–11.5 mouse hearts and identified muskelin (MKLN1) as an interacting protein. In this study, the TBX20b-MKLN1 interaction was confirmed as a novel isoform-specific interaction occurring in the perinuclear region of mammalian cells. Expression analysis determined that TBX20 and MKLN1 are coexpressed in regions involved in development of the interventricular septum and the valvuloseptal regions of the AVC.
2. Materials and Methods
Plasmid construction and yeast two-hybrid library screening
The human TBX20b isoform cDNA (BC120946.1) was obtained from Open Biosystems and was cloned into the DNA-binding domain vector, pGBTK–T7 (Clontech). To create the yeast two-hybrid screening library, cDNA fragments were fused with the Gal4 activation domain in the pGAD-T7 vector (Clontech). First, mRNA was isolated from embryonic hearts of ICR mice between E9.5 and E11.5, and primed with the NotI-oligo-d(T) primer to generate cDNAs using the SuperScript Plasmid System for cDNA Synthesis and Cloning (Invitrogen). pGAD-T7 was modified by digestion with PvuII and self-ligation to remove the NotI site. Then, SalI and NotI sites were inserted into the polylinker region. Finally, the cDNA fragments were linked with a SalI adaptor, digested with NotI, and directionally cloned into the SalI and NotI sites of the modified pGAD-T7 vector to generate the library. At least 5×106 independent clones were included in the primary library and all 16 randomly picked clones contained inserts with an average size of 1.9 kilobases (data not shown). The yeast two-hybrid screening assay was done in AH109 cells following Clontech’s Matchmaker protocol. One of the resultant clones was the full ORF of Mkln1 (RefSeq NM_013791.2).
Copurification studies
For the in vitro copurification studies, TBX20b was cloned into pGEX-2T (GEHealthcare). BL21 cells were transformed with either GST or GST-TBX20b and protein production was induced with IPTG treatment. GST and GST-TBX20b proteins were purified with glutathione sepharose beads (Invitrogen) and incubated with in vitro translated radiolabeled MKLN1 generated with the TNT-coupled transcription/translation system (Promega). Protein complexes were isolated by centrifugation, eluted, and analyzed by SDS-PAGE and radiography.
For mammalian cell overexpression copurification studies, MKLN1 was cloned into the pCMV-HA expression vector (Clontech) and TBX20b was cloned into the pCMV-GST vector [15]. The copurification studies were done as previously described [16]. Briefly, COSM6 cells were transfected according to the Lipofectamine 2000 protocol (Invitrogen). After 48 hours, lysates were collected and copurified with glutathione sepharose beads rotating at 4°C for 4–6 hours. The beads were collected by centrifugation and the interacting proteins were eluted. Whole cell lysate and copurification elution fractions were analyzed by SDS-PAGE and western blot analysis. Antibodies included GST (GEHealthcare, 27457701V) and HA (HA1.1 16B12). HRP-conjugated secondary antibodies were used for visualization of the antibody complexes with enzyme-linked chemiluminescence (Millipore).
Immunohistochemistry and immunocytochemistry
ICR mice matings were timed to collect E9.5–12.5 embryos. After dissection, the whole embryo or embryonic hearts were fixed in 4% paraformaldehyde, dehydrated with ethanol washes, cleared with Histo-clear (National Diagnostics) and embedded in paraffin wax. The tissues were sectioned at 7um and processed for immunohistochemistry. Antibodies included TBX20 at 1:1000 (Sigma, HPA008192) and MKLN1 at 1:50 (Sigma, HPA022817). Signals were visualized through a DAB chromogen system (Dako) and slides were counterstained with hematoxylin.
For cellular immunostaining, cells were plated on glass coverslips in a 24-well plate overnight. Cells were transfected using Lipofectamine 2000. After 48 hours, cells were stained overnight with primary antibody at 4°C. The cells were washed, stained with fluorophore-conjugated secondary antibody and mounted onto glass microscope slides with DAPI counterstaining mounting solution (Vectashield).
Subcellular localization analysis
Subcellular localization of TBX20 and MKLN1 was determined with the nuclear/cytoplasmic fractionation kit from ThermoScientific. Cells were transfected as described previously. TBX20b was cloned into the pCMV-Tag3 construct to create a myc-tagged fusion protein (Stratagene). Protein lysates were quantified and analyzed by SDS-PAGE and western analysis. Antibodies for cytoplasmic and nuclear fraction controls are MEK1/2 (Cell Signaling, L38C12) and LSD1 (Cell Signaling, C69G12), respectively.
3. Results
3.1 Yeast two-hybrid assay identified MKLN1 as a novel interacting partner to TBX20b
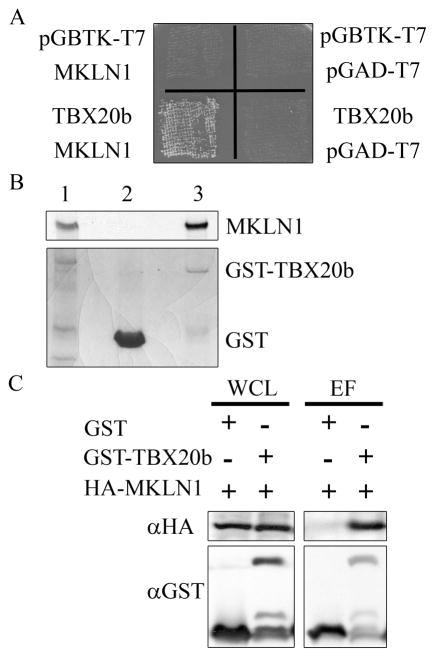
A novel yeast two-hybrid library was constructed to facilitate the identification of protein interactions relevant to mouse heart development. The library contains cDNAs isolated from the hearts of E9.5–11.5 mice. To avoid interference from the activation/repression domains of the longer human TBX20a isoform [14], the shorter human TBX20b isoform was used as bait. In this study, the yeast two-hybrid assay yielded 1.5 × 105 transformants as determined by growth on Tryptophan/Leucine (Trp/Leu) double dropout selection plates. After selection for protein interaction on quadruple (Q) dropout plates, fifteen colonies were established. Sequence analysis showed that 13 colonies contained 3 unique clones of MKLN1. The interaction of TBX20b and MKLN1 was confirmed with a yeast cotransformation assay as shown in Figure 1a. A single colony from each cotransformation plate was re-plated as a patch on a Trp/Leu dropout plate, with growth confirming cotransformation. The yeast patches were replica-plated into a Q dropout plate. Growth of yeast cotransformed with pGBTK-TBX20b and pGAD-MKLN1 on the Q plate confirmed the protein-protein interaction.
Fig. 1. Identification of MKLN1 as a novel interacting partner to TBX20b.
(A) AH109 yeast cells were cotransformed with pGBTK-T7 and pGAD-T7 vector controls, pGBTK-TBX20b with the pGAD-T7 control, pGAD-MKLN1 with the pGBTK-T7 control, and pGBTK-TBX20b with pGAD-MKLN1. Each cotransformation was replated as a patch on a Trp/Leu plate, and replica-plated onto a Q plate. Growth with pGBTK-TBX20b and pGAD-MKLN1 indicates protein interaction. (B) GST and GST-TBX20b proteins were isolated from bacterial lysates and copurified with in vitro radiolabeled MKLN1. The bottom panel shows the coomassie-stained SDS-PAGE gel with radioactive MKLN1 input (lane 1), and the GST-pulldown elution fractions of GST (lane 2) and GST-TBX20 (lane 3). The top panel is the radiography film revealing presence of MKLN1. (C) COSM6 cells were transfected with HA-MKLN1 and GST or GST-TBX20b as indicated. Interacting proteins were isolated by copurification. Whole cell lysate (WCL) and elution fractions (EF) were analyzed by SDS-PAGE and western blot analysis probing for GST and HA1.1.
An in vitro copurification assay using a GST pull-down method with a GST-TBX20b fusion protein and radiolabeled MKLN1 confirmed direct interaction. In vitro translated, radiolabeled MKLN1 was generated and incubated with GST or GST-TBX20b proteins isolated from bacterial cultures. Protein complexes were isolated by GST pull-down copurification and analyzed by SDS-PAGE. The presence of the GST and GST-TBX20b proteins are shown on the coomassie-stained gel, which was then exposed on radiography film to reveal the presence of MKLN1. MKLN1 was found in the input lane and the GST-TBX20b elution lane, confirming the direct interaction between the radiolabeled MKLN1 and GST-TBX20b (Figure 1b).
To confirm the TBX20b-MKLN1 interaction within mammalian cells, a copurification assay using transfected cell lysates was done. COSM6 cells were cotransfected with HA-MKLN1 and GST or GST-TBX20b. Lysates were collected and whole cell lysates were saved. The remaining lysates were incubated with GST-binding beads to isolate the GST fusion proteins and interacting proteins. The copurification assay resulted in the whole cell lysate and GST-bound elution fractions which were analyzed by SDS-PAGE and western blot analysis. HA-MKLN1 was found in both whole cell lysates, but it only copurified with GST-TBX20b, confirming an interaction between TBX20b and MKLN1 within mammalian cells (Figure 1c).
3.2 Characterization of the interaction domains between TBX20 and MKLN1
While the TBX20b isoform was used in the yeast two-hybrid screening, the TBX20a isoform is also of interest in studying the genetic regulation of cardiogenesis and was included in the following experiment. A copurification assay using transfected mammalian cells was done as previously stated. COSM6 cells were cotransfected with HA-MKLN1 in addition to GST, GST-TBX20b, or GST-TBX20a. Interestingly, HA-MKLN1 did not copurify with the GST-TBX20a isoform (Figure 2a). To identify the domain of interaction, truncated TBX20b constructs were cloned into the GST vector to separate the N-terminal portion and the T-box domain. The copurification assay revealed that the T-box domain of TBX20 (aa 102–297) is responsible for the interaction with HA-MKLN1, whereas the N-terminal portion of TBX20 does not bind to HA-MKLN1. (Figure 2a).
Fig. 2. Identification of the interaction domains of TBX20b and MKLN1.
(A) Top: Schematic of TBX20 isoforms. Bottom: COSM6 cells were transfected with HA-MKLN1 and GST, GST-TBX20a, GST-TBX20b, and truncations of GST-TBX20b. Copurification assays and western blot analyses compared whole cell lysate (WCL) and the elution fraction (EF). The blots were probed with GST and HA1.1 antibodies as before. (B) Top: Schematic of MKLN1 protein. Bottom: Similar copurification analyses were performed with truncated HA-tagged MKLN1 constructs. Gels were loaded as in panel (A) comparing WCL and EF with the GST and the GST-TBX20b transfected cells. Western blot analysis of the truncated HA-MKLN1 constructs (top-to-bottom) are listed left-to-right.
The MKLN1 domain required for interaction with TBX20b was determined by creating HA-tagged truncated constructs of MKLN1 and using the same copurification assay with GST-TBX20b. MKLN1 was truncated from N- and C-terminal ends deleting domains listed in Figure 2b. The first 259 amino acids of MKLN1 consist of a discoidin-like domain and a LiSH/CTLH domain. The C-terminal 476 amino acids of MKLN1 consist of a kelch motif with 6 kelch repeats followed by a C-terminal tail with no functional characteristics. The removal of the kelch motif and C-terminal tail abolished the interaction with GST-TBX20b. The interaction was narrowed to the kelch repeats domain with the removal of the last 3 repeats resulting in no interaction (Figure 2b).
3.3 TBX20b and MKLN1 colocalize in the perinuclear cytoplasm
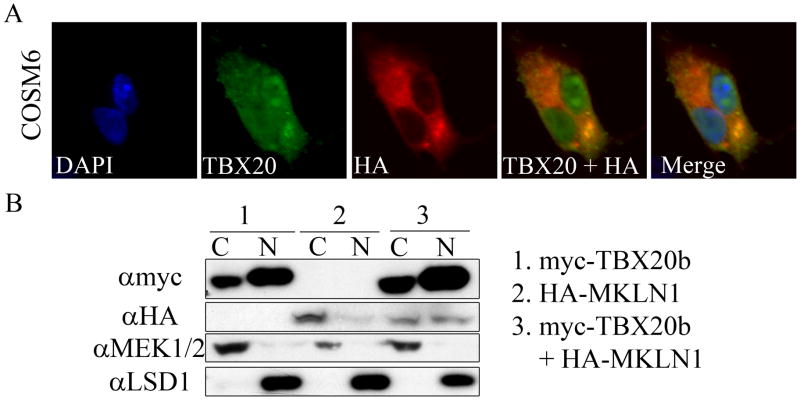
To further explore the TBX20b-MKLN1 interaction, the subcellular localizations of the proteins were determined in multiple cell types. As a transcription factor, the primary subcellular localization for TBX20 is in the nucleus. However, with previous studies placing the majority of MKLN1 in the cytoplasm [17,18,19,20], it was of interest to determine where the TBX20b-MKLN1 interaction was localized. COSM6 cells were cotransfected with myc-TBX20b and HA-MKLN1 and analyzed by immunofluorescent staining. HA-MKLN1 was localized primarily in the cytoplasm and myc-TBX20b was localized primarily in the nucleus with some cytoplasm protein. Colocalization was seen in the cytoplasm with perinuclear intensity (Figure 3a). These localization results were verified by using a nuclear/cytoplasmic fractionation kit on transfected cell lysates with western blot analysis confirming the immunostaining result (Figure 3b). MKLN1 has been identified within protein complexes involved in nucleocytoplasmic shuttling and an interaction with a transcription factor, such as TBX20b, could indicate a negative regulation by sequestering the transcription factor from the nucleus. However, the overexpression of HA-MKLN1 in the transfected COSM6 cells did not result in any identifiable changes in subcellular localization of TBX20.
Fig. 3. Colocalization of TBX20b and MKLN1.
(A) COSM6 cells were transfected with myc-TBX20b and HA-MKLN1. Fluorescent immunocytochemistry was performed with antibodies for TBX20, HA1.1, and DAPI for counterstain. (B) COSM6 cells were transfected with 1. myc-TBX20b, 2. HA-MKLN1, and 3. myc-TBX20b and HA-MKLN1. Cells were lysed and fractionated into cytoplasmic (C) and nuclear (N) fractions and analyzed by western blotting. MEK1/2 and LSD1 are cytoplasmic and nuclear controls, respectively.
3.4 Expression studies of TBX20 and MKLN1 using immunohistochemistry
To determine where and when the TBX20b-MKLN1 interaction takes place within cardiogenesis, the expression patterns of TBX20 and MKLN1 were analyzed in developing mice from E9.5–12.5 by immunohistochemistry. TBX20 expression analysis confirmed previously published data with expression in the ventricular myocardium wall, atrial myocardium, and endocardial cushions [14,21,22]. TBX20 expression correlates to its functions necessary for the development of the chamber myocardium in the ventricles and the valvuloseptal structures that develop from the endocardial cushion regions. Expression patterns of MKLN1 have not been examined throughout mouse heart development. From E9.5–10.5, MKLN1 expression is seen in numerous embryonic tissues including the heart. Within the heart, MKLN1 expression is found throughout the developing myocardium. From E11.5–12.5, MKLN1 expression is restricted and enriched within the endocardial cells in the AVC and in the myocardial cells of the IVS (Figure 4). Regions of overlapping expression between TBX20 and MKLN1 can be seen in the endocardial cushion cells of the developing atrioventricular region as well as in the myocardial cells in the interventricular region. Development of these regions is critical for proper cardiogenesis, failure of which leads to valvuloseptal defects
Fig. 4. Immunohistochemistry in the developing mouse heart.
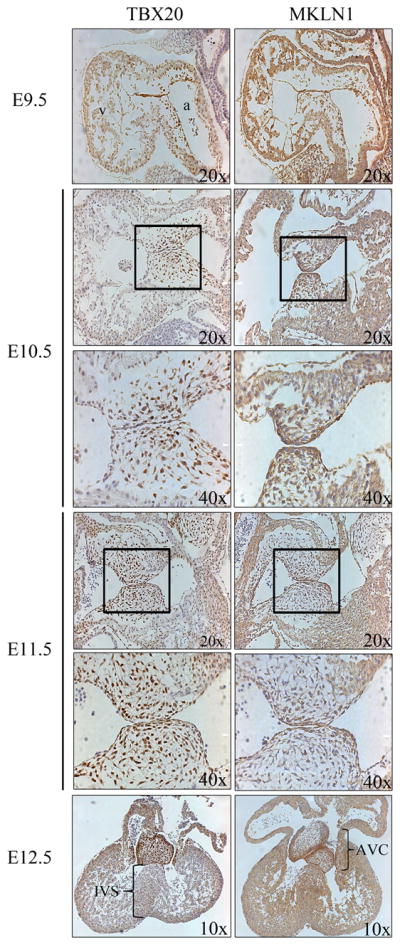
ICR mice matings were timed to collect embryos at the stated time points. Embryos were dissected and processed for immunohistochemistry on sagittal sections (E9.5–11.5) and frontal sections (E12.5). Anti-TBX20 and anti-MKLN1 were visualized with the DAB chromogen system. Slides were counterstained with hematoxylin. Boxed areas of developing endocardial cushion region indicate higher magnification panel below. (v), ventricles; (a), atrium; (IVS), interventricular septum; (AVC), atrioventricular cushions.
4. Discussion
Identifying novel regulators of critical cardiac transcription factors is crucial to better define the regulatory networks involved in cardiogenesis and provide insights into the mechanisms of CHD. In this study, a novel interaction between TBX20b and MKLN1 was discovered. Interestingly, this is an isoform-specific interaction with MKLN1 only interacting with the shorter TBX20b isoform. The TBX20 locus is alternatively transcribed into multiple TBX20 isoforms, mainly TBX20a and TBX20b [14]. Both isoforms are expressed in the heart throughout development and into adulthood with the TBX20a being the major transcript in both mice and humans [14,23]. Both isoforms can interact with cardiac transcription cofactors and activate transcription of critical cardiac genes and, in some cases, TBX20b can activate transcription more effectively than the longer isoform [14]. Both isoforms have identical T-box DNA-binding domains which suggest that they activate the same target genes. An isoform-specific protein interaction adds further complexity to the activities of TBX20 transcription factors.
TBX transcription factors are characterized by a highly conserved DNA-binding domain designated the T-box, which is typically 180 amino acids and recognizes a 24 base pair sequence called the T-site [3,14,24]. TBX20 binds to the T/2-site (half of the typical T-site) as a monomer in a molten globule state at 37°C, which increases conformation flexibility and potentially allows for more protein-protein interactions [24]. The T-box of TBX20 is also important for the binding of cofactors such as NKX2-5, GATA4 and GATA5 [14]. The binding of these proteins to the T-box implies cooperative or competitive binding with DNA and/or cofactors. Since MKLN1 binds to TBX20b at the T-box domain, determining if MKLN1 can alter DNA-binding or cofactor binding abilities of TBX20b would be of interest in future studies.
MKLN1 is a 735 aa protein with discoidin-like and LisH/CTLH domains in the N-terminal half. The C-terminal portion contains a kelch motif with 6 repeats [19,25]. The kelch motif typically contains 5–7 repeats that each form a 4-stranded β sheet, or blade, in a β-propeller and is most often involved in protein-protein interactions within protein complexes [25,26]. Proteins containing a kelch motif have a variety of functions, ranging from cytoskeleton organization to gene regulation. For instance, the kelch motif-containing protein KEAP1 regulates gene expression by sequestering the transcription factor NRF2 [26]. The binding of TBX20b to the kelch motif of MKLN1 suggests the presence of a protein complex with other binding partners in the cytoplasm. Identification of other protein partners in the TBX20b-MKLN1 interaction complex would be of interest to further explore regulatory roles of MKLN1 on TBX20b activity.
To better understand the localization of the TBX20b-MKLN1 interaction throughout heart development, the expression patterns of TBX20 and MKLN1 were determined by immunohistochemistry. In this study, TBX20 expression supports previous reports [14,22,27,28]. MKLN1 is expressed in a variety of tissues in developing and adult mice including the skeletal muscle, liver, heart, and parts of the central nervous system [17,20,29]. In this study, MKLN1 expression was detailed in heart development of E9.5–12.5 mice. While MKLN1 expression is broadly expressed in E9.5–10.5 hearts, in later stages there is strong expression in the endocardial cells of the developing AVC and the myocardial cells in the IVS. The coexpression of TBX20 and MKLN1 correlate to regions important for the developing valves and septa.
While the functions of MKLN1 have been largely unexplored, the colocalization between TBX20b and MKLN1 at the perinuclear region suggests that the potential nucleocytoplasmic shuttling roles of MKLN1 regulate TBX20b activity [30]. However, any nuclear function of MKLN1 has yet to be determined and future studies could reveal new methods of transcription factor regulation. In addition, TBX20 does have a significant cytoplasmic localization which opens the possibility of a cytoplasmic, DNA-independent role for the transcription factor. Instead of a decrease of TBX20 activity in the nucleus due to sequestering, perhaps MKLN1 and TBX20b play a DNA-independent role in heart development. Recent publications have spotlighted DNA-independent functions of TBX transcription factors TBX1 and TBX20. TBX20 reduced the BMP/Smad signaling through the binding and inhibiting the Smad1/5 signaling that is required to activate Tbx2 expression [31]. This DNA-independent regulation of critical signaling pathways is a relatively unexplored function of transcription factors. Another potential regulatory role for the TBX20b-MKLN1 interaction involves the development of the AVC which will form the heart valves. Valvulogenesis requires changes in cellular proliferation and ECM properties. TBX20 has been identified as regulating proliferation and the expression of different ECM components [6,7,8,9,13]. MKLN1 was first identified having a role in the regulation of cell attachment through ECM signaling [17]. Perhaps TBX20 and MKLN1 regulate the changes in the ECM during valvuloseptal development. Understanding how TBX20 function is regulated in different developmental processes throughout cardiogenesis would provide key information in how CHD occur and are identified.
Acknowledgments
We thank the members of the Jiao and Wang labs for their assistance on this project. We thank Dr. B. Hogan (Duke Univ. Med. Ctr.) for supporting the construction of the yeast two-hybrid library. We thank Dr. M. Kneussel (University of Hamburg Medical School, Germany) for providing materials.We acknowledge the services of the UAB Heflin Center Genomics Core Facility for all sequencing. This work is supported by a UAB faculty development grant and a NHLBI R01 grant (HL095783-01A1) awarded to K.J.
Abbreviations
- aa
amino acids
- AVC
atrioventricular canal
- CHD
congenital heart defects
- E
embryonic day
- ECM
extracellular matrix
- EF
elution fraction
- EMT
epithelial-to-mesenchymal transition
- GST
glutathione s-transferase
- HA
Hemagglutinin
- MKLN1
muskelin1
- OFT
outflow tract
- Q
quadruple
- TBX
T-box
- Trp/Leu
tryptophan and leucine
- WCL
whole cell lysate
Footnotes
Present/Permanent address: Dr. Kai Jiao. Kaul 768. 720 20th St. South, Birmingham, AL 35294 USA
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wilson V, Conlon F. The T-box family. Genome Biol. 2002;3:REVIEWS3008. doi: 10.1186/gb-2002-3-6-reviews3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Packham E, Brook J. T-box genes in human disorders. Hum Mol Genet. 2003;12(Spec No 1):R37–44. doi: 10.1093/hmg/ddg077. [DOI] [PubMed] [Google Scholar]
- 3.Bollag R, Siegfried Z, Cebra-Thomas J, Garvey N, Davison E, Silver L. An ancient family of embryonically expressed mouse genes sharing a conserved protein motif with the T locus. Nat Genet. 1994;7:383–389. doi: 10.1038/ng0794-383. [DOI] [PubMed] [Google Scholar]
- 4.Barnett J, Desgrosellier J. Early events in valvulogenesis: a signaling perspective. Birth Defects Res C Embryo Today. 2003;69:58–72. doi: 10.1002/bdrc.10006. [DOI] [PubMed] [Google Scholar]
- 5.Stennard F, Harvey R. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development. 2005;132:4897–4910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- 6.Cai C, Zhou W, Yang L, Bu L, Qyang Y, Zhang X, Li X, Rosenfeld M, Chen J, Evans S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh M, Christoffels V, Dias J, Trowe M, Petry M, Schuster-Gossler K, Bürger A, Ericson J, Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132:2697–2707. doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]
- 8.Stennard F, Costa M, Lai D, Biben C, Furtado M, Solloway M, McCulley D, Leimena C, Preis J, Dunwoodie S, Elliott D, Prall O, Black B, Fatkin D, Harvey R. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi J, Mileikovskaia M, Koshiba-Takeuchi K, Heidt A, Mori A, Arruda E, Gertsenstein M, Georges R, Davidson L, Mo R, Hui C, Henkelman R, Nemer M, Black B, Nagy A, Bruneau B. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 10.Posch M, Gramlich M, Sunde M, Schmitt K, Lee S, Richter S, Kersten A, Perrot A, Panek A, Al Khatib I, Nemer G, Mégarbané A, Dietz R, Stiller B, Berger F, Harvey R, Ozcelik C. A gain-of-function TBX20 mutation causes congenital atrial septal defects, patent foramen ovale and cardiac valve defects. J Med Genet. 2010;47:230–235. doi: 10.1136/jmg.2009.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Shen A, Li X, Jiao W, Zhang X, Li Z. T-box transcription factor TBX20 mutations in Chinese patients with congenital heart disease. Eur J Med Genet. 2008;51:580–587. doi: 10.1016/j.ejmg.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Kirk E, Sunde M, Costa M, Rankin S, Wolstein O, Castro M, Butler T, Hyun C, Guo G, Otway R, Mackay J, Waddell L, Cole A, Hayward C, Keogh A, Macdonald P, Griffiths L, Fatkin D, Sholler G, Zorn A, Feneley M, Winlaw D, Harvey R. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet. 2007;81:280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shelton E, Yutzey K. Tbx20 regulation of endocardial cushion cell proliferation and extracellular matrix gene expression. Dev Biol. 2007;302:376–388. doi: 10.1016/j.ydbio.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stennard F, Costa M, Elliott D, Rankin S, Haast S, Lai D, McDonald L, Niederreither K, Dolle P, Bruneau B, Zorn A, Harvey R. Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262:206–224. doi: 10.1016/s0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]
- 15.Tsai RY, Reed RR. Using a eukaryotic GST fusion vector for proteins difficult to express in E. coli. Biotechniques. 1997;23:794–796. 798, 800. doi: 10.2144/97235bm06. [DOI] [PubMed] [Google Scholar]
- 16.Jiao K, Zhou Y, Hogan BL. Identification of mZnf8, a mouse Krüppel-like transcriptional repressor, as a novel nuclear interaction partner of Smad1. Mol Cell Biol. 2002;22:7633–7644. doi: 10.1128/MCB.22.21.7633-7644.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams J, Seed B, Lawler J. Muskelin, a novel intracellular mediator of cell adhesive and cytoskeletal responses to thrombospondin-1. EMBO J. 1998;17:4964–4974. doi: 10.1093/emboj/17.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa H, Katoh H, Fujita H, Mori K, Negishi M. Receptor isoform-specific interaction of prostaglandin EP3 receptor with muskelin. Biochem Biophys Res Commun. 2000;276:350–354. doi: 10.1006/bbrc.2000.3467. [DOI] [PubMed] [Google Scholar]
- 19.Prag S, Collett G, Adams J. Molecular analysis of muskelin identifies a conserved discoidin-like domain that contributes to protein self-association. Biochem J. 2004;381:547–559. doi: 10.1042/BJ20040253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prag S, De Arcangelis A, Georges-Labouesse E, Adams J. Regulation of post-translational modifications of muskelin by protein kinase C. Int J Biochem Cell Biol. 2007;39:366–378. doi: 10.1016/j.biocel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Carson C, Kinzler E, Parr B. Tbx12, a novel T-box gene, is expressed during early stages of heart and retinal development. Mech Dev. 2000;96:137–140. doi: 10.1016/s0925-4773(00)00376-2. [DOI] [PubMed] [Google Scholar]
- 22.Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene tbx20. Mech Dev. 2001;100:87–91. doi: 10.1016/s0925-4773(00)00499-8. [DOI] [PubMed] [Google Scholar]
- 23.Hammer S, Toenjes M, Lange M, Fischer J, Dunkel I, Mebus S, Grimm C, Hetzer R, Berger F, Sperling S. Characterization of TBX20 in human hearts and its regulation by TFAP2. J Cell Biochem. 2008;104:1022–1033. doi: 10.1002/jcb.21686. [DOI] [PubMed] [Google Scholar]
- 24.Macindoe I, Glockner L, Vukasin P, Stennard F, Costa M, Harvey R, Mackay J, Sunde M. Conformational stability and DNA binding specificity of the cardiac T-box transcription factor Tbx20. J Mol Biol. 2009;389:606–618. doi: 10.1016/j.jmb.2009.04.056. [DOI] [PubMed] [Google Scholar]
- 25.Adams J. Characterization of a Drosophila melanogaster orthologue of muskelin. Gene. 2002;297:69–78. doi: 10.1016/s0378-1119(02)00887-9. [DOI] [PubMed] [Google Scholar]
- 26.Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 27.Ahn D, Ruvinsky I, Oates A, Silver L, Ho R. tbx20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech Dev. 2000;95:253–258. doi: 10.1016/s0925-4773(00)00346-4. [DOI] [PubMed] [Google Scholar]
- 28.Meins M, Henderson D, Bhattacharya S, Sowden J. Characterization of the human TBX20 gene, a new member of the T-Box gene family closely related to the Drosophila H15 gene. Genomics. 2000;67:317–332. doi: 10.1006/geno.2000.6249. [DOI] [PubMed] [Google Scholar]
- 29.Tagnaouti N, Loebrich S, Heisler F, Pechmann Y, Fehr S, De Arcangelis A, Georges-Labouesse E, Adams J, Kneussel M. Neuronal expression of muskelin in the rodent central nervous system. BMC Neurosci. 2007;8:28. doi: 10.1186/1471-2202-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valiyaveettil M, Bentley A, Gursahaney P, Hussien R, Chakravarti R, Kureishy N, Prag S, Adams J. Novel role of the muskelin-RanBP9 complex as a nucleocytoplasmic mediator of cell morphology regulation. J Cell Biol. 2008;182:727–739. doi: 10.1083/jcb.200801133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh R, Horsthuis T, Farin H, Grieskamp T, Norden J, Petry M, Wakker V, Moorman A, Christoffels V, Kispert A. Tbx20 interacts with smads to confine tbx2 expression to the atrioventricular canal. Circ Res. 2009;105:442–452. doi: 10.1161/CIRCRESAHA.109.196063. [DOI] [PubMed] [Google Scholar]